Summary
We compared clinical symptoms, laboratory findings, radiographic signs and outcomes of COVID‐19 and influenza to identify unique features. Depending on the heterogeneity test, we used either random or fixed‐effect models to analyse the appropriateness of the pooled results. Overall, 540 articles included in this study; 75,164 cases of COVID‐19 (157 studies), 113,818 influenza type A (251 studies) and 9266 influenza type B patients (47 studies) were included. Runny nose, dyspnoea, sore throat and rhinorrhoea were less frequent symptoms in COVID‐19 cases (14%, 15%, 11.5% and 9.5%, respectively) in comparison to influenza type A (70%, 45.5%, 49% and 44.5%, respectively) and type B (74%, 33%, 38% and 49%, respectively). Most of the patients with COVID‐19 had abnormal chest radiology (84%, p < 0.001) in comparison to influenza type A (57%, p < 0.001) and B (33%, p < 0.001). The incubation period in COVID‐19 (6.4 days estimated) was longer than influenza type A (3.4 days). Likewise, the duration of hospitalization in COVID‐19 patients (14 days) was longer than influenza type A (6.5 days) and influenza type B (6.7 days). Case fatality rate of hospitalized patients in COVID‐19 (6.5%, p < 0.001), influenza type A (6%, p < 0.001) and influenza type B was 3%(p < 0.001). The results showed that COVID‐19 and influenza had many differences in clinical manifestations and radiographic findings. Due to the lack of effective medication or vaccine for COVID‐19, timely detection of this viral infection and distinguishing from influenza are very important.
Keywords: coronavirus, COVID‐19, influenza, meta‐analysis, SARS‐Cov‐2
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- ARDS
acute respiratory distress syndrome
- CDC
Centre for Disease Controls
- CI
confidence interval
- COVID‐19
coronavirus disease 2019
- CRP
C‐reaction protein
- CT scan
computed tomography scan
- ESR
erythrocyte sedimentation rate
- GGO
ground‐glass opacity
- ICU
intensive care unit
- IL
interleukin
- IQR
interquartile range
- MCP‐I
monocyte chemoattractant protein I
- N
number
- NA
not known
- PRISMA
preferred reporting items for systematic reviews and meta‐analysis statement
- RT‐PCR
real‐time polymerase chain reaction
- SARS‐Cov‐2
severe acute respiratory syndrome coronavirus‐2
- TNF‐α
tumour necrosis factor‐α
- WBCs
white blood cells
- WHO
World Health Organization
1. INTRODUCTION
Influenza outbreaks are seen every year during the colder months of the year. Until now, four influenza pandemics have been reported, including the H1N1 (1918), the H2N2 (1957), the H3N2 (1967) and the H1N1 pandemic in 2009. 1 In addition, seasonal flu is reporting in different countries every year. Influenza mortality is associated with age, underlying disease and pregnancy. 2 , 3 The most important clinical findings in patients with influenza are fever, cough and runny nose. 4 , 5 According to the Centre for Disease Controls (CDC), the influenza virus has infected 35.5 million people, with 490,600 people hospitalized and about 34,200 have died in the United States between 2018 and 2019. The influenza virus is divided into four types (A, B, C and D) by antigenic differences in their core proteins, including matrix protein (M1) and nucleoprotein (NP). 6 Types A and B are the dominant types of circulating influenza virus, and the most influenza epidemics are related to type A. Influenza type A viruses are subdivided into subtypes according to glycoproteins on the surface of the virus: hemagglutinin (HA) and neuraminidase (NA). There are 18 different HA subtypes and 11 different NA subtypes (H1–H18 and N1–N11, respectively). 6 Current subtypes of influenza A viruses that routinely circulate in people include flu A (H1N1) and A (H3N2)6. Type A subtypes often cause mild and symptomatic respiratory illness, but some subtypes such as H5N1 are highly pathogenic and have a higher mortality rate. 6
In the last 20 years, in addition to the influenza virus, other respiratory viruses belonging to coronavirus families such as severe acute respiratory syndrome (SARS) (in 2002) and Middle East respiratory syndrome (in 2012) were among the most severe respiratory pathogens. 6 In late 2019, the SARS‐Cov‐2 virus caused the COVID‐19 infection, and it became a pandemic disease in a short time. Between December 2019 and 26 April 2020, about three million people were infected with COVID‐19, and about 300,000 died from the disease. Both influenza and SARS‐Cov‐2 viruses cause respiratory disease with a wide range of asymptomatic or mild to severe disease. 7 Also, both viruses are transmitted via contact, droplets and contaminated surfaces. 8 Moreover, basic reproduction number R zero (R0) for COVID–19 (1.5–5.7) is more significant than influenza (0.9–2.1), which shows that the SARS‐Cov‐2 virus can infect more people than influenza. 8 , 9 , 10 , 11 , 12
Children are the primary group of virus carriers in influenza infections. 13 Moreover, children, older adults, pregnant women, individuals with underlying chronic disease and immunosuppressed patients are at high risk of severe influenza infection. 13 , 14 , 15 While, in the COVID‐19, children are less affected than adults and the older age with underlying conditions, and are associated with severe symptoms of the disease. 16 Among COVID‐19 patients, the primary treatment is mostly supportive, although multiple experimental antiviral medications are being evaluated. 17 , 18 Thus, prevention and rapid diagnosis of infected patients are crucial. Most of the clinical symptoms of COVID‐19 patients are similar to influenza infections. Therefore, in this study, we attempted to distinguish the clinical symptoms, laboratory findings, radiographic signs and outcomes of confirmed COVID‐19 and influenza patients. All findings are compared to determine the unique features among each virus. These data could be helpful in the early diagnosis and prevention of infection as well as providing more reliable epidemiological data on a large scale for healthcare policies and future studies.
