Abstract
The deadly pandemic, coronavirus disease 2019 (COVID‐19), caused due to the severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), has paralyzed the world. Although significant methodological advances have been made in the field of viral detection/diagnosis with 251 in vitro diagnostic tests receiving emergency use approval by the US‐FDA, little progress has been made in identifying curative or preventive therapies. This review discusses the current trends and potential future approaches for developing COVID‐19 therapeutics, including repurposed drugs, vaccine candidates, immune‐modulators, convalescent plasma therapy, and antiviral nanoparticles/nanovaccines/combinatorial nanotherapeutics to surmount the pandemic viral strain. Many potent therapeutic candidates emerging via drug‐repurposing could significantly reduce the cost and duration of anti‐COVID‐19 drug development. Gene/protein‐based vaccine candidates that could elicit both humoral and cell‐based immunity would be on the frontlines to prevent the disease. Many emerging nanotechnology‐based interventions will be critical in the fight against the deadly virus by facilitating early detection and enabling target oriented multidrug therapeutics. The therapeutic candidates discussed in this article include remdesivir, dexamethasone, hydroxychloroquine, favilavir, lopinavir/ritonavir, antibody therapeutics like gimsilumab and TJM2, anti‐viral nanoparticles, and nanoparticle‐based DNA and mRNA vaccines.
Keywords: anti‐inflammatory drugs, antivirals, convalescent plasma, COVID‐19, immune therapy, nanomedicines, vaccines
In view of what is known so far about the SARS‐CoV‐2 structure, its pathophysiology, life cycle, and related immunological responses, key steps are envisaged where nanotechnologists and nanotechnology could play a pivotal part in designing advanced biomimetic approaches and engineered nanomaterials having versatile chemical functionalities against COVID‐19 prophylaxis, diagnosis, and treatment.
1. Introduction
The entire world is facing the pandemic coronavirus disease of 2019 (COVID‐19) due to the outbreak of 2019 novel coronavirus (2019‐nCoV; SARS‐CoV‐2) and an international health emergency has been declared. The disease has touched nearly every corner of the world. 2019‐nCoV infection is highly contagious and containment efforts mostly deal with identification and quarantine of exposed and asymptomatic suspects, contact tracing, detection, and strict isolation of infected patients to limit its spread.[ 1 , 2 , 3 , 4 ]
World Health Organization (WHO), on 30th January 2020, declared the disease caused by 2019‐nCoV as the sixth international public health emergency. Due to its rapid escalation across the globe, the WHO declared the 2019‐nCoV outbreak a global pandemic on the 11th of March 2020. So far, i.e., from 31st December 2019 to 21st September 2020, more than 30 949 804 individuals were afflicted by 2019‐nCoV, and 959 116 deaths have occurred worldwide.[ 3 ] On the 11th of February 2020, the WHO officially declared a name for the ongoing 2019‐nCoV related disease pandemic as coronavirus disease 2019 (COVID‐19) whereas, the International Committee on Taxonomy of Viruses (ICTV) has renamed 2019‐nCoV as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) based on its close genetic resemblance with another coronavirus SARS‐CoV.[ 2 , 5 ]
SARS‐CoV‐2 can transfer from an infected person with or without symptoms to another individual, with a high basic reproduction number (R 0) value. Although the initial R 0 was estimated to be between 2.2 and 2.7 for SARS‐CoV‐2, one estimate published in February 2020 indicated that the R 0 value is as high as 4.7 to 6.6, with the infected individual doubling time of 2.4 d.[ 6 , 7 ] The R 0 value indicates that each individual can spread the infection to 4.7–6.6 individuals on average. More recent studies published in August 2020 indicated that COVID‐19 has a higher transmission rate than severe acute respiratory syndrome (SARS) and middle east respiratory syndrome (MERS) with a basic reproduction number of 3.32 and an incubation period of 5.24 d.[ 7 ] Globalization and the convenience of country to country travel had additionally fueled the worldwide spread of the disease.[ 2 , 8 , 9 , 10 ]
This article discusses SARS‐CoV‐2 nanostructure, the virus biology in connection to its epidemiology, clinical manifestations, and potential and future therapeutic options including repurposed drugs, vaccine/protein therapies, immune therapies, and nanotherapeutics. Additionally, the recommended preventive and protective measures are presented (Figure 1 ).
Figure 1.
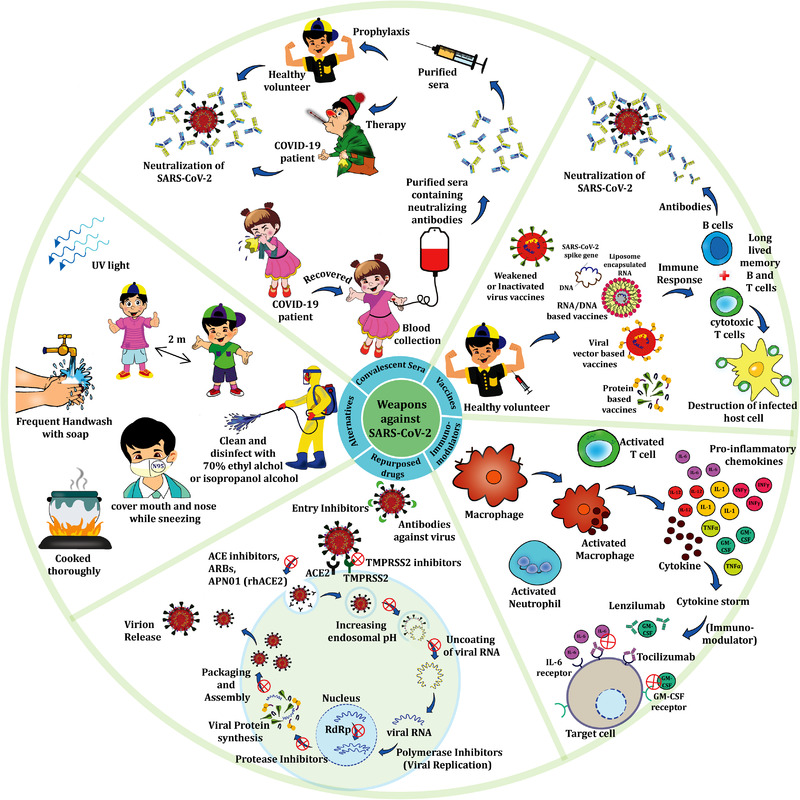
Scheme showing various strategies to tackle COVID‐19 pandemic.
2. SARS‐CoV‐2 and COVID‐19
Coronaviruses (CoVs) are a large family of viruses and are known causative agents for respiratory, hepatic, intestinal, and neurological diseases of altering severity in several animal species. Tyrell and Bynoe in 1966 were the first to cultivate and describe the CoVs from patients having common colds.[ 11 ] CoVs are single‐stranded ribonucleic acid (RNA) viruses. Morphologically, they are spherical virions, having a core of RNA that is being surrounded by a membrane decorated with protein projections (called spike proteins) like a ring/crown (Latin: corona = crown). CoVs are currently classified into four genera, alphacoronaviruses (α‐CoV), betacoronaviruses (β‐CoV), gammacoronaviruses (γ‐CoV), and deltacoronaviruses (δ‐CoV). α‐CoV and β‐CoV originate particularly from bats and other mammals, whereas γ‐CoV and δ‐CoV mostly originate from animals like pigs and birds. The genus β‐CoV is further classified into lineages A to D.[ 1 , 10 , 12 , 13 , 14 , 15 ]
SARS‐CoV‐2 comes under the subgenus sarbecovirus, belongs to lineage B of the genus betacoronavirus (β‐CoV), family coronaviridae, and the order nidovirales.[ 10 ] Before the discovery of SARS‐CoV‐2, other known lineage B β‐CoVs were severe acute respiratory syndrome CoV (SARS‐CoV), SARS‐related to palm civet CoV (SARSr‐CiCoV), and SARS‐belonging to rhinolophus bat CoV HKU3 (SARSr‐Rh‐BatCoV HKU3).[ 16 ]
Coronaviruses (CoVs) that can infect humans are HCoV‐NL63 and HCoV‐229E belonging to α‐CoVs, HCoV‐OC43 and HCoV‐HKU1 belonging to β‐CoVs lineage A, SARS‐CoV belonging to β‐CoVs lineage B, and MERS‐CoV belonging to β‐CoVs lineage C. SARS‐CoV‐2 is the seventh CoV known to cause human infection, second in the lineage B β‐CoVs, and the third in the past three decades to cross interspecies barriers to infect humans (animal‐to‐human transmission).[ 13 , 15 , 16 , 17 ]
Bats are major reservoir of diverse α‐CoVs and β‐CoVs of lineage B, C, and D; but not β‐CoVs lineage A.[ 18 ] The close resemblance of bat CoVs to MERS‐CoV and SARS‐CoV indicates that bats can be the natural reservoir hosts of both the viral pandemics (SARS and MERS). At the same time, there could be other animal based intermediate or middle hosts for the virus facilitating animal‐to‐human transmission, e.g., palm civets and raccoon dogs in the instance of SARS,[ 19 , 20 ] and dromedary camels in the instance of MERS.[ 21 ] However, the evolutionary paths of the virus transferring from bats to intermediary hosts and then to humans have not been fully elucidated.[ 13 ] Among the seven CoVs carrying the ability to infect humans, the β‐CoVs can cause mild to severe illness and higher fatalities, whereas α‐CoVs mostly cause asymptomatic infections or those with mild symptoms.[ 1 , 16 ]
Two immensely pathogenic viruses emanating from animal reservoirs, SARS‐CoV in 2002 affecting 8096 patients globally with 774 deaths[ 22 , 23 ] and MERS‐CoV in 2012 affecting 2519 patient globally (as of January 2020) with 866 deaths, resulted in global pandemics in the last two decades with high mortality rates.[ 22 , 24 ] According to the WHO, SARS‐CoV and MERS‐CoV have mortality rates of 9.6% and 34.3%, respectively.
SARS‐CoV‐2, the pathogen behind the current pandemic, is a positive‐sense, single‐stranded RNA virus (+ssRNA virus) with a lipid‐based envelope. CoVs contain the largest known RNA genome, with SARS‐CoV‐2 genome size being around 30 000 bases long. SARS‐CoV‐2 poses two notable genomic features; first, its receptor‐binding domain (RBD) binds with a very strong affinity to angiotensin‐converting enzyme 2 (ACE2) receptors in humans, cats, and other species carrying inflated receptor homology, second, incorporation of a polybasic cleavage site at S1/S2 junction. S1 and S2 are the two spike protein subunits. These genome‐based features of SARS‐CoV‐2 make them unique among the known β‐CoVs and may explain the reason behind heightened infectivity and transmission ability of the virus in humans.[ 25 , 26 , 27 ]
Viruses with their size in nanometers are nature's own prefabricated smart and capable nanovectors, associated with several diseases and carry a negative connotation. These are natural nanomaterials with very complex structures. They are efficient nanovectors capable of transferring their genetic materials to the infected host cells, followed by hijacking host‐cell machinery to express their proteins.[ 28 ] Many viruses have proven their worth in the field of drug, gene, and vaccine delivery.[ 29 , 30 ] Each SARS‐CoV‐2 virion has an average diameter of 50–150 nm (Figure 2 ).[ 14 , 31 , 32 ] Similar to other CoVs, SARS‐CoV‐2 carries four major structural genes encoding major structural proteins, known as the spike protein (S), an envelope protein (E), the membrane protein (M), and the nucleocapsid protein (N); N protein embraces the genome consisting of ssRNA and the proteins S, E, and M constitute the envelope of the virus (Figure 3 ). The S protein enables the virus to bind to and amalgamate with the host cell membrane. It is the key immunogenic antigen and has a decisive role in defining virulence, tissue tropism, host range, and protective immunity.[ 25 ] The trimeric S protein belongs to a class I fusion protein consisting of S1 (receptor binding) and S2 (membrane fusion) subunits. The subunit S1 consists of a C domain carrying the RBD, and an N domain. S2 contains fusion peptide, the heptad repeat (HR‐1 and HR‐2), transmembrane, and cytoplasmic domains. The locking of the RBD to the ACE2 receptor of the host cell initiates conformational modifications in the S2 subunit that further facilitates fusion among viral and host cell membranes. The S1/S2 juncture is the cleavage site for proteases and is also required to trigger membrane fusion, viral entry, and in the formation of syncytium.[ 16 , 27 ]
Figure 2.

Illustration of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2).
Figure 3.

A scheme showing different SARS‐CoV‐2 proteins.
Other structural proteins, like E, M, and N, are involved in the viral assembly. Alternatively, nonstructural proteins constituting viral cysteine proteases such as papain‐like protease (PL pro), 3‐chymotrypsin like protease (3CL pro), RNA‐dependent RNA polymerase (RdRp), helicase, and other accessory proteins, participate in the viral transcription followed by replication. To counter the host immune response, the M protein and PL pro, antagonize the host interferon (IFN) response and help the virus to control in vivo replication efficacy and pathogenesis.[ 16 ]
Alike SARS‐CoV, SARS‐CoV‐2 ingress into target cells is also aided by the S protein. Viral entry depends on i) the locking of S1 unit of S protein to host cell ACE2 receptors; ii) the S protein priming by host cellular protease TMPRSS2, which facilitates S protein cleavage right at the S1/S2 juncture and the S2’ site, facilitating the merger between viral and cellular membranes.
Following cellular entry, CoVs disassemble to release their genome (nucleocapsid and viral RNA) into the cytoplasm of the infected host cell for initiating the translation of viral polyproteins and subsequent replication of genomic RNA (Figure 4 ).[ 33 ] Replicase polyproteins of SARS‐CoV‐2 are processed by PL pro and 3CL pro to form nonstructural proteins (helicase/RdRp).[ 16 ] Various 3CL pro inhibitors can be expected to repress viral replication by obstructing the cleavage function of the protein. RdRp is a fundamental enzyme of the RNA synthesizing machinery of RNA viruses, and it is highly conserved in all HCoVs. Therefore, it served as a prime target in various viral infections to halt genome replication and can be inhibited by various polymerase inhibitors (favilavir, remdesivir).[ 16 , 33 , 34 ]
Figure 4.
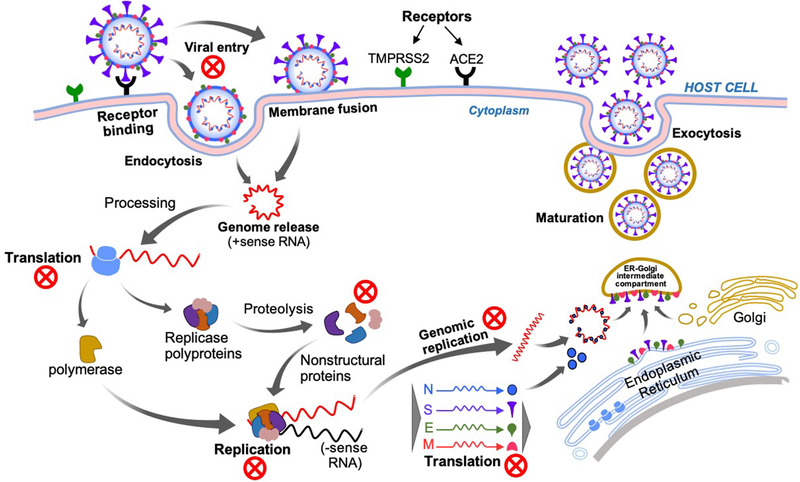
SARS‐CoV‐2 virus and host cell interaction and replication mechanism. The red‐colored cross marks represent potential therapeutic targets.
SARS‐CoV‐2 utilizes the ACE2 receptors to gain entry into the host cells.[ 35 ] ACE2 is a counteractive component of the renin angiotensin system (RAS) that is responsible for maintaining a balance between fluid volume and pressure by utilizing the cleavage products of angiotensin (AGT) and their receptors.[ 36 , 37 , 38 ] Among these peptides, angiotensin II (Ang‐II) causes vasoconstriction and helps in sodium retention by means of AGTR1 receptor and results in vasodilation and natriuresis by binding to the AGTR2 receptor. The enzyme ACE is responsible for producing Ang‐II. After infecting host cells, the virus also uses its protease 3CL pro to suppress NFkappaB by degrading the activating factor IKK‐gamma.[ 39 ] Since, NFkappaB is involved in the induction of ACE by attaching to its promoter and enhancing transcription.[ 40 ] The virus thus reduces the expression of ACE. ACE further downregulates the expression of ACE2 in‐part by Ang‐II, which is its catalytic product.[ 41 ] Thus, the viral infection results in upregulation of ACE2 and downregulation of ACE.
It has been shown that, bradykinin, another important molecule that forms an important component of vasosuppressor system involved in inducing hypotension and vasodilation, is being regulated by ACEs. Bradykinin is degraded by ACE and its concentration is enhanced in presence of angiotensin1‐9 a peptide produced by ACE2. Thus, the enhanced ACE2 expression caused by the virus leads to increased levels of bradykinin causing “bradykinin storm.” This is because SARS‐CoV‐2 increases the amount of bradykinin in the infected tissues. Bradykinin causes vasodilation leading to swelling and inflammation of the tissue and induces pain. It also increases the production of hyaluronic acid. The bradykinin storm induced leakage of fluid and hyaluronic acid induced formation of hydrogel into the lungs may be the reason behind low oxygen uptake in severe COVID‐19 patients and generation of severe symptoms like hypokalemia associated with arrhythmia and sudden cardiac death.[ 42 , 43 , 44 , 45 ] Hence, the drugs which can take care of the bradykinin storm can also be considered as potential therapeutics for COVID‐19.
