Abstract
Rice production and sustainability are challenged by its most dreadful pest, the brown planthopper (Nilaparvata lugens Stål, BPH). Therefore, the studies on rice-BPH interactions and their underlying mechanisms are of high interest. The rice ontogenetic defense, such as the role of microRNAs (miRNAs) has mostly been investigated against the pathogens, with only a few reports existing against the insect infestations. Thus, revealing the involvement of rice miRNAs in response to BPH infestations will be beneficial in understanding these complex interactions. In this study, the small RNA profiling of the IR56 rice in response to separate BPH infestations of varied virulence levels identified the BPH-responsive miRNAs and revealed the differential transcript abundance of several miRNAs during a compatible and incompatible rice-BPH interaction. The miRNA sequence analysis identified 218 known and 28 novel miRNAs distributed in 54 miRNA families. Additionally, 138 and 140 numbers of differentially expressed (DE) miRNAs were identified during the compatible and incompatible interaction, respectively. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed the target gene candidates of DE miRNAs (including osa-miR2871a-3p, osa-miR172a, osa-miR166a-5p, osa-miR2120, and osa-miR1859) that might be involved in the IR56 rice defense responses against BPH infestation. Conversely, osa-miR530-5p, osa-miR812s, osa-miR2118g, osa-miR156l-5p, osa-miR435 and two of the novel miRNAs, including novel_16 and novel_52 might negatively modulate the IR56 rice defense. The expressional validation of the selected miRNAs and their targets further supported the IR56 rice defense regulatory network. Based on our results, we have proposed a conceptual model depicting the miRNA defense regulatory network in the IR56 rice against BPH infestation. The findings from the study add further insights into the molecular mechanisms of rice-BPH interactions and will be helpful for the future researches.
Subject terms: Entomology, Herbivory
The normal human dietary uptakes depend largely on cereals, including rice (Oryza sativa L.), which is a staple food for more than half of the world population fulfilling more than 20% of the daily calorie needs1. It’s been estimated that by the year 2050, the global crop productions need to be increased at least 50% to satisfy the demand2. However, the current trend of rice production indicates only a 1.0% per year increase, which can be extrapolated to be a ~ 42% global increase by 2050, below par of the need2. In addition, rice productivity is severely challenged by several insect infestations3. Among them, the brown planthopper (Nilaparvata lugens Stål, BPH) is the most dreadful rice pest causing hopperburn, a fatal drying of rice plants that results in huge economic losses in Asia1. Although, the use of different insecticides is most common practice employed to control the BPH infestations, the abuse of these chemicals has resulted in many adversities, including insecticide resistance, insect resurgence, the elimination of natural enemies and other environmental hazards. Therefore, the identification and molecular breeding of rice germplasms continaing the BPH-resistance genes are considered to be the most suitable strategy for the control and management of BPH1. To date, 39 BPH resistance loci (Bph/bph genes) have been reported from different rice cultivars and from wild-rice species4. Amongst them, bph2/Bph265, Bph3/Bph176, Bph67, Bph98, Bph149, Bph1510, Bph1811, bph2912, and Bph3213 have been isolated by map-based cloning. On one hand, the Bph3 (a cluster of four plasma-membrane-localized lectin receptor kinases, OsLecRK1-4) has been considered for molecular breeding of rice cultivars with broad-spectrum and durable insect resistance6,14. On the other hand, it is of evident that these cultivars carrying the Bph genes can lose their resistance to BPH due to the evolution of new biotypes or populations1,15,16. Recently, a newly established virulent BPH population (IR56-BPH population) was discovered that could successfully break down the Bph3-mediated resistance in IR56 rice16. However, the underlying mechanisms for rice-BPH interactions is still unclear.
MicroRNAs (miRNAs) are of 21–24 nucleotides long endogenous regulatory non-coding small RNAs (sRNA) in plants having vital roles in the post-transcriptional regulation of gene expression during plant defense responses17–19. The early reports of their involvements in plant defense against herbivory were revealed from the sRNA transcriptome analysis of Nicotiana attenuate20. The silencing of RNA-directed RNA polymerase 1 (RdR1) and Dicer-like 3 (Dcl3) or Dicer-like 4 (Dcl4), important proteins from the miRNA biogenesis pathway, impaired the N. attenuate resistance against insect attacks. Further, the RdR1 expression was induced either by application of jasmonic acid (JA) or salicylic acid (SA) or caterpillar oral secretions, but not by mechanical wounding itself. In Cucumis melo, the resistant Vat+ near isogenic lines and the susceptible Vat− exhibited distinct miRNA profiles under the infestation of Aphis gossypii21. Likewise, in Camellia sinensis infestation of Ectropis oblique resulted in the differential expression of 150 miRNAs, supporting the role of miRNA in plant–insect interactions22. To our knowledge, the first study on the roles of miRNAs in rice-BPH interactions was reported in 2017, revealing the differential miRNA responses in a resistant (BPH15 introgression line) and susceptible rice (recurrent parent 9311) in response to the infestation of BPH biotype 123. However, to the best of our knowledge, no report exists on the role of miRNAs in a resistant rice variety in response to BPH infestations of variable virulence levels. Thus, the current scenario offers good opportunity to study the involvement of miRNA and their subsequent defense modulatory roles during the rice-BPH interactions.
In this study, the miRNA profiling of the resistant IR56 rice (carrying Bph3) have been performed under the independent infestations of a virulent IR56-BPH and an avirulent TN1-BPH. The IR56 rice and IR56-BPH interaction (hereafter referred as IR-IR56-BPH) is considered to be compatible, whereas the IR56 rice and TN1-BPH interaction (hereafter referred as IR-TN1-BPH) is considered to be incompatible in nature24. Small RNA sequencing data from three different libraries, i.e. IR-IR56-BPH, IR-TN1-BPH, and control (no BPH) were analyzed to find out the conserved and novel miRNAs involved in rice-BPH interactions. Further, the identification of the differentially expressed (DE) miRNAs in IR-IR56-BPH and IR-TN1-BPH suggested their roles in specific rice-BPH interactions. Additionally, the validation of some selected DE miRNA and their targets by qPCR analysis further strengthened their involvement in IR56 rice defense. The miRNA target predictions and their functional annotations by GO and KEGG enrichments indicated the defense modulatory roles of some DE miRNAs and their targets. Lastly, based on the findings of this study, a conceptual model depicting the miRNA defense regulatory network in IR56 rice has been proposed.
Result
Small RNAs sequencing in the IR56 rice
To reveal the involvement of miRNAs during the rice-BPH interactions, the small RNA sequencing was performed from the IR-IR56-BPH, IR-TN1-BPH, and control after 24 h of BPH feedings. Total raw reads of control, IR-IR56-BPH, and IR-TN1-BPH were 15296583, 12601079, and 16575853, respectively (Table 1). Raw reads of the three libraries were filtered to remove low quality reads, poly A, incorrect adaptors and sequences shorter than 18 nt. After sequence filtering, 14761791 (control), 12259165 (IR-IR56-BPH), and 15644891 (IR-TN1-BPH) clean reads were obtained (Supplementary Figure S1, Table 1). Subsequently, the alignment of all clean sequences with sRNAs in GenBank and Rfam databases resulted in the removal of sequences other than the unique reads. The obtained unique sequences were then mapped to the rice genome to exclude any matches with the exons or introns and repeat sequences. The remaining unique sequences were then aligned with the miRNA database in miRBase (release 21) to find out the known miRNAs. In total, these clean reads were mapped to 218 known miRNAs. The known miRNA length distributions of all three libraries were found to be mostly concentrated at 21 and 24 nt as reported in several plant species, including rice. Additionally, most of the 21 nt long miRNAs contained the 5′U as the first base (Fig. 1).
