Abstract
SARS‐CoV‐2, the causative agent of COVID‐19, has been responsible for a million deaths worldwide as of September 2020. At the time of this writing, there are no available US FDA−approved therapeutics for the treatment of SARS‐CoV‐2 infection. Here, we describe a detailed protocol to generate recombinant (r)SARS‐CoV‐2 using reverse‐genetics approaches based on the use of a bacterial artificial chromosome (BAC). This method will allow the production of mutant rSARS‐CoV‐2—which is necessary for understanding the function of viral proteins, viral pathogenesis and/or transmission, and interactions at the virus‐host interface—and attenuated SARS‐CoV‐2 to facilitate the discovery of effective countermeasures to control the ongoing SARS‐CoV‐2 pandemic. © 2020 Wiley Periodicals LLC.
Basic Protocol: Generation of recombinant SARS‐CoV‐2 using a bacterial artificial chromosome
Support Protocol: Validation and characterization of rSARS‐CoV‐2
Keywords: bacterial artificial chromosome, coronavirus, COVID‐19, infectious clone, reverse genetics, SARS‐CoV‐2, virus rescue
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the causative agent of coronavirus disease 2019 (COVID‐19), a life‐threatening respiratory illness in humans that originated in December 2019 in Wuhan, China (Andersen, Rambaut, Lipkin, Holmes, & Garry, 2020; Lu et al., 2020; Wu et al., 2020). In only 9 months since its first identification, SARS‐CoV‐2 has caused approximately 16 million confirmed COVID‐19 cases and more than 1 million deaths in over 200 countries (https://www.who.int/emergencies/diseases/novel‐coronavirus‐2019). SARS‐CoV‐2 is one of three zoonotic coronaviruses that have recently caused outbreaks in the human population, together with severe acute respiratory syndrome coronavirus (SARS‐CoV) in 2002‐2003 (Ksiazek et al., 2003; Peiris et al., 2003) and Middle East respiratory syndrome coronavirus (MERS‐CoV) in 2012‐2013 (van Boheemen et al., 2012; Zaki, van Boheemen, Bestebroer, Osterhaus, & Fouchier, 2012). Although SARS‐CoV and MERS‐CoV have higher fatality rates (Alfaraj et al., 2019; Gudbjartsson et al., 2020; Petersen et al., 2020; Sikkema et al., 2019), their outbreaks were successfully controlled. SARS‐CoV‐2, however, displays asymptomatic spread (Gudbjartsson et al., 2020; Mizumoto, Kagaya, Zarebski, & Chowell, 2020) and is more easily transmissible, as evidenced by the current magnitude of the ongoing pandemic. Despite intensive efforts, there are currently no United States (US) Food and Drug Administration (FDA)−approved vaccines (prophylactics) and/or antivirals (therapeutics) available to protect against SARS‐CoV‐2 infections and associated COVID‐19 disease (Liu et al., 2020; Williamson et al., 2020).
SARS‐CoV‐2 is a member of the Coronaviridae family, which are positive‐sense, single‐stranded RNA viruses with an ∼30 kb viral genome (Wu et al., 2020). As with SARS‐CoV and MERS‐CoV, the virion membrane is decorated with three structural proteins (Fig. 1A): the membrane (M), the envelope (E), and the spike (S). The unusually large viral (v)RNA genome is found within the viral particle encapsidated by the viral nucleocapsid (N) protein (Fig. 1A). These structural proteins are encoded at the 3′ end of the viral genome (Fig. 1B) and are essential for viral entry, replication and transcription, assembly, and budding. Scattered between the structural genes are the accessory genes that are important for virus propagation (Wu et al., 2020). Two‐thirds of the 5′ end of the viral genome encodes the replicase gene, containing two open reading frames (ORFs), ORF1a and ORF1b, in which the latter is translated via a ribosomal frameshift (Ziebuhr, 2005). Two polyproteins (PP1a and PP1ab) are synthesized by the translation of these two ORFs, and are subsequently cleaved into multiple nonstructural proteins (nsp) by viral proteases. Among these nsp are components of the replication‐transcription complex (nsp7, nsp8, and nsp12), helicase, endoribonuclease, 3′‐to‐5′ exonuclease, and 2′‐O‐ribose methyltransferase (Table 1).
Figure 1.
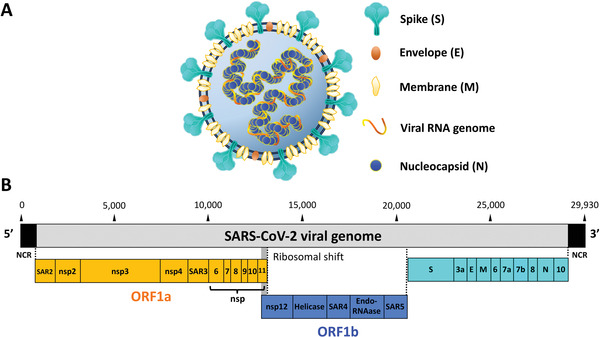
Virion structure and genomic organization of SARS‐CoV‐2. (A) Schematic representation of SARS‐CoV‐2 virion: The lipid bilayer surface of SARS‐CoV‐2 is decorated by the envelope (E), membrane protein (M), and spike (S) glycoproteins. S is the viral protein responsible for attachment and entry. Inside the virion is the positive‐sense, single‐stranded RNA viral genome that is encapsulated by the viral nucleocapsid (N) protein. (B) Schematic representation of SARS‐CoV‐2 genome organization: SARS‐CoV‐2 has a genome of ∼29,930 nucleotides. At the end of the viral genome are located the 5′ and 3′ untranslated non‐coding regions (NCR). The SARS‐CoV‐2 genome encodes for the viral ORF1a (orange) and ORF1b (blue). ORF1a polyprotein is cleaved into leader protein (SAR2), nsp2, nsp4, nsp4, 3C‐like proteinase (SAR3), nsp6, nsp7, nsp8, nsp9, nsp10, and nsp11. ORF1b polyprotein is generated through a ribosomal frameshift, and cleaved into the RNA‐dependent RNA polymerase, RdRp (nsp12), helicase, 3′‐to‐5′ exonuclease (SAR4), endoRNase, and 2′‐O‐ribose methyltransferase (SAR5). After the ORF1ab, SARS‐CoV‐2 genome encodes for the viral structural S, E, M and N proteins, along with the accessory proteins 3a, 6, 7a, 7b, 8, and 10.
Table 1.