SARS‐CoV‐2 phylogenetic analysis showed that it is meticulously related to two bat‐derived SARS‐like CoVs (bat‐SL‐CoVZC45 and bat‐SL‐CoVZXC21) found in horseshoe bats (Rhinolophus) with ≈89% nucleotide sequence resemblance with bat‐SL‐CoVZC45. It exhibits ≈79% nucleotide sequence resemblance with SARS‐CoV and ≈50% with MERS‐CoV. Further, a 98.7% nucleotide sequence resemblance to the partial RdRp gene of the bat‐CoV strain BtCoV/4991 was also observed.[ 2 , 10 ]
3. Epidemiology
Gaining in‐depth insight into the SARS‐CoV‐2 virus epidemiology, and characterizing its possible impact, is a pressing need of the current time in order to expand health care measures to tackle the pandemic. The overall impact of a pandemic is governed by the total number of infected persons, transmissibility, and its medical severity (asymptomatic, mild‐to‐severe symptomatic, requiring hospitalization, or fatal).[ 46 ]
COVID‐19 pandemic is continuously evolving, with massive number of cases and deaths each day. Based on data from the initial outbreak and considering worldwide incidence, the trend of an increasing incidence of COVID‐19 principally follows exponential growth in the number of reporting cases.[ 2 , 47 ]
The present mean incubation time for COVID‐19 is 5.5 d, and the median incubation time is 5.1 d, with the potential asymptomatic transmission. Following SARS‐CoV‐2 infection, most patients (≈97.5%) develop symptoms within 11.5 d and almost every patient shows symptoms within 14 d. Very few (≈2.5%) SARS‐CoV‐2 infected patients develop symptoms within 2.2 d.[ 1 , 2 , 31 , 48 ]
As of 21st September 2020, 216 countries/areas/territories/regions have reported confirmed cases of the disease. USA has reported the largest number of confirmed COVID‐19 patients (6 703 698), followed by India (5 487 580), Brazil (4 528 240), Russian Federation (1 109 595), Peru (762 865), Colombia (758 398), Mexico (694 121), South Africa (661 211), Spain (640 040), Argentina (622 934), Chile (446 274), and France (432 424) surpassing China's total number of COVID‐19 patients (90 876). This highlights how rapidly the virus can spread throughout the world. As of 21st September 2020, a total of 959 116 mortalities have been confirmed globally, showing 3.09% mortality rate, with 530 373 deaths in the American region, the highest being reported from USA (198 094) and Brazil (136 532).[ 49 ]
SARS‐CoV‐2 can affect all age groups especially the higher risk groups which include: i) children <59 months, ii) pregnant women, iii) elderly, iv) people with chronic medical ailments (cardiac, pulmonary, hepatic, renal, metabolic, hematologic, neurodevelopmental diseases, or weak immune system), v) people with immunosuppressive conditions (HIV/AIDS, under chemotherapy or steroids), and vi) health care professionals.[ 4 , 47 , 50 ] Lower risk groups should self‐isolate themselves at home, drink plenty of fluids, and follow general good‐health guidelines to keep their immune system strong and healthy.
4. Clinical Manifestations
SARS‐CoV‐2 infection is highly contagious, and containment efforts mostly emphasize on quarantine of exposed and asymptomatic suspects and strict isolation of infected patients during the incubation period to limit the disease spread. Preliminary clinical feature of the SARS‐CoV‐2 infection in humans is pneumonia, which formed the basis of identification, detection as well as isolation of patients. SARS‐CoV‐2 infection manifests varied clinical features, extending from asymptomatic to a condition with acute respiratory distress syndrome (ARDS).[ 51 ]
In symptomatic patients so far, the clinical manifestations consist of fever >39.1 °C (most common symptom), dry cough, nasal congestion, sore throat, dyspnea, headache, myalgia, fatigue, upper respiratory tract infections, smell/taste dysfunctions, and diarrhea. Very few patients are reported with rhinorrhea. Pneumonia mostly occurs by 2–3 weeks following the initial infection with prominent signs of hypoxemia and deviations in blood gas.[ 1 , , 2 , 47 , 50 , 52 ]
Visible changes observed in chest X‐rays and computed tomography demonstrating deterioration in lung tissue are observed to be the ground glass irregularities of the disease. Pulmonary consolidation, alveolar exudates, and interlobular involvements have also been observed. Patients also show low white blood cell count (leukopenia), and elevation in inflammatory markers (C‐reactive proteins, proinflammatory cytokines, etc.) and formation of blood clots in some cases. COVID‐19 patients seeking intensive care unit (ICU) are particularly older and more likely to carry pre‐existing comorbid conditions like hypertension and related heart diseases followed by diabetes. Although, certain epidemiological features were identified, additional studies are still warranted.[ 1 , 2 , 46 ]
Recent reports have also revealed the existence of asymptomatic infections and gastrointestinal symptoms, specifically among youngsters.[ 53 ] The occurrence of asymptomatic individuals with the ability to spread the infection may be a great hurdle in the control and containment of the disease and are posing significant public health threats. However, detailed studies on the extent of asymptomatic persons, their viral loads, and share in transmission need further evaluation. This allows a better understanding of the viral pathogenesis and will aid policy makers to develop scientifically sound guidelines.[ 8 ]
5. Strategies for Infection Control and Prevention
Currently, there is no specific approved therapeutic agent available to treat SARS‐CoV‐2. Treatment is supportive and based on the patient's medical condition to alleviate symptoms. Proper public health measures to check the viral infections are of an urgent need to tackle the growing pandemic. Additionally, stringent measures must be taken to check the human‐to‐human transmission to minimize secondary infections and prevent community transmission among near contacts, and health care professionals, and to prevent further spread.
Based on earlier knowledge of the MERS and SARS outbreak, the WHO recommends adaptation of various infection control measures to lessen the overall risk of the disease transmission and prevent overall spread. This includes avoiding close proximity with people showing symptoms of acute respiratory infections, washing hands with soap frequently (minimum for a span of 20 s), using 60–95% alcohol‐based hand sanitizers after contacting infected people, and avoiding contact with infected inanimate surfaces and pet/wild animals without having proper protection.[ 54 ]
The practice of cough etiquettes by people suffering from acute respiratory tract infection, maintaining distance from these patients, proper covering of cough or sneezes, and washing hands frequently are few other disease preventive and control measures being recommended. The WHO further recommends proper and consistent adoption of environmental disinfection and cleaning methods like cleaning of supposedly infected surfaces with water, soaps, disinfectants, and detergents. People are recommended to stay at home, avoiding social contact (stay home stay safe). While the governments have imposed lockdown measures, travel bans, and ban on people gathering in parks and public places. People should leave their homes putting facemasks and only for essential/basic commodities or medical needs. People should avoid unnecessary journeys and travel to or from work only when they cannot work from home. People going outside should maintain a social distancing of more than 2 m apart from anyone other than members of their own family.
A person showing SARS‐CoV‐2 symptoms (fever of ≥37.8 °C, a persistent cough or breathing problems) should take extra precautions and self‐isolate (quarantine) himself/herself for 10–14 d including his family members.[ 55 ]
6. Potential Therapeutics/Vaccines in Pipeline for COVID‐19
Although, significant methodological advances have been made in the field of viral detection of COVID‐19 with many companies receiving emergency use approvals for their in vitro diagnostic tests, molecular‐based high complexity laboratory tests, or antibody tests as mentioned by the US‐FDA website (a total of 251 as of September 21, 2020), little progress has been made in identifying curative or preventive therapies.[ 56 ] Thus, there is a definite need to find therapeutic agents directly targeting SARS‐CoV‐2, and several scientists around the world are focusing on the development of fruitful prevention/treatment strategies and finding new drugs/antivirals/vaccines against SARS‐CoV‐2, to halt the ongoing outbreak. The US‐FDA website lists dozens (a total of 100 companies as of September 21, 2020) of companies involved in developing antiviral or vaccine therapies for COVID‐19.[ 57 ]
Antivirals can be broadly categorized into two classes, i.e., 1) virus targeting antivirals, which target the viral life cycle, machinery and pathways or directly inactivate viral structural proteins; and 2) host targeting antivirals, which either target the host cellular machinery important for viral infection or target the host's immune response pathways and cascades elicited toward the viral infection.[ 12 , 58 ] In the course of viral infection, various events in the viral life cycle and virus‐host protein–protein interactions have been identified as the potential targets for antivirals.[ 59 ] Cellular events such as virion adsorption, intracellular transport, uncoating, genome and protein synthesis, and assembly inside infected host cells play a decisive role in the viral pathogenesis and targeting these can be a good strategy in the development of current therapies for tackling viral infections. The existence of a potentially small number of viral targets and fast mutating genes sometimes make the business of finding and developing novel and potent antivirals a challenging task. Viruses offer limited intrinsic targets for engineering antivirals.[ 60 ] As viruses are greatly dependent on the host cellular machinery for replication,[ 61 ] targeting various host factor(s)[ 62 ] and molecular pathways hijacked by the virus opens a pandora of opportunities to construct novel antivirals.[ 12 , 62 , 63 ]
Potential therapeutic targets in SARS‐CoV‐2: Mostly various genes and their encoded proteins of SARS‐CoV‐2 can act as favorable diagnostic, therapeutic, or vaccine targets for the virus (Figures 4 and 5 ).
Figure 5.

The replication of SARS‐CoV‐2 inside lower respiratory tract cells and potential target sites for antivirals.
Mechanisms such as inhibition of viral enzymes (DNA and RNA polymerases, 3CL pro, TMPRSS2, reverse transcriptase, neuraminidase, endonucleases, and other proteases) or processes such as ACE2 cellular receptor inhibitors and endosomal acidification mediators prohibiting viral fusion; molecules interfering with glycosylation of the viral protein, viral assembly, new viral particle transport, and release, and immunomodulation of cytokine release can be potential targets in developing various antiviral drugs for the SARS‐CoV‐2.[ 64 , 65 , 66 ]
TMPRSS2 inhibitors (camostat, nafamostat) and furin inhibitors can abrogate the S1/S2 proteolytic cleavage, thereby blocking the viral entry.[ 35 ] In addition, the viral entry can be curbed by using carbohydrate‐binding protein inhibitors such as griffithsin, which bind to the spike glycoprotein.[ 67 ] Endosomal cell entry and S protein activation inside endosomes depend on the pH‐dependent endosomal protease cathepsins and might be clogged using lysosomotropic agents like ammonium chloride, chloroquine, and cathepsin inhibitors.[ 68 ] Antibodies against the RBD and other viral proteins can effectively inhibit virus entry into the host cell. The enzyme 3CL pro, one of the essential proteases of the SARS‐CoV life cycle, plays a crucial role in the proteolysis of various viral polyproteins, controlling viral replication. The 3CL pro enzyme is considered as a key drug target in the case of SARS‐CoV and MERS.[ 69 ] Recent studies revealed that SARS‐CoV‐2 3CL pro is conserved and carries 99% nucleotide sequence identity with SARS‐CoV 3CL pro. Hence, 3CL pro inhibitors that impede the cleaving ability of 3CL pro might repress virus replication, rendering this enzyme an attractive therapeutic target against COVID‐19.[ 69 , 70 ]
However, most of the potential drugs/vaccines in the spotlight as potential COVID‐19 therapeutics are only experimental candidates, and vigorous clinical studies are warranted to verify their efficacy and safety. The antivirals being investigated may be toxic and many of them can merely alleviate certain conditions.
To date, apart from the emergency use approval of the antiviral drug favilavir in China, India, Russia, and parts of the Middle East and the emergency use approval of remdesivir by the US‐FDA and Japan in COVID‐19 patients, there are no approved therapeutic molecules to treat the COVID‐19 pandemic.[ 56 , 71 ]
Thus far, therapeutic modalities including various western,[ 72 ] natural product‐based (NCT04382040), and traditional Chinese medicines[ 73 , 74 ] with some potential activity against COVID‐19 have been briskly tested clinically, and they exhibited preliminary efficacy against COVID‐19.[ 65 ]
6.1. Repurposed Drugs for SARS‐CoV‐2
Drug repurposing (repositioning, or retasking) expedites drug product development by identifying new uses for existing approved or experimental drugs in the current global crisis. This strategy could significantly reduce the cost and duration of drug development compared to discovering and developing entirely new therapeutics.[ 75 ]
The following are the lists of various drugs/therapeutics under consideration and testing to be repositioned against COVID‐19.
6.1.1. Favilavir
Favilavir (also known as favipiravir, T‐705) a guanosine nucleotide analogue, is an antiviral with broad‐spectrum activity. It is a pyrazine carboxamide derivative and is currently being marketed in China and Japan as Avigan for treating influenza. It became the first antiviral drug to gain approval from the National Medical Products Administration of China for clinical trials in treating SARS‐CoV‐2. Favilavir is highly effective in treating RNA virus infections by inhibiting the RdRp. Favilavir, originally was formulated to battle catarrhal (inflammation in nose and throat), has shown efficacy in clinical trials carried out in COVID‐19 patients. ChiCTR2000029600 and ChiCTR2000029544 are ongoing clinical studies evaluating the safety profile and efficacy of favilavir. It has been approved in Italy by Italian Medicines Agency (AIFA) for experimental use against COVID‐19 and clinical trials are underway (NCT04336904). It is being administered to moderate COVID‐19 patients at 1800 mg twice a day on the first day, and thereafter 600 mg thrice a day up to 14 d; however, the dose may vary based on indications. Favilavir is one of the potential treatment options currently being tried for possible use in the treatment of patients suffering from COVID‐19. Despite its potential effectiveness and mass production in China, favilavir is not yet approved as a drug product for COVID‐19. NCT04310228 is another ongoing clinical trial assessing the usefulness and safety of favilavir in combination with tocilizumab. Vero E6 cells infected with nCoV2019BetaCoV/Wuhan/WIV04/2019 exhibited a half‐maximal effective concentration value (EC50) of 61.9 × 10−6 m against favilavir.[ 72 ]
6.1.2. Remdesivir
Remdesivir has been labeled as the most promising antiviral by the WHO for the ongoing SARS‐CoV‐2 caused COVID‐19 pandemic.[ 76 ] Remdesivir (development code GS‐5734), an adenosine nucleotide analogue, is a broad‐spectrum antiviral drug. A study reported the EC50 value of remdesivir against Vero E6 cells infected with nCoV2019BetaCoV/Wuhan/WIV04/2019 to be around 0.77 × 10−6 m and also showed its antiviral effect against 2019‐nCoV infected human liver cancer Huh‐7 cell line.[ 72 ] It is a monophosphoramidate prodrug and is metabolized to the active form, GS‐441524, which disguises and gets incorporated in new RNA strand by viral RNA polymerase, thereby escapes proofreading by viral exonuclease, halting genome replication.[ 77 , 78 , 79 ]
Originally, remdesivir was developed and synthesized to battle Ebola and was reported to have treated an American COVID‐19 patient, who has now fully recovered after receiving the drug.[ 80 ] However, more clinical data are required before the drug can be approved and is considered as an effective and official drug to treat either SARS‐CoV‐2 or Ebola virus. Warren and group reported the EC50 of remdesivir against Ebola virus infected macrophages, human endothelial and liver cells to be around 0.01 × 10−6 to 0.20 × 10−6 m, and demonstrated its therapeutic efficacy in an Ebola virus infected monkey model of the disease.[ 81 ]
Initially China planned Phase III clinical studies to estimate the safety profile and efficacy of remdesivir in COVID‐19 patients based on its promising preclinical data in SARS‐CoV and MERS‐CoV infections,[ 82 , 83 ] evaluating remdesivir in severe COVID‐19 (NCT04257656) and in mild/moderate COVID‐19 patients (NCT04252664). However, these trials were either terminated or suspended due to the unavailabilty of eligible COVID‐19 patients.
Similarly, in February 2020, the U.S. National Institute of Allergy and Infectious Diseases (NIAID) carried out a Phase III adaptive COVID‐19 treatment trial (ACTT) globally to assess the efficacy of various investigational molecules compared to the control arm (NCT04280705). Preliminary results from the trial indicated that remdesivir was superior as compared to placebo in shortening the recovery time in hospitalized COVID‐19 patients.[ 84 ]
Another multicentric trial for remdesivir compared the drug with the standard of care treatment, which includes supplementary oxygen and ventilator support when indicated, in severe COVID‐19 (NCT04292899) patients and patients with moderate COVID‐19 (NCT04292730). In the trial, along with the standard care, a 200 mg loading dose of remdesivir was administered on day 1, followed by 100 mg dose administered as intravenous (IV) infusions every 24 h for 2–5 or 2–10 d.[ 85 ] Results of the trials indicated no significant difference in clinical status of COVID‐19 patients compared to the standard of care.[ 86 , 87 ]
6.1.3. Chloroquine
Other drugs like chloroquine or hydroxychloroquine are also under consideration in the global hunt to discover an effective COVID‐19 therapy[ 88 ] after their approval for limited and emergency use for COVID‐19 by the US‐FDA.