2. METHODS
2.1. Search strategy
This study was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta‐Analyses Statement (PRISMA) guidelines. 19 We searched all studies published up to 26 April 2020, from the following databases: Embase, Scopus, PubMed, Web of Science and the Cochrane library. Search medical subject headings (MeSH) terms used were 'COVID‐19', 'SARS Cov‐2', 'severe acute respiratory syndrome coronavirus 2', 'coronavirus disease 2019 virus', '2019 novel coronavirus', 'COVID‐19 virus', 'influenza virus,' 'influenza,' 'Human flu', and all their synonyms like 'SARS‐CoV‐2' and '2019‐nCoV'. Moreover, we searched for unpublished and grey literature with Google scholar, CDC and World Health Organization (WHO) databases. We also examined references of included articles to find additional relevant studies. There was no language restriction, and all included studies were written in English or Chinese languages; the letter was translated by https://translate.google.com/. Additional search strategy details are provided in Table S1.
2.2. Study selection
Duplicate studies were removed using EndNote X7 (Thomson Reuters). Records were initially reviewed by title and abstract by independent five authors (AP, SG, MR, AK and EA). The full text of potentially eligible records was retrieved and examined, and any disagreement was resolved by consensus.
2.3. Eligibility and inclusion criteria
Studies to be eligible for inclusion in our meta‐analysis had to have following pre‐determined criteria. All case‐control, cross‐sectional, cohort studies, case reports and case series peer‐reviewed studies were included if they reported the number of confirmed cases of patients with demographic data [AND] [OR] clinical data [AND] [OR] radiology data [AND] [OR] laboratory data [AND] [OR] risk factor data. Also, influenza virus studies from 2000 to 2020 were included.
2.4. Exclusion criteria
Studies without the number of confirmed cases, letters to editor, review articles, individual case reports and news reports were excluded. Duplicate data from the same patients were combined and counted as a single case when the data were reported more than one.
2.5. Data extraction
All COVID‐19 included literature that was published in 2020, and all influenza studies were from 2000 to 2020. The following items were extracted from each article: first author, centre and study location, countries, sample collection time, patient follow‐up time, the reference standard for infection confirmation, number of confirmed cases, study type, and all demographic, clinical, radiological, laboratory data and risk factor data. Four of our authors (Saied Ghorbani, Mohammad Hossein Razizadeh, Ehsan Alborzi and Alireza Khatami) independently extracted data, and all extracted data were checked randomly by another author (Ali Pormohammad); the differences were resolved by consensus.
2.6. Quality assessment
Quality assessments of studies were performed by two reviewers independently according to the critical appraisal checklist recommended by the Joanna Briggs Institute, and disagreements were resolved by consensus. The checklist is composed of nine questions that reviewers addressed for each study. The 'Yes' answer to each question received one point. Thus, the final scores for each study could range from 0 to 9 (Table S2).
2.7. Analysis
Data cleaning and preparation were done in Microsoft Excel 2010 (Microsoft©), and further analyses were carried out via Comprehensive Meta‐Analysis Software Version 2.0 (Biostat). Determination of heterogeneity among the studies was undertaken using the chi‐squared test (Cochran's Q) to assess the appropriateness of pooling data. Depending on the heterogeneity test, we used either random‐ or fixed‐effect model for pooled results. In the case of high heterogeneity (I 2 >50%), a random‐effect model (M‐H heterogeneity) was applied, while in low heterogeneity cases (I 2 < 50%), a fixed‐effect model was used. 20 Percentages and means ± SDs were calculated to describe the distributions of categorical and continuous variables, respectively. p‐Values reflect study heterogeneity with <0.05 being significant. We also used the funnel plot, Begg's and Egger's tests based on the symmetry assumption to detect publication bias (Figure S1).
3. RESULTS
3.1. Characteristics of included studies
The process of study selection is represented in Figure 1. A total of 194,092 reports were screened for the analysis of patients with COVID‐19 and influenza; 363,827 of them were excluded after the duplicate removing, title and abstract screening, and the full text of 611 reports were reviewed in full text. We excluded studies that did not report sufficient data. Out of 540 included studies, 157 studies met the inclusion criteria for COVID‐19 and 383 for influenza. The characteristics of the selected articles are summarized in Table S5. Of the 157 COVID‐19 studies that were included in the analysis, 155 studies were in English and 2 of them were in Chinese languages. 21 , 22 All COVID‐19 studies were retrospective, published in 2020, and 150 studies were from China, 2 from the United States, 1 from Italy, 1 from Japan, 1 from the United Kingdom, 1 from Iran and 1 from Taiwan. All influenza studies were from 2000 to 2020, and out of 383 influenza studies, 251 studies were influenza A and 47 studies were influenza B.
FIGURE 1.