List of SARS‐CoV‐2 Gene Products
| Gene | Locus | Protein product | Start | End |
|---|---|---|---|---|
| 5′ NCR | 5′ NCR | None | 1 | 177 |
| ORF1a | SAR2 | Leader protein | 178 | 717 |
| ORF1a | nsp2 | nsp2 | 718 | 2631 |
| ORF1a | nsp3 | nsp3 | 2632 | 8466 |
| ORF1a | nsp4 | nsp4 | 8467 | 9966 |
| ORF1a | SAR3 | 3C‐like proteinase | 9967 | 10,884 |
| ORF1a | nsp6 | nsp6 | 10,885 | 11,754 |
| ORF1a | nsp7 | nsp7 | 11,755 | 12,003 |
| ORF1a | nsp8 | nsp8 | 12,004 | 12,597 |
| ORF1a | nsp9 | nsp9 | 12,598 | 12,936 |
| ORF1a | nsp10 | nsp10 | 12,937 | 13,353 |
| ORF1b | nsp11 | nsp11 | 13,354 | 13,392 |
| ORF1b | nsp12 | RNA‐dependent RNA‐polymerase (RdRp) | 13,352 | 16,148 |
| ORF1b | helicase | Helicase | 16,149 | 17,951 |
| ORF1b | SAR4 | 3′‐to‐5′ exonuclease | 17,952 | 19,532 |
| ORF1b | endoRNAse | endoribonuclease | 19,533 | 20,570 |
| ORF1b | SAR5 | 2′‐O‐ribose methyltransferase | 20,571 | 21,464 |
| S | S | Spike (S) | 21,475 | 25,296 |
| ORF3a | 3a | 3a | 25,305 | 26,132 |
| E | E | Envelope (E) | 26,157 | 26,384 |
| M | M | Membrane (M) | 26,435 | 27,103 |
| ORF6 | 6 | 6 | 27,114 | 27,299 |
| ORF7a | 7a | 7a | 27,306 | 27,671 |
| ORF7b | 7b | 7b | 27,668 | 27,799 |
| ORF8 | 8 | 8 | 27,806 | 28,171 |
| N | N | Nucleocapsid (N) | 28,186 | 29,445 |
| ORF10 | 10 | 10 | 29,470 | 29,586 |
| 3′ NCR | 3′ NCR | None | 29,587 | 29,930 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Reverse genetics have allowed the manipulation of viral genomes by introducing predetermined mutations to study fundamental features of the virus life cycle, such as viral entry, replication and transcription, assembly, budding, and/or the functions of viral proteins. Recently, the rescue of recombinant (r)SARS‐CoV‐2 has been reported, involving the assembly of the entire SARS‐CoV‐2 genome by in vitro ligation (Xie et al., 2020) or a yeast‐based synthetic genome recombination platform (Thi Nhu Thao et al., 2020). However, these methods require in vitro transcription for the production of vRNA, which is not convenient compared to DNA for efficient virus rescue by transfection in susceptible cells.
Here, we report a detailed protocol for the rescue of rSARS‐CoV‐2 by transfection of a full‐length cDNA clone of the SARS‐CoV‐2 USA‐WA1/2020 strain (accession no. MN985325) using a bacterial artificial chromosome (BAC) in Vero E6 cells. The BAC is used to assemble the SARS‐CoV‐2 cDNA under the control of the cytomegalovirus (CMV) promoter, which allows the expression of the vRNA in the nucleus by cellular RNA polymerase II. This system has previously been shown to overcome the difficulties associated with undesirable expression of toxic viral proteins during the propagation of cDNA clones in bacteria (Almazán et al., 2006; Hotard et al., 2012; Pu et al., 2011), which permits the production of infectious virus without requiring an in vitro ligation and transcription step (Ávila‐Pérez et al., 2020; Ávila‐Pérez, Park, Nogales, Almazán, & Martínez‐Sobrido, 2019). The rescued rSARS‐CoV‐2 is easily detectable by cytopathic effect (CPE) and immunofluorescence (IFA) with a monoclonal antibody (MAb) against the N protein. Furthermore, rSARS‐CoV‐2 displays similar viral fitness to the wild‐type (WT) parental SARS‐CoV‐2, as determined by plaque assay and growth kinetics. The use of BAC‐based reverse genetics to generate rSARS‐CoV‐2 represents an excellent tool to study fundamental viral processes, pathogenesis, and transmission, as well as for development of attenuated forms of the virus for implementation as live‐attenuated vaccines (LAVs) for the prophylactic treatment of SARS‐CoV‐2 infection, in addition to the identification and characterization of antivirals for the therapeutic treatment the SARS‐CoV‐2 in humans.
GENERATION OF RECOMBINANT SARS‐CoV‐2 USING A BAC
In this article, we describe a detailed protocol to generate rSARS‐CoV‐2, USA‐WA1/2020 strain, using reverse genetics techniques based on the use of a BAC. The SARS‐CoV‐2 BAC can be stably propagated in bacteria and, upon transfection of Vero E6 cells, generates infectious rSARS‐CoV‐2. The rSARS‐CoV‐2 is easily detected in infected Vero E6 cells by cytopathic effect (CPE) and immunofluorescence, and displays plaque sizes and growth kinetics similar to its wild‐type counterpart. Since the reverse genetics system is based on a single BAC plasmid, the viral genome can be directly manipulated to produce mutant rSARS‐CoV‐2, which is necessary to understand the function of specific viral proteins, mechanisms of viral pathogenesis and/or transmission, and interactions at the virus‐host interface, as well as to generate attenuated forms of the virus, which will facilitate the discovery of effective countermeasures to control the ongoing SARS‐CoV‐2 pandemic.
Biosafety Recommendations
All of our experiments involving infectious wild‐type or recombinant SARS‐CoV‐2 were conducted under appropriated Biosafety Level 3 (BSL‐3) laboratories and approved by the Texas Biomed Institutional Biosafety (IBC) committee. Individuals working with SARS‐CoV‐2 had proper biosafety training before entering BSL‐3 laboratories. Cell culture procedures involving no infectious virus was conducted at BSL‐2 and moved to BSL‐3 for viral infections. The development of plaque assays and viral titrations were conducted at BSL‐2 after complete inactivation of the virus in BSL‐3 with established inactivation procedures.
Materials
SARS‐CoV‐2: SARS‐CoV‐2 USA‐WA1/2020 strain was obtained from BEI Resources (cat. no. NR‐52281) and amplified on Vero E6 cells. This strain was selected because it was isolated from an oropharyngeal swab from a patient with respiratory illness in January 2020 in Washington, US. The SARS‐CoV‐2 USA‐WA1/2020 sequence to generate the rSARS‐CoV‐2 was available from Genbank (accession no. MN985325)
pBeloBAC11 plasmid (NEB)
Vero E6 cells (see recipe)
Other reagents and materials needed to generate pBeloBAC11‐SARS‐CoV‐2 (Ye et al., 2020)
DH10B E. coli competent cells (Thermo Fisher Scientific, cat. no. 18290015)
SOC medium (see recipe)
Luria‐Bertani (LB) medium (see recipe) and LB agar plates (see recipe)
Chloramphenicol (Thermo Fisher Scientific, cat. no. 56‐75‐7): prepared as working stock of 12.5 mg/ml in 100% ethanol
E.Z.N.A Fast Filter Plasmid DNA Maxi kit (Promega, cat. no. D6924‐04)
Cell culture medium for Vero E6 cells: Dulbecco's Modified Eagle Medium (DMEM; Corning, cat. no. 15‐013‐CV) containing 10% fetal bovine serum (FBS; Avantor Seradigm, cat. no. 1500‐500) and 1× penicillin‐streptomycin‐l‐glutamine (PSG; Corning, cat. no. 30‐009‐CI)
Opti‐MEM serum‐free medium (Gibco, cat. no. 31985‐070)
Lipofectamine 2000 (LPF2000) transfection reagent (Thermo Fisher Scientific, cat. no. 11668019)
Transfection medium for Vero E6 cells: Dulbecco's Modified Eagle Medium (DMEM; Corning, cat. no. 15‐013‐CV) containing 10% fetal bovine serum (FBS; Avantor Seradigm, cat. no. 1500‐500)
Post‐infection medium for Vero E6 cells: Dulbecco's Modified Eagle Medium (DMEM; Corning, cat. no. 15‐013‐CV) containing 2% fetal bovine serum (FBS; Avantor Seradigm, cat. no. 1500‐500) and 1× penicillin‐streptomycin‐l‐glutamine (PSG; Corning, cat. no. 30‐009‐CI)
Phosphate‐buffered saline (PBS; see recipe)
0.5% (10×) trypsin‐EDTA solution (Gibco, cat. no. 15400‐054): dilute to 10× in 1× PBS prior to use
Gene Pulser Electroporation System (Bio‐Rad)
Gene Pulser/MicroPulser Electroporation Cuvettes, 0.2 cm gap (Bio‐Rad, cat. no. 1652086)
10‐cm culture tubes
Incubator (PHCbi, model cat. no. MCO‐170AICUVDL)
MaxQ Large Incubated Benchtop Orbital Shaker (Thermo Scientific, MaxQ 4000, cat. no. SHKE4000)
2‐L Erlenmeyer flasks
Spectrophotometer
Tissue culture plates: 6‐, 12‐, and 96‐well cell culture plates (Greiner Bio‐One, catalog #657160, #665180, and #655180, respectively).