Chloroquine a “4‐aminoquinoline” is an old drug used in the prevention and therapy of malaria. It is further prescribed in systemic lupus erythematosus (SLE) and rheumatoid arthritis (RA) due to its anti‐inflammatory activity. In preliminary studies, the drug has shown efficacy and tolerable safety profile against COVID‐19 associated pneumonia.[ 89 ]
Wang and group reported effective in vitro inhibition of the virus, using chloroquine. Vero E6 cells infected with nCoV2019BetaCoV/Wuhan/WIV04/2019 and treated with chloroquine exhibited an EC50 of 1.13 × 10−6 m.[ 72 ]
Chloroquine also acts as zinc ionophore, and allows passage of extracellular zinc to the cytoplasm and inhibits viral RdRp.[ 90 , 91 , 92 ] It also interferes with viral entry by inhibiting host receptor glycosylation. Administered to a patient orally at a dose of 500 mg in 12 to 24 h for 5 to 10 d.[ 85 ] Hydroxychloroquine, a chloroquine derivative carrying a similar mechanism of action as well as therapeutic activity as chloroquine[ 93 ] but with minimal adverse effects, has also been evaluated as an anti‐COVID‐19 therapeutic.[ 94 ]
In a study by Yao and group, hydroxychloroquine inhibited SARS‐CoV‐2 infected Vero cells in vitro after 48 h of growth with a lower EC50 value (0.72 × 10−6 m) as compared to chloroquine (5.47 × 10−6 m).[ 93 ] Multicentric clinical trials in China have also revealed that the drug shows a potent broad‐spectrum antiviral effect. The mechanism of action may be increased endosomal pH, thereby interfering with the fusion process of the virus with the cell membrane. Thus, the virus is incapable of releasing its genetic payload inside the cell and replicate further.
Unlike the US‐FDA, European regulators restricted the general use of chloroquine for COVID‐19 without significant data and limited their use to clinical trials only. Therefore, clinical research at a large‐scale is still looked‐for to elucidate its mode of action and potential prophylactic/therapeutic efficacy against COVID‐19.
A study (ChiCTR2000029559) in China has tested the drug in 62 COVID‐19 patients showing mild to moderate symptoms. All the patients received treatments like antiviral and antibacterial drugs, oxygen therapy, immunoglobulin as standard care, whereas only half of them received hydroxychloroquine (400 mg per day) together with standard care up to 5 d. Clinical symptoms like the rate of recovery of the body temperature, the time required for cough remission were significantly shortened in patients receiving hydroxychloroquine. Larger percentage of patients (80.6%) with improved pneumonia symptoms in the treatment group compared to the control was reported (54.8%).[ 95 ]
Another Phase II clinical study (NCT04335084) testing the efficacy of hydroxychloroquine in combination with vitamin C, D, and zinc toward the prevention of COVID‐19 infection is underway. A report by Dr. Raoult from France further demonstrated significant improvements in COVID‐19 patients administered with hydroxychloroquine at a dose of 600 mg kg‐1 for 6 d along with azithromycin.[ 88 ]
However, WHO has pulled out the drugs from their Solidarity trial due to lack of efficacy in treating COVID‐19.[ 96 ]
6.1.4. NP‐120
NP‐120 (Ifenprodil—brand name Cerocal), a potential therapeutic option for idiopathic pulmonary fibrosis (IPF), acute lung injury (ALI), and persistent coughs may be repurposed for COVID‐19. NP‐120 is N‐methyl‐d‐aspartate (NDMA) receptor glutamate receptor antagonist, which targets the NMDA‐type subunit 2B (Glu2NB).[ 97 ] Ifenprodil is a vasodilator, originally developed by Sanofi as an oral medication to treat blood circulation disorders in French and Japanese markets. It is no longer sold in France but is still marketed in Japan. Ifenprodil is an approved drug in countries like Japan and South Korea to treat certain neurological conditions. An independent animal study showing a considerable reduction in ALI and enhanced survivability in Avian H5N1 infected mice encourages researchers to expand clinical programs to ALI and ARDS associated with COVID‐19 infection.[ 97 ] An adaptive Phase IIb/III study assessing the safety and efficacy of ifenprodil (20/40 mg three times a day) together with standard of care in comparison to standard of care alone in the treatment of hospitalized COVID‐19 patients is underway (NCT04382924). Ifenprodil is already being tested in clinical trials in patients suffering from IPF and its associated cough (NCT04318704).
6.1.5. Lopinavir/Ritonavir and Others
A repurposed drug combination treatment against SARS‐CoV‐2, lopinavir and ritonavir (protease inhibitor combination; Kaletra), is currently used as both first‐ and second‐line antiretroviral medication for HIV. Lopinavir is given in conjunction with ritonavir to increase its half‐life.[ 98 , 99 ]
Several clinical trials of lopinavir/ritonavir, either alone or with various combinations, are underway. China launched a controlled trial (ChiCTR2000029308) to test the efficacy of lopinavir/ritonavir and IFNα‐2b combination in patients hospitalized with COVID‐19. The most commonly studied dosing regimen of lopinavir/ritonavir for COVID‐19 infection is 200 mg/50 mg orally twice daily for 14 d.[ 98 ]
Some other antiretrovirals were also screened for anti‐SARS‐CoV‐2 activity.[ 85 ] A randomized controlled Phase III trial (NCT04252274) assessing the efficacy and safety of darunavir/cobicistat combination (PREZCOBIX) for treatment of COVID‐19 is underway. However, due to the lack of efficacy in hospitalized COVID‐19 patients, WHO withdrew lopinavir/ritonavir from their Solidarity trial.[ 96 ]
6.1.6. Viral Entry Protein Inhibitors
Hoffmann and co‐workers showed that SARS‐CoV‐2 infection relies on the host cellular factors, such as ACE2 and TMPRSS2, and could be successfully blocked by using protease inhibitors.[ 35 ] ACE2 enzyme is a protein recognized by various CoVs (SARS‐CoV and SARS‐CoV‐2) to gain cell entry.[ 27 , 100 ] ACE2 receptor is expressed by the epithelial cells covering organs like lung, intestine, and blood vessels.[ 101 ] In addition to ACE2 as the entry receptor, the virus also requires cellular proteases for the priming of S protein to enter the host cells. TMPRSS2 is a serine protease employed by CoVs for S protein priming.[ 35 ] Therefore, it is anticipated that the viral entry inside host cells can be obstructed by serine protease TMPRSS2 inhibitors.
The marketed TMPRSS2 inhibitors, nafamostat and camostat have been demonstrated to be effective in blocking the SARS‐CoV‐2 cellular entry.[ 102 ] A Phase IIa trial studying the impact of camostat mesilate (Foipan) on 180 COVID‐19 patients was initiated during March 2020 (NCT04321096). Nafamostat exhibits inhibitory effect against nCoV2019BetaCoV/Wuhan/WIV04/2019 infected Vero E6 cells with an EC50 value of 22.5 × 10−6 m.[ 72 ]
6.1.7. Leronlimab
Leronlimab (PRO 140) is used to treat breast cancer and HIV. Leronlimab is a humanized monoclonal antibody (MAb) capable of blocking CCR5 (CCR5 antagonist). It is in clinical trials for mild to moderate COVID‐19 patients (NCT04343651) and it is expected to benefit patients showing respiratory complications by diminishing the cytokine storm.
Currently, leronlimab has received fast track approval by the US‐FDA as a combination therapy with antiretroviral therapy for HIV and triple‐negative metastatic breast cancer and has completed nine clinical trials, including a Phase III trial in HIV‐infected patients.[ 103 ]
6.1.8. Umifenovir
Another promising repurposed antiviral drug is umifenovir, also known as Arbidol which inhibits the virion membrane fusion to host cell by targeting the interaction between S protein/ACE2 receptors. Arbidol is currently approved in China and Russia as prophylactic and treatment drug against influenza. In vitro cellular models showed antiviral activity against SARS and therefore it is anticipated to have promising results against SARS‐CoV‐2. The most studied dosing regimen of umifenovir for COVID‐19 infection is 200 mg orally every 8 h. Ongoing clinical trials are further evaluating the efficacy of umifenovir.[ 85 ]
A randomized, placebo‐controlled, Phase IV clinical trial assessing the safety and efficacy of umifenovir as an adjuvant therapy to the combined therapeutic regimen of IFNβ1a, lopinavir/ritonavir and hydroxychloroquine in moderate to severe COVID‐19 patients (NCT04350684) is underway.
6.2. Vaccines/Protein Therapeutics for SARS‐CoV‐2 Prophylaxis
Our immune system relies on two vital pillars: 1) the innate/general immunity; and 2) the adaptive/specialized immunity. Recent literature suggests that innate immunity can also influence the nature of adaptive responses apart from protecting the host against viruses during early infection when the initial adaptive immune responses take effect.[ 104 ] The synchronized activities of innate and adaptive immunity are vital for gaining overall protection against viruses.
Innate immunity gives early reactions by enabling the detection and destruction of pathogens quickly within hours and consists of the skin and mucous membranes in the body openings forming external barriers, phagocytic cells, natural killer (NK) cells, various substances in the blood and body fluids.[ 105 ] Adaptive immunity takes more time in the detection and destruction of pathogens, but it targets the pathogen more precisely and can patrol the body against antigens for months/years by producing memory cells. Adaptive immunity consists of T cells, B cells, antibodies, and cytokines as soluble proteins in the blood and tissue.[ 105 ]
Upon virus entry inside host cells, here most specifically the airway epithelial cells for SARS‐CoV‐2, two types of immune responses are being observed (Figure 6 ). One is the positive and healthy immune response and the other is the negative and defective immune response.
Figure 6.

The immune responses generated against SARS‐CoV‐2. Mainly two types of immune responses are generated in the host against the virus. One is the normal or positive immune response, which leads to virus neutralization and ceasing of the disease progression and the other one is the abnormal or aggressive immune response that gives rise to disease associated complications like the ARDS during a severe COVID‐19 infection.
In the case of RNA viruses, after the virus entry and replication and further release inside the host cells, the infection is detected by a set of pattern/pathogen recognition receptors (PRRs) of innate immune system comprising first‐line of defense against viral infection. Inside the cell, PRRs like toll‐like receptors (TLRs) and retinoic acid‐inducible gene I (RIG‐I)‐like receptors (RLRs) sense the viral ss/dsRNA genome and its replication intermediates. Upon cellular entry, the virus is recognized by the endosomal ssRNA sensor (TLR7/8), the cytosolic dsRNA sensor (RIG‐I, MDA‐5), and the cytosolic inflammasome sensor (NLRP3). Subsequently, these different sensors recruit adaptor proteins including myeloid differentiation primary response 88 (MyD88) and mitochondrial antiviral signaling protein (MAVS) respectively to further activate downstream signaling pathways. This causes activation of the transcription factors like nuclear factor kappa B (NF‐κB) and IFN regulatory factors (i.e., IRF3 and IRF7), and subsequently triggers the production of type I/III interferons (IFN‐α and IFN‐β) and proinflammatory cytokines like interleukin 1 beta (IL‐1β), IL‐6, and tumor necrosis factor alpha (TNF‐α), respectively.[ 106 ]
The antiviral activity of IFN‐α and IFN‐β is further amplified by the expression of IFN stimulated genes (ISGs) such as ribonuclease L (RNAse L) and the proinflammatory chemokine (CXCL10) and is vital in limiting the spread and replication of the virus and modulating the innate/adaptive immune responses. Cytokines released by infected cells further modulate the adaptive immune response by recruiting and activating immune cells in eliminating the virus. This comprises the positive immune response, wherein the infection attracts T cells specific to the virus, and destroys the virus checking the infection. Neutralizing antibodies are also produced against the viruses, which block the viral infection, and alveolar macrophages further clear the viruses by phagocytosis.[ 107 ]
The virus additionally activates the inflammasome sensor, NLRP3, that lead to the secretion of highly inflammatory cytokine IL‐1β and the initiation of pyroptosis (highly inflammatory form of programmed cell death). This comprises the negative immune response, wherein the host cells undergo pyroptosis, elicited against cytopathic viruses, in response to the viral release. Then pathogen‐associated molecular patterns (PAMPs) like viral m‐RNA and damage‐associated molecular patterns (DAMPs) like ATP, nucleic acids, and ASC oligomers are released from the host cells in reaction to the virus. These molecular patterns are then recognized by other neighboring cells, which include macrophages, epithelial and endothelial cells causing them to release various chemokines and proinflammatory cytokines like IFN gamma‐induced protein 10 (IP‐10), macrophage inflammatory protein 1α (MIP‐1α), MIP‐1β, and MIP‐1γ, and IL‐10. These chemokines and cytokines attract immune cells like macrophages, monocytes, and T‐cells to the infection site leading to further inflammation and additional production of IFNγ by T cells. In a defective response, higher accumulation of immune cells causes overproduction of the proinflammatory cytokines causing a cytokine storm that damages the organ being infected, and then it circulates to other organs causing multiorgan damage. It has also been observed that non‐neutralizing antibodies produced against the virus by the activated B cells further enhance the damage by SARS‐CoV‐2 infection by a phenomenon called antibody‐dependent enhancement.[ 108 ]
Downregulation of the host IFN response can cause an unbalanced immune response producing high levels of proinflammatory cytokines and infiltration of inflammatory cells leading to hyperinflammation causing more severe clinical symptoms of COVID‐19. CoVs have evolved many mechanisms to stop type I IFN induction and signaling. Severe COVID‐19 patients demonstrated unusually impaired type I IFN signatures in comparison to mild or moderate cases.[ 107 ]
Therefore the category of host immune response generated against the virus (Figure 7 ) defines the disease severity in the SARS‐CoV‐2 infection, and aggressive immune response against the virus causes significant damage to the lung airways.[ 109 ] So, the adoption of immune therapeutic strategy against the virus could be either based on fortifying the positive immune response or minimizing the dysfunction immune response by means of immune‐suppressive therapies. The process of vaccine development against the virus should be dealt with caution. Following are some of the list of therapeutic molecules/drug candidates that are meant to target the immune response generated against the virus and under consideration in the present scenario.
Figure 7.
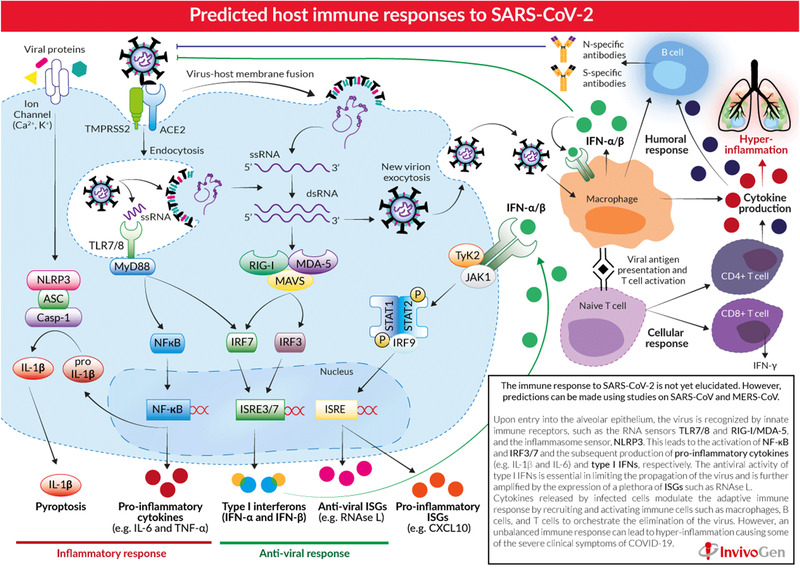
Predicted host immune responses to SARS‐CoV‐2. Reproduced with permission.[ 107 ] Copyright 2020, InvivoGen.
6.2.1. Fusogenix DNA Vaccine
One promising vaccine candidate for COVID‐19 is the Fusogenix DNA vaccine (Covigenix). Fusogenix is a proteo‐lipid vehicle (PLV), formulated with well‐tolerated neutral lipids and Entos proprietary fusion‐associated small trans‐membrane proteins (FAST proteins), which have a novel fusion mechanism to deliver therapeutic nucleic acid payloads directly into target cells, intact and unmodified.
Fusogenix DNA vaccine utilizes plasmid DNA, which encodes several protein epitopes (antigen) from key immunogenic SARS‐CoV‐2 proteins to develop an advanced therapeutic payload with maximum protection. These protein epitopes will help activate the body's inherent antibody production and elicitation of a protective immune response against COVID‐19. Further, in comparison to traditional vaccines, the DNA‐based vaccine avoids the use of the infectious agent, shows enhanced stability, easy large‐scale production, and is expected to stimulate both B‐ and T‐cell responses.
In preclinical in vivo studies, Covigenix showed high immunogenicity, efficacy, and safety. In Phase I/II human clinical trials, Covigenix will be further evaluated for its safety, tolerability, immunogenicity, and efficacy.[ 103 ]
6.2.2. mRNA‐1273 Vaccine
Another vaccine candidate developed against SARS‐CoV‐2 is mRNA‐1273. It is a lipid nanoparticle (LNP) loaded with an mRNA that encodes for a complete, prefusion stabilized form of the S protein of the SARS‐CoV‐2. mRNA‐1273 is in Phase I human clinical trial to examine the safety and immunogenicity of its five dose levels (10, 25, 50, 100, and 250 µg) on a double‐dose vaccination schedule (NCT04283461).
On 12th May 2020, mRNA‐1273 was granted fast track designation by US‐FDA. Data from the Phase 1 study (first clinical batch), with 25 µg and 100 µg dose levels, have shown the presence of well‐tolerated neutralizing antibodies titers at or above convalescent sera. Safety data obtained from the Phase 1 study led to the discontinuation of the 250 µg dose level in the subsequent Phase II studies.[ 110 ] Phase III trial was next launched to assess the efficacy, safety, as well as immunogenicity of the vaccine at a dose of 100 µg in adult participants (NCT04470427).