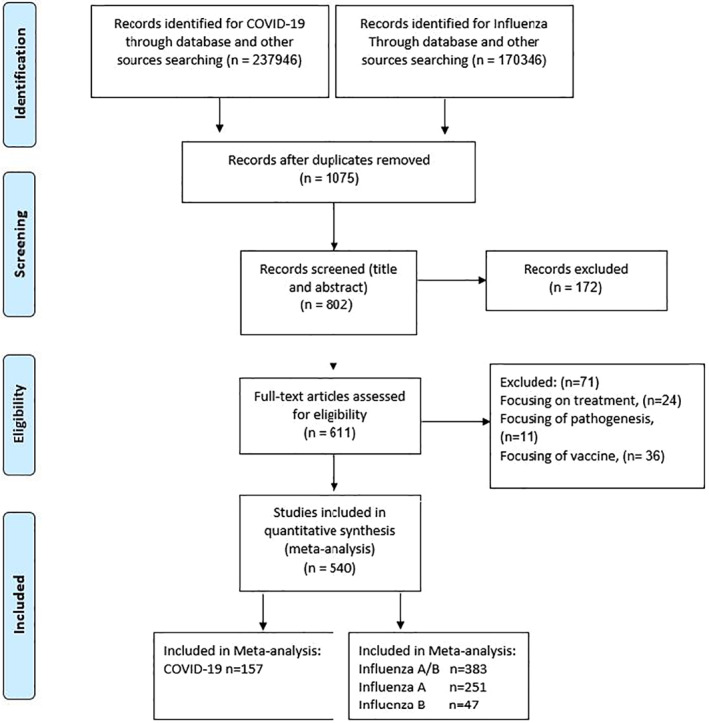
Flow diagram of literature search and study selection (Preferred Reporting Items for Systematic Reviews and Meta‐Analyses Statement [PRISMA] flow chart)
3.2. Quality assessment
Quality assessment of included studies was performed based on the critical appraisal checklist, and the final quality scores of the included studies are represented in Table S2. In brief, studies by Chen, 23 Wang, 24 Huang, 25 Guan, 26 Zhang, 27 Cheng, 28 Li, 29 Wei Xu 30 and Song 31 had the highest quality of the COVID‐19 studies available to date in the purpose of this study.
3.3. Demographics, baseline characteristics and clinical characterization
Overall, 75,164 confirmed patients with COVID‐19 infection, 113,818 with influenza type A and 9266 with influenza type B were included in the meta‐analysis, of which 51% (95% CI 50–52.2, p < 0.001) of COVID‐19, 54% (95% CI 53–54.5, p < 0.001) of influenza type A and 52% (95% CI 48–55.5, p < 0.001) of influenza type B included patients who were male. Funnel plots for included studies did not detect significant publication bias (Figure S1). Table 1 shows that most patients of COVID‐19 (76% [95% CI 72.5–79, p < 0.001]), influenza type A (87.5% [95% CI 85–90, p < 0.001]) and influenza type B (89.5% [95% CI 83–93, p < 0.001]) had fever. Cough was the second most common symptom presenting in the patients of COVID‐19 (54% [95% CI 50–58, p < 0.001]), influenza type A (83.5% [95% CI 81–85, p < 0.001]) and influenza type B (79% [95% CI 75–84, p < 0.001]). Runny nose was the third most common symptom presenting in the patients of influenza type A (70% [95% CI 71–72, p < 0.001]) and influenza type B (74% (95% CI 72–75, p < 0.001]) of patients. While runny nose was less common symptom in COVID‐19 and it is presented in 14% (95% CI 7.3–25, p < 0.001) of patients. Also, fatigue was the fourth most common symptom in influenza type A (60% [95% CI 59–61, p < 0.001]), while it was less common in COVID‐19 (27% [95% CI 23–31.5, p < 0.001]) and influenza type B (21% [95% CI 18–24, p < 0.001]) patients. Dyspnea was less common in COVID‐19 patients (15% [95% CI 12–19, p < 0.001]), in comparison to influenza type A (45.5% [95% CI 41–50, p < 0.001]) and influenza type B (33% [95% CI 23.5–45, p < 0.001]). Likewise, sore throat was less common in COVID‐19 patients (11.5% [95% CI 9–14, p < 0.001]), in comparison to influenza type A (49% [95% CI 48–51, p < 0.001]) and influenza type B (38% [95% CI 36–39, p < 0.001]). Also, rhinorrhea was less common in COVID‐19 patients (9.3% [95% CI 6–14, p < 0.001]), in comparison to influenza type A (44.5% [95% CI 39–49.5, p < 0.001]) and influenza type B (49% [95% CI 42–56, p < 0.001]). There was no information about coryza in COVID‐19 patients, while it is presented in influenza type A by 47% (95% CI 43–52, p < 0.001) and influenza type B by 32% (95% CI 10–67, p < 0.001) of the patients (Figure 2).
TABLE 1.