Table 1.
The summary of sRNA sequencing result data.
| Sequence type | Control | IR56-BPH | TN1-BPH | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Raw counts | Unique counts | Total counts | Raw counts | Unique counts | Total counts | Raw counts | Unique counts | Total counts | |
| Raw reads | 9,128,932 | 417,277 | 9,546,209 | 7,400,347 | 439,295 | 7,839,642 | 8,812,250 | 336,072 | 9,148,322 |
| Clean reads | 3,032,174 | 159,462 | 3,191,636 | 2,325,929 | 175,454 | 2,501,383 | 2,763,080 | 134,601 | 2,897,681 |
| rRNA | 8,520,462 | 160,027 | 8,680,489 | 6,720,834 | 165,447 | 6,886,281 | 8,448,361 | 157,756 | 8,606,117 |
| tRNA | 26,782 | 2566 | 29,348 | 39,360 | 3136 | 42,496 | 25,390 | 2412 | 27,802 |
| snRNA | 2853 | 1054 | 3907 | 3809 | 1250 | 5059 | 2146 | 911 | 3057 |
| snoRNA | 17,043 | 3966 | 21,009 | 21,350 | 4378 | 25,728 | 11,999 | 3394 | 15,393 |
| Repeats | 205,796 | 94,997 | 300,793 | 202,490 | 100,147 | 302,637 | 47,939 | 29,365 | 77,304 |
| Exons | 164,544 | 76,430 | 240,974 | 167,697 | 79,725 | 247,422 | 126,311 | 73,424 | 199,735 |
| Introns | 19,312 | 9384 | 28,696 | 23,323 | 11,159 | 34,482 | 11,660 | 6766 | 18,426 |
| Others | 172,140 | 68,853 | 240,993 | 221,484 | 74,053 | 295,537 | 138,444 | 62,044 | 200,488 |
Figure 1.
The first nucleotide bias (uridine, U; adenine, A; cytosine, C; guanine, G) at the 5′ end position of different lengths of the known miRNA in control (A), IR-IR56-BPH (B), and IR-TN1-BPH (C) libraries. The numbers of miRNA for each condition were denoted in the stacked histogram.
Identification and expression of known and novel miRNAs
A total of 218 known rice miRNAs belonging to 54 rice miRNA families, and 28 novel miRNAs were discovered in the three libraries (Supplementary Table S1). Among all identified known and novel miRNAs, six miRNAs were detected only in IR-TN1-BPH, 24 miRNAs were found only in IR-IR56-BPH, and 31 miRNAs were only found in the control (Supplementary Table S2), while 185 remaining miRNAs were found in all three libraries.
The miRNAs having more than 1000 transcripts per million (TPM) were considered as abundantly expressed miRNAs, while those with less than 10 TPM were classified as rarely expressed miRNAs. The 20 most abundant miRNAs in each of the libraries (accounting for ~ 90% of total miRNA reads) are listed in Table 2. Two of them (novel_117, and novel_121, shown in bold) were novel miRNAs. The number of rare miRNAs in TN1-BPH (98) was twice the number of IR56-BPH (43), while showed no significant difference between IR56-BPH (43) and control (35), indicating that many miRNAs may be down-regulated in incompatible rice-BPH interaction (Supplementary Table S3).
Table 2.
Top 20 most abundant miRNAs expressed in the four libraries (TPM were shown).
| miRNA | IR56_CK | miRNA | IR56_IR | miRNA | IR56_TN |
|---|---|---|---|---|---|
| osa-iR1861b | 395,432.94 | osa-miR1861b | 249,283.49 | osa-miR1861b | 173,834.54 |
| osa-iR396f.-5p | 62,370.48 | osa-miR396f.-5p | 97,201.05 | osa-miR396f.-5p | 121,963.23 |
| osa-iR396e-5p | 62,207.96 | osa-miR396e-5p | 96,835.17 | osa-miR396e-5p | 121,963.23 |
| osa-miR166a-3p | 57,657.15 | osa-miR166a-3p | 78,968.23 | osa-miR166a-3p | 106,040.71 |
| osa-miR166k-3p | 41,891.84 | osa-miR162a | 46,466.25 | osa-miR159a.1 | 43,663.82 |
| osa-miR167d-5p | 35,756.37 | osa-miR167d-5p | 36,099.76 | osa-miR166k-3p | 40,052.53 |
| novel_121 | 32,830.85 | osa-miR166k-3p | 35,977.80 | osa-miR166g-3p | 39,067.63 |
| osa-miR1861a | 30,311.65 | osa-miR159a.1 | 35,977.80 | osa-miR167d-5p | 36,277.08 |
| osa-miR162a | 27,508.02 | osa-miR1862e | 30,245.75 | osa-miR162a | 24,130.01 |
| osa-miR159a.1 | 23,607.33 | osa-miR166g-3p | 25,428.38 | osa-miR1861a | 17,564.02 |
| osa-miR1861h | 17,959.45 | osa-miR812k | 23,355.08 | osa-miR156a | 14,937.62 |
| osa-miR166g-3p | 17,918.82 | osa-miR1861a | 19,147.51 | osa-miR1862d | 14,773.47 |
| osa-miR1862e | 14,018.12 | osa-miR1862d | 18,354.78 | osa-miR166j-5p | 14,445.17 |
| osa-miR1862d | 11,864.61 | osa-miR166j-5p | 14,513.08 | osa-miR396g | 13,788.58 |
| osa-miR812k | 11,458.29 | osa-miR396c-5p | 8110.25 | osa-miR166m | 12,967.83 |
| osa-miR166j-5p | 10,523.75 | osa-miR166m | 7500.46 | osa-miR812k | 11,654.63 |
| novel_117 | 8085.82 | osa-miR5794 | 7195.56 | osa-miR168a-5p | 8699.93 |
| osa-miR396c-5p | 8004.55 | osa-miR168a-5p | 6829.68 | osa-miR396c-5p | 8207.49 |
| osa-miR166m | 6907.48 | osa-miR820a | 6768.71 | osa-miR11337-5p | 7222.59 |
| osa-miR399d | 6541.79 | osa-miR156a | 5671.08 | osa-miR1862e | 7058.44 |
Identification of DE miRNAs in response to BPH infestations
The differential expression and Pearson correlation analysis of the miRNAs revealed the DE miRNAs in IR56 rice in response to BPH infestations. The correlation analysis indicated a positive correlation among all the samples used in the sRNA sequence analysis (Supplementary Figure S2). The differential expression analysis of all identified miRNAs (known and novel) revealed that BPH infestations have significant effects on the transcript abundance of the IR56 rice miRNAs. Out of the 246 identified miRNAs, 138 for IR-IR56-BPH and 140 for IR-TN1-BPH were found to be deferentially expressed, compared with the control (no BPH) (Supplementary Table S3). In the IR-IR56-BPH, 69 numbers of miRNAs were found to be upregulated, whereas 69 miRNAs were down regulated in comparison to the control (Fig. 2A). On the other hand, 36 miRNAs were found to be upregulated in the IR-TN1-BPH, while 104 miRNAs were downregulated (Fig. 2B). Additionally, some DE miRNA families were found to be exclusive in the IR-IR56-BPH or IR-TN1-BPH, respectively. For instance, miRNAs from the families like MIR397 and MIR398 were found exclusive to the IR-IR56-BPH but not in IR-TN1-BPH. Conversely, miRNAs from the families like MIR172 and MIR435 were found exclusively in the IR-TN1-BPH but not in IR-IR56-BPH. Collectively, the DE miRNA analysis results in the IR-IR56-BPH and IR-TN1-BPH libraries suggested that different miRNAs are involved in the rice-BPH interactions and the kind of interaction (compatible or incompatible) affects the number (how many), kind (which family/novel), and nature (up or downregulation) of the miRNA expressions in rice.