6.2.3. Ad5‐nCOV Vaccine
Another vaccine for COVID‐19 is based on an adenovirus and is named as Ad5‐nCOV. This vaccine is being developed by the Chinese company CanSino Biologics in partnership with China's Institute of Biology at the country's Academy of Military Medical Sciences. Results from a Phase I safety (NCT04313127) and Phase II (NCT04341389) trials conducted for the vaccine, demonstrated strong immune response against SARS‐CoV‐2. Therefore, in an unprecedented move, the Chinese military approved the vaccine as a “specially needed drug.”[ 111 , 112 ] Its Phase III trial in Saudi Arabia is underway (NCT04526990).
6.2.4. BNT162b2 Vaccine
Another RNA based vaccine candidate developed against SARS‐CoV‐2 is BNT162b2. It is based on LNPs loaded with nucleoside‐modified messenger RNA (modRNA) that encodes for an optimized SARS‐CoV‐2 full length S protein and RBD.
Based on preclinical and Phase I/II studies (NCT04380701) data and in consultation with the US‐FDA's Center for Biologics Evaluation and Research (CBER), BNT162b2 has received fast‐track designation by the US‐FDA and will be tested at a 30 µg dose level (in a two dose regimen) in a global Phase II/III study (NCT04368728).[ 113 ]
6.2.5. CoronaVac Vaccine
Another vaccine candidate against COVID‐19 is a purified inactivated SARS‐CoV‐2 virus vaccine called CoronaVac (formerly called as PiCoVacc). Preclinical studies for the vaccine showed the induction of specific neutralizing antibodies against SARS‐CoV‐2 in rats, mice, and macaques (nonhuman primates) those were able to neutralize 10 representative SARS‐CoV‐2 strains. Three immunizations with the vaccine at two different doses (3/6 µg per dose) provided partial or complete protection in non‐human primates (macaques) against SARS‐CoV‐2 infection.[ 114 ]
Preliminary results of Phase I/II clinical trials (NCT04383574 and NCT04352608), which was tested at three/two different doses in a two doses regimen either scheduled on day 0/28 or on day 0/14 or both. (antigen content of 300, 600, 1200 SU/0.5 mL) showed promising immunogenicity and no severe adverse events. CoronaVac got an emergency approval for limited use by the Chinese government. Phase III clinical trial (NCT04456595) is underway to assess its safety and tolerability. The immunization schedule is two doses IM (600 SU/0.5ml) with a 14 d interval.
6.2.6. INO‐4800 DNA Vaccine
INO‐4800 DNA vaccine is designed in such a way to directly deliver optimized DNA plasmids into cells using Inovio's trademarked device called CELLECTRA. CELLECTRA utilizes a transitory electric pulse to reversibly open tiny cellular pores, enabling the plasmids to enter. Once inside the cell, the plasmids begin replicating along with the cell's DNA and are translated into proteins (MAbs), producing a specific immune response. This methodology has the potential to generate therapeutic MAbs in vivo. Clinical trials are underway to assess its safety, tolerability, and immunogenicity (NCT04336410).
Smith and co‐workers in a preclinical study testing humoral immunogenicity in both mice and guinea pigs have shown the successful generation of neutralizing antibodies and T cell immune responses against SARS‐CoV‐2. These encouraging preclinical results further support its translation to large randomized clinical trials.[ 115 ]
6.2.7. ChAdOx1 nCoV‐19 Vaccine
An adenoviral vaccine candidate AZD1222 (provisionally named ChAdOx1 nCoV‐19) based on the SARS‐CoV‐2 S protein that was originally developed to target MERS is also a potential vaccine candidate for COVID‐19. It is a weakened and safer form of a common cold adenovirus (ChAdOx1) that cannot reproduce within the body but can produce CoV S protein after vaccination resulting in the formation of antibodies against the S proteins. The vaccine's seed stock was in production at Oxford University's Biomanufacturing Facility, UK, and Phase I/II clinical trials (NCT04324606/ISRCTN15281137 and NCT04444674) assessing its safety, efficacy, and immunogenicity against SARS‐CoV‐2 are underway. ChAdOx1 nCoV‐19 is being administered intramuscularly (IM) either at a single dose of 5 × 1010 viral particles (vp) or a single dose of 5 × 1010 vp followed by a booster dose.
The preliminary results from Phase I/II trial (NCT04324606) showed an acceptable safety profile, and a second vaccine dose (boosting dose) further enhanced the antibody responses inducing both humoral and cellular immune responses.[ 116 ]
Doremalen and co‐workers in a study successfully demonstrated a robust humoral and cell‐mediated response from a single dose of an investigational vaccine, AZD1222. A single vaccination has prevented pneumonia caused by SARS‐CoV‐2 in six rhesus macaques by halting SARS‐CoV‐2 replication.[ 117 ]
The vaccine has progressed to Phase II/III (NCT04400838) as well as Phase III trials (ISRCTN89951424). On September 6, 2020 AstraZeneca stopped global trials of the vaccine after finding a volunteer, who developed a type of inflammation called transverse myelitis. The British trial resumed on September 12, 2020, but trials in other countries are still on hold.
6.2.8. Plant‐Based Coronavirus Vaccine
Another vaccine in progress is based on virus‐like particles (VLP). It is a protein‐based vaccine developed using Medicago's proprietary plant‐based technology that uses VLPs grown in Nicotiana Benthamiana. VLP mimic viruses without genetic material and enables the body's immune system to generate an immune response. VLPs cannot replicate and are noninfectious.
In this plant‐based approach, the genetic sequence of a virus is inserted into agrobacterium, which then enters into the plant tissues. The plant begins to produce the protein that is used as a vaccine. In case the virus begins to mutate, the production may be updated in new plants. Using plants and genetically engineered agrobacteria, the mass‐production of vaccines is easy, faster, and inexpensive. The Phase I trial (NCT04450004) evaluating safety, tolerability, and immunogenicinity at dosages of 3.75, 7.5, or 15 µg of the vaccine candidate alone or followed by a booster dose in healthy human volunteers is underway. Medicago is also planning to initiate its Phase II/III trials.[ 118 ]
6.2.9. Avian Coronavirus Infectious Bronchitis Virus (IBV) Vaccine
A genetically modified infectious bronchitis virus (IBV) vaccine is also in the pipeline to treat COVID‐19. IBV was originally developed to treat avian coronavirus. It is available in oral form and has shown efficacy in preclinical trials. The new vaccine is anticipated to turn SARS‐CoV‐2 infection into a very mild cold.[ 119 ]
6.2.10. COVID‐19 S‐Trimer
Another potential protein‐based vaccine candidate in the pipeline is COVID‐19 S‐Trimer (SCB‐2019). It is a recombinant subunit vaccine developed using Clover's patented Trimer‐Tag technology via a prompt mammalian cell expression system. S‐Trimer vaccine is based on the S protein of the SARS‐CoV‐2. The company also identified the antibodies against the trimeric S protein in the serum of patients fully recovered from COVID‐19. A randomized, placebo‐controlled Phase I trial of SCB‐2019 has been launched and will be evaluating the safety, reactogenicity, as well as immunogenicity at multiple dose levels (3, 9, and 30 µg) with and without the adjuvant (NCT04405908).[ 103 , 118 ]
6.2.11. Oral Coronavirus Vaccine
VAAST, oral recombinant vaccine tablets based on the genome COVID‐19 causative virus using Vaxart's proprietary oral vaccine platform, are in preclinical trials. Each vaccine constructs is a nonreplicating viral vector based on a different SARS‐CoV‐2 antigen combination.[ 103 , 118 ]
6.2.12. AdCOVID
AdCOVID is another potent single dose, intranasal vaccine candidate in preclinical trials to protect against COVID‐19. It is an adenovirus‐based vaccine expressing SARS‐CoV‐2 S protein.[ 103 ]
6.2.13. Sputnik V
Sputnik V a COVID‐19 vaccine was registered on 11th August 2020 and approved for early use by the Russian Ministry of Health under the adopted emergency rules during the pandemic. It is a dual adenovirus vectors (rAd26 and rAd5) based vaccine loaded with a fragment of gene coding S protein of the SARS‐CoV‐2. The use of two dissimilar forms of adenovirus vectors, for the first and second vaccination doses is a unique technology of the Gamaleya National Center for ensuring long lasting immunity.
On 1st August 2020, Phase I/II clinical trials of Sputnik V (NCT04437875 and NCT04436471) have been completed and results suggested the induction of strong antibody and cellular immune responses. Postregistration Phase III clinical trial comprising ≥ 40000 people in Russia was expected to get started 31st August, 2020 onwards (NCT04530396). Other countries (UAE, Saudi Arabia, Philippines, India, and Brazil) may also join the Phase III clinical trial locally.[ 118 ]
6.3. Monoclonal Antibodies (MAbs) for SARS‐CoV‐2
The key focus of developing therapeutics against the virus has been revolving around finding antivirals and vaccines. However, many reports point toward various patients associated complications such as cytokine storm syndrome (CRS) and macrophage activation syndrome (MAS), which lead to ARDS during severe COVID‐19 infection.
COVID‐19 patients developing CRS secondary to COVID‐19 pneumonia show increased production of various proinflammatory cytokines, chemokines, and other growth factors. Therefore, treatment of hyperinflammation or the aggressive immune dysfunctions raised in response to the viral infection using immunosuppression mechanism can be advantageous and aid in reducing the mortality.[ 52 , 120 ]
Although immunomodulatory therapy is not recommended in COVID‐19 pneumonia in general, the compassionate use of immunomodulators might be advantageous in COVID‐19 patients exhibiting CRS complications, decreasing the high levels of proinflammatory cytokines, and thereby preventing multiorgan failure.[ 121 ] The following are some of the immunomodulators under consideration as anti‐COVID‐19 therapeutics.
6.3.1. Gimsilumab
Gimsilumab (KIN 1901), a human MAb, working by targeting granulocyte macrophage colony stimulating factor (GM‐CSF), is under clinical trials to prevent and treat ARDS in COVID‐19 patients. As GM‐CSF elevated levels in blood further augment the expression of other proinflammatory cytokines causing the progression of ARDS and serum from COVID‐19 patients have shown high levels of GM‐CSF; therefore, gimsilumab is anticipated to reduce the mortality rate by reducing lung damage. A multicenter, Phase II trial (NCT04351243) is underway to assess gimsilumab's efficacy and safety profile in patients with ARDS/lung injury secondary to COVID‐19.[ 103 ]
6.3.2. TJM2
TJM2 (TJ003234) is a neutralizing antibody against human GM‐CSF. TJM2 shows a high affinity towards human GM‐CSF, thereby blocking GM‐CSF binding to its receptor. This will further prevent downstream signaling cascades resulting in response to GM‐CSF binding, halt target cell activation, resulting in inhibition of inflammatory responses causing reduced tissue inflammation and death. TJM2 is in clinical trials to study its efficacy in reducing the severity of complications associated with COVID‐19.[ 103 ] A Phase I/II, randomized, multicenter trial (NCT04341116) will assess the efficacy of TJM2 under supportive care when administered as an IV infusion (3 mg kg‐1 or 6 mg kg‐1) in subjects with severe COVID‐19.
6.3.3. Lenzilumab
Lenzilumab, another GM‐CSF neutralizing immunotherapy, is being studied in minimizing and treating the cytokine storm and associated lung dysfunction/ARDS in COVID‐19 patients. Lenzilumab is Humanigen's proprietary Humaneered MAb targeting GM‐CSF and is being approved by US‐FDA for sympathetic use in COVID‐19 patients.[ 103 ] Currently, it is being studied in a multicentric, Phase III trial in comparison to the current standard of care for taking care of respiratory failure and prevention of death in hospitalized COVID‐19 patients (NCT04351152).
6.3.4. Namilumab
Namilumab (IZN‐101, AMG203) is another fully human immunoglobulin G1 MAb targeting GM‐CSF, which has received compassionate use approval from US‐FDA in the treatment of critical as well as hospitalized COVID‐19 patients before their admission to ICU.103 The double center compassionate use study of the drug will be conducted in Bergamo and Milan cities in Italy. It is currently in later stages of clinical development programs against RA and ankylosing spondylitis.[ 122 ]
6.3.5. TZLS‐501
TZLS‐501 is another antibody in progress as a potential treatment for COVID‐19. TZLS‐501 is a humanized MAb targeting interleukin‐6 receptor (anti‐IL‐6R). Early clinical studies channeled in China suggest that anti‐IL‐6R MAbs might be used clinically for treating COVID‐19 patients. TZLS‐501 binds to both forms of IL‐6R (membrane‐bound or soluble), thereby speedily depleting the levels of IL‐6 in blood. Overproduction of IL‐6 leads to the elicitation of long‐lasting inflammation, causing lung damage following SARS‐CoV‐2 infection.[ 103 ]
6.3.6. AT‐100
Another immunotherapeutic candidate explored for COVID‐19 is AT‐100. AT‐100 is a human recombinant surfactant protein D (rhSP‐D). Initially, AT‐100 was developed for broncho‐pulmonary dysplasia (BPD) in premature infants who requisite mechanical ventilation and oxygen therapy.[ 103 ]
6.3.7. Tocilizumab
Tocilizumab (Actemra) is a recombinant version of a humanized MAb targeting the IL‐6 receptor, which binds to IL‐6R, thereby inhibiting signal transduction. US‐FDA has approved it for the treatment of CRS, rheumatoid arthritis, giant cell arthritis, polyarticular juvenile idiopathic arthritis, and systematic juvenile idiopathic arthritis.[ 121 , 123 ]
Xu and group from China in their pilot study used tocilizumab (single dose of 400 mg IV) to treat a total of 21 COVID‐19 patients. Tocilizumab treated patients showed a significant reduction in fever and IL‐6 levels. Lymphocyte counts in the patients returned to normal after treatment and they demonstrated an improvement in lung function.[ 123 ] A multicentered randomized Phase III trial of tocilizumab is underway in China to assess its efficacy and safety in COVID‐19 patients with pneumonia and elevated IL‐6 (ChiCTR2000029765). Another proof of concept study (TOSCA) in Italy is underway for its efficacy and safety profile assessment in the treatment of patients with ARDS and CRS secondary to COVID‐19 (NCT04332913). Another multicentric Phase III placebo‐controlled clinical trial, called COVACTA (NCT04320615), is underway to evaluate tocilizumab (maximum single dose of 800 mg IV and one more dose only if clinical symptoms deteriorate/or exhibits no improvements) along with standard of care versus placebo along with standard of care in severe COVID‐19 patients globally.
6.3.8. Sarilumab
Sarilumab (Kevzara) is another IL‐6 receptor blocker under clinical trials for its ability to lessen the effect of the inflammatory immune response against SARS‐CoV‐2 infection. It is a fully human MAb against the IL‐6 receptor. It has been approved by US‐FDA toward the treatment of rheumatoid arthritis, but its use to treat COVID‐19 patients is investigational. Sarilumab was developed using proprietary VelocImmune technology (Regeneron's) using a genetically engineered mouse equipped with a genetically humanized immune system to produce fully human antibodies. A multicenter, double blind, Phase II/III trial (NCT04327388) is underway to assess the efficacy and safety of a single IV dose of sarilumab compared to supportive care plus placebo in severe/critical COVID‐19 patients. In Phase II, patients were distributed into three groups at random, with two doses of sarilumab (200 and 400 mg) and placebo. Preliminary data from the trial led to its immediate amendment to enroll only critically ill COVID‐19 patients (either on ICU or requiring mechanical ventilation/high flow oxygenation) receiving a higher dose of the drug (400 mg) or placebo.[ 103 ]
6.3.9. Cytosorb
CytoSorb is an extracorporeal blood purification technology based on hemo‐adsorbent polymer to treat uncontrolled inflammation in patients those have undergone cardiac surgery. It consists of polymer beads with high porosity those can successfully adsorb and remove toxic substances and inflammatory mediators from blood and other bodily fluids. CytoSorb has received US‐FDA's emergency use authorization in the US for use in severely ill COVID‐19 patients approaching or confirmed respiratory/multiple organ failure and are admitted to ICU. It is approved in the European regions as an adjuvant therapy designed to lessen the complications associated with CRS by absorbing a wide range of cytokines, PAMPs and DAMPs, thereby reducing their levels in circulation.[ 124 ]
6.3.10. Convalescent Sera
The use of convalescent sera is a promising treatment strategy against SARS‐CoV‐2. It consists of patient derived serum enriched with passive neutralizing antibodies against the virus, obtained from COVID‐19 recovered patients.[ 125 ] Many reports from China suggest the use of convalescent serum as therapy for patients with COVID‐19. China has offered convalescent plasma therapy to 200 COVID‐19 patients (ChiCTR2000029757). Convalescent plasma therapy is promising, but its safety and efficacy as a treatment modality for COVID‐19 are yet to be established. Therefore, it is important to study and establish the safety as well as efficacy of convalescent plasma therapy in COVID‐19 patients in clinical trials. The US‐FDA based on encouraging results from early trials has given it the emergency use authorization against COVID‐19.