Meta‐analysis on demographics, baseline characteristics and clinical outcomes of patients with confirmed COVID‐19, influenza type A and influenza type B
| COVID‐19 (total of 157 studies, 75,164 patients) | Influenza type A (total 251 studies, 113,818 patients) | Influenza type B (total 47 studies, 9266 patients) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | |
| Age, years | 49.7 (mean) (43–54) | 123 | 24,360 | 99 | <0.001 | 36.5 (26–48) | 164 | 64,602 | 99 | <0.001 | 38.5 (32–42) | 27 | 4179 | 99 | <0.001 |
| Sex (male) | 51 (50–52.2) | 144 | 71,778 | 72.5 | <0.001 | 54 (53–54.5) | 212 | 98,403 | 81 | <0.001 | 52 (48–55.5) | 28 | 4513 | 77 | <0.001 |
| Fever | 76 (72.5–79) | 113 | 15,537 | 94 | <0.001 | 87.5 (85–90) | 171 | 61,212 | 97 | <0.001 | 89.5 (83–93) | 31 | 8388 | 97 | <0.001 |
| Cough | 54 (50–58) | 114 | 15,162 | 93 | <0.001 | 83.5 (81–85) | 164 | 59,840 | 96 | <0.001 | 79 (75–84) | 26 | 7735 | 94 | <0.001 |
| Fatigue | 27 (23–31.5) | 82 | 12,645 | 94 | <0.001 | 60 (59–61) | 40 | 13,637 | 96 | <0.001 | 21 (18–24) | 6 | 750 | 86 | <0.001 |
| Sputum production | 21 (18–24) | 42 | 4506 | 81 | <0.001 | 34 (33.5–35) | 56 | 22,107 | 96 | <0.001 | 26 (23–27) | 6 | 4546 | 97 | <0.001 |
| Myalgia | 20 (16–24) | 41 | 5077 | 87 | <0.001 | 32 (27–36) | 90 | 39,018 | 98 | <0.001 | 22.5 (11–40) | 15 | 6397 | 98 | <0.001 |
| Dyspnoea | 15 (12–19) | 51 | 7761 | 93 | <0.001 | 45.5 (41–50) | 105 | 34,079 | 98 | <0.001 | 33 (23.5–45) | 9 | 973 | 90 | <0.001 |
| Chill | 14 (9–21) | 18 | 2577 | 91 | <0.001 | 34 (28–40) | 36 | 25,479 | 98 | <0.001 | 25.5 (12–26) | 7 | 4706 | 98 | <0.001 |
| Sore throat | 11.5 (9–14) | 46 | 7737 | 82 | <0.001 | 49 (48–51) | 116 | 47,041 | 96 | <0.001 | 38 (36–39) | 20 | 6475 | 95 | <0.001 |
| Headache | 10.5 (9–12) | 59 | 9311 | 75 | <0.001 | 28 (18–30) | 96 | 36,580 | 99 | <0.001 | 13.4 (8.5–20) | 14 | 3643 | 95 | <0.001 |
| Chest pain | 10.5 (8–13.5) | 41 | 6759 | 91 | <0.001 | 9 (7–12) | 41 | 1,6670 | 95 | <0.001 | 11 (6–20) | 7 | 1245 | 89 | <0.001 |
| Diarrhoea | 8.5 (6.6–11) | 78 | 11,421 | 91 | <0.001 | 12 (10–14) | 112 | 27,360 | 94 | <0.001 | 9 (5.5–15) | 16 | 3942 | 96 | <0.001 |
| Rhinorrhoea | 9.3 (6–14) | 16 | 879 | 60 | 0.001 | 44.5 (39–49.5) | 70 | 29,166 | 98 | <0.001 | 49 (42–56) | 17 | 2190 | 89 | <0.001 |
| Nausea and vomiting | 6 (4–8) | 36 | 5063 | 86 | <0.001 | 20 (17–22.5) | 107 | 28,728 | 96 | <0.001 | 20 (16–26) | 18 | 3872 | 91 | <0.001 |
| Runny nose | 14 (7.3–25) | 14 | 1758 | 91 | <0.001 | 70 (71–72) | 19 | 12,814 | 97 | <0.001 | 74 (72–75) | 4 | 3618 | 96 | <0.001 |
| Pneumonia | NA | NA | NA | NA | NA | 28 (24–34) | 94 | 43,192 | 98 | <0.001 | 14.5 (10–20) | 23 | 4571 | 93 | <0.001 |
| Croup | NA | NA | NA | NA | NA | 4 (2–5) | 13 | 6712 | 84 | <0.001 | 4 (2–4.5) | 6 | 2120 | 0 | 0.4 |
| Coryza | NA | NA | NA | NA | NA | 47 (43–52) | 16 | 6819 | 88 | <0.001 | 32 (10–67) | 3 | 1857 | 98 | <0.001 |
| Asthma | NA | NA | NA | NA | NA | 12 (10–14) | 89 | 61,655 | 96 | <0.001 | 15 (7–29) | 4 | 570 | 88 | <0.001 |
| Comorbid conditions | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID‐19 | Influenza type A | Influenza type B | |||||||||||||
| Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | |
| Travel historya | 51 (44–58) | 52 7 | 54,154 | 98 | <0.001 | – | – | – | – | – | – | – | – | – | – |
| Contact history b | 43.5 (35–52) | 48 | 7126 | 96 | <0.001 | – | – | – | – | – | – | – | – | – | – |
| Hypertension | 20 (16–25) | 66 | 54,151 | 97 | <0.001 | 20 (16–25) | 42 | 15,915 | 96 | <0.001 | 7 (4–12) | 1 | 194 | 0 | 1 |
| ARDS | 26.6 (18–38) | 5 | 3826 | 95 | <0.001 | 31.5 (26–38) | 51 | 17,762 | 96 | <0.001 | 0.8 (0.1–6) | 1 | 55 | 0 | 1 |
| Diabetes | 10.5 (7.5–14.5) | 61 | 46,232 | 97 | <0.001 | 11 (10–13) | 107 | 66,761 | 94 | <0.001 | 19 (11–29) | 6 | 850 | 88 | <0.001 |