Polystyrene T‐75 tissue culture flask (Corning, cat. no. 431081)
Polypropylene sterile conical tubes, 15‐ and 50‐ml (Greiner Bio‐One, cat. no. 188261 and 227270, respectively)
Centrifuge
Nalgene® System 100 Cryogenic Vials (Thermo Fisher Scientific, cat. no. 5000‐1020)
Brightfield microscope
Ziploc bag (1 gallon)
Preparation of pBeloBAC11‐SARS‐CoV‐2 for the rescue of rSARS‐CoV‐2
The generation and validation of the BAC containing the full‐length genome of SARS‐CoV‐2 is detailed in Ye et al. (2020). Briefly, unique restriction sites were selected within the SARS‐CoV‐2 viral genome based on their distribution and spacing, and by their absence within the pBeloBAC11 plasmid. The BstBI and Mlul restriction sites were removed from the S and M gene, respectively, via silent mutation to ensure that these restriction sites were unique, and as a molecular marker to distinguish the rescued rSARS‐CoV‐2 from the natural viral isolate. Five cDNA fragments covering the entire 29,930‐bp viral genome of SARS‐CoV‐2 USA‐WA1/2020 (accession no. MN985325) were synthesized de novo by Bio Basic (Ontario, Canada). Then, the fragments were sequentially assembled into the pBeloBAC11 plasmid (NEB) using unique restriction enzymes and standard molecular biology techniques. The full‐length SARS‐CoV‐2 genome was cloned under the control of the cytomegalovirus (CMV) promoter and flanked at the 3′ end by the Hepatitis Delta Virus (HDV) Ribozyme (Rz) and the bovine growth hormone (bGH) termination and polyadenylation sequences (Fig. 2), and is designated as pBeloBAC11‐SARS‐CoV‐2.
Figure 2.
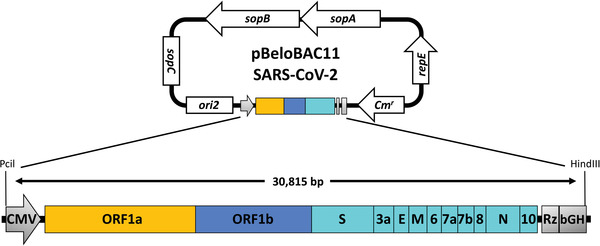
Schematic representation of the pBAC plasmid to rescue rSARS‐CoV‐2: The full‐length cDNA of the SARS‐CoV‐2 genome is flanked at the 5′ end by the cytomegalovirus (CMV) polymerase II−driven promoter and at the 3′ end by the hepatitis delta ribozyme (Rz) and bovine growth hormone (bGH) polyadenylation signal. The entire ∼30,815‐bp construct was inserted into the pBeloBAC11 plasmid using Pcil and HindIII restriction sites. The pBeloBAC11 plasmid is an E. coli vector commonly used to generate BACs because it can support the insertion of large DNA fragments as a single copy in cells. The pBeloBAC11 contains a chloramphenicol resistance (CmR) gene as a selective marker. Viral proteins are those previously described in Figure 1.
-
1
Transform 100 µl of DH10B E. coli competent cells with 100 ng of pBeloBAC11‐SARS‐CoV‐2 by electroporation (2.5 kV, 600 Ω and 10 µF) in a Gene Pulser® Cuvette with 0.2‐cm gap.
-
2
Transfer bacteria to a 10‐ml culture tube containing 1 ml of SOC medium and incubated at 37°C for 1 hr with shaking at 200‐250 rpm.
-
3
Spread 100 µl of the bacteria onto LB agar plates supplemented with 12.5 µg/ml of chloramphenicol and place in an incubator at 37°C for 16 hr.
-
4
After 16 hr incubation, transfer and grow a single bacterial colony transformed with pBeloBAC11‐SARS‐CoV‐2 in 1 ml of LB medium supplemented with 12.5 µg/ml of chloramphenicol in a 37°C shaking incubator at 200‐250 rpm for 16 hr.
We recommend growing three bacterial colonies individually.
-
5
Add 1 ml of bacterial culture to a 2‐L flask containing 500 ml of LB liquid medium supplemented with 12.5 µg/ml of chloramphenicol, and grow the bacteria in a 37°C shaking incubator at 200‐250 rpm for 16 hr, or until an OD600 of 0.8 is reached.
-
6
Purify the pBeloBAC11‐SARS‐CoV‐2 using a Plasmid DNA Maxi kit according to the manufacturer's instructions. Elute plasmid DNA in 500 µl of ddH2O or elution buffer from the kit. This purification method usually yields approximately 20‐30 µg of purified pBeloBAC11‐SARS‐CoV‐2. Store the pBeloBAC11‐SARS‐CoV‐2 at 20°C.
Avoid vortexing or vigorously shaking the BAC, as this may cause DNA shearing due to its large size.
Rescue of rSARS‐CoV‐2
For the rescue of rSARS‐CoV‐2 from pBeloBAC11‐SARS‐CoV‐2, we recommend three independent transfections for each recombinant virus to increase the probability of successful rescue. If one is attempting rescue of multiple recombinant viruses, scale up the following steps accordingly. We also recommend transfecting the empty pBeloBAC11 plasmid as an internal control for these viral rescues. A schematic representation of the transfection protocol is detailed in Figure 3.
Figure 3.
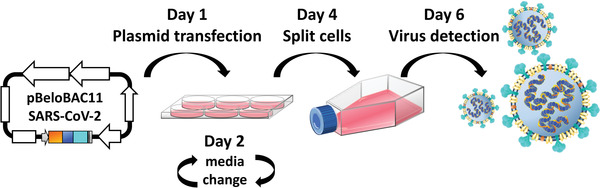
Generation of rSARS‐CoV‐2: Vero E6 cells (6‐well plate format, 1.2 × 106 cells/well, triplicates) are transfected, using LPF2000, with 2 µg of pBeloBAC11‐SARS‐CoV‐2 (Fig. 2) overnight in a tissue culture incubator at 37°C with 5% CO2. At 6‐8 hr post‐transfection, the transfection medium is replaced by post‐infection medium. At day 4 post‐transfection, Vero cells are scaled up into T‐75 flasks. After an additional 48 hr, CPE should be detected, and tissue culture supernatants are collected to evaluate the presence of recombinant virus (Fig. 4).
-
7
Plate Vero E6 cells (6‐well plate format, 5 × 105 cells/well, triplicates) in cell culture medium the day prior to transfection.
-
8
Prepare Opti‐MEM‐LPF2000 mixture: Combine 250 µl of Opti‐MEM medium with 4‐8 µl of LPF2000 per transfection. Gently invert to mix and incubate at room temperature for 5‐10 min.