6.4. Anti‐Inflammatory Drugs for SARS‐CoV‐2
6.4.1. Dexamethasone
Glucocorticoids may modulate inflammation‐mediated lung injury in COVID‐19 patients and thereby reduce progression to respiratory failure and the resulting death. However, there is uncertainty about the effectiveness of glucocorticoids in COVID‐19. Dexamethasone is a corticosteroid used in a wide range of conditions for its anti‐inflammatory along with immunosuppressant effects.
Preliminary findings of the Phase II/III RECOVERY trial (NCT04381936), a randomized, controlled, open‐label study conducted in the UK, noted a survival benefit with the use of dexamethasone (oral or IV at a dose of 6 mg per day for up to 10 d) as compared to standard care alone (invasive mechanical ventilation or oxygen) in hospitalized COVID‐19 patients. Patients on ventilators showed one third and patients requiring only oxygen showed one fifth reductions in the 28 day mortality. Dexamethasone can also be used in both children and elderly, but in pregnancy/breast‐feeding cases, the recovery trial used prednisolone (40 mg orally) or hydrocortisone (80 mg twice daily IV) instead of dexamethasone.[ 126 ]
6.4.2. N‐acetylcysteine
Among several drugs employed for the treatment of COVID‐19, N‐acetylcysteine (NAC) is a potential therapeutic/preventive/adjuvant against SARS‐CoV‐2.
NAC is a precursor of reduced glutathione (GSH). It has anti‐inflammatory, antioxidant, antiviral, and anticoagulant/platelet‐inhibiting properties. It is approved as an over‐the‐counter dietary supplement and is in the WHO list of essential medications. The US‐FDA has approved the usage of NAC to treat acetaminophen overdose associated liver side effects, and also as mucolytic agent (due to its free sulfhydryl group) in cystic fibrosis or chronic obstructive pulmonary disease (COPD). It has been also used as an antioxidant in a variety of disorders involving GSH depletion and oxidative stress. Thiols can block ACE2 receptors thereby blocking penetration of SARS‐CoV‐2 into cells. NAC can act as a potential therapeutic agent in the treatment and/or attenuating the risk associated with COVID‐19 through a variety of potential mechanisms, including increasing glutathione level, improving T cell response, and modulating inflammation thereby preventing COVID‐19 patients from moving to ICUs.[ 127 ]
An ongoing Phase II clinical trial (NCT04374461) is assessing the number of successful COVID‐19 patients discharged/transferred from critical care units after receiving NAC (6 g per day IV) in addition to supportive or other treatments as prescribed against COVID‐19. Other Phase III (NCT04455243) and Phase IV (NCT04419025) clinical trials of NAC are assessing NAC's anti‐inflammatory effect and its efficacy in minimizing the severity associated with COVID‐19, respectively.[ 128 ]
6.4.3. Cyclosporine A
Cyclosporine A (CsA) is a calcineurin inhibitor and is used to avert organ rejection and for treating T‐cell associated autoimmune diseases. Anti‐inflammatory and immunosuppressive effects of CsA are exerted due to its bonding to cyclophilin‐A (Cyp‐A), thereby preventing the activation of nuclear factor of activated T‐cell (NFAT) and production of IL‐2 (a cytokine crucial for the proliferation of T‐cells). CsA has shown potent antiviral activity at noncytotoxic micromolar concentrations against many CoVs including SARS‐CoV in cell cultures. CsA may be successful in COVID‐19 due to the close resemblance between SARS‐CoV‐2 and SARS‐CoV. This antiviral property might have resulted due to the inhibition of Cyp‐A‐dependent viral assembly and inhibition of the NFAT pathway.[ 129 , 130 , 131 ]
Ongoing Phase I safety study of CsA (oral or IV) is assessing the tolerability and clinical effects in COVID‐19 patients requiring oxygen supplementation (NCT04412785). A Phase IIa randomized clinical trial (NCT04492891) is underway for the treatment of non‐ICU hospitalized COVID‐19 patients. Patients will receive CsA orally for up to 7 d at 2.5 mg kg‐1 body weight twice a day along with standard of care. Another ongoing Phase IV clinical trial (NCT04392531) is assessing the efficacy and safety of CsA plus standard treatment in comparison to standard treatment in hospitalized COVID‐19 patients.
6.4.4. Budesonide
Another corticosteroid in pipeline is inhaled budesonide. It may directly inhibit the viral replication or may exert the inflammatory effect. Steroids reduce the effect of cytokines and thereby modulate immune response. Budesonide may prevent the ARDS associated with COVID‐19. Inhaled corticosteroid's therapeutic effect against COVID‐19 is limited and still need to be explored. Various clinical trials assessing their efficacy in COVID‐19 are ongoing (Phase IV: NCT04355637).
6.4.5. Vitamin D
Vitamin D is a steroid hormone. Vitamin D is being evaluated as an adjuvant therapy for COVID‐19 based on its immunomodulatory, anti‐inflammatory, antifibrotic, and antioxidant effects. Vitamin D exerts direct effect on immune cell proliferation and activity, and ACE2 expression.[ 132 ]
The aim of one of its ongoing randomized Phase III trial (NCT04385940) in COVID‐19 patients is to assess its efficacy at daily low dose (1000 IU) versus weekly high dose (50000 IU) for 3 weeks and to determine the correlation between vitamin D deficiency and clinical characteristics. Another Phase III trial (NCT04344041) assessing the efficacy of vitamin D3 at high dose (400000 IU) in comparison to standard dose (50000 IU) is underway.
7. Future Potential Therapeutics
Apart from the currently available drugs, immunomodulators, and vaccine candidates, there are many other potent molecules like novel protein or peptide therapeutics and even nanoparticulate based strategies yet to be tested against the virus. These new therapeutics, once proven, can be the game changers in tackling the pandemic. Below we list some of the potential future therapeutics for treating COVID‐19 patients.
7.1. Peptides Based Therapeutics for COVID‐19
In the last decades, peptides as therapeutics and diagnostic molecules have gained massive interest in drug development due to the dynamic research conducted by pharmaceutical and biotech companies.[ 133 ] Currently, a large number of both natural as well as synthetic therapeutic peptides are in clinical development for various diseases ranging from cancer, neurodegenerative, cardiovascular, metabolic disorders, autoimmune disorders, hormonal deficiencies, to infectious diseases.[ 133 , 134 ] Peptide based therapeutics offer numerous benefits over other molecules such as high selectivity and specificity, high efficacy and safety, tolerability, less immunogenicity, biocompatibility, low toxicity, easy, and cost‐effective production. According to their source and method of production, peptide based therapeutics can be classified as: 1) natural peptides (derived from peptide hormones, plant proteins, animals, and human‐derived peptides); 2) recombinant peptides; and 3) synthetic peptides.[ 133 , 134 , 135 ]
Peptides are relatively easy to modify structurally. Improved therapeutic performances can be achieved by utilization of modified amino acids, cyclization, etc. Many antiviral peptides are being explored for viral diseases like hepatitis, influenza, and HIV.[ 136 ] Peptides exhibit a reduced possibility of developing resistance during the treatment. Antiviral activities of the peptides can be attributed to three major mechanisms: 1) inhibition of viral attachment and penetration into host cells; 2) inhibition of replication by interacting with viral polymerases, and 3) disruption of the viral envelope. They can be presented as a new generation of antiviral agents with broad‐spectrum activities.[ 136 , 137 ]
Potential peptide‐based therapeutics which are in the pipeline against COVID‐19 include FlowVax peptide vaccine from Flow Pharma,[ 103 ] Corvax signal peptide vaccine from Vaxil Bio, (VaxHit platform),[ 103 ] epitope‐based peptide vaccine from OncoGen (Vaccinomics strategy),[ 103 ] Ii‐Key peptide vaccine from Generex Biotechnology (EpiVax's computational tools and li‐Key technology),[ 118 , 138 ] etc. Peptide‐based therapeutics can be effective alternatives for COVID‐19 prophylaxis.
7.2. Nanoparticle Based Therapeutics for COVID‐19
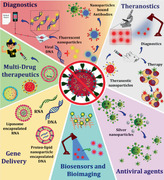
Nanotechnology, as one of the ground‐breaking approaches, can be really promising in offering remedial solutions toward fighting the on‐going COVID‐19 outbreak and future pandemics. Nanotechnology allows a number of tactics to fabricate highly advanced therapeutic options to cope with SARS‐CoV‐2 both outside and inside the host. The scope of nanotechnology against COVID‐19 includes conventional as well as advanced biomimetic approaches and engineered nanomaterials with versatile chemical functionalities. Therefore, nano intervention will be extremely relevant in advancing COVID‐19 prophylaxis, diagnosis, and treatment (Figure 8 ). In view of what we know so far about the SARS‐CoV‐2 structure, its pathophysiology, life cycle, and related immunological response, we envisage key steps where nanotechnologist and nanotechnology could play a pivotal part.
Figure 8.

Schematic showing the areas where nanotechnology can significantly benefit the fight against COVID‐19.
Herein, nanotechnology is discussed in terms of fabricating engineered nanocarriers to overcome the limitations associated with conventional antiviral and other biological therapeutics, nanomaterials as novel therapeutics, risk free immunization techniques, engineered nanomaterials in the development of point‐of‐care diagnostics, and alternative advanced disinfection methodologies. Nanotechnology aims toward safe, effective, and targeted delivery of existing as well as future therapeutics, fabricating risk‐free vaccines for safe and effective immunization, curbing the interactions between viruses and host cells, and permanent disruption of viral particles.[ 139 , 140 , 141 ]
In the recent past, many nanoparticulate systems have been tested for their efficacy and made their way to preclinical studies against several pathogenic viruses such as HIV, herpes simplex, human papilloma virus, and respiratory viruses.[ 140 , 141 , 142 ]
7.3. Nanocarrier‐Based Delivery Systems
In the dearth of a specified/broad spectrum antiviral agent against SARS‐CoV‐2, current therapy focusses on repurposing existing molecules. Therefore, to make the repurposed therapeutics more effective and safer, an appropriate nanocarrier delivery system might be useful. Successful delivery of therapeutics especially peptide/protein, DNA/RNA, and other biologicals to the target site is most often hampered due to their poor aqueous solubility, degradation, fast‐clearance, low‐bioavailability leading to subtherapeutic concentrations. Nanomaterials with large surface area to volume ratios, versatile functionalities and surface modification, unique physicochemical properties, and suitable for various routes of administration can easily address the challenges associated with conventional therapeutics. Biocompatible organic/inorganic nanoparticles can be potentially tuned to encapsulate therapeutics with large payloads, control release, minimize drug resistance, and improve pharmacokinetic/pharmacodynamic properties and patient compliance. Further actively targeted nanocarriers can cross the biological barriers thereby reaching the protected target sites with higher specificity excluding the unwanted exposure to nontargeted areas resulting in dosage reduction, reduced systemic toxicity, improved bioavailability, and enhanced efficacy. Nanotechnology further makes it possible to tailor the therapy according to individual needs.[ 140 ]
Itani and group have demonstrated the potential role of different nanoparticles as efficient delivery agents for potential therapeutics or immunomodulators against COVID‐19.[ 142 ] Nanoparticle based delivery of therapeutics (drugs, vaccines, RNA/DNA, and peptides) ensures targeted and optimized concentration in the desired site while minimizing adverse effects and unwanted degradation enhancing efficacy. Donalisio et al. demonstrated nanotechnological approach for the tropical treatment of herpes simplex virus (HSV‐1 and HSV‐2) by encapsulating acyclovir in chitosan nanoparticles. The obtained gel formulation showed enhanced penetration across the porcine skin as compared to the commercial cream product, and revealed increased efficacy as compared to its free form.[ 143 ] LaBauve and group used a lipid‐coated mesoporous silica nanocarrier system to deliver antiviral molecule ML336 for inhibiting Venezuelan equine encephalitis virus (VEEV), in order to improve its solubility and stability. The designed ML336 loaded nanosystem showed enhanced circulation time, biocompatibility, and viral inhibition in vivo in a murine model of VEEV infection.[ 144 ] Kumar et al. demonstrated the fabrication of zidovudine loaded lactoferrin nanoparticles. Zidovudine is a highly bioavailable antiretroviral drug, which shows serious side effects and toxicity. Results revealed higher efficacy and reduced toxicity of nanocarrier based formulation as compared to their free soluble counterparts.[ 145 ] Liu et al. demonstrated the fabrication of liposomes encapsulating cholesterol modified hydroxychloroquine for the treatment of bleomycin induced pulmonary fibrosis. The engineered nanosystem inhibited the proliferation of rat lung fibroblasts and reduced any toxicity associated with hydroxychloroquine thereby, providing evidence as a possible therapeutic agent for pulmonary fibrosis.[ 146 ]
Encapsulation in a nanosystem enhances nucleic acid‐based vaccine stability and offers a newer fusion mechanism to transport the genetic material loaded inside them directly to the cells. Similarly, the efficacy of other mRNA/siRNA vaccines can be significantly enhanced via complexing them with lipid/polymer‐based nanoparticles. These lipid/polymer‐based nanoparticles are based on ionizable monomers, which complex negatively charged RNA/DNA and thus facilitate escape of the entrapped RNA/DNA from endosomes into the cytosol where the genetic material can be translated.[ 30 ] The successful inhibition of viruses achieved with nanoplatforms could have extensive advantage in combatting various viral infections including SARS‐CoV‐2.
7.4. Nanoparticle‐Based Combinatorial Therapy
Combinatorial therapy aims in suppressing more than one pathway simultaneously, thereby, producing synergistic effect. Therefore, combinatorial therapy may represent another therapeutic approach against SARS‐CoV‐2. It displays numerous advantages such as dose reduction of the individual therapeutics, leading to fewer adverse events. It may also overcome drug resistance. Nanocarriers are very promising systems for the co‐delivery of multiple drugs with different physicochemical properties. They enable encapsulation and sequential release of both hydrophobic and hydrophilic drugs in a single platform. Nanoparticles can also be instrumental in attaining combinatorial multidrug therapeutics for controlling uncontrolled inflammation and manage disease severity. A very recent work by Dormont et al. developed targeted multidrug nanoparticles composed of an endogenous lipid squalene, an immunomodulator adenosine and an antioxidant α‐tocopherol for treating acute viral inflammation (Figure 9 ).[ 147 ] Freeling et al. fabricated a lipid‐drug nanoparticle system comprising of three antiretroviral drugs (lopinavir, ritonavir, and tenofovir) to overcome lymphatic drug insufficiency and reported 50‐fold higher and sustained intracellular concentrations in lymph nodes compared to current oral therapy of free drugs. This combinatorial nanotherapy can be used to target SARS‐CoV‐2.[ 148 ] Bearing in mind the risk of random mutations in SARS‐CoV‐2 genome, which may result in variation in antigenic protein, nanoparticle functionalization with a varied number of antigenic moieties/proteins concurrently will enhance the efficacy of vaccines against viral infections. These next‐generation nanocarrier‐based vaccines will not only help in guarding the antigenic proteins against metabolism/degradation but will also help in greater exposure of antigenic proteins to antigen‐presenting cells, thereby improving their delivery and efficacy.
Figure 9.

Fabrication and characterization of multidrug SQAd/VitE nanoparticles. A) Schematic illustration of SQAd bio‐conjugation and VitE encapsulation to form SQAd/VitE nanoparticles. B) VitE encapsulation efficiency (weight%) in SQAd/VitE nanoparticles as measured by HPLC. C) Hydrodynamic size (diameter, nanometers) of SQAd/VitE nanoparticles measured by dynamic light scattering (DLS). D) Measurement of surface zeta potential of SQAd/VitE and SQAd nanoparticles. E) cryo‐TEM images of SQAd/VitE nanoparticles. F) Stability of SQAd/VitE nanoparticles in 50% fetal bovine serum (FBS) over 5 days as measured by DLS. G) Release profile of Ad from SQAd/VitE nanoparticles in 50% FBS. Reproduced with permission.[ 147 ] Copyright 2020, The Authors.