| Current smoker | 10.5 (6.5–16) | 28 | 6643 | 96 | <0.001 | 20 (16–24) | 53 | 20,914 | 92 | <0.001 | 14 (10–20) | 4 | 395 | 22 | 0.3 |
| Chronic liver disease | 5.4 (4–7) | 34 | 5269 | 79 | <0.001 | 3 (2–4.5) | 55 | 53,532 | 97 | <0.001 | 2 (7–6.5) | 5 | 1920 | 85dia | <0.001 |
| Digestive system disease | 6 (4–10) | 28 | 5807 | 93 | <0.001 | 15 (13–18) | 39 | 22,105 | 95 | <0.001 | 15 (10–20) | 10 | 4463 | 86 | <0.001 |
| Healthcare worker | 23 (10–43) | 10 | 46,509 | 95 | <0.001 | 4 (105–10) | 8 | 3753 | 97 | <0.001 | NA | NA | NA | NA | NA |
| Past smoker | 6.5 (5–9) | 5 | 1307 | 33 | 0.2 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Cardiovascular and cerebrovascular diseases | 10.5 (7–15) | 56 | 54,270 | 98 | <0.001 | 9.5 (8–11) | 134 | 72,509 | 96 | <0.001 | 9 (3.5–20) | 8 | 2407 | 95 | <0.001 |
| Chronic respiratory disease | 9.5 (6–14) | 33 | 49,731 | 95 | <0.001 | 12 (10–12) | 112 | 67,817 | 96 | <0.001 | 16 (10–24) | 8 | 988 | 85 | <0.001 |
| Nervous system disease | 3 (0.1–6) | 2 | 314 | 92 | <0.001 | 7 (6–9) | 90 | 63,704 | 97 | <0.001 | 3 (2–7) | 12 | 2614 | 2614 | 87 |
| HIV | 2 (0.2–14) | 2 | 232 | 70 | 0.06 | 3.5 (2.5–4.5) | 13 | 26,637 | 85 | <0.001 | NA | NA | NA | NA | NA |
| Cancer | 3 (2–5) | 41 | 52,749 | 93 | <0.001 | 7 (5.5–8.5) | 52 | 39,954 | 93 | <0.001 | 10 (5–18) | 6 | 850 | 85 | <0.001 |
| Renal failure | 7 (4–11) | 39 | 5676 | 91 | <0.001 | 5 (4–6) | 9 | 64,775 | 95 | <0.001 | 7 (2–17) | 8 | 1077 | 94 | <0.001 |
| Bacteria co‐infection | 4.5 (1.5–12) | 10 | 1196 | 84 | <0.001 | 12 (9–15) | 62 | 14,326 | 85 | <0.001 | 15 (7–29) | 9 | 740 | 83 | <0.001 |
| Fungi co‐infection | 4.8 (2–11) | 2 | 109 | 0 | 0.4 | 1.5 (1–2) | 9 | 3580 | 38 | 0.1 | 20 (6–42) | 3 | 95 | 74 | 0.03 |
| Virus co‐infection | 4 (1.5–10) | 10 | 2499 | 88 | <0.001 | 8.5 (5.5–13) | 47 | 33,680 | 97 | <0.001 | 24 (8–54) | 10 | 2027 | 96 | <0.001 |
| Chest x‐ray and CT scan findings | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID‐19 | Influenza type A | Influenza type B | |||||||||||||
| Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | |
| Abnormal chest x‐ray | 84 (78–8.5) | 12 | 1706 | 85 | <0.001 | 57 (50–64) | 65 | 19,500 | 89 | <0.001 | 33 (6–80) | 5 | 651 | 86 | <0.001 |
| Bilateral involvement | 76.8 (62.5–87) | 12 | 46,270 | 94 | <0.001 | 37.5 (27.5–48.5) | 35 | 6003 | 88 | <0.001 | 16.5 (7–35) | 4 | 545 | 87 | <0.001 |
| Consolidation | 75.5 (50.5–91) | 6 | 1378 | 92 | <0.001 | 27 (17–40) | 25 | 8730 | 98 | <0.001 | 27.5 (8–62) | 3 | 95 | 78 | 0.09 |
| Ground‐glass opacity | 71 (40–90) | 12 | 46,270 | 87 | <0.001 | 47 (30–65) | 7 | 529 | 90 | <0.001 | 6.5 (0.3–65) | 2 | 73 | 80 | <0.001 |
| Unilateral involvement of chest radiography | 15.5 (10.5–22.5) | 29 | 4615 | 94 | <0.001 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Outcome | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COVID‐19 | Influenza type A | Influenza type B | |||||||||||||
| Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | Clinical presentation* (CI 95%) | Included studies number | Included patients number | I‐squared** | p‐value** | |
| Incubation period (mean) (day) | 6.4 (5.8–7) | 53 | 12,609 | 98 | <0.001 | 3.4 (2.25–4.5) | 8 | 784 | 97 | <0.001 | NA | NA | NA | NA | NA |
| Discharged | 57.5 (49.5–62.5) | 57 | 7906 | 96 | <0.001 | 82 (77–86) | 122 | 68,373 | 98 | <0.001 | 87.5 (63–97) | 14 | 2646 | 97 | <0.001 |
| Duration of hospitalization (mean) (day) | 14 (12–16) | 32 | 3674 | 97 | <0.001 | 6.5 (6–8) | 82 | 23,510 | 98 | <0.001 | 6.7 (5.3–8) | 15 | 2227 | 97 | <0.001 |
| Mortality | 6.2 (6–6.5) | 64 | 56,269 | 97 | <0.001 | 6 (5–6.5 | 133 | 78,648 | 97 | <0.001 | 3 (2–4) | 13 | 2912 | 85 | <0.001 |
Abbreviations: ARDS, acute respiratory distress syndrome; CI, confidence interval; CT scan, computed tomography scan; ICU, intensive care unit; NA, not available.