-
9
Prepare plasmid transfection mixture: Combine 50 µl of Opti‐MEM and 2 µg of pBeloBAC11‐SARS‐CoV‐2 per transfection and invert the tubes to mix.
-
10
Prepare Opti‐MEM‐LPF2000‐DNA plasmid mixture: Add the 250 µl Opti‐MEM‐LPF2000 mixture (step 8) to the 50 µl plasmid transfection mixture (step 9). Allow the mixture to incubate at room temperature for 20‐30 min. Meanwhile, remove the cell culture medium from the Vero E6 cells and add 1 ml of transfection medium.
The cells should be 90% confluent (~1.2 × 106 cells/well) on the day of the transfection.
-
11
Slowly add the 300 µl Opti‐MEM‐LPF2000‐DNA plasmid mixture (step 10) to the Vero E6 cells in a dropwise fashion. Gently rock the plate back and forth and place in an incubator at 37°C with 5% CO2 for 16 hr.
-
12
Approximately 6‐8 hr post‐transfection, remove the transfection medium (step 11) and add 2 ml of post‐infection medium.
-
13
Place the cells back in the 37°C, 5% CO2 incubator.
-
14At 72 hr post‐transfection (day 4), scale up transfected Vero E6 cells to a T‐75 flask.
- Wash the cells with 1× PBS twice and then add 1 ml of 1× trypsin‐EDTA solution.
- When the cells detach, resuspend the cells in 10 ml of cell culture medium and transfer to a 15‐ml conical tube.
- Centrifuge the cells 10 min at 226 × g room temperature.
-
Remove the medium and resuspend the cells in 12 ml of post‐infection medium. Transfer the cells to a T‐75 flask.As additional internal controls, two additional T‐75 flasks with confluent monolayers of Vero E6 cells are prepared and are either mock‐infected or infected with WT SARS‐CoV‐2 at a multiplicity of infection (MOI) of 0.01 plaque forming units (PFU).
-
15
Incubate the T‐75 flasks in a 37°C incubator with 5% CO2 for 2 days. Check the cells daily for cytopathic effect (CPE) to assess the presence of rSARS‐CoV‐2 (Fig. 4A).
Figure 4.
Detection and in vitro characterization of rSARS‐CoV‐2. (A) CPE: At 48 hr after scaling up transfected Vero E6 cells into T‐75 flasks (Fig. 3), CPE can be already observed. Representative images of mock‐infected, SARS‐CoV‐2‐infected, and transfected Vero E6 are shown. Scale bars are 400 µm. (B) IFA: Vero E6 cells (12‐well plate format, 0.5 × 106 cells/well, triplicates) were infected with tissue culture supernatants from transfected Vero E6 cells. Mock‐infected and SARS‐CoV‐2‐infected Vero E6 cells were included as internal controls. At 24 hr post‐infection, cells were fixed with 10% neutral buffered formalin. After fixation for 16 hr, cells were permeabilized with 0.5% Triton X‐100 for 10 min. Cells were then washed three times with 1× PBS and incubated with 1 μg/ml of an anti‐SARS NP MAb 1C7 at 37°C. After 1 hr incubation with the primary NP 1C7 MAb, cells are washed three times with 1× PBS and incubated with an anti‐mouse FITC‐conjugated secondary antibody at 37°C. After 1 hr, cells are washed three times with 1× PBS and observed under a fluorescent microscope. DAPI was used to stain the nucleus. Scale bars are 100 µm. (C) Plaque assay: Confluent monolayers of Vero E6 cells (6‐well plate format, 1.2 × 106 cells/well, triplicates) were infected with SARS‐CoV‐2 or rSARS‐CoV‐2 at 37°C for 1 hr and overlaid with agar. After 72 hr in a 37°C incubator with 5% CO2, cells were fixed in 10% neutral buffered formalin for 16 hr before agar was removed. Next, cells were permeabilized with 0.5% Triton X‐100 for 10 min and prepared for immunostaining as previously described using the anti‐NP MAb (1C7) and vector kits (Vectastain ABC kit and DAB HRP substrate kit; Vector Laboratories). (D) Viral growth kinetics: Vero E6 cells (12‐well plate format, 0.5 × 106 cells/well, triplicates) were infected (MOI of 0.01) with SARS‐CoV‐2 (black) or rSARS‐CoV‐2 (red) and placed in a 37°C incubator with 5% CO2 for 4 days. At 12, 24, 48, 72, and 96 hr post‐infection, viral titers in supernatants were determined by plaque assay (PFU/ml). Error bars indicate the standard deviations from three separate experiments. The dashed black line indicates the limit of detection (10 PFU/ml).
-
16
Once CPE is observed, place the T‐75 flasks into ziploc bags and freeze at −80°C. Thaw the cells at room temperature and transfer the tissue culture supernatant containing SARS‐CoV‐2 to a 50‐ml conical tube. Centrifuge 10 min at 226 × g, 4°C, to pellet cell debris.
-
17
Collect the tissue culture supernatants and aliquot them into cryogenic tubes (0.5 ml/tube) before storage at −80°C.
VALIDATION AND CHARACTERIZATION OF rSARS‐CoV‐2
In order to validate the successful rescue of rSARS‐CoV‐2, fresh Vero E6 cells are infected with tissue culture supernatants containing rSARS‐CoV‐2. The presence of rSARS‐CoV‐2 can be determined by immunofluorescence by expression of SARS‐CoV‐2 NP using a mouse anti‐SARS NP monoclonal antibody (MAb) 1C7 (Fig. 4B). WT SARS‐CoV‐2 and mock infected cells should be included as internal controls in the IFA.