7.5. Nanomaterials as Risk‐Free Immunization Techniques
Nanomaterials have emerged as a favorable tool both as immune stimulators and immune suppressors for immune modulation. Nanomaterial based nonviral vectored vaccines can successfully overcome two major limitations associated with conventional vaccines of weak immunogenicity and reversion risk associated with live/attenuated viral vectors, and can be a reliable alternative to viral vaccines. Nanosystems can imitate the genome transfer ability of viral vaccines which show high pathogenicity and can also deliver molecular adjuvants simultaneously.[ 139 ] Recombinant protein and inactivated vaccines in general necessitate the use of adjuvants to enhance their immunogenicity. Many nanomaterials depending on their functionalization possess an intrinsic adjuvant property and can amplify the immune response in a host. Therefore, fabricating nanomaterials with intrinsic ability to amplify host's immune response will be very helpful in enhancing the efficacy of vaccines especially in patients with impaired immune system. Nanosystems can also enhance the targeted delivery of immunosuppressants to both adaptive and innate immune systems.[ 139 ]
7.6. Nanomaterials with Intrinsic Antiviral Properties
Various nanoparticles like graphene oxide, silver, gold, copper, and zinc nanoparticles show intrinsic antiviral/antibacterial properties and can act as potential therapeutics against the disease.[ 149 , 150 , 151 , 152 , 153 , 154 , 155 , 156 , 157 , 158 , 159 , 160 ] They may offer alternative methods to traditional disinfection protocols used in healthcare sites. Nanoparticles can potentially sustain release, enhance cell entry, and reduce efflux of toxic metal ions. The use of metal nanoparticles would be particularly advantageous as antiviral agents because metals may attack a wide range of viral targets. Also, there is a lower risk of developing drug resistance in the case of metals as compared to conventional antivirals.[ 149 ]
Silver nanoparticles (AgNPs) have been investigated for their antimicrobial potency against bacteria, but many reports point toward their antiviral activities against viruses, which includes HIV, herpes simplex virus, respiratory syncytial virus, hepatitis B virus, etc.[ 150 , 151 , 152 ] Li and group have demonstrated efficient in vitro inhibition of H1N1 infection utilizing AgNPs based codelivery of oseltamivir (neuraminidase inhibitor) by inhibiting viral attachment and release.[ 153 ] The fact that AgNPs are less toxic and are nontoxic at lower concentrations to humans, vouch for their possible usage for therapeutic purposes. Given the fact that silver ion itself attacks a broad range of targets in the microbial entry, microorganisms generally do not develop resistance against silver.[ 152 ] Nanocolloid AgNPs have also been shown to be effective in controlling viral proliferation in the respiratory tract after inhalation delivery, thus controlling the disease progression.[ 154 ]
Another study by Levina and group demonstrated in vitro antiviral activity of DNA decorated titanium dioxide (TiO2) nanosystem against different influenza A virus subtypes infected MDCK cells. The nanosystem was found to be highly efficient in delivering nucleic acids directly inside the cells without the aid of any transfection agent with an IC50 value of 1.5 µg mL‐1 (30 × 10−9 m for DNA).[ 155 ]
Another nanostructured material, graphene, has shown promising antimicrobial/antiviral properties.[ 156 , 157 , 158 ] Recently, the efficacy of Cu to inactivate HuCoV‐229E was demonstrated. It was found that the HuCoV‐229E (103 plaque forming units) was inactivated irreversibly in less than 60 min on brasses/alloys containing at least 70–90% Cu, whereas the virus was shown to persist on surfaces like glass, rubber, and stainless steel in an infectious state for at least 5 d.[ 159 ] Dhanasezhian et al. fabricated biogenic gold and AgNPs using the seaweed Sargassum wightii. Their cytotoxic and antiviral activity against HSV‐1 and HSV‐2 strains on Vero cells showed that functionalized metallic nanoparticles act as antiviral agents by hindering virion attachment and cellular entry, depending on their particle size.[ 160 ]
7.7. Nanomaterials as Diagnostic Agents
Nanotechnology can be influential in early viral detection, disease monitoring, and targeted destruction of the viruses. Early and efficient detection using nanoparticle enabled technologies will accelerate patient management and identification and containment of infection hotspots. Nanoparticle based disease monitoring mediated by various metallic nanoparticles carrying magnetic, fluorescence, and surface‐enhanced Raman scattering features would aid in tracking treatment response.[ 161 , 162 ]
Moitra and group reported the development of gold nanoparticles (AuNPs) based colorimetric assay for rapid, selective, and visual diagnosis of positive COVID‐19 patients within 10 min after the isolation of viral RNA. In the presence of targeted RNA from the virus, thiol‐modified antisense oligonucleotides against N protein of SARS‐CoV‐2 capped AuNPs exhibited selective agglomeration leading to altered surface plasmon resonance. Subsequent addition of RNaseH further cleaved the RNA from the RNA‐antisense oligonucleotides complex leading to the formation of a precipitate (Figure 10 ).[ 163 ] Graphene sheets conjugated to antibodies can rapidly detect targeted viral proteins and viral targeting antibody functionalized graphenes can further be useful in the development of biosensors against COVID‐19.[ 164 ] Similarly, utilizing theranostic nanoparticles, both therapy and diagnosis can be combined in order to detect and neutralize the virus in a single go, halting the virus to survive and reproduce within the host. The theranostic nanoparticles can efficiently complex with SARS‐CoV‐2, detecting and disrupting their structure.[ 142 ]
Figure 10.
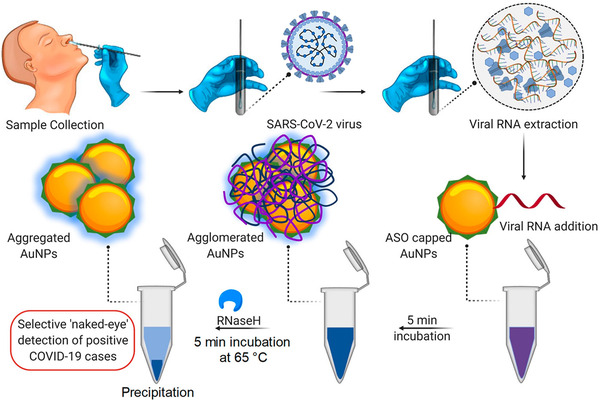
Schematic illustration for the selective naked‐eye detection of SARS‐CoV‐2 RNA facilitated by the suitably designed ASO‐capped Au nanoparticles. Reproduced with permission.[ 163 ] Copyright 2020, American Chemical Society (ACS). https://pubs.acs.org/doi/10.1021/acsnano.0c03822
Potential nanoparticle based therapeutics which are in the pipeline against the current COVID‐19 outbreak include proteo‐lipid nanovehicle based fusogenix DNA vaccine, lipid nanoparticle‐loaded mRNA vaccine (mRNA‐1273), plant‐based virus‐like particles (VLP), recombinant protein nanoparticle vaccine (NVX‐CoV2373),[ 103 , 165 ] and nanomicelles based antiviral.[ 138 ]
Analogous to cancer nanotherapeutics, COVID‐19 can also be managed successfully by exercising 3D nanotechnology‐ based approaches that include efficient detection, diagnosis, and delivery. The tissue and cell targeting principle achieved by targeting antibodies and peptides in handling other dreaded diseases like cancer and neurodegeneration can be adopted in the case of the antiviral nanoparticles for achieving targeted and effective therapy of infected patients. Furthermore, theranostic nanoparticles can be developed to detect and kill the virus simultaneously. Nanoparticle based kits can be developed to detect the virus at an early stage in asymptomatic patients with lesser viral loads. Nanoparticle based antiviral material can be developed for fabricating superfine filters for face masks, personal protective equipment, and surface coatings. Further, nanotechnology may assist in large‐scale production of therapeutics and equipment addressing COVID‐19 and similar pandemics.
Thus, there are varieties of therapeutic molecules against COVID‐19. The list of these potential therapeutic molecules under consideration for COVID‐19 is summarized in Table 1 .
Table 1.
List of potential therapeutic molecules under consideration for COVID‐19
| Potential candidates | Platform | Type of candidate | Company/ Developer | Current stage of clinical evaluation/ regulatory status | Same platform targeting other viruses | Refs. |
|---|---|---|---|---|---|---|
| Repurposed drugs | ||||||
| Favilavir (favipiravir, T‐705) | Antiviral | Guanosine analogue; RNA‐dependent RNA polymerase (RdRp) inhibitor | Fujifilm Toyama Chemical's |
Phase 0 ChiCTR2000029600 ChiCTR2000029544 Phase III |
Influenza, Catarrhal | [ 71 , 72 ] |
| Remdesivir (GS‐5734) | Antiviral | Adenosine analogue; RNA‐dependent RNA polymerase (RdRp) inhibitor | Gilead Sciences |
Phase II Phase III |
Ebola | [ 72 , 77 , 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 , 86 , 87 ] |
| Chloroquine/hydroxychloroquine | Anti‐malarial/ Antiviral | Quinine derivative | Bayer/Sanofi |
Phase II Phase III Phase IV ChiCTR2000029559 |
Malaria, SLE, and RA | [ 72 , 85 , 88 , 89 , 90 , 91 , 92 , 93 , 94 , 95 , 96 ] |
| NP‐120 (Ifenprodil) | Anti‐fibrotic | N‐methyl‐d‐aspartate (NDMA) receptor glutamate receptor antagonist | Algernon |
Phase II Phase IIb/III |
Blood circulation disorders, idiopathic pulmonary fibrosis | 0[ 97 ] |
| Lopinavir and ritonavir | Antiretrovirals | Protease inhibitors (3CLpro inhibitor) | Fujifilm Toyama Chemical's |
Phase 0 ChiCTR2000029308 |
HIV, SARS‐CoV and MERS‐CoV | [ 85 , 96 , 98 , 99 ] |
| Camostat and nafamostat | Serine protease inhibitors | TMPRSS2 inhibitors | Ono Pharmaceutical |
Phase IIa |
Pancreatitis and postoperative reflux esophagitis | [ 72 , 102 ] |
| Leronlimab (PRO 140) | Humanized IgG4 monoclonal antibody | CCR5 antagonist | CytoDyn/ AGC Biologics |
Phase II |
Cancer and HIV | [ 103 ] |
| Umifenovir (Arbidol) | Antiviral | Targets S protein/ACE2 interaction and inhibits membrane fusion of the viral envelope | Pharmstandard |
Phase IV |
Influenza | [ 85 ] |
| APN01 | Mimics the ACE2 enzyme | Recombinant human ACE2 (rhACE2) | Apeiron Biologics/ University of British Columbia |
Phase II EudraCT: 2020‐001172‐15 |
Acute lung injury (ALI), ARDS, and PAH | [ 103 , 138 ] |
| EIDD‐2801 | Antiviral | Ribonucleoside analogue | Ridgeback Biotherapeutics |
Phase I Phase II |
MERS and SARS | [ 57 , 103 ] |
| Vaccines | ||||||
| Fusogenix DNA vaccine (Covigenix) | DNA | Proteo‐lipid vehicle (PLV) encapsulated DNA plasmid that encodes several protein epitopes for SARS‐CoV‐2 | Entos Pharmaceuticals | Preclinical | – | [ 103 ] |
| mRNA‐1273 | RNA | Lipid nanoparticle carrying mRNA for (S) protein | Moderna/ Vaccine Research Center (NIAID) |
Phase I NCT04283461 Phase III NCT04470427 |
– | [ 110 ] |
| Ad5‐nCOV | Nonreplicating viral vector | Recombinant adenovirus type 5 vector | CanSino Biologics/ Beijing Institute of Biotechnology |
Phase I ChiCTR2000030906 Phase II Phase III |
Ebola | [ 111 , 112 ] |
| BNT162b2 | RNA | Lipid nanoparticle carrying nucleoside‐modified mRNA (modRNA) for S protein and RBD | BioNTech in collaborations with Pfizer, and Fosun Pharma |
Phase I/II NCT04380701 Phase II/III NCT04368728 |
– | [ 113 ] |
| CoronaVac (PiCoVacc) | Inactivated virus | Purified inactivated SARS‐CoV‐2 vaccine | Sinovac Biotech |
Phase 1/II Phase III |
– | [ 114 ] |
| INO‐4800 | DNA | Optimized DNA plasmids delivery directly into cells using device called CELLECTRA | Inovio/ Advaccine Biotechnology |
Preclinical Phase I |
– | [ 115 ] |
| AZD1222 (ChAdOx1) | Nonreplicating adenoviral vector | Adenovirus vector and the SARS‐CoV‐2 spike protein | AstraZeneca/University of Oxford/Advent SRL |
Phase I/II Phase II/III Phase III ISRCTN89951424 |
MERS, Influenza, TB, Chikungunya, Zika, MenB, plague | [ 116 , 117 ] |
| Plant‐based vaccine | Protein | Virus‐like particles (VLP) | Medicago/ Laval University |
Phase I |
– | [ 118 ] |
| Modified avian coronavirus vaccine | Protein | Genetically modified infectious bronchitis virus vaccine (IBV) | Migal/ Volcani Institute | Preclinical | Avian coronavirus infectious bronchitis virus (IBV) | [ 119 ] |
| COVID‐19 S‐trimer | Protein | Recombinant subunit on the trimeric S protein (S‐trimer) of the SARS‐CoV‐2 | Clover Biopharmaceuticals/ GlaxoSmithKline (GSK)/ Dynavax |
Phase I |
– | [ 103 , 118 ] |
| Oral recombinant vaccine (VAAST) | Nonreplicating adenovirus vector | Based on a different SARS‐CoV‐2 antigen combination | Vaxart/ Emergent BioSolutions | Preclinical | – | [ 103 , 118 ] |
| AdCOVID | Nonreplicating viral vector | Adenovirus‐based vaccine expressing SARS‐CoV‐2 spike protein | Altimmune/ University of Alabama | Preclinical | – | [ 103 ] |
| Sputnik V (Gam‐Covid‐Vac) | Nonreplicating adenoviral vector | Two different types of adenovirus‐based vaccine expressing SARS‐CoV‐2 spike protein | Gamaleya National Research Institute of Epidemiology and Microbiology |
Phase I/II Phase III |
– | [ 118 ] |
| Linear DNA vaccine | DNA | Based on SARS‐CoV‐2 spike (S) protein | Applied DNA Sciences/ Takis Biotech | Preclinical | – | [ 118 ] |
| ZyCoV‐D | DNA | Plasmid DNA vaccine | Zydus Cadila | Phase I/II CTRI/2020/07/026352 | [ 103 , 118 ] | |
| Bacillus Calmette‐Guerin (BCG) vaccine | Live‐attenuated vaccine | Weakened strain of bovine tuberculosis bacillus, Mycobacterium bovis | University of Melbourne / Murdoch Children's Research Institute and Radboud University |
Phase III |
Tuberculosis (TB) | [ 118 ] |
| Inactivated novel coronavirus pneumonia (COVID‐19) vaccine | Inactivated virus | Inactivated SARS‐CoV‐2 virus (Vero cell) | Sinopharm/ Wuhan Institute of Biological Products |
Phase III ChiCTR2000034780 |
– | [ 118 ] |
| BBIBP‐CorV | Inactivated virus | Inactivated SARS‐CoV‐2 vaccine | Sinopharm/ Beijing Institute of Biological Products | Phase III | – | [ 103 , 118 ] |
| Covaxin | Inactivated coronavirus | Whole‐Virion inactivated SARS‐CoV‐2 Vaccine (BBV152) | Bharat Biotech |
Phase I/II |
– | [ 118 ] |
| NVX‐CoV2373 | Protein | Recombinant SARS‐CoV‐2 nanoparticle vaccine consisting of trimeric full‐length SARS‐CoV‐2 spike glycoproteins and saponin based Matrix‐M1 adjuvant | Novavax |
Phase I/II |
– | [ 103 , 165 ] |
| FlowVax peptide vaccine | Adjuvanted microsphere peptide vaccine targeting SARS‐CoV‐2 nucleocapsid | Adjuvanted microsphere peptide vaccine targeting SARS‐CoV‐2 nucleocapsid | Flow Pharma | Preclinical | Ebola, Marburg, HIV, Zika, Influenza, HPV | [ 103 ] |
| Signal peptide vaccine (VXL‐301, 302, 303) | Peptide | Signal peptide vaccine using VaxHit bioinformatics platform | Vaxil Bio | Preclinical | – | [ 103 ] |
| Epitope‐based peptide vaccine | Peptide | Synthetic long peptide vaccine candidate for S and M proteins (Vaccinomics strategy) | OncoGen | Preclinical | – | [ 103 ] |
| Ii‐Key peptide | Peptide | Synthetic peptides that mimic the epitopes of the SARS‐CoV‐2 using EpiVax's computational tools and li‐Key technology | Generex/ EpiVax | Preclinical | Influenza, HIV, SARS‐CoV | [ 138 ] |
| Monoclonal antibodies | ||||||
| Gimsilumab (KIN 1901) | Human Mab | Targets GM‐CSF | Roivant Sciences |
Phase II |
– | [ 103 ] |
| TJM2 (TJ003234) | Neutralizing antibody | Neutralizing antibody against human GM‐CSF | I‐Mab |
Phase I/II |
– | [ 103 ] |
| Lenzilumab | Neutralizing antibody | Neutralizing antibody against human GM‐CSF | Humanigen |
Phase III Compassionate use |
– | [ 103 ] |
| Namilumab (IZN‐101, AMG203) | Human Mab | Targets GM‐CSF | Izana Bioscience | Compassionate use | RA and ankylosing spondylitis | [ 103 , 122 ] |
| TZLS‐501 | Human monoclonal antibody | Mab against IL‐6 receptor | Tiziana Life Sciences | Preclinical | Autoimmune diseases | [ 103 ] |
| AT‐100 | Protein | Human recombinant surfactant protein D (rhSP‐D) | Airway Therapeutics | Preclinical | Bronchopulmonary dysplasia (BPD) in premature infants | [ 103 ] |
| Tocilizumab (Actemra) | Human Mab | Mab against IL‐6 receptor | Roche |
Phase III ChiCTR2000029765 |
RA, giant cell arthritis | [ 121 , 123 ] |
| Sarilumab (Kevzara) | Human Mab | IL‐6 receptor blocker | Sanofi and Regeneron |
Phase II/III |
RA | [ 103 ] |
| Convalescent sera | Passive neutralizing antibody | Passive neutralizing antibody developed in COVID‐19 recovered patients | ‐ |
Phase 0 ChiCTR2000029757 Phase II |
– | [ 125 ] |
| Anti‐inflammatory molecules | ||||||
| Dexamethasone | Glucocorticoid | Anti‐inflammatory and immunosuppressant | – |
Phase II/III |
– | [ 126 ] |
| N‐acetylcysteine | Amino acid (Glutathione precursor) | Anti‐inflammatory, antioxidant, antiviral, and anticoagulant | – |
Phase II Phase III Phase IV |
– | [ 127 , 128 ] |
| Cyclosporine A | Calcineurin inhibitor | Anti‐inflammatory and immunosuppressant |
Phase IIa Phase IV |
[ 129 , 130 , 131 ] | ||
| Budesonide | Corticosteroid | Anti‐inflammatory | – |
Phase IV |
||
| Vitamin D | Steroid | Anti‐inflammatory, antioxidant, immunomodulatory, | – |
Phase III |
– | [ 132 ] |
8. Conclusion
The exceptional speed, with which the SARS‐CoV‐2 pandemic is escalating across the world, is causing global anxiety to attain new heights. COVID‐19 has become the reason for the globally prevalent fear, stress, and concern. All these are natural and common reactions to the fast‐changing and undefined situation that all of us find ourselves in. Further, there is a noticeable degree of fear, stress, and apprehension in certain groups with the higher risk factor, such as children, older people, health care professionals, frontline workers, and people with pre‐existing maladies (such as asthma, diabetes, heart diseases, and hypertension). The impact of the crisis on people's mental health is also of major concern. As the new measures to prevent the infection are being introduced, such as quarantine (social isolation), cases of depression and loneliness are expected to rise.