Figure 2.
The volcano plots showing the DE miRNAs in the comparisons of IR-IR56-BPH versus control (A) and IR-TN1-BPH versus control (B). The significantly upregulated, and downregulated miRNAs were shown in red and green, respectively (adjusted P value < 0.01). No differential expression between the two groups was shown in blue (adjusted P value > 0.01). The number of genes in each group was parenthesized. The randomly selected 20 miRNAs in following qRT-PCR were labelled. The miRNAs that were not expressed in both libraries were not shown in the figure.
Prediction of the targets of the DE miRNAs and their functional annotations
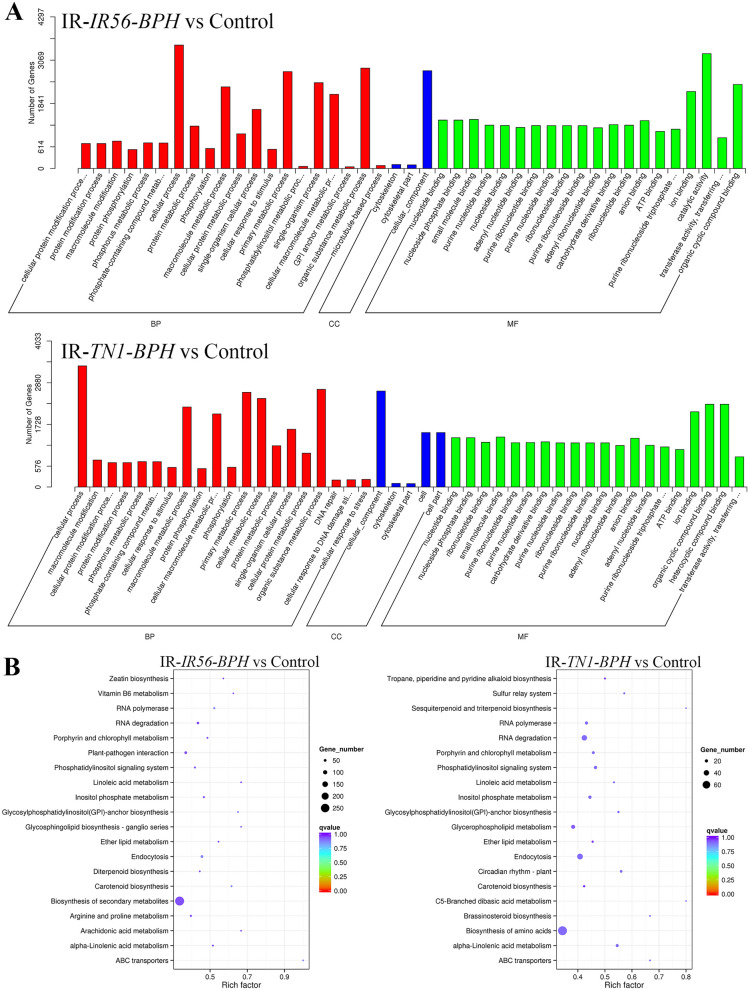
The prediction of the targets of the DE miRNAs provided a means to understand the possible defense modulatory roles of the miRNAs. Also, the analysis revealed that a single miRNA can have multiple targets in rice, whereas a single mRNA transcript can also be targeted by multiple miRNAs. To functionally characterize the miRNA targets, GO and KEGG enrichments of the predicted targets were carried out. The GO term annotations revealed similar biological process and molecular functions for the targets in both IR-IR56-BPH and IR-TN1-BPH, such as metabolic process and protein modifications, and nucleotide-binding and transferase activity (Fig. 3A). However, the targets associated with IR-TN1-BPH DE miRNAs have crucial stress-responsive GO enrichments indicating their possible roles in rice defense responses. Similarly, the KEGG pathway enrichments revealed that some of the target genes of the miRNAs from IR-IR56-BPH and IR-TN1-BPH samples are commonly involved in the physiological processes, including RNA degradation, glycosylphosphatidylinositol (GPI)-anchor biosynthesis, lipid metabolisms, carotenoid biosynthesis inositol metabolism and phosphatidylinositol signaling, and endocytosis (Fig. 3B). However, the IR-IR56-BPH and IR-TN1-BPH targets had some different and exclusive KEGG enrichments such as, zeatin biosynthesis, plant-pathogen interaction, and vitamin B6 metabolism for the IR-IR56-BPH, while brassinosteroid biosynthesis, circadian rhythm, and sulfur relay system for the IR-TN1-BPH. Thus, having the common metabolic pathway enriched targets in IR-IR56-BPH and IR-TN1-BPH suggest that the BPH-feeding response in IR56 rice share several common features immaterial with the type of BPH population, whereas exclusive pathway enrichments to IR-IR56-BPH or IR-TN1-BPH miRNA targets suggest the differential metabolic response in IR56 rice to BPH feedings depending on the infested population.
Figure 3.
Gene ontology annotations (A) and top 20 KEGG pathways (B) enriched in the DE miRNA targets in IR-IR56-BPH and IR-TN1-BPH.
Expression validation of selected DE miRNAs and their targets
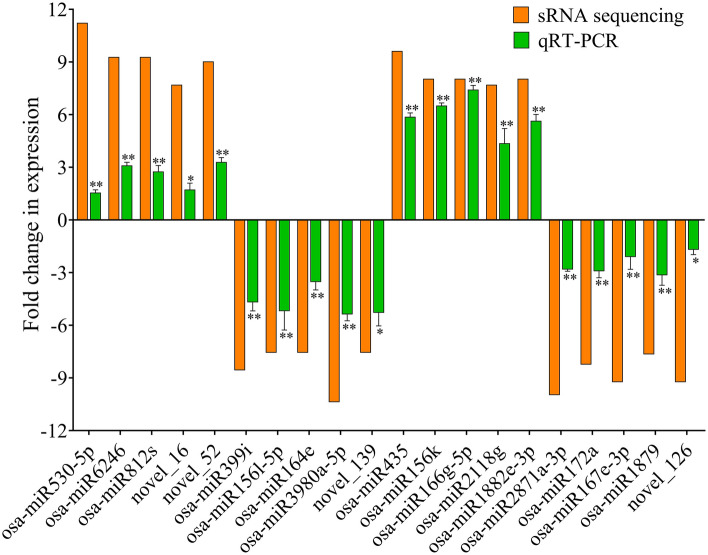
To validate the transcript abundance of the identified miRNAs in the sRNA sequencing, we have randomly selected 20 miRNAs (10 each from IR-IR56-BPH and IR-TN1-BPH) and their relative expressions were evaluated by real-time quantitative PCR (qPCR, primers listed in Supplementary Table S4). The qPCR results were found to be in accordance with the sRNA sequencing results indicating a similar and significant trend of relative expression levels in the BPH infested rice samples as compared to the control (Fig. 4). However, the degree of the relative fold changes of the DE miRNAs obtained via sRNA sequencing and qPCR analysis did differ. Thus, the similar expression profiles of the miRNAs between the qPCR and sRNA sequencing indicated the sequencing results to be reliable and suitable for further analyses.