Materials
Vero E6 cells (see recipe in Reagents and Solutions)
Cell culture medium for Vero E6 cells: Dulbecco's Modified Eagle Medium (DMEM; Corning, cat. no. 15‐013‐CV) containing 10% fetal bovine serum (FBS; Avantor Seradigm, cat. no. 1500‐500) and 1× penicillin‐streptomycin‐l‐glutamine (PSG; Corning, cat. no. 30‐009‐CI)
rSARS‐CoV‐2 (see Basic Protocol)
Wild‐type (WT) SARS‐CoV‐2: SARS‐CoV‐2 USA‐WA1/2020 strain was obtained from BEI Resources (cat. no. NR‐52281) and amplified on Vero E6 cells. This strain was selected because it was isolated from an oropharyngeal swab from a patient with respiratory illness in January 2020 in Washington, US. The SARS‐CoV‐2 USA‐WA1/2020 sequence to generate the rSARS‐CoV‐2 is available from Genbank (accession no. MN985325)
Post‐infection medium for Vero E6 cells: Dulbecco's Modified Eagle Medium (DMEM; Corning, cat. no. 15‐013‐CV) containing 2% fetal bovine serum (FBS; Avantor Seradigm, cat. no. 1500‐500) and 1× penicillin‐streptomycin‐l‐glutamine (PSG; Corning, cat. no. 30‐009‐CI)
Phosphate‐buffered saline (PBS; see recipe)
10% neutral buffered formalin solution (Sigma‐Aldrich, cat. no. HT501128–4 liters)
Permeabilization solution: 0.5% (v/v) Triton X‐100 (Sigma‐Aldrich, cat. no. X100−500 ML) in 1× PBS (see recipe)
Blocking solution: 2.5% bovine serum albumin (BSA; Sigma‐Aldrich, cat. no. A9647) in 1× PBS (see recipe)
Mouse anti‐SARS NP MAb 1C7 cross‐reactive with SARS‐CoV‐2 (Millipore Sigma, cat. no. ZMS1075) was generated at the Center for Therapeutic Antibody Development at the Icahn School of Medicine at Mount Sinai (ISMMS) and kindly provided by Dr. Thomas Moran
Polyclonal rabbit anti‐mouse FITC (Dako, cat. no. F0261)
4′,6′−diamidino‐2‐phenylindole (DAPI; Research Organics, cat. no. 2039D)
DMEM/F12 agar medium (see recipe)
Vectastain ABC‐HRP kit, peroxidase, mouse IgG (Vector laboratories, cat. no. P‐4002)
Biotinylated anti‐mouse secondary antibody (from Vectastain ABC‐HRP kit)
DAB substrate kit, peroxidase (HRP), with nickel (Vector Laboratories, cat. no. SK‐4100)
Avicel PH‐101 (Sigma‐Aldrich, cat. no. 11365‐1KG)
Tissue culture plates: 6‐, 12‐, and 96‐well cell culture plates (Greiner Bio‐One, cat. no. 657160, 665180, and 655180, respectively)
Polypropylene sterile conical tubes, 15‐ and 50‐ml (Greiner Bio‐One, cat. no. 188261 and 227270, respectively)
Serological pipette, 5, 10, and 25 ml (Greiner Bio‐One, cat. no. 606180, 607180, and 760180, respectively)
Universal pipette tips, 20, 200, and 1000 µl (VWR, cat. no. 76322‐134, 76322‐150, and 16466‐008, respectively)
Incubator (PHCbi, model cat. no. MCO‐170AICUVDL)
Fluorescence microscope
-
1
Prepare Vero E6 cells (12‐well plate format, 0.1 × 106 cells/well, triplicates) the day before infection. Infect cells when they have reached ~90%‐100% confluency (0.5 × 106 cells/well)
-
2
Thaw aliquots of rSARS‐CoV‐2 and wild‐type (WT) SARS‐CoV‐2 on ice. Prepare undiluted and serially diluted 1:10 and 1:100 viral stocks in post‐infection medium.
-
3
Wash Vero E6 cells three times with 1× PBS and infect each well, in triplicate, with 500 µl of virus. For column A of a 12‐well plate, add 500 µl of undiluted rSARS‐CoV‐2. For columns B, C, and D, add 500 µl of 10‐fold‐diluted rSARS‐CoV‐2, 100‐fold diluted rSARS‐CoV‐2, and appropriately diluted WT SARS‐CoV‐2, respectively.
-
4
Incubate the plates in a 37°C, 5% CO2 incubator for 1 hr.
-
5
After viral absorption, remove the viral inoculum, wash once with 1× PBS, and then add 1 ml of fresh post‐infection medium to each well. Return the plates back to the 37°C, 5% CO2 incubator. Incubate the plates for 24 hr.
-
6
Remove the tissue culture supernatants from the infected Vero E6 cells and fix the cells by completely submerging plates in 10% neutral buffered formalin for 16 hr.
The process of fixation for 16 hr with 10% neutral buffered formalin will inactivate the virus and permit it to be safely removed from the BSL‐3 laboratory to continue the characterization of the virus at BSL‐2. Biocontainment procedures to remove samples from BSL‐3 must still be strictly followed.
-
7
Gently rinse the plates with tap water to remove remaining 10% neutral buffered formalin. Do not place the cells directly under the faucet.
-
8
Tap off all remaining water from the plates and wash with 1× PBS three times.
-
9
Add 500 µl/well of permeabilization solution for 10 min at room temperature.
-
10
Remove the permeabilization solution and wash the cells three times with 1× PBS.
-
11
Add 500 µl/well of blocking solution for 1 hr at room temperature.
-
12
Remove the blocking solution and incubate the cells with 500 µl/well of mouse anti‐SARS NP MAb 1C7 (1 µg/ml) diluted in blocking solution in a 37°C incubator for 1 hr.
If desired, other MAb or polyclonal antibodies (PAb) for the detection of SARS‐CoV‐2 may be used in place of 1C7.
-
13
After 1 hr, remove the solution and wash the cells three times with 1× PBS.
-
14
Remove the 1× PBS and add 500 µl/well of anti−mouse IgG‐FITC‐conjugated antibody (1:200 dilution) and 4′,6′‐diamidino‐2‐phenylindole (DAPI; 1 mg/ml) in blocking solution. Place the plates in a 37°C incubator for 1 hr.
At this point, plates should be shielded from light with aluminum foil.
-
15
Remove the previous solution containing secondary antibody and DAPI and wash the cells three times with 1× PBS. Leave 1 ml of 1× PBS in each well. Samples can be stored at 4°C while protected from light with aluminum foil.
-
16
Analyze the samples under a fluorescence microscope.
-
17
In immunofluorescence (Fig. 4B), the detection of SARS‐CoV‐2 NP is shown in green (FITC), while the nucleus is stained in blue (DAPI). As expected, only Vero E6 cells treated with tissue culture supernatants containing WT; SARS‐CoV‐2 or rSARS‐CoV‐2 were positive in FITC signal.
Plaque assay of rSARS‐CoV‐2 for titration and visualization of plaque sizes
In order to quantitatively titrate the amount of rSARS‐CoV‐2 from our rescue tissue culture supernatants and to assess their plaque phenotype, we recommend the use of plaque assays. WT SARS‐CoV‐2 and mock infected cells should be included as internal controls in the plaque assay.
-
18
A day prior to conducting plaque assays, seed 6‐well plates with 0.5 × 106 Vero E6 cells/well in cell culture medium. Vero E6 cells should be ∼90%‐100% confluency (∼1.2 × 106 cells/well) at the time of viral infection. Prepare enough 6‐well plates to titrate each sample in triplicate.
-
19
Thaw an aliquot of the rSARS‐CoV‐2‐containing supernatant and the WT SARS‐CoV‐2‐containing supernatant, and make 10‐fold serial dilutions in post‐infection medium.
-
20
Wash Vero E6 cells with 1× PBS three times before infection with 1 ml/well of the 10‐fold serial dilutions of rSARS‐CoV‐2.
We used 100 (10‐2) to 10,000,000 (10‐7)−fold serial dilutions of the tissue culture supernatants.
-
21
Incubate the plates at 37°C in a 5% CO2 incubator for 1 hr. Gently rock the plates back and forth every 15 min.
-
22
After 1 hr of viral absorption, remove the inoculum and add 3 ml of prewarmed DMEM/F12/agar medium. Let the agar solidify at room temperature.
-
23
Once the agar has solidified, invert the plates and place them in a 37°C incubator with 5% CO2 for 3‐4 days. Observe the plates daily for the formation of viral plaques.
Note that viral plaques may be difficult to observe and may need to be held upright under a light source for visibility.
-
24
When viral plaques are visible, fix the cells by completely submerging the plates in 10% neutral buffered formalin for 16 hr.
-
25
Gently rinse the plates with tap water to remove remaining 10% neutral buffered formalin. Do not place the cells directly under the faucet.
-
26
Carefully remove the agar and wash the plates three times with 1× PBS.
-
27
Next, permeabilize the cells with permeabilization solution for 10 min at room temperature. Then, wash the plate with 1× PBS three times.
-
28
Remove the 1× PBS and add 1 ml of blocking solution for 1 hr at room temperature.