Recent travel or contact with endemic people resident of Wuhan.
Contact with another person with respiratory symptoms.
*Age is an exception, presented in mean age in years.
**Greater than 50% is considered high heterogeneity, less than 50% is considered low heterogeneity. A low p‐value (<0.05) is consistent with high heterogeneity.
FIGURE 2.
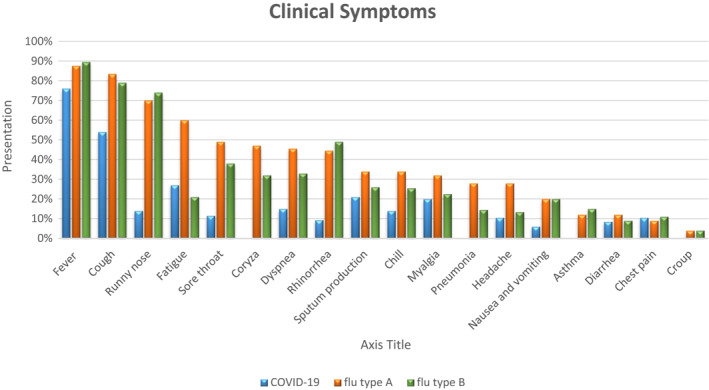
Meta‐analysis on symptoms presentation of patients with confirmed COVID‐19, influenza type A and influenza type B
3.4. Risk factors and common comorbid of patients infected with COVID‐19
Up to 26 April 2020, 51% (95% CI 44–58, p < 0.001) of COVID‐19 patients had a history of recent travel or contact with endemic people, and 43.5% (95% CI 35–52, p < 0.001) had contacted with another person with respiratory symptoms. Another risk factor for COVID‐19 was healthcare worker by 23% (95% CI 10–43, p < 0.001). The most common comorbid chronic condition for COVID‐19 and influenza type A is hypertension by 20% (95% CI 16–25, p < 0.001) and diabetes for influenza type B by 19% (95% CI 11–29, p < 0.001). Acute respiratory syndrome (ARDS) occurred more frequently presented in influenza type A by 31.5% (95% CI 26–38, p < 0.001) compared to COVID–19 by 26.6% (95% CI 18–38, p < 0.001) and influenza type B by 0.8% (95% CI 0.1–6, p < 0.001). Virus co‐infection occurred more frequently in influenza type B by 24% (95% CI 8–54, p < 0.001), in comparison to influenza type A by 8.5% (95% CI 5.5–13, p < 0.001) and COVID–19 by 4% (95% CI 1.5–10, p < 0.001) patients (Figure 3).
FIGURE 3.
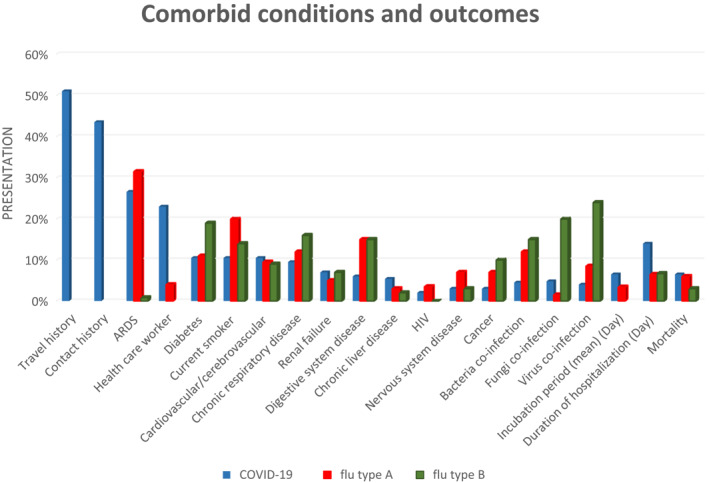
Meta‐analysis on presentation of comorbid conditions and outcomes of patients with confirmed COVID‐19, influenza type A, and influenza type B. ARDS, acute respiratory distress syndrome
3.5. Chest x‐ray and CT scan findings in patients infected with COVID–19
Analysis showed that 84% (95% CI 78–8.5, p < 0.001) of COVID–19 patients, 57% (95% 50–64, p < 0.001) of influenza type A patients and 33% (95% 6–80, p < 0.001) of influenza type B patients had abnormal radiological findings on chest x‐ray and CT scans. The most common radiological abnormalities in COVID–19 patients were bilateral involvement of chest x‐ray by 76.8% (95% CI 62.5–87, p < 0.001), consolidation by 75.5% (95% CI 50.5–91, p < 0.001) and ground‐glass opacity (GGO) by 71% (95% CI 40–90, p < 0.001) (Table 1).