Figure 4.
Expression validation of the selected miRNAs by qPCR. The fold changes (log2) in the expression of the miRNAs were calculated and compared to the sRNA sequencing data. Bars represent the mean ± SE of three biological replicates for the qPCR data. Asterisks * and ** indicate the significant difference in the expression levels of miRNAs in IR-IR56-BPH or IR-TN1-BPH as compared to control at P < 0.05 and P < 0.01, respectively (Student’s t‐test).
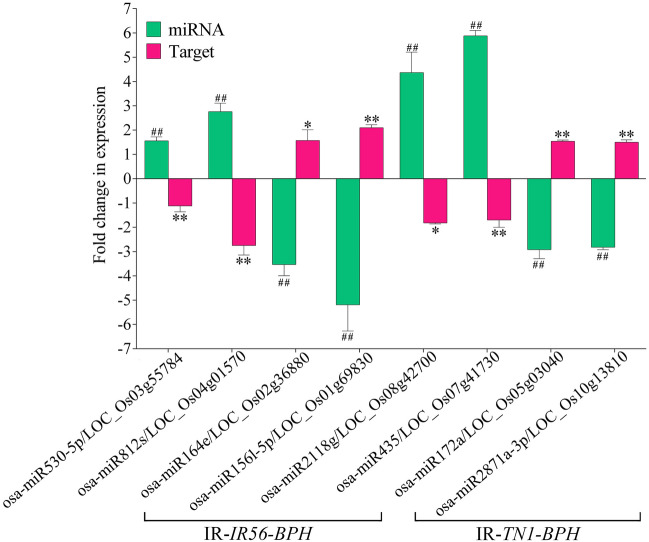
The target gene predictions of the DE miRNAs have resulted in identifying several genes in the IR56 rice associated with defense responses and plant protection (Supplementary Table S5). Additionally, the GO and KEGG enrichments further strengthen their roles in rice defense. From the predicted targets, we selected 8 genes and validated their relative expressions by qPCR. All the selected targets exhibited a negative correlation to their corresponding miRNA expression levels (Fig. 5). In the IR-IR56-BPH, strong downregulation (log2 fold change > 2) was observed in the target of osa-miR812s (LOC_Os04g01570) that encodes a pectin methylesterase inhibitor (PEMI) protein. Significant downregulation in its transcript accumulation in IR-IR56-BPH was also observed in the target of osa-miR530-5p (LOC_Os03g55784) that encodes an allene oxide synthase (AOS). Conversely, the targets of the downregulated miRNAs in IR-IR56-BPH, including osa-miR3980a-5p and osa-miR156l-5p were found to show significant upregulated expression levels as compared to the control. These targets include LOC_Os01g69830 and LOC_Os02g36880, encoding a squamosa promoter binding protein (SBP) and a no apical meristem (NAM) protein, respectively. In the IR-TN1-BPH, the targets of the upregulated miRNAs osa-miR2118g and osa-miR435 were found to be significantly downregulated, including LOC_Os08g42700 (encoding a NB-ARC domain containing protein) and LOC_Os07g41730 (encoding an alpha/beta hydrolase domain containing protein), respectively. On the other hand, the targets of the downregulated miRNAs osa-miR2871a-3p and osa-miR172a in IR- TN1-BPH were found to show significant upregulations, including LOC_Os10g13810 (encodes a glycosyltransferase family protein) and LOC_Os05g03040 (encodes an AP2/EREBP family transcription factor), respectively. The negative correlations in between the DE miRNAs and their targets suggest the existence of a well-orchestrated post-transcriptional regulation in response to the BPH infestation in rice. Moreover, the transcript dynamics of different target genes depending upon the nature of rice BPH interactions (IR-IR56-BPH or IR-TN1-BPH) indicate the possible existence of a differential repertoire of defense-related transcription factors/proteins for the compatible or incompatible interactions.
Figure 5.
Expression validation of the selected miRNAs targets by qPCR. The fold changes (log2) in the expression of the target genes were calculated and compared to those of the miRNAs in IR56 rice. Bars represent the mean ± SE of three biological replicates for the qPCR data. Hashtags # and ## represent the significant difference in the expression levels of miRNAs and asterisks * and ** indicate the significant difference in the expression levels of target genes in IR-IR56-BPH or IR-TN1-BPH as compared to control at P < 0.05 and P < 0.01, respectively.
Discussion
Extensive researches on understanding the roles of miRNAs in plant physiology and protection have resulted in the discovery and characterization of more numbers of miRNAs than ever before. Although, many works depicting the role of miRNAs in the plant-aphid interactions have been reported25, only a few reports indicating the role of rice miRNA in insect defense response is available23,26,27. To the best of our knowledge, this is the first report briefing the miRNA dynamics in a resistant rice in response to the infestations of a virulent and an avirulent BPH population. In the current work, the comparative sRNA sequencing of IR56 rice infested separately by the virulent IR56-BPH (IR-IR56-BPH, compatible interaction) and the avirulent TN1-BPH (IR-TN1-BPH, incompatible interaction) revealed the differential involvement of miRNAs during BPH feeding. Besides, comparisons among the sRNA libraries of the BPH-infested and the control (no BPH) revealed that BPH feeding reprogrammed the miRNA expressions in the IR56 rice. A total of 278 DE miRNAs were found in response to the BPH infestations, out of which we have identified 28 mature novel and 218 mature conserved miRNAs. Further, the exclusive nature of the presence of some miRNA families in either IR-IR56-BPH or IR-TN1-BPH samples indicated the interaction-specific nature of those miRNA families in IR56 rice. The exclusive nature of tissue-specific expression of several miRNA families has been reported in a rice cultivar in response to drought stress28. In addition, several miRNA families showed exclusive differential expressions during the interactions between two rice cultivars (Bph15 IL and 9311) and BPH biotype 123. On the contrary, 71 DE miRNAs (18 up and 53 downregulated) were found to be in common in between IR-IR56-BPH and IR-TN1-BPH suggesting that several miRNAs act in a similar pattern in response to BPH feeding irrespective of their virulence levels. The relative transcript abundances of some of the selected DE miRNAs were validated by qPCR analysis, which further strengthened the reliability of the sRNA sequencing data.