-
29
Detect viral plaques by immunostaining using mouse anti‐SARS NP MAb 1C7 as follows. Remove the blocking solution and add 500 µl/well of anti‐SARS NP MAb 1C7 (1 µg/ml) in blocking solution before returning the plates to a 37°C incubator for 1 hr.
If desired, other MAbs or PAbs against SARS‐CoV‐2 may be used in place of 1C7 for immunostaining of the viral plaques. Alternatively, viral plaques can be visualized by crystal violet staining.
-
30
Wash the plate three times with 1× PBS and add 500 µl/well of biotinylated anti‐mouse secondary antibody diluted in blocking solution before returning the plates to the 37°C incubator for 30 min.
-
31
Wash the cells three times with 1× PBS and add 400 µl/well Vectastain ABC reagent, then move the plates back to the 37°C incubator for 30 min.
-
32
Remove the medium in each well, wash with 1× PBS three times, and visualize the viral plaques using a DAB peroxidase kit in accordance to the manufacturer's instructions.
-
33
To determine the viral titer, count the number of PFU in each well, average the triplicates, then multiply by the dilution factor and divide by the volume used during infection.
A representative result of a viral plaque assay is shown in Figure 4C.
Growth kinetics
In order to assess viral fitness of the rSARS‐CoV‐2 and to compare it to that of its natural counterpart, we recommend doing growth kinetics (Fig. 4D).
-
34
Prepare confluent monolayers of Vero E6 cells (12‐well plate format, 0.1 × 106 cells/well, triplicates) the day before infection. Infect cells when they have reached ~90%‐100% confluency (0.5 × 106 cells/well)
-
35
Infect the Vero E6 cells with WT SARS‐CoV‐2 or rSARS‐CoV‐2 at a MOI of 0.01, in triplicate, and place in a 37°C, 5% CO2 incubator for 1 hr.
-
36
After 1 hr viral absorption, wash the plate once with 1× PBS, then remove the viral inoculum and add 1.5 ml of post‐infection medium to each well.
-
37
Incubate the plates at 37°C in a 5% CO2 incubator. At 12, 24, 48, 72, and 96 hr post‐infection, collect 200 µl of the tissue culture supernatants and store at −80°C in cryogenic tubes.
-
38
For viral titration, prepare confluent monolayers of Vero E6 cells (96‐well plate format, 4 × 104 cells/well, triplicates) the day before infection.
-
39
Thaw virus‐containing tissue culture supernatants and prepare 10‐fold viral dilutions. To that end, add at least 80 µl of each sample, in triplicate, to row A of a 96‐well plate (dilution plates).
-
40
Add 90 µl of post‐infection medium to the other wells (row B to H).
-
41
Serially dilute the samples down the dilution plates by transferring 10 µl from row A to row B. Continue transferring 10 µl down the plate until row H.
-
42
Remove the cell culture medium from the 96‐well plates containing Vero E6 cells (from step 38), and wash once with 1× PBS. Remove the 1× PBS before infection.
-
43
Transfer 50 µl of virus from the dilution plates (step 41) to corresponding wells of the 96‐well plates containing Vero E6 cells. Allow viral absorption for 1 hr at 37°C in a 5% CO2 incubator.
-
44
Remove the viral inoculum, wash once with 1× PBS, and overlay the cells with 1% Avicel in post‐infection medium. Place the plates in an incubator set to 37°C with 5% CO2 for 24 hr.
Avicel overlays assist in the formation of localized viral plaques, and due to their low viscosity, produce significantly larger plaques compared to agar or methylcellulose overlay media. Moreover, Avicel overlays are easier to remove from 96‐well plates, and, if desired, can be applied without removal of the viral inoculum.
-
45
Fix the cells by completely submerging the plates in 10% neutral buffered formalin for 16 hr.
-
46
Gently wash the plates with tap water, but not directly under a faucet. Then, wash the wells three times with 1× PBS.
-
47
Remove the 1× PBS and permeabilize the cells with permeabilization solution for 10 min, then wash the cells with 1× PBS three times.
-
48
After removing the 1× PBS, incubate the cells with blocking solution at room temperature for 1 hr.
-
49
Detect viral plaques using mouse MAb against SARS NP 1C7, Vectastain ABC kit, and DAB reagent, as described for plaque assays above.
-
50
To determine the viral titer, count the number of PFU in each well, average the triplicates, then multiply by the dilution factor and divide by the volume used during infection.
-
51
A representative result of a viral growth kinetic assay is shown in Figure 4D.
REAGENTS AND SOLUTIONS
DMEM/F‐12, 2×
Dissolve a packet of DMEM/F‐12 (Gibco, cat. no. 12400‐024) in 450 ml of cell culture−grade water, along with:
10 ml of 100× PSG (Corning, cat. no. 30‐009‐CI)
6 ml of 35% bovine serum albumin (BSA; Sigma‐Aldrich, cat. no. A9647)
10 ml 1M HEPES buffer solution (Gibco, cat. no. 15630‐080)
24 ml of 5% sodium bicarbonate (NaHCO3; Sigma, cat. no. S‐5761)
Store up to 3 weeks at 4°C
DMEM/F‐12/agar
Mix 25 ml of 2× DMEM/F‐12 medium with:
0.5 ml of 1% DEAE‐Dextran (MP Biomedicals, cat. no. 195133)
1 ml of 5% sodium bicarbonate (NaHCO3; Sigma, cat. no. S‐5761)
8.5 ml of cell culture−grade water
Just before use, add 15 ml of pre‐warmed 2% (w/v) agar (Oxoid, cat. no. LP0028) for a final concentration of 0.5%.
LB agar plates
Prepare 1 L of LB medium (see recipe), and add 15 g of Bacto agar (BD, cat. no. 214010). Autoclave, cool to ∼50°C, add 12.5 µg/ml of chloramphenicol, and pour into 100‐mm bacterial culture plates. Store up to 3 weeks at 4°C.
Luria‐Bertani (LB) medium
For 1 L of medium, add the following to 1 L of ddH2O:
10 g Bacto Tryptone (BD, cat. no. 211699)
5 g Bacto Yeast Extract (BD, cat. no. 212720)
10 g of sodium chloride (NaCl, JT Baker, cat. no. 4058‐01)
Autoclave to sterilize
Store up to 6 weeks at 4°C
Phosphate‐buffered saline (PBS)
Prepare 10× PBS:
80 g NaCl
2 g KCl
11.5 g Na2HPO4·7H2O
2 g KH2PO4
ddH2O to 1 L
Adjust pH to 7.3
Sterilize by autoclaving
Store 10× PBS up to 6 weeks at room temperature
To prepare 1× PBS, dilute 10× PBS 1:10 in ddH2O, autoclave, and store up to 6 weeks at room temperature.
Vero E6 cells
Vero E6 cells (African green monkey kidney epithelial cells; ATCC, cat. no. CRL‐1586) are used in all experimental procedures, including viral rescue, plaque assay, titration, and growth kinetics. This cell line was selected because they are readily infected with SARS‐CoV‐2 (Ogando et al., 2020), produce high viral titers, and have moderate transfection efficiencies.
Cells are cultured in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin‐streptomycin‐l‐glutamine (PSG) and maintained in a 37°C incubator with 5% CO2. DMEM supplemented with 2% FBS and 1% PSG is used as post‐infection medium. Opti‐MEM medium (Invitrogen) is used for transfection of the BAC to rescue rSARS‐CoV‐2. Lipofectamine 2000 (LPF2000, Invitrogen) is used at a 2:1 ratio to DNA for BAC transfection into Vero E6 cells.