3.6. Outcome
Based on the available data, the mean incubation period was 6.4 days (95% CI 5.8–7, p < 0.001) in 12,609 COVID‐19 cases and 3.4 days (95% CI 2.25–4.5, p < 0.001) in 784 influenza type A, while there is no available data about the incubation period of influenza type B in the included articles. The mean duration of hospitalization among 3674 COVID‐19 confirmed cases was 14 days (95% CI 12–16, p < 0.001), 6.5 days (95% CI 6–8, p < 0.001) in 23,510 influenza type A cases and 6.7 days (95% CI 5.3–8, p < 0.001) in 2227 influenza type B cases . The hospital discharged rate of COVID–19 was 57.5% (95% CI 49.5–62.5, p < 0.001), which was lower in comparison to influenza type A (82% [95% CI 77–86, p < 0.001]) and influenza type B (87.5% [95% CI 63–97, p < 0.001]) patients.
Case fatality rate of COVID‐19 hospitalized patients was 6.5% (95% CI 4.5–9, p < 0.001) (Figure S2), influenza type A was 6% (95% 5–6.5, p < 0.001) and influenza type B was 3% (95% CI 2–4, p < 0.001).
3.7. The influenza mortality rate in different subtypes and countries
The mortality rate in the influenza A subtypes are subtypes H1N1 (5.5% [95% CI 4–7, p < 0.001]), H3N2 (1.7% [95% CI 0.2–15, p < 0.001]), H5N1 (42% [95% CI 29–56, p < 0.001]), H7N9 (30% [95% CI 25.6–35, p < 0.001]) and non‐H1N1 (2% [95% CI 1–5, p < 0.001]). Influenza mortality rate was associated with age, and the high mortality rate was in ≥50‐year‐old ages (12% [95% CI 6–22.5, p < 0.001]) (Table 2). Influenza H1N1 mortality rate in ICU cases was 25% (95% CI 15.5–37.6, p < 0.001) (Figure 4).
TABLE 2.
Meta‐analysis: mortality rate of COVID‐19 patients in different age ranges
| Age groups (year) | Mortality rate (%) (CI 95%) | Included studies number | Included patients number | Heterogeneity test, I‐squared (%)** | Heterogeneity test, p‐value** |
|---|---|---|---|---|---|
| <8 | 0.6 (0–9) | 1 | 82 | 0 | 1 |
| >50 | 38 (35–40) | 14 | 1935 | 85 | <0.001 |
| All range | 4.3 (4–4.5) | 49 | 54,252 | 92 | <0.001 |
| Overall | 6.2 (6–6.5) | 64 | 56,269 | 91 | <0.001 |
FIGURE 4.
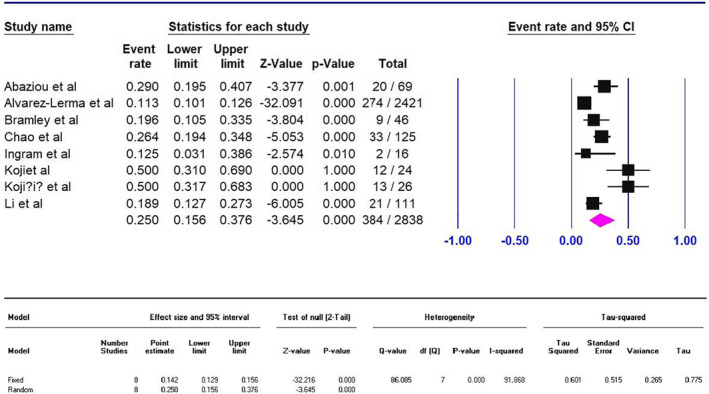
Meta‐analysis forest plot on mortality rate of influenza type A (subtype H1N1) in intensive care unit. All patients in two Kojiret studies (from Bosnia and Herzegovina) were mechanically ventilated patients
Figure 5 showed influenza type A, B and A/B mortality rates in different countries based on the reported data from these countries. The highest mortality by influenza type A/B was reported in Indonesia at 77% (95% 48–92, p 0.9) and the lowest mortality rate in Rwanda at 0.1% (95% CI 0–1.5, p 0.9) (Table S3). Vietnam had the highest mortality in influenza type A by 33% (95% CI 20–48, p < 0.17), and Kuwait had the lowest mortality rate by 0.4% (95% CI 0.1–2.5, p 0.9) (Table S4). Moreover, Spain had the highest mortality in influenza type B by 12.5% (95% CI 0.7–73, p < 0.9), and France had the lowest mortality rate by 0.9% (95% CI 0.6–1.5, p 0.9) (Figure S3).
FIGURE 5.