In response to the IR56-BPH infestation, osa-miR530-5p and osa-miR812s were found to be highly upregulated in the IR-IR56-BPH samples. The MIR530 family members, including osa-miR530-5p have been reported to be involved in stress responses29–31. The target prediction results revealed that the osa-miR530-5p target LOC_Os03g55784 encodes an AOS that participate in the JA signaling in rice32. JA has been reported to be associated with insect defense responses in rice, and also might act as an early event in rice-BPH interaction24,33. Thus, the elevated transcript abundance of osa-miR530-5p in the IR-IR56-BPH during the feedings of the virulent IR56-BPH might have tampered the JA signaling pathway, a common signaling event to insect attacks in rice. Manipulation of JA signaling in rice in response to BPH infestation has also been reported to be achieved by other miRNA family members, including miR160f-3p, miR166c-5p and miR169r-3p23. In addition to this, the target of osa-miR812s, LOC_Os04g01570, encodes a PEMI protein reported to be involved in plant growth and stress responses34. Although, PEMIs have been reported to play significant roles in the growth and development in rice35,36, several recent reports have revealed their importance in biotic stress tolerance in plants, including Arabidopsis and cotton37,38. Thus, the upregulated expression of osa-miR812s during the compatible rice-BPH interaction, resulting in the downregulation of PEMI proteins in the IR56 rice might have aided in the successful rice resistance breakdown. In addition, in the IR-IR56-BPH plants the elevated transcript abundances of two novel miRNAs, novel_16 and novel_52, were observed that target a serine/threonine (S/T) kinase and a lectin kinase protein, respectively. The roles of S/T kinases and lectin kinases in rice resistance response have already been discovered10,39. Further, the OsLecRK1-4 have been identified to confer broad-spectrum resistance in rice6. Our previous study revealed that in IR56 rice OsLecRK3 and OsLecRK4 displayed induced expressions during the incompatible rice-BPH interactions24. Thus, the downregulation of the S/T kinase or the lectin kinases, possibly by the miRNA-mediated transcript cleavage might have favored a continuous feeding and resistance breakdown in the IR-IR56-BPH plants. Conversely, downregulations of several miRNAs were also observed during the compatible IR-IR56-BPH interactions, including the osa-miR156l-5p. The osa-miR156 is a conserved miRNA having a usual target of the SPB transcription factors40,41. Target mimic-based silencing of the miR156 that targets multiple SBP proteins in rice has resulted in the enhanced resistance response against BPH infestation26. Further, downregulation of OsSPL2 by the overexpression of osa-miR529 had conferred enhanced stress resistance in rice by the elevated expression of superoxide dismutase (SOD) and peroxidase (POD) genes42. However, the lower transcript abundance of osa-miR156l-5p in the IR-IR56-BPH plants resulted in the upregulated expression of OsSPL2 (LOC_Os01g69830), which might have suppressed the POD and SOD transcription, thereby decreasing the reactive oxygen species (ROS)-signaling events. ROS-signaling might act as a basal defense response and an early event in rice-BPH interactions24,43. Thus, these consequences of the upregulated expression of osa-miR156l-5p might have helped IR56-BPH to overcome the rice resistance.
In the TN1-BPH infested IR56 rice (IR-TN1-BPH), the elevated transcript accumulations of osa-miR2118g and osa-miR435 were observed and verified by qPCR. LOC_Os08g42700, encoding a NB-ARC domain containing protein was predicted as a target of osa-miR2118g, whereas LOC_Os07g41730, encoding an α/β-hydrolase was predicted to be the target of osa-miR435. The α/β-hydrolase has been reported to serve as the core structure for phytohormones and ligand receptors, including that of gibberellins (GA)44. GA has been reported to positively regulate rice defense against BPH infestations, as the overexpression of GA receptor OsGID1 enhanced BPH resistance in rice45. But the downregulation of α/β-hydrolase that is associated with the GA pathway might be induced by the TN1-BPH feedings as an attempt to disrupt rice defenses. Further, the NB-ARC or NB-LLR proteins have been extensively studied in plants, including rice, specifically for their roles in defense responses46. Also, the phytopathogen-induced miRNA-mediated suppression of rice defense genes has been reported during the rice blast disease47. Thus, the suppression of the NB-ARC protein in rice mediated by the upregulated expression of osa-miR2118g could be an attempt by the TN1-BPH to outrun the IR56 rice defense. Ouyang and colleagues proposed that due to the transcript abundance of miRNAs that targets the NB-domain genes in tomato, the susceptible cultivars express insufficient resistance proteins48. Besides, as the NB-ARC family forms a vital class of R-genes, interacting with the pathogen/insect effectors, osa-miR2118g might participate in the ETI response to channel the defense response to BPH. On the other hand, more numbers of miRNAs exhibited a reduced transcript abundance in the IR-TN1-BPH plants. For instance, osa-miR2871a-3p, osa-miR172a, osa-miR166a-5p, osa-miR2120, and osa-miR1859 were found to be downregulated many folds as compared to the control. A glycosyltransferase family protein (LOC_Os10g13810) was predicted to be the target of osa-miR2871a-3p has been associated with multiple functions in rice, including growth, development, and stress responses49,50. Two glycosyltransferase genes UGT73B3 and UGT73B5 were identified to be necessary for the pathogen defense in Arabidopsis51. In addition, UDP-glycosyltransferase was reported to facilitate the modifications and storage of secondary metabolites in rice and thereby defending against stress50. Thus, the transcript accumulation of LOC_Os10g13810 in IR-TN1-BPH might have boosted the rice resistance, possibly by regulating the secondary metabolite pool. Dai et al. has reported that by sequestering osa-miR396 the rice defense response against BPH can be enhanced via the increased biosynthesis of flavonoids27. Our previous findings also supported the positive role of secondary metabolites, such as phenylpropanoids in the rice-BPH interactions24. Additionally, upregulation of LOC_Os01g23530 (the predicted target of osa-miR1859) encoding a terpene synthase further supports this hypothesis. Another gene LOC_Os05g03040 (target of osa-miR172a), encoding an AP2/EREBP family transcription factor and participating in starch biosynthesis was found to exhibit upregulated expressions in IR-TN1-BPH plants. It is evident that during a compatible rice-BPH interaction, rapid starch breakdown occurs to produce more sucrose in rice plants as a large amount of sucrose is consumed by the BPH feeding1. On the contrary, during the incompatible rice-BPH interaction the resistant rice plants produce more starch indicating lesser loss of sucrose, showing resistance to the BPH feedings1,52. Thus, our results from this study are in accordance with the previous report, as in the IR-TN1-BPH plants the starch biosynthesis is upregulated, supporting the IR56 rice resistance to the TN1-BPH. In addition to this, downregulation of osa-miR2120 and thus, the upregulation of its target LOC_Os02g32680, encoding a lectin receptor-type protein kinase was observed in the IR-TN1-BPH plants. The upregulation of the lectin receptor kinase are believed to be crucial for the rice defense responses against BPH infestation during an incompatible interactions6,24. Thus, these results further support the incompatible nature of IR56 rice and TN1-BPH interactions and the vital regulatory roles of miRNAs. On the basis of the miRNA results obtained in this study, their target prediction and functional annotations, and the validation of some miRNA and target gene negative correlations, a conceptual model depicting the miRNA regulatory network of the IR56 rice in response to BPH feedings from two different populations of varied virulence levels have been proposed (Fig. 6). The findings from this study added new information about the involvement of miRNAs and their defense modulatory roles in response to BPH feedings on the resistance IR56 rice.
Figure 6.