COMMENTARY
Background Information
Since the identification of SARS‐CoV‐2 in December 2019 in China, the virus has rapidly dispersed across the world and caused over 16 million confirmed cases and over 1 million deaths (Andersen et al., 2020; Wu et al., 2020). The World Health Organization (WHO) officially declared a pandemic of SARS‐CoV‐2 in March of 2020.
Currently, no FDA‐approved therapeutic vaccines or prophylactic antivirals are available for the treatment of SARS‐CoV‐2 infection and associated COVID‐19. Reverse genetics systems to rescue recombinant viruses represent an excellent experimental tool for the development of attenuated forms of SARS‐CoV‐2 for their implementation as live attenuated vaccines (LAVs) and for the rapid identification of drugs with antiviral activity against SARS‐CoV‐2. To date, two different approaches to generate rSARS‐CoV‐2 have been described (Thi Nhu Thao et al., 2020; Xie et al., 2020). However, both of these reverse genetics techniques to generate rSARS‐CoV‐2 are laborious and require in vitro ligation and transcription for successful viral rescue.
Here, we describe the experimental procedure to generate rSARS‐CoV‐2 using a BAC‐based reverse genetics approach (Ye et al., 2020). After transfection of Vero E6 cells with the pBeloBAC11‐SARS‐CoV‐2 plasmid, infectious rSARS‐CoV‐2 can be generated in the tissue culture supernatants of transfected cells. The rSARS‐CoV‐2 is easily detectable by determining CPE or by IFA using an anti‐NP MAb. Importantly, the rSARS‐CoV‐2 generated using our BAC‐based reverse genetics approach exhibit plaque phenotype and growth kinetics similar to a WT SARS‐CoV‐2 natural isolate. Many problems, including the large size of the SARS‐CoV‐2 genome and the instability of some viral sequences in bacteria, can be circumvented by the use of the BAC platform, since pBeloBAC11 is maintained as a single copy per cell.
The development of a BAC‐based reverse genetic system for SARS‐CoV‐2 is a powerful tool that enables researchers to study multiple aspects of SARS‐CoV‐2 biology and pathogenesis in vitro and in vivo. This approach permits easy manipulation of the SARS‐CoV‐2 genome for the generation of recombinant viruses containing mutations or deletions of viral proteins, allowing investigators to ask important questions regarding SARS‐CoV‐2 replication, virulence, and pathogen‐host interactions in cultured cells or validated animal models of SARS‐CoV‐2 infection (Golden et al., 2020). Also, these reverse genetics approaches will allow the generation of attenuated forms of SARS‐CoV‐2 for their potential implementation as LAVs for the prevention of COVID‐19 disease. Finally, with these reverse genetics in place, rSARS‐CoV‐2‐expressing reporter genes such as fluorescent or luciferase proteins could be rescued. Reporter‐expressing rSARS‐CoV‐2 represents an excellent option for the rapid identification of compounds with antiviral activity using high‐throughput settings that could be employed for the treatment of SARS‐CoV‐2 infections.
Critical Parameters
SARS‐CoV‐2 is a Biosafety Level 3 (BSL‐3) pathogen, and all steps prior to inactivation must be done in the proper biocontainment laboratory. The success of the experiments described in this article is dependent on many critical parameters. First, a large quantity of highly purified pBeloBAC11‐SARS‐CoV‐2 is required, which can be generated as described in the Basic Protocol. The rescue of rSARS‐CoV‐2 requires transfection with Lipofectamine 2000 (LPF2000) in Opti‐MEM containing no antibiotics. The inclusion of antibiotics may hinder transfection efficiency and the production of infectious rSARS‐CoV‐2. It is important to maintain healthy Vero E6 cells and check them for potential contamination (e.g., mycoplasma). Vero E6 cells should be passaged a day prior to transfection to ensure optimal cell conditions. The proper amount of Vero E6 should also be used in each of the experiments. We recommend conducting all transfections in triplicate to ensure the successful rescue of rSARS‐CoV‐2. If attempting to rescue a mutant rSARS‐CoV‐2, the researcher must determine whether the mutation is deleterious to the virus. Mutations that are too detrimental to the virus will prevent successful viral rescue. Another consideration that must be addressed is the optimal temperature for viral rescues. For WT rSARS‐CoV‐2, the optimal temperature is 37°C; however, mutant rSARS‐CoV‐2 may require lower temperatures. In plaque assays, the DMEM/F‐12 agar must be overlaid over the cells at approximately 39°C, by maintaining the DMEM/F‐12 mixture in a 39°C water bath before adding warm agar. We recommend sequencing the rescued rSARS‐CoV‐2 to identify potential mutations that could arise from passaging the virus in Vero E6 cells.
Troubleshooting
If viral rescue is not achieved, there are several possible causes. Vero E6 cells may be at a high passage number that decreases transfection efficacy. A new batch of fresh Vero E6 cells with a low passage number should be used. We also recommend using Vero E6 cells that were passaged the day before transfection to improve transfection efficiency. A poor preparation of pBeloBAC11‐SARS‐CoV‐2 might also result in low virus rescue efficiency. It is recommended to generate and verify the purity of the pBeloBAC11‐SARS‐CoV‐2 preparation for successful viral rescue. Note that the pBeloBAC11‐SARS‐CoV‐2 is a low‐copy‐number plasmid, at approximately 1‐2 copies per cell. Unexpected contamination can occur during transfection of the pBeloBAC11‐SARS‐CoV‐2 without antibiotics. To resolve this, use fresh media and be sure all materials are sterile.
Understanding Results
CPE in transfected Vero E6 cells is an indication of successful viral rescue. However, full validation is conducted by immunofluorescence and/or plaque assays. In the immunofluorescence assay, the presence of SARS‐CoV‐2 in infected cells is detected with the 1C7 anti‐NP monoclonal antibody and a secondary anti‐mouse FITC‐antibody using fluorescent microscopy. We recommend including mock‐transfected cells or cells transfected with empty pBAC as control. In the plaque assay, the presence of virus is determined by immune staining using the same 1C7 anti‐NP monoclonal antibody. Viral plaque sizes are dependent on the incubation time of the plaque assay. We recommend an incubation time of 48‐96 hr post‐infection for the plaque assays, to allow the formation of visible plaques.
Time Considerations
After transfection of Vero E6 cells with pBeloBAC11‐SARS‐CoV‐2, yielding rSARS‐CoV‐2, we have been able to observe CPE as early as 48‐72 hr post‐transfection. However, we recommend maintaining the transfection for 3‐4 days. The validation and characterization of rSARS‐CoV‐2 (IFA and/or plaque assays) require approximately 24 and 48‐72 hr, respectively. An extra 16‐hr overnight fixation of samples in 10% neutral buffered formalin for complete viral inactivation before removing the plates from BSL‐3 conditions is also required. Immunostaining of the IFA and plaque assays requires an additional ∼5‐6 hr. So, from the transfection of the Vero E6 cells until verification of virus, the total protocol requires approximately 7 days for completion.
Conflict of Interest Statement
The authors declare not conflict of interest.
Author Contributions
Luis Martinez‐Sobrido: Conceptualization; formal analysis; funding acquisition; resources; supervision; writing‐review & editing.