The disruptive effects of the COVID‐19 outbreak are dominating our daily lives; therefore, it is of prime importance that we should manage and react properly to this challenging situation unfolding fast in our lives. The world is in total chaos, lives have frozen, and everything around us has come to a standstill situation. Nature has forced us to stay indoors. Let us hope, together; we will be able to find a cure and curb the grave epidemic as soon as possible.
Many drug candidates are under investigation for the disease. Initial reports suggest some of them are promising at this juncture. However, whether these drugs will act following virus dynamics and host response is a big question. How they will behave in a clinical trial is yet another long‐term debate. Besides, their affordability in developing countries is a significant concern. Antiviral therapy shows a wide margin of ineffectiveness as many viruses possess extraordinary genetic adaptability, which endows them with the ability to escape antiviral repression. In some instances, the viruses reclaim advantage over the host by online mutagenesis, which creates novel strains with additional acquired resistance to the existing antivirals. Therefore, finding new and multifaceted antiviral agents is the unmet need of the hour. An approach analogous to cancer nanotechnology, including early diagnosis, targeted delivery, and combinatorial approaches can be followed to tackle this deadly viral nanovector.
US‐FDA's extraordinary speed in reviewing emergency use authorization (EUAs) and investigational new drug (IND) applications and their prompt approval to expedite anti‐COVID‐19 therapeutics into the clinical development is highly appreciable. WHO and partners had launched an international clinical trial named Solidarity, for comparing potential options against standard of care to find an effective treatment for COVID‐19. The trial's aim is to expedite the discovery of any potential drugs that can slow down the disease progression or improve patient's survival rate. On 4th July 2020, WHO has withdrawn hydroxychloroquine and lopinavir/ritonavir from their Solidarity trial. The Solidarity Trial's committee found that these drugs were ineffective in treating the infection. Similar findings were also reported from the UK's Recovery trial. Further, US‐FDA's approval for the emergency use of remdesivir and other drugs, and other medical countermeasures (MCMs) and facilitating their availability is another noteworthy move in combating COVID‐19 and is an example of using science and technology at its best in protecting and rescuing the world from emerging chemical, biological, radiological and nuclear (CBRN) threats. With the current focused, rapid, worldwide discoveries in basic science, translational research, and clinical studies, and the expedited regulatory scrutiny, it is anticipated that preventive and curative therapies for COVID‐19 will be a reality in the near future.
Author Contributions
T.D: writing–original draft preparation, conceptualization, visualization. A.G: Writing—original draft preparation. J.M.: Writing—reviewing and editing, visualization, investigation. U.B.K.: Writing—reviewing and editing, conceptualization, investigation, supervision. J.J.P.: Writing—original draft preparation, reviewing and editing, conceptualization, investigation, supervision.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
J.J.P. thanks ICMR‐DHR for fellowship.
Biographies
Taru Dube is a senior research fellow and pursuing her Ph.D. at the Institute of Nano Science and Technology (INST, Punjab, India). Her research work is focused on the development of peptide‐based nanostructures/theranostics for brain delivery. Before joining the Ph.D. programme at INST, she earned her Masters degree in Pharmaceutics (M. Pharm.) from the University of Pune, India. She is also a recipient of University medal in her graduation in pharmacy from Bundelkhand University, U.P., India.

Amrito Ghosh Majumdar is a final year student of integrated B.Tech‐M.Tech dual degree in Biotechnology at the KIIT School of Biotechnology, KIIT University, Odisha, India. He is currently pursuing a research internship at INST, India. He completed his Secondary Education in 2012 and Higher Secondary Education in 2014 from New Barrackpur Colony Boys High School, New Barrackpur, Kolkata, India.

Jibanananda Mishra is an experienced cell biologist and biomedical scientist currently working as an associate professor in the School of Bioengineering and Biosciences, Lovely Professional University (Punjab, India). Besides academic research and teaching in India and at the University of Colorado Denver (USA), Dr. Mishra has years of experience in the R&D divisions of renowned pharmaceutical industries such as Dr. Reddy's Laboratories Ltd. India, etc. He has significantly contributed to the development of several biopharmaceuticals and antibiotics. His research interest focuses on understanding the pathogenesis of difficult‐to‐treat diseases such as neurological disorders and cancers, and exploring the possibilities of effective therapeutics delivery to restore normal physiological functions.

Uday B. Kompella is a professor of Pharmaceutical Sciences, Ophthalmology, and Bioengineering at the University of Colorado, Anschutz Medical Campus, with research interests in the areas of drug discovery, drug delivery, and nanotechnology for treating a variety of degenerative, neovascular, and inflammatory disorders including retinitis pigmentosa, age‐related macular degeneration (AMD), diabetic retinopathy, and cancers. He is a codirector of the Colorado Center for Nanomedicine and Nanosafety.

Jiban Jyoti Panda is a scientist at the Institute of Nano Science and Technology (INST, Govt. of India), (Punjab, India). She pursued Ph.D. in Biotechnology at ICGEB, New Delhi. She carried out her postdoctoral research from Skagg's School of Pharmacy and Pharmaceutical Sciences, University of Colorado, Denver. Her research interest involves developing drug delivery systems across various physiological barriers, especially the blood‐brain barrier and developing theranostic nanosystems for different neural disorders. Her group also works in the field of the development of peptide‐based nanostructures for targeted drug delivery applications. She has extensive experience in developing nanoformulations for various diseases.

Contributor Information
Uday B. Kompella, Email: Uday.Kompella@CUAnschutz.edu.
Jiban Jyoti Panda, Email: jyoti@inst.ac.in.
References
- 1. Velavan T. P., Meyer C. G., Trop. Med. Int. Health 2020, 25, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lai C.‐C., Shih T.‐P., Ko W.‐C., Tang H.‐J., Hsueh P.‐R., Int. J. Antimicrob. Agents 2020, 55, 105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization , 2020. Coronavirus disease (COVID‐19) weekly epidemiological update and weekly operational update. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/, (accessed: September 2020).
- 4. World Health Organization , 2020. COVID‐19 strategy update. https://www.who.int/publications-detail/covid-19-strategy-update-14-april-2020 (accessed: September 2020).
- 5. World Health Organization , 2020. Naming the coronavirus disease (COVID‐19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus- 2019/technical‐guidance/naming‐the‐coronavirus‐disease‐(covid‐2019)‐and‐the‐virus‐ that‐causes‐it/, (accessed: September 2020).
- 6. Sanche S., Lin Y. T., Xu C., Romero‐Severson E., Hengartner N. W., Ke R., medRxiv. Preprint 2020. 10.1101/2020.02.07.20021154 [DOI] [Google Scholar]
- 7. Xie Y., Wang Z., Liao H., Marley G., Wu D., Tang W., BMC Infect. Dis. 2020, 20, 640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Al‐Tawfiq J. A., Travel Med. Infect. Dis. 2020, 35, 101608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., N. Engl. J. Med. 2020, 382, 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan J. F.‐W., Yuan S., Kok K.‐H., To K. K.‐W., Chu H., Yang J., Xing F., Liu J., Yip C. C.‐Y., Poon R. W.‐S., Lancet 2020, 395, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tyrrell D., Bynoe M., Lancet 1966, 287, 76. [DOI] [PubMed] [Google Scholar]
- 12. Zumla A., Chan J. F., Azhar E. I., Hui D. S., Yuen K.‐Y., Nat. Rev. Drug Discovery 2016, 15, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lau S. K., Feng Y., Chen H., Luk H. K., Yang W.‐H., Li K. S., Zhang Y.‐Z., Huang Y., Song Z.‐Z., Chow W.‐N., J. Virol. 2015, 89, 10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fehr A. R., Perlman S., Methods Mol. Biol. 2015, 1282, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui J., Li F., Shi Z.‐L., Nat. Rev. Microbiol. 2019, 17, 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan J. F., Lau S. K., To K. K., Cheng V. C., Woo P. C., Yuen K.‐Y., Clin. Microbiol. Rev. 2015, 28, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Forni D., Cagliani R., Clerici M., Sironi M., Trends Microbiol. 2017, 25, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J. H., Wang H., Crameri G., Hu Z., Zhang H., Zhang J., McEachern J., Field H., Daszak P., Eaton B. T., Zhang S., Wang L.‐F., Science 2005, 310, 676. [DOI] [PubMed] [Google Scholar]
- 19. Song H.‐D., Tu C.‐C., Zhang G.‐W., Wang S.‐Y., Zheng K., Lei L.‐C., Chen Q.‐X., Gao Y.‐W., Zhou H.‐Q., Xiang H., Zheng H.‐J., Chern S.‐W. W., Cheng F., Pan C.‐M., Xuan H., Chen S.‐J., Luo H.‐M., Zhou D.‐H., Liu Y.‐F., He J.‐F., Qin P.‐Z., Li L.‐H., Ren Y.‐Q., Liang W.‐J., Yu Y.‐D., Anderson L., Wang M., Xu R.‐H., Wu X.‐W., Zheng H.‐Y., Chen J.‐D., et al., Proc. Natl. Acad. Sci. USA 2005, 102, 2430.15695582 [Google Scholar]
- 20. Lau S. K., Woo P. C., Li K. S., Huang Y., Tsoi H.‐W., Wong B. H., Wong S. S., Leung S.‐Y., Chan K.‐H., Yuen K.‐Y., Proc. Natl. Acad. Sci. USA 2005, 102, 14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haagmans B. L., Al Dhahiry S. H., Reusken C. B., Raj V. S., Galiano M., Myers R., Godeke G.‐J., Jonges M., Farag E., Diab A., Lancet Infect. Dis. 2014, 14, 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. De Wit E., Van Doremalen N., Falzarano D., Munster V. J., Nat. Rev. Microbiol. 2016, 14, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. World Health Organization , 2020. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003 (Based on data as of the 31 December 2003). https://www.who.int/csr/sars/country/table2004_04_21/en/, (accessed: September 2020).
- 24. World Health Organization , 2020. Epidemic and pandemic‐prone diseases (MERS situation update, January 2020). http://www.emro.who.int/pandemic-epidemic- diseases/mers‐cov/mers‐situation‐update‐january‐2020.html, (accessed: September 2020).
- 25. Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N., Decroly E., Antiviral Res. 2020, 176, 104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., Nat. Med. 2020, 26, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walls A. C., Park Y.‐J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Cell 2020, 181, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kostarelos K., Nat. Nanotechnol. 2020, 15, 343. [DOI] [PubMed] [Google Scholar]
- 29. Steinmetz N. F., Mol. Pharmaceutics 2013, 10, 1. [DOI] [PubMed] [Google Scholar]
- 30. Rauch S., Jasny E., Schmidt K. E., Petsch B., Front. Immunol. 2018, 9. 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bar‐On Y. M., Flamholz A., Phillips R., Milo R., Elife 2020, 9, 57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Prasad S., Potdar V., Cherian S., Abraham P., Basu A., Team I.‐N. N., Indian J. Med. Sci. 2020, 151, 241. 10.4103/ijmr.IJMR_577_20. [DOI] [Google Scholar]
- 33. Antibodies‐online.com, 2020 . SARS‐CoV‐2 life cycle: stages and inhibition targets. https://www.antibodies-online.com/resources/18/5410/sars-cov-2-life-cycle-stages-and-inhibition-targets/ (accessed: September 2020).
- 34. Elfiky A. A., Life Sci. 2020, 253, 117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoffmann M., Kleine‐Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.‐H., Nitsche A., Cell 2020, 181, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carey R. M., Curr. Hypertens. Rep. 2017, 19, 21.28271418 [Google Scholar]
- 37. Arendse L. B., Danser A. J., Poglitsch M., Touyz R. M., Burnett J. C., Llorens‐ Cortes C., Ehlers M. R., Sturrock E. D., Pharmacol. Rev. 2019, 71, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Flores‐Munoz M., Smith N., Haggerty C., Milligan G., Nicklin S., J. Physiol. 2011, 589, 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang D., Fang L., Shi Y., Zhang H., Gao L., Peng G., Chen H., Li K., Xiao S., J. Virol. 2016, 90, 2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garcia V., Shkolnik B., Milhau L., Falck J. R., Schwartzman M. L., J. Pharmacol. Exp. Ther. 2016, 356, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patel V. B., Zhong J.‐C., Grant M. B., Oudit G. Y., Circ. Res. 2016, 118, 1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang D.‐D., Gao Z.‐X., Vio C. P., Xiao Y., Wu P., Zhang H., Guo X.‐W., Meng X.‐X., Gu L., Wang J.‐L., Hypertension 2018, 72, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kjeldsen K., Exp. Clin. Cardiol. 2010, 15, e96. [PMC free article] [PubMed] [Google Scholar]
- 44. Bielecka‐Dabrowa A., Mikhailidis D. P., Jones L., Rysz J., Aronow W. S., Banach M., Int. J. Cardiol. 2012, 158, 12. [DOI] [PubMed] [Google Scholar]
- 45. Skogestad J., Aronsen J. M., Front. Physiol. 2018, 9, 1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lipsitch M., Swerdlow D. L., Finelli L., N. Engl. J. Med. 2020, 382, 1194. [DOI] [PubMed] [Google Scholar]
- 47. Rothan H. A., Byrareddy S. N., J. Autoimmun. 2020, 109, 102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lauer S. A., Grantz K. H., Bi Q., Jones F. K., Zheng Q., Meredith H. R., Azman A. S., Reich N. G., Lessler J., Ann. Intern. Med. 2020, 172, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. World Health Organization , 2020. WHO Coronavirus Disease (COVID‐19) Dashboard. https://covid19.who.int/?gclid=CjwKCAjwwab7BRBAEiwAapqpTIXx50 DJR‐ApRDEFiT5cqX7RYSY8‐xExRAYcmwGzPLJPZXTNHrGQYxoC‐ rIQAvD_BwE, (accessed: September 2020).
- 50. He F., Deng Y., Li W., J. Med. Virol. 2020, 92, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., a. Xia J., Yu T., Zhang X., Zhang L., Lancet 2020, 395, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Lancet 2020, 395, 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu Y., Li X., Zhu B., Nat. Med. 2020, 26, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Centers for Disease Control and Prevention , 2020. Infection control guidance for healthcare professionals about coronavirus (COVID‐19). https://www.cdc.gov/coronavirus/2019-ncov/hcp/infection-control.html, (accessed: September 2020).
- 55. World Health Organization , 2020. Coronavirus disease (COVID‐19) advice for the public. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice- for‐public, (accessed: September 2020).
- 56. U.S. Food & Drug Administration , 2020. Emergency Use Authorization (EUA) information, and list of all current EUAs. https://www.fda.gov/emergency-preparedness‐and‐response/mcm‐legal‐regulatory‐and‐policy‐framework/emergency‐ use‐authorization#abouteuas, (accessed: September 2020).
- 57. Reagan Udall Foundation , 2020. COVID‐19 Company Directory. https://reaganudall.org/companies-developing-covid-19-therapies, (accessed: September 2020).
- 58. Lou Z., Sun Y., Rao Z., Trends Pharmacol. Sci. 2014, 35, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Loregian A., Marsden H. S., Palu G., Rev. Med. Virol. 2002, 12, 239. [DOI] [PubMed] [Google Scholar]
- 60. Schroder M., Bowie A., Biochem. Soc. Trans. 2007, 35, 1512. [DOI] [PubMed] [Google Scholar]
- 61. Konig R., Stertz S., Zhou Y., Inoue A., Hoffmann H.‐H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., Nature 2010, 463, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ullah H., Hou W., Dakshanamurthy S., Tang Q., Oncotarget 2019, 10, 3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin K., Gallay P., Antiviral Res. 2013, 99, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Smith T., Bushek J., LeClaire A., Prosser T., Clinical Drug Information, Clinical Solutions 2020. https://www.elsevier.com/__data/assets/pdf_file/0016/1004173/COVID-19-Drug-Therapy_5.28.2020.pdf, (accessed: August 2020).