A conceptual model depicting the miRNA regulatory network of the IR56 rice against the infestation of IR56-BPH and TN1-BPH. AOS: allene oxide synthase; PEMI: pectin methylesterase inhibitor; ERP: ethylene-responsive protein; STK: serene/threonine protein kinase; LPK: lectin protein kinase; SBP: sqamosa promoter-binding protein; NAM: no apical meristem; ET: ethylene; SA: salicylic acid; ETS: effector-triggered susceptibility; GA: gibberellins; NB-ARC: NB-ARC domain containing proteins; GTF: glycosyltransferase; AP2/ERE: AP2/ERE domain containing transcription factor; LecRK: lectin receptor kinase; CtP450: cytochrome P450; Sec.Metabol: secondary metabolites. Arrows indicate a positive correlation, whereas blunt ended lines represent a negative correlation.
Conclusion
In conclusion, the comparative miRNA profiling of the IR56 rice infested separately by the virulent IR56-BPH and the avirulent TN1-BPH revealed the dynamics of miRNA pool. Identification of the DE miRNA indicated that BPH feeding reprogramed the miRNA transcriptions in the IR56 rice. Although, the total numbers of DE miRNAs were found to be similar in IR-IR56-BPH and IR-TN1-BPH, but more numbers of miRNAs were found to be downregulated during the incompatible interactions, suggesting the defense modulatory roles of their targets. Prediction of targets and their functional enrichments by GO and KEGG analysis added more insights to their putative functionality in rice-BPH interactions. Validation of the expression profiles of selected miRNAs and their predicted targets further strengthened their involvements in rice defense against BPH attacks. Finally, based on the findings of this study, a conceptual model depicting the regulatory network of IR56 rice in response to the BPH infestations has been proposed. Furthermore, to the best of our knowledge, this is the first report on the differential rice miRNA roles in a resistance rice in response to BPH infestations of different virulence level. In depth analysis experiments, such as overexpression or selective mutation of a miRNA or the miRNA target MIMIC studies will further validate the functional roles in rice resistance of the identified candidate miRNA in this study. Moreover, the findings from this study will add new insights to the defense regulatory roles of rice miRNA in response to BPH attacks and will help to understand the rice-BPH interactions.
Materials and methods
Ethics statement
All animal work has been conducted according to the relevant national and international guidelines.
Plant material and growth
An indica rice variety ‘IR56’ was used in this experiment as the plant material. In a net house, pre-germinated IR56 rice seeds were planted in mud beds and grown under natural light and temperature conditions. After 14 days old seedlings were then transplanted into mud-filled cups (diameter 12 cm, height 15 cm) and put in the China National Rice Research Institute (CNRRI) greenhouse with following growth conditions: temperature was set to 28 ± 2 °C with an 80 ± 5% relative humidity RH. Ten days post transplantation, the IR56 rice plants were used for conducting the experiments.
Insect materials
Two BPH populations, i.e. TN1-BPH and IR56-BPH of different virulence were used to perform the BPH bioassays in this study24. BPH colonies initially collected from rice fields in Hangzhou, China, were maintained on Taichung Native 1 (TN1) rice (TN1-BPH) or IR56 rice (IR56-BPH) in a climate-controlled chamber (26 ± 2 °C, 80 ± 5% RH) for more than 7 years at the CNRRI24.
BPH bioassays and sample collections
Individual IR56 rice plants were infested with 4 newly emerged adult females BPH, and were confined in a transparent plastic cage (diameter 10 cm, height 60 cm) equipped with a net (with holes of diameter 0.5 mm)24. As per the aforementioned procedures, both TN1-BPH and IR56-BPH populations were infested onto the separate IR56 rice plants. The interactions in between IR56-BPH with IR56 rice (IR-IR56-BPH) was considered as the compatible, whereas the interaction in between TN1-BPH and IR56 rice (IR-TN1-BPH) was considered to be the incompatible rice-BPH interaction24. IR56 rice plants with no BPH treatment put inside a plastic cage served as a control for this experiment. The BPH assay experiments were performed with 3 independent biological replicates. After 1 day of the BPH infestations, the stem portions of the plants (control and treated) were collected and immediately frozen in liquid nitrogen. The replicates of each sample were combined as one and stored at − 80 °C until further use.
Small RNA library construction and sequencing
Total RNA was isolated from each sample (Control, no BPH; IR56-BPH infested, IR-IR56-BPH; TN1-BPH infested, IR-TN1-BPH) by using the TransZol Up reagent (Transgen, Beijing, China) according to the manufacturer’s instructions. The purity, concentration and integrity of isolated RNA were determined by a NanoPhotometer spectrophotometer (IMPLEN, CA, USA), Qubit RNA Assay Kit in Qubit 2.0 Flurometer (Life Technologies, CA, USA), and Agilent 2100 (Agilent Technologies, CA, USA), respectively. The RNA degradation and contamination was monitored on 1% (w/v) agarose gel electrophoresis. Total RNA (> 3 μg) of good quality was used to construct an sRNA library for each sample by using NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, USA.) following manufacturer’s recommendations. To the enriched small RNA pool, 5′ and 3′ adapters were ligated by T4 RNA ligase. Complementary first strand cDNAs were PCR amplified to generate cDNA libraries. Subsequently, the libraries were sequenced (single-end) on an Illumina Hiseq2500 at the Novegene Company (Beijing, China) following the vendor’s recommended protocol. The sequencing data have been submitted to the NCBI’s GEO database.
Data analysis and identification of the differentially expressed miRNAs
After Illumina sequencing, raw data were processed using Novogene’s Perl and Python scripts. Clean data were screened to remove reads containing more than three N (undetermined bases), reads with 5′ adapter contaminants, reads without 3′ adapter or the insert tags, those containing poly A, T, G, or C and low quality reads obtained from the raw data. Then, sRNA sequences of 18–35 nt were selected to conduct all downstream analyses. To prevent every unique sRNA mapping to multiple non-coding RNA (ncRNAs), we used the following priority rule: known miRNA > rRNA > tRNA > snRNA > snoRNA > repeat > gene > novel miRNA so that every unique sRNA mapped to only one annotation. The Bowtie v1.2.353 was used to map the sRNA tags to the indica rice ShuHui498 (R498) genome (https://www.mbkbase.org/R498/)54 without mismatch to analyze their expression and distribution on the reference sequence. Next, the mappable sRNA tags were aligned to the miRNA precursor of O. sativa in the miRNA database (miRbase v. 22.1) to obtain the known miRNA count. Then, rRNAs, tRNAs, snRNAs, and snoRNAs were removed by mapping the remained sRNA tags to Rfam release 1455. Repeat sequences were filtered by using a repeat sequences database56, and tags originating from protein coding genes were discarded by mapping to the exon and intron of mRNAs of O. sativa. Finally, novel miRNAs were predicted by exploring the secondary structure, the Dicer cleavage site and the minimum free energy of the former unannotated sRNA tags which could be mapped to the reference sequence by integrating two available software miREvo57 and mirdeep258.
Since the biological replicates of each samples were combined as one bulk, therefore, when analyzing differentially expressed miRNAs between libraries, we first transformed the raw read count matrix of miRNAs into TPM (transcript per million)59, then used the DEGseq R package60 to analyze the differences. The adjusted P value (q Value) < 0.01 and absolute value of log2 (fold change) > 1 was set as the threshold for significant differential expression by default61. We compared the expression level of miRNAs between IR-IR56-BPH versus control, and IR-TN1-BPH versus control.