Acknowledgment
We want to thank Dr. Thomas Moran (Icahn School of Medicine at Mount Sinai) for providing us with the SARS‐CoV cross‐reactive NP 1C7 MAb. We also want to thank BEI Resources for providing the SARS‐CoV‐2 USA‐WA1/2020 isolate (NR‐52281). Additionally, we thank members at our institute for their efforts in keeping them fully operational during the COVID‐19 pandemic, and the BSC and IACUC committees for reviewing our protocols in a time‐efficient manner. We would like to dedicate this manuscript to all COVID‐19 victims and to all heroes battling this disease.
Chiem, K. , Ye, C. , & Martinez‐Sobrido, L. (2020). Generation of recombinant SARS‐CoV‐2 using a bacterial artificial chromosome. Current Protocols in Microbiology, 59, e126. doi: 10.1002/cpmc.126
Literature Cited
- Alfaraj, S. H. , Al‐Tawfiq, J. A. , Assiri, A. Y. , Alzahrani, N. A. , Alanazi, A. A. , & Memish, Z. A. (2019). Clinical predictors of mortality of Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) infection: A cohort study. Travel Medicine and Infectious Disease, 29, 48–50. doi: 10.1016/j.tmaid.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almazán, F. , Dediego, M. L. , Galán, C. , Escors, D. , Alvarez, E. , Ortego, J. , … Enjuanes, L. (2006). Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. Journal of Virology, 80(21), 10900–10906. doi: 10.1128/JVI.00385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen, K. G. , Rambaut, A. , Lipkin, W. I. , Holmes, E. C. , & Garry, R. F. (2020). The proximal origin of SARS‐CoV‐2. Nature Medicine, 26(4), 450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ávila‐Pérez, G. , Nogales, A. , Park, J. G. , Vasquez, D. M. , Dean, D. A. , Barravecchia, M. , … Martínez‐Sobrido, L. (2020). In vivo rescue of recombinant Zika virus from an infectious cDNA clone and its implications in vaccine development. Scientific Reports, 10(1), 512. doi: 10.1038/s41598-020-57545-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ávila‐Pérez, G. , Park, J. G. , Nogales, A. , Almazán, F. , & Martínez‐Sobrido, L. (2019). Rescue of recombinant Zika virus from a bacterial artificial chromosome cDNA clone. Journal of Visualized Experiments, 148, 59537. doi: 10.3791/59537. [DOI] [PubMed] [Google Scholar]
- Golden, J. W. , Cline, C. R. , Zeng, X. , Garrison, A. R. , Carey, B. D. , Mucker, E. M. , … Hooper, J. W. (2020). Human angiotensin‐converting enzyme 2 transgenic mice infected with SARS‐CoV‐2 develop severe and fatal respiratory disease. Journal of Clinical Investigation Insight, 5(19), 142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson, D. F. , Helgason, A. , Jonsson, H. , Magnusson, O. T. , Melsted, P. , Norddahl, G. L. , … Stefansson, K. (2020). Spread of SARS‐CoV‐2 in the Icelandic population. New England Journal of Medicine, 382(24), 2302–2315. doi: 10.1056/NEJMoa2006100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotard, A. L. , Shaikh, F. Y. , Lee, S. , Yan, D. , Teng, M. N. , Plemper, R. K. , … Moore, M. L. (2012). A stabilized respiratory syncytial virus reverse genetics system amenable to recombination‐mediated mutagenesis. Virology, 434(1), 129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ksiazek, T. G. , Erdman, D. , Goldsmith, C. S. , Zaki, S. R. , Peret, T. , Emery, S. , … Group, S. W. (2003). A novel coronavirus associated with severe acute respiratory syndrome. New England Journal of Medicine, 348(20), 1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- Liu, J. , Cao, R. , Xu, M. , Wang, X. , Zhang, H. , Hu, H. , … Wang, M. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS‐CoV‐2 infection in vitro. Cell Discovery, 6, 16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. , Wu, H. , … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet, 395(10224), 565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizumoto, K. , Kagaya, K. , Zarebski, A. , & Chowell, G. (2020). Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID‐19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveillance, 25(10), 2000180. doi: 10.2807/1560-7917.ES.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogando, N. S. , Dalebout, T. J. , Zevenhoven‐Dobbe, J. C. , Limpens, R. W. A. L. , van der Meer, Y. , Caly, L. , … Snijder, E. J. (2020). SARS‐coronavirus‐2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. Journal of General Virology, 101(9), 925‐940. doi: 10.1099/jgv.0.001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris, J. S. , Lai, S. T. , Poon, L. L. , Guan, Y. , Yam, L. Y. , Lim, W. , … SARS study group . (2003). Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet, 361(9366), 1319–1325. doi: 10.1016/s0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, E. , Koopmans, M. , Go, U. , Hamer, D. H. , Petrosillo, N. , Castelli, F. , … Simonsen, L. (2020). Comparing SARS‐CoV‐2 with SARS‐CoV and influenza pandemics. The Lancet Infectious Diseases, 20(9), e238‐e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu, S. Y. , Wu, R. H. , Yang, C. C. , Jao, T. M. , Tsai, M. H. , Wang, J. C. , … Yueh, A. (2011). Successful propagation of flavivirus infectious cDNAs by a novel method to reduce the cryptic bacterial promoter activity of virus genomes. Journal of Virology, 85(6), 2927–2941. doi: 10.1128/JVI.01986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikkema, R. S. , Farag, E. A. B. A. , Islam, M. , Atta, M. , Reusken, C. B. E. M. , Al‐Hajri, M. M. , & Koopmans, M. P. G. (2019). Global status of Middle East respiratory syndrome coronavirus in dromedary camels: A systematic review—CORRIGENDUM. Epidemiology and Infection, 147, e198. doi: 10.1017/S0950268819000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thi Nhu Thao, T. , Labroussaa, F. , Ebert, N. , V'kovski, P. , Stalder, H. , Portmann, J. , … Thiel, V. (2020). Rapid reconstruction of SARS‐CoV‐2 using a synthetic genomics platform. Nature, 582(7813), 561–565. doi: 10.1038/s41586-020-2294-9. [DOI] [PubMed] [Google Scholar]
- van Boheemen, S. , de Graaf, M. , Lauber, C. , Bestebroer, T. M. , Raj, V. S. , Zaki, A. M. , … Fouchier, R. A. (2012). Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio, 3(6). doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, B. N. , Feldmann, F. , Schwarz, B. , Meade‐White, K. , Porter, D. P. , Schulz, J. , … de Wit, E. (2020). Clinical benefit of remdesivir in rhesus macaques infected with SARS‐CoV‐2. Nature, 585(7824), 273‐276. doi: 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. , Yu, B. , Chen, Y. M. , Wang, W. , Song, Z. G. , … Zhang, Y. Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, X. , Muruato, A. , Lokugamage, K. G. , Narayanan, K. , Zhang, X. , Zou, J. , … Shi, P. Y. (2020). An infectious cDNA clone of SARS‐CoV‐2. Cell Host Microbe, 27(5), 841–848.e843. doi: 10.1016/j.chom.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, C. , Chiem, K. , Park, J. G. , Oladunni, F. , Platt, R. N. , Anderson, T. , … Martinez‐Sobrido, L. (2020). Rescue of SARS‐CoV‐2 from a single bacterial artificial chromosome. Mbio 11(5), e02168‐20. doi: 10.1128/mBio.02168-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki, A. M. , van Boheemen, S. , Bestebroer, T. M. , Osterhaus, A. D. , & Fouchier, R. A. (2012). Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. New England Journal of Medicine, 367(19), 1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- Ziebuhr, J. (2005). The coronavirus replicase. Current Topics in Microbiology and Immunology, 287, 57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]