- 65. Duan Y., Zhu H.‐L., Zhou C., Drug Discovery Today 2020, 25, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu C., Liu Y., Yang Y., Zhang P., Zhong W., Wang Y., Wang Q., Xu Y., Li M., Li X., Acta Pharm. Sin. B 2020, 10, 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lee C., Mar. Drugs 2019, 17, 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ashfaq U. A., Javed T., Rehman S., Nawaz Z., Riazuddin S., Virol. J. 2011, 8, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sisay M., Pharmacol. Res. 2020, 156, 104779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. ul Qamar M. T., Alqahtani S. M., Alamri M. A., Chen L.‐L., J. Pharm. Anal. 2020, 10, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. The Science Times , 2020. Favilavir: First approved drug to possibly treat coronavirus. https://www.sciencetimes.com/articles/25053/20200317/favilavir-first-approve-drug-treat-coronavirus.htm, (accessed: April 2020).
- 72. Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G., Cell Res. 2020, 30, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yang Y., Islam M. S., Wang J., Li Y., Chen X., Int. J. Biol. Sci 2020, 16, 1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang D., Zhang B., Lv J.‐T., Sa R.‐N., Zhang X.‐M., Lin Z.‐J., Pharmacol. Res. 2020, 157, 104882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F., Cell Discovery 2020, 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. World Health Organization , 2020. Outline of trial designs for experimental therapeutics. https://www.who.int/publications-detail/outline-of-trial-designs-for-experimental-therapeutics, (accessed: September 2020).
- 77. Al‐Tawfiq J. A., Al‐Homoud A. H., Memish Z. A., Travel Med. Infect. Dis. 2020, 34, 101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gordon C. J., Tchesnokov E. P., Woolner E., Perry J. K., Feng J. Y., Porter D. P., Gotte M., J. Biol. Chem. 2020, 295, 6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Costanzo M., De Giglio M., Roviello G., Curr. Med. Chem. 2020, 27, 4536. [DOI] [PubMed] [Google Scholar]
- 80. Holshue M. L., DeBolt C., Lindquist S., Lofy K. H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S. I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M. A., Weldon W. C., Biggs H. M., Uyeki T. M., Pillai S. K., N. Engl. J. Med. 2020, 382, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Warren T., Jordan R., Lo M., Soloveva V., Ray A., Bannister R., Mackman R., Perron M., Stray K., Feng J., Xu Y., Wells J., Stuthman K., Welch L., Doerffler E., Zhang L., Chun K., Hui H., Neville S., Lew W., Park Y., Babusis D., Strickley R., Wong P., Swaminathan S., Lee W., Mayers D., Cihlar T., Bavari S., Open Forum Infect. Dis. 2015, 2, 67. [Google Scholar]
- 82. Sheahan T. P., Sims A. C., Graham R. L., Menachery V. D., Gralinski L. E., Case J. B., Leist S. R., Pyrc K., Feng J. Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M. O., Mackman R. L., Spahn J. E., Palmiotti C. A., Siegel D., Ray A. S., Cihlar T., Jordan R., Denison M. R., Baric R. S., Sci. Transl. Med. 2017, 9, eaal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Sheahan T. P., Sims A. C., Leist S. R., Schäfer A., Won J., Brown A. J., Montgomery S. A., Hogg A., Babusis D., Clarke M. O., Nat. Commun. 2020, 11, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Beigel J. H., Tomashek K. M., Dodd L. E., Mehta A. K., Zingman B. S., Kalil A. C., Hohmann E., Chu H. Y., Luetkemeyer A., Kline S., N. Engl. J. Med. 2020. 10.1056/NEJMoa2007764. [DOI] [Google Scholar]
- 85. Sanders J. M., Monogue M. L., Jodlowski T. Z., Cutrell J. B., J. Am. Med. Assoc. 2020, 323, 1824. [DOI] [PubMed] [Google Scholar]
- 86. Goldman J. D., Lye D. C., Hui D. S., Marks K. M., Bruno R., Montejano R., Spinner C. D., Galli M., Ahn M.‐Y., Nahass R. G., N. Engl. J. Med. 2020. 10.1056/NEJMoa2015301. [DOI] [Google Scholar]
- 87. Spinner C. D., Gottlieb R. L., Criner G. J., Lopez J. R. A., Cattelan A. M., Viladomiu A. S., Ogbuagu O., Malhotra P., Mullane K. M., Castagna A., J. Am. Med. Assoc. 2020, 324, 1048. [Google Scholar]
- 88. Gautret P., Lagier J.‐C., Parola P., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V. E., Dupont H. T., Honore S., Colson P., Chabriere E., Scola B. L., Rolain J.‐M., Brouqui P., Raoult D., Int. J. Antimicrob. Agents 2020, 56, 105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89. Gao J., Tian Z., Yang X., BioSci. Trends 2020, 14, 72. [DOI] [PubMed] [Google Scholar]
- 90. Al‐Bari M. A. A., Pharmacol. Res. Perspect. 2017, 5, e00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Xue J., Moyer A., Peng B., Wu J., Hannafon B. N., Ding W.‐Q., PLoS One 2014, 9, e109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Te Velthuis A. J., van den Worm S. H., Sims A. C., Baric R. S., Snijder E. J., van Hemert M. J., PLoS Pathog. 2010, 6, e1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Clin. Infect. Dis. 2020, 71, 732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M., Cell Discovery 2020, 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chen Z., Hu J., Zhang Z., Jiang S., Han S., Yan D., Zhuang R., Hu B., Zhang Z., MedRxiv 2020. 10.1101/2020.03.22.20040758 [DOI] [Google Scholar]
- 96. World Health Organization , 2020. “Solidarity” clinical trial for COVID‐19 treatments. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments, (accessed: September 2020).
- 97. Zhang C., Zhang Y., Qin Y., Zhang Q., Liu Q., Shang D., Lu H., Li X., Zhou C., Huang F., Msystems 2019, 4, e00431.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., N. Engl. J. Med. 2020, 382, 1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Martinez M. A., Antimicrob. Agents Chemother. 2020, 64, e00399. 10.1128/AAC.00399-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Jia H. P., Look D. C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford‐Lenane C., Perlman S., McCray P. B., J. Virol. 2005, 79, 14614. 10.1128/JVI.79.23.14614-14621.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Hamming I., Timens W., Bulthuis M., Lely A., Navis G., van Goor H., J. Pathol. 2004, 203, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Hoffmann M., Schroeder S., Kleine‐Weber H., Muller M. A., Drosten C., Pohlmann S., Antimicrob. Agents Chemother. 2020, 64, e00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. BioWorld , 2020. Biopharma products in development for COVID‐19. https://www.bioworld.com/COVID19products#vac, (accessed: September 2020).
- 104. Bell E., Nat. Rev. Immunol. 2002, 2, 381. [DOI] [PubMed] [Google Scholar]
- 105. Janeway C. A. Jr, Travers P., Walport M., Shlomchik M. J., in Immunobiology: The Immune System in Health and Disease, 5th ed., Garland Science, New York: 2001. [Google Scholar]
- 106. Vabret N., Britton G. J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M. D., Immunity 2020, 52, 910.32505227 [Google Scholar]
- 107. InvivoGen , 2020. Predicted immune responses to SARS‐CoV‐2. https://www.invivogen.com/spotlight-covid-19-predicted-immune-responses. (accessed: September 2020).
- 108. Tay M. Z., Poh C. M., Rénia L., MacAry P. A., Ng L. F., Nat. Rev. Immunol. 2020, 20, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Wong C., Lam C., Wu A., Ip W., Lee N., Chan I., Lit L., Hui D., Chan M., Chung S., Clin. Exp. Immunol. 2004, 136, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Jackson L. A., Anderson E. J., Rouphael N. G., Roberts P. C., Makhene M., Coler R. N., McCullough M. P., Chappell J. D., Denison M. R., Stevens L. J., N. Engl. J. Med. 2020. 10.1056/NEJMoa2022483. [DOI] [Google Scholar]
- 111. Zhu F.‐C., Li Y.‐H., Guan X.‐H., Hou L.‐H., Wang W.‐J., Li J.‐X., Wu S.‐P., Wang B.‐S., Wang Z., Wang L., Lancet 2020, 395, 1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zhu F.‐C., Guan X.‐H., Li Y.‐H., Huang J.‐Y., Jiang T., Hou L.‐H., Li J.‐X., Yang B.‐F., Wang L., Wang W.‐J., Lancet 2020, 396, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Mulligan M. J., Lyke K. E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K. A., Nature 2020, 1. 10.1038/s41586-020-2639-4. [DOI] [Google Scholar]
- 114. Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Science 2020, 369, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Smith T. R. F., Patel A., Ramos S., Elwood D., Zhu X., Yan J., Gary E. N., Walker S. N., Schultheis K., Purwar M., Xu Z., Walters J., Bhojnagarwala P., Yang M., Chokkalingam N., Pezzoli P., Parzych E., Reuschel E. L., Doan A., Tursi N., Vasquez M., Choi J., Tello‐Ruiz E., Maricic I., Bah M. A., Wu Y., Amante D., Park D. H., Dia Y., Ali A. R., Zaidi F. I., et al., Nat. Commun. 2020, 11, 2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Folegatti P. M., Ewer K. J., Aley P. K., Angus B., Becker S., Belij‐Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E. A., Lancet 2020, 396, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. van Doremalen N., Lambe T., Spencer A., Belij‐Rammerstorfer S., Purushotham J. N., Port J. R., Avanzato V., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E. R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade‐ White K., Pérez‐Pérez L., Bissett C., Gilbride C., Williamson B. N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P. W., Scott D., Saturday G., de Wit E., Gilbert S. C., Munster V. J., BioRxiv 2020. 10.1101/2020.05.13.093195 [DOI] [Google Scholar]
- 118. New York Times , 2020. Coronavirus Vaccine Tracker. https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html (accessed: September 2020).
- 119. The Times of Israel , 2020. Israeli‐made oral vaccine for coronavirus on track, but testing will take months. https://www.timesofisrael.com/israeli-made-oral-vaccine-for-coronavirus-on-track-but-testing-will-take-months/, (accessed: September 2020).
- 120. Mehta P., McAuley D. F., Brown M., Sanchez E., Tattersall R. S., Manson J. J., Lancet 2020, 395, 1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Zhang S., Li L., Shen A., Chen Y., Qi Z., Clin. Drug Invest. 2020, 40, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Taylor P. C., Saurigny D., Vencovsky J., Takeuchi T., Nakamura T., Matsievskaia G., Hunt B., Wagner T., Souberbielle B., N. S. G. for the , Arthritis Res. Ther. 2019, 21, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Xu X., Han M., Li T., Sun W., Wang D., Fu B., Zhou Y., Zheng X., Yang Y., Li X., Proc. Natl. Acad. Sci. USA 2020, 117, 10970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gruda M. C., Ruggeberg K.‐G., O'Sullivan P., Guliashvili T., Scheirer A. R., Golobish T. D., Capponi V. J., Chan P. P., PLoS One 2018, 13, e0191676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Casadevall A., Pirofski L.‐A., J. Clin. Invest. 2020, 130, 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Horby P., Lim W. S., Emberson J. R., Mafham M., Bell J. L., Linsell L., Staplin N., Brightling C., Ustianowski A., Elmahi E., N. Engl. J. Med. 2020. 10.1056/NEJMoa2021436. [DOI] [Google Scholar]
- 127. De Flora S., Balansky R., La Maestra S., FASEB J. 2020, 34, 13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Jorge‐Aaron R.‐M., Rosa‐Ester M.‐P., Future Med. 2020, 15, 959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Pfefferle S., Schopf J., Kogl M., Friedel C. C., Muller M. A., Carbajo‐Lozoya J., Stellberger T., von Dall'Armi E., Herzog P., Kallies S., PLoS Pathog. 2011, 7, e1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. de Wilde A. H., Pham U., Posthuma C. C., Snijder E. J., Virology 2018, 522, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Cour M., Ovize M., Argaud L., BioMed Cent. 2020, 24, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Martineau A. R., Forouhi N. G., Lancet Diabetes Endocrinol. 2020, 8, 735.32758429 [Google Scholar]
- 133. Baig M. H., Ahmad K., Saeed M., Alharbi A. M., Barreto G. E., Ashraf G. M., Choi I., Biomed. Pharmacother. 2018, 103, 574. [DOI] [PubMed] [Google Scholar]
- 134. Rastogi S., Shukla S., Kalaivani M., Singh G. N., Drug Discovery Today 2019, 24, 148. [DOI] [PubMed] [Google Scholar]
- 135. Fisher E., Pavlenko K., Vlasov A., Ramenskaya G., Pharm. Med. 2019, 33, 9. [DOI] [PubMed] [Google Scholar]
- 136. Skalickova S., Heger Z., Krejcova L., Pekarik V., Bastl K., Janda J., Kostolansky F., Vareckova E., Zitka O., Adam V., Viruses 2015, 7, 5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Vilas Boas L. C. P., Campos M. L., Berlanda R. L. A., de Carvalho Neves N., Franco O. L., Cell. Mol. Life Sci. 2019, 76, 3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Genetic Engineering & Biotechnology News , 2020. Catching Up to Coronavirus: Top 60 Treatments in Development. https://www.genengnews.com/virology/coronavirus/catching-up-to-coronavirus-top-60-treatments-in-development/, (accessed: September 2020).
- 139. Fries C. N., Curvino E. J., Chen J.‐L., Permar S. R., Fouda G. G., Collier J. H., Nature Nanotechnology 2020, 1. [DOI] [PubMed] [Google Scholar]
- 140. Singh L., Kruger H. G., Maguire G. E., Govender T., Parboosing R., Therapeutic advances in infectious disease 2017, 4, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Weiss C., Carriere M., Fusco L., Capua I., Regla‐Nava J. A., Pasquali M., Scott J. A., Vitale F., Unal M. A., Mattevi C., ACS Nano 2020, 14, 6383. [DOI] [PubMed] [Google Scholar]
- 142. Itani R., Tobaiqy M., Al Faraj A., Theranostics 2020, 10, 5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Donalisio M., Leone F., Civra A., Spagnolo R., Ozer O., Lembo D., Cavalli R., Pharmaceutics 2018, 10, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. LaBauve A. E., Rinker T. E., Noureddine A., Serda R. E., Howe J. Y., Sherman M. B., Rasley A., Brinker C. J., Sasaki D. Y., Negrete O. A., Sci. Rep. 2018, 8, 13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Kumar P., Lakshmi Y. S., Bhaskar C., Golla K., Kondapi A. K., PLoS One 2015, 10, e0140399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Liu L., Ren J., He Z., Men K., Mao Y., Ye T., Chen H., Li L., Xu B., Wei Y., Sci. Rep. 2017, 7, 10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Dormont F., Brusini R., Cailleau C., Reynaud F., Peramo A., Gendron A., Mougin J., Gaudin F., Varna M., Couvreur P., Sci. Adv. 2020, 6, eaaz5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Freeling J. P., Koehn J., Shu C., Sun J., Ho R. J., AIDS 2014, 28, 2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Aderibigbe B. A., Molecules 2017, 22, 1370. [Google Scholar]
- 150. Baram‐Pinto D., Shukla S., Perkas N., Gedanken A., Sarid R., Bioconjugate Chem. 2009, 20, 1497. [DOI] [PubMed] [Google Scholar]
- 151. Burdusel A.‐C., Gherasim O., Grumezescu A. M., Mogoanta L., Ficai A., Andronescu E., Nanomaterials 2018, 8, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Galdiero S., Falanga A., Vitiello M., Cantisani M., Marra V., Galdiero M., Molecules 2011, 16, 8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Li Y., Lin Z., Zhao M., Xu T., Wang C., Hua L., Wang H., Xia H., Zhu B., ACS Appl. Mater. Interfaces 2016, 8, 24385. [DOI] [PubMed] [Google Scholar]
- 154. Zachar O., ScienceOpen Prepr. 2020. 10.14293/S2199-1006.1.SOR-.PPHBJEO.v1 [DOI] [Google Scholar]
- 155. Levina A. S., Repkova M. N., Zarytova V. F., Mazurkova N. A., in IEEE 15th Int. Conf. on Nanotechnology (IEEE‐NANO), IEEE, Piscataway, NJ: 2015, p. 1509. [Google Scholar]
- 156. Ye S., Shao K., Li Z., Guo N., Zuo Y., Li Q., Lu Z., Chen L., He Q., Han H., ACS Appl. Mater. Interfaces 2015, 7, 21571. [DOI] [PubMed] [Google Scholar]
- 157. Kumar P., Huo P., Zhang R., Liu B., Nanomaterials 2019, 9, 737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Statnano , 2020. Antibacterial and Antiviral Graphene‐based Coating for Public Settings. https://statnano.com/news/67560/Antibacterial-and-Antiviral-Graphene-based-Coating-for-Public-Settings#:~:text=Nano%20Graphene%20Inc.%2C%20also%20known,surfaces%20of%20public%20settings%20to, (accessed: May 2020).
- 159. Warnes S. L., Little Z. R., Keevil C. W., mBio 2015, 6, e01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Dhanasezhian A., Srivani S., Govindaraju K., Parija P., Sasikala S., Kumar M., Indian J. Geo‐Mar. Sci. 2019, 48, 1252. [Google Scholar]
- 161. Colino C. I., Millán C. G., Lanao J. M., Int. J. Mol. Sci. 2018, 19, 1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Doria G., Conde J., Veigas B., Giestas L., Almeida C., Assuncao M., Rosa J., Baptista P. V., Sensors 2012, 12, 1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Moitra P., Alafeef M., Dighe K., Frieman M. B., Pan D., ACS Nano 2020, 14, 7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Palmieri V., Papi M., Nano Today 2020, 33, 100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J. S., Zhu M., Cloney‐Clark S., Zhou H., N. Engl. J. Med. 2020. 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]