Prediction and functional annotations of the miRNA targets
The targets of the identified DE miRNAs in the O. sativa genome were predicted using the TargetFinder software62. To further reveal functions related to the putative target genes, GO (https://geneontology.org/) and KEGG (www.kegg.jp/kegg) enrichment analysis of the predicted target genes was performed using the clusterProfiler R package63.
Validations of the selected DE miRNAs and their targets by qRT-PCR
To validate some of the selected DE miRNAs and their targets, their relative expressions were determined by performing qPCR analysis. From the collected samples, miRNA and total RNA was isolated using the Easy pure miRNA kit (Transgen, Beijing, China) and the TransZol Up reagent (Transgen), respectively, according to the manufacturer’s instructions. First strand cDNA from the miRNA and the total RNA was amplified by using the miRcute Plus miR-first strand cDNA kit (Tiangen, Shenzhen, China) and the Transcript one-step gDNA removal and cDNA synthesis supermix kit (Transgen), respectively. The miRNA-specific forward primers and a universal reverse primer, and gene-specific primer pairs were used to determine the miRNA and target expression analysis, along with the SYBR Green PCR mix (Transgen) on the ABI 7500 real-time PCR system (Applied Biosystems, CA, USA). Three independent biological samples for each reaction, and three technical replicates for each biological sample, were used for the qPCR analysis. The U6 gene was used as the internal reference gene for evaluating the miRNA relative expression levels, whereas the constitutively expressed housekeeping gene OsUbq from was used as an endogenous control for the targets64. The relative expression was evaluated using the 2−ΔΔCt method65.
Statistical analysis
The statistical analyses of the relative expressions were carried out using Data Processing System software66. Data are reported as mean ± SE. Expressions of miRNAs and the targets were analyzed by student’s t-test. The statistical significance level was set for P values < 0.05 or 0.01.
Supplementary information
Acknowledgements
This research was supported by grants from the China Agriculture Research System (CARS-01-35), the National Key Research and Development Program of China, the National Key R&D Program of China (2016YFD0200801), and the Rice Pest Management Research Group of the Agricultural Science and Technology Innovation Program of China Academy of Agricultural Science.
Author contributions
S.N., P.J.W., and Q.F. conceived and designed the research. S.N. performed the research work and analyzed the data. P.J.W., and Q.F. participated in the data analysis. S.Y.Y. helped in rice transplantation and insect maintenance. F.X.L. and W.X.W. established the insect populations. S.N., P.J.W., and Q.F. wrote the manuscript. The manuscript has been read and approved by all authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Qiang Fu, Email: fuqiang@caas.cn.
Pin-Jun Wan, Email: wanpinjun@caas.cn.
Supplementary information
is available for this paper at 10.1038/s41598-020-76198-9.
References
- 1.Cheng X, Zhu L, He G. Towards understanding of molecular interactions between rice and the brown planthopper. Mol. Plant. 2013;6:621–634. doi: 10.1093/mp/sst030. [DOI] [PubMed] [Google Scholar]
- 2.Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS ONE. 2013;8:e66428. doi: 10.1371/journal.pone.0066428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jahn GL, Litsinger JA, Chen Y, Barrion AT, et al. Integrated pest management of rice: ecological concepts. In: Koul O, et al., editors. Ecologically Based Integrated Pest Management. Wallingford: CAB International; 2007. pp. 315–366. [Google Scholar]
- 4.Zhang Y, et al. Identification of major locus Bph35 resistance to brown planthopper in rice. Rice Sci. 2020;27:237–245. doi: 10.1016/j.rsci.2020.04.006. [DOI] [Google Scholar]
- 5.Tamura Y, et al. Map-based cloTning and characterization of a brown planthopper resistance gene BPH26 from Oryza sativa L. ssp. indica cultivar ADR52. Sci. Rep. 2014;4:5872. doi: 10.1038/srep05872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat. Biotechnol. 2014;33:301. doi: 10.1038/nbt.3069. [DOI] [PubMed] [Google Scholar]
- 7.Guo J, et al. Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 2018;50:297–306. doi: 10.1038/s41588-018-0039-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Y, et al. Allelic diversity in an NLR gene BPH9 enables rice to combat planthopper variation. Proc. Natl. Acad. Sci. USA. 2016;113:12850–12855. doi: 10.1073/pnas.1614862113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du B, et al. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. USA. 2009;106:22163–22168. doi: 10.1073/pnas.0912139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng X, et al. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013;76:687–698. doi: 10.1111/tpj.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji H, et al. Map-based cloning and characterization of the BPH18 gene from wild rice conferring resistance to brown planthopper (BPH) insect pest. Sci. Rep. 2016;6:34376. doi: 10.1038/srep34376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, et al. Map-based cloning and characterization of BPH29, a B3 domain-containing recessive gene conferring brown planthopper resistance in rice. J. Exp. Bot. 2015;66:6035–6045. doi: 10.1093/jxb/erv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ren J, et al. Bph32, a novel gene encoding an unknown SCR domain-containing protein, confers resistance against the brown planthopper in rice. Sci. Rep. 2016;6:37645. doi: 10.1038/srep37645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Marker assisted pyramiding of two brown planthopper resistance genes, Bph3 and Bph27 (t), into elite rice cultivars. Rice (New York, N.Y.) 2016;9:27–27. doi: 10.1186/s12284-016-0096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sogawa, K. In Rice Planthoppers: Ecology, Management, Socio Economics and Policy (eds. Heong, K. L. et al.) Ch. Chapter 2, 33–63 (Springer Netherlands, 2015).
- 16.Zheng Y, He J, Wan P, Lai F, Sun Y, Lin J, Fu Q. Virulence characteristics of Nilaparvata lugens(Stål) reared on resistant rice variety IR56. Chin. J. Rice Sci. 2016;30:552–558. doi: 10.16819/j.1001-7216.2016.6016. [DOI] [Google Scholar]
- 17.Baldrich P, San Segundo B. MicroRNAs in rice innate immunity. Rice. 2016;9:6. doi: 10.1186/s12284-016-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staiger D, Korneli C, Lummer M, Navarro L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013;197:394–404. doi: 10.1111/nph.12022. [DOI] [PubMed] [Google Scholar]
- 19.Brant EJ, Budak H. Plant small non-coding RNAs and their roles in biotic stresses. Front. Plant Sci. 2018;9:1038. doi: 10.3389/fpls.2018.01038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pandey SP, Shahi P, Gase K, Baldwin IT. Herbivory-induced changes in the small-RNA transcriptome and phytohormone signaling in Nicotiana attenuata. Proc. Natl. Acad. Sci. 2008;105:4559–4564. doi: 10.1073/pnas.0711363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sattar S, Song Y, Anstead JA, Sunkar R, Thompson GA. Cucumis melo microRNA expression profile during aphid herbivory in a resistant and susceptible interaction. Mol. Plant Microbe Interact. 2012;25:839–848. doi: 10.1094/MPMI-09-11-0252. [DOI] [PubMed] [Google Scholar]
- 22.Jeyaraj A, et al. Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L.) Sci. Rep. 2017;7:13634. doi: 10.1038/s41598-017-13692-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu Y, et al. Identification and analysis of brown planthopper-responsive microRNAs in resistant and susceptible rice plants. Sci. Rep. 2017;7:8712. doi: 10.1038/s41598-017-09143-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nanda S, et al. Differential responses of OsMPKs in IR56 rice to two BPH populations of different virulence levels. Int. J. Mol. Sci. 2018;19:4030. doi: 10.3390/ijms19124030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sattar S, Thompson GA. Small RNA regulators of plant-hemipteran interactions: micromanagers with versatile roles. Front. Plant Sci. 2016;7:1241. doi: 10.3389/fpls.2016.01241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge Y, et al. Silencing of miR156 confers enhanced resistance to brown planthopper in rice. Planta. 2018;248:813–826. doi: 10.1007/s00425-018-2942-6. [DOI] [PubMed] [Google Scholar]
- 27.Dai Z, et al. The OsmiR396–OsGRF8–OsF3H-flavonoid pathway mediates resistance to the brown planthopper in rice (Oryza sativa) Plant Biotechnol. J. 2019 doi: 10.1111/pbi.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Awasthi JP, et al. Identification and characterization of drought responsive miRNAs in a drought tolerant upland rice cultivar KMJ 1–12-3. Plant Physiol. Biochem. 2019;137:62–74. doi: 10.1016/j.plaphy.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 29.Lu S, Sun Y-H, Chiang VL. Stress-responsive microRNAs in Populus. Plant J. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- 30.Xu D, Mou G, Wang K, Zhou G. MicroRNAs responding to southern rice black-streaked dwarf virus infection and their target genes associated with symptom development in rice. Virus Res. 2014;190:60–68. doi: 10.1016/j.virusres.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, Xie K, Xiong L. Global expression profiling of rice microRNAs by one-tube stem-loop reverse transcription quantitative PCR revealed important roles of microRNAs in abiotic stress responses. Mol. Genet. Genom. 2010;284:477–488. doi: 10.1007/s00438-010-0581-0. [DOI] [PubMed] [Google Scholar]
- 32.Tamaoki D, et al. Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal. Behav. 2013;8:e24260. doi: 10.4161/psb.24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J, et al. Induced jasmonate signaling leads to contrasting effects on root damage and herbivore performance. Plant Physiol. 2015;167:1100. doi: 10.1104/pp.114.252700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An SH, et al. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta. 2008;228:61–78. doi: 10.1007/s00425-008-0719-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen HP, Jeong HY, Kim H, Kim YC, Lee C. Molecular and biochemical characterization of rice pectin methylesterase inhibitors (OsPMEIs) Plant Physiol. Biochem. 2016;101:105–112. doi: 10.1016/j.plaphy.2016.01.021. [DOI] [PubMed] [Google Scholar]
- 36.Nguyen HP, Jeong HY, Jeon SH, Kim D, Lee C. Rice pectin methylesterase inhibitor28 (OsPMEI28) encodes a functional PMEI and its overexpression results in a dwarf phenotype through increased pectin methylesterification levels. J. Plant Physiol. 2017;208:17–25. doi: 10.1016/j.jplph.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 37.Lionetti V, et al. Three pectin methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol. 2017;173:1844–1863. doi: 10.1104/pp.16.01185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu N, et al. A pectin methylesterase inhibitor enhances resistance to Verticillium wilt. Plant Physiol. 2018;176:2202–2220. doi: 10.1104/pp.17.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin H-Y, You MK, Jeung JU, Shin JS. OsMPK3 is a TEY-type rice MAPK in Group C and phosphorylates OsbHLH65, a transcription factor binding to the E-box element. Plant Cell Rep. 2014;33:1343–1353. doi: 10.1007/s00299-014-1620-9. [DOI] [PubMed] [Google Scholar]
- 40.Xie K, Wu C, Xiong L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006;142:280–293. doi: 10.1104/pp.106.084475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preston JC, Hileman LC. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 2013;4:80–80. doi: 10.3389/fpls.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue E, et al. Overexpression of miR529a confers enhanced resistance to oxidative stress in rice (Oryza sativa L.) Plant Cell Rep. 2017;36:1171–1182. doi: 10.1007/s00299-017-2146-8. [DOI] [PubMed] [Google Scholar]
- 43.Lu J, et al. An EAR-motif-containing ERF transcription factor affects herbivore-induced signaling, defense and resistance in rice. Plant J. 2011;68:583–596. doi: 10.1111/j.1365-313X.2011.04709.x. [DOI] [PubMed] [Google Scholar]
- 44.Mindrebo JT, Nartey CM, Seto Y, Burkart MD, Noel JP. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016;41:233–246. doi: 10.1016/j.sbi.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 45.Chen L, Cao T, Zhang J, Lou Y. Overexpression of OsGID1 enhances the resistance of rice to the brown planthopper Nilaparvata lugens. Int. J. Mol. Sci. 2018;19:2744. doi: 10.3390/ijms19092744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee H-A, Yeom S-I. Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief. Funct. Genom. 2015;14:233–242. doi: 10.1093/bfgp/elv012. [DOI] [PubMed] [Google Scholar]
- 47.Zhang X, et al. Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 2018;177:352–368. doi: 10.1104/pp.17.01665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ouyang S, et al. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014;10:e1004464. doi: 10.1371/journal.ppat.1004464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yin Y, Chen H, Hahn MG, Mohnen D, Xu Y. Evolution and function of the plant cell wall synthesis-related glycosyltransferase family 8. Plant Physiol. 2010;153:1729–1746. doi: 10.1104/pp.110.154229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ko JH, Kim BG, Hur H-G, Lim Y, Ahn J-H. Molecular cloning, expression and characterization of a glycosyltransferase from rice. Plant Cell Rep. 2006;25:741–746. doi: 10.1007/s00299-006-0119-4. [DOI] [PubMed] [Google Scholar]
- 51.Langlois-Meurinne M, Gachon CMM, Saindrenan P. Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 2005;139:1890–1901. doi: 10.1104/pp.105.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, et al. Responses of two contrasting genotypes of rice to brown planthopper. Mol. Plant Microbe Interact. 2007;21:122–132. doi: 10.1094/MPMI-21-1-0122. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Du H, et al. Sequencing and de novo assembly of a near complete indica rice genome. Nat. Commun. 2017;8:15324. doi: 10.1038/ncomms15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kalvari I, et al. Rfam 13.0: shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2017;46:D335–D342. doi: 10.1093/nar/gkx1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mobile DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wen M, Shen Y, Shi S, Tang T. miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012;13:140–140. doi: 10.1186/1471-2105-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2011;40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou L, et al. Integrated profiling of microRNAs and mRNAs: MicroRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE. 2011;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2009;26:136–138. doi: 10.1093/bioinformatics/btp612%JBioinformatics. [DOI] [PubMed] [Google Scholar]
- 61.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. Ann. Stat. 2003;31:2013–2035. doi: 10.1214/aos/1074290335. [DOI] [Google Scholar]
- 62.Fahlgren N, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li C, et al. Gene expression and plant hormone levels in two contrasting rice genotypes responding to brown planthopper infestation. BMC Plant Biol. 2017;17:57. doi: 10.1186/s12870-017-1005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Tang Q-Y, Zhang C-X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2012;20:254–260. doi: 10.1111/j.1744-7917.2012.01519.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.