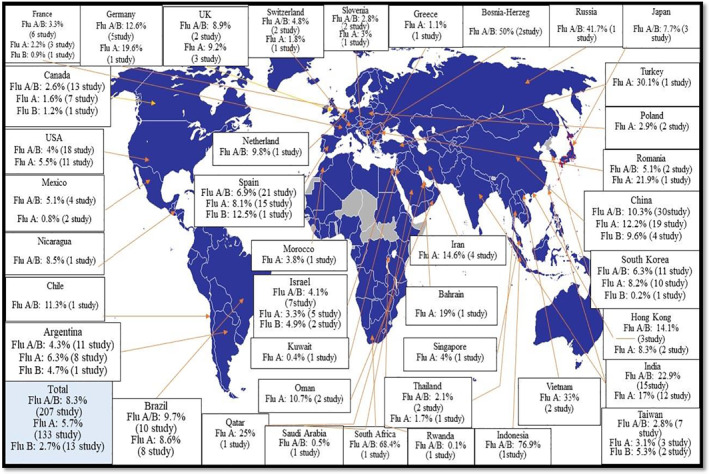
Meta‐analysis; global estimate of influenza type A, B and A/B hospitalized patients' case fatality rate (all patients in two Bosnia and Herzegovina studies were mechanically ventilated patients)
3.8. Laboratory findings of patients infected with COVID‐19
The laboratory findings showed that among 10,185 COVID‐19 cases where data were available, lymphopenia was 62.5% (95% CI 45–72, p < 0.001), which is more than influenza type A (8820 cases) at 49% (95% CI 35–56.4, p < 0.001) (Table 3). In addition, C‐reactive protein increased in 1054 COVID‐19 patients by 81% (95% CI 68–89, p < 0.001), in 5237 influenza type A patients by 62% (95% CI 55–73, p < 0.001), and in 287 influenza type B patients by 43% (95% CI 37–49, p < 0.001). In COVID‐19 confirmed patients, 80% (95% CI 75–85, p < 0.001) had decreased albumin and 1783 patients had increased LDH at 70.3% (95% CI 65–76, p < 0.001) (Table 4).
TABLE 3.
Meta‐analysis; mortality rate of influenza type and subtypes in different age ranges
| Age groups (year) | Mortality rate (%) (CI 95%) | Included studies number | Included patients number | Heterogeneity test, I‐squared (%) a | Heterogeneity test, p‐value a | |
|---|---|---|---|---|---|---|
| Type A/B | All ages | 7 (6–8) | 207 | 194,931 | 97 | <0.001 |
| <1 year | 3 (4–18) | 5 | 3291 | 85 | <0.001 | |
| <5 year | 0.8 (0.1–5) | 4 | 962 | 92 | <0.001 | |
| <18 year | 2 (0.8–4) | 24 | 15,229 | 56 | 0.05 | |
| ≥18 year | 8 (6–10) | 38 | 20,824 | 84 | <0.001 | |
| ≥50 year | 12 (6–22.5) | 12 | 10,152 | 56 | 0.05 | |
| Type A (all subtypes) | All ages | 6 (5–6.5) | 133 | 98,545 | 94 | <0.001 |
| <1 year | 5 (0.7–25) | 2 | 18 | 91 | <0.001 | |
| <5 year | 1 (0.1–8) | 3 | 775 | 89 | <0.001 | |
| <18 year | 1 (0.1–8) | 3 | 775 | 87 | <0.001 | |
| ≥18 year | 10 (8–13.5) | 21 | 5735 | 92 | <0.001 | |
| ≥50 year | 16 (3–51) | 6 | 7186 | 56 | 0.05 | |
| Type A (subtype H1N1) | All ages | 5.5 (4–7) | 84 | 88,603 | 97 | <0.001 |
| In ICU cases | 25 (15.5–37.6) | 8 | 2838 | 91 | <0.001 | |
| <1 year | 4.5 (0.3–45) | 1 | 10 | 0 | 1 | |
| <5 year | – | – | – | – | – | |
| <18 year | 1.5 (0.6–40) | 10 | 3945 | 94 | <0.001 | |
| ≥18 year | 5.5 (3–8) | 9 | 1105 | 40 | <0.01 | |
| ≥50 year | 17.5 (12–25.5) | 2 | 120 | 96 | <0.001 | |
| Type A (subtype H3N2) | All ages | 1.7 (0.2–15) | 4 | 68,579 | 87 | <0.001 |
| <1 year | 5 (0.7–28) | 2 | 17 | 8 | 0.1 | |
| <5 year | – | – | – | – | – | |
| <18 year | 1.5 (0.6–4) | 10 | 3945 | 9 | <0.001 | |
| ≥18 year | 2.5 (4–14) | 4 | 825 | 65 | 0.003 | |
| ≥50 year | – | – | – | 0 | 0.4 | |
| Type A (subtype H5N1) | All ages | 42 (29–56) | 5 | 146 | 56 | 0.05 |
| <1 year | – | – | – | – | – | |
| <5 year | – | – | – | – | – | |
| <18 year | 77 (48–92) | 1 | 13 | 87 | <0.001 | |
| ≥18 year | 40 (28–51) | 1 | 67 | 87 | <0.001 | |
| ≥50 year | – | – | – | – | – | |
| All ages | 30 (25.6–35) | 11 | 1018 | 52 | 0.02 | |
| Type A (subtype H7N9) | All ages | 2 (1–5) | 4 | 291 | 0 | 0.5 |
| Type A (non‐H1N1) | All ages | 3 (2–4) | 13 | 2812 | 87 | <0.001 |
| Type B | <1 year | – | – | – | – | – |
| <5 year | – | – | – | – | – | |
| <18 year | 2.5 (0.7–7.6) | 3 | 1550 | 53 | 0.1 | |
| ≥18 year | – | – | – | – | – | |
| ≥50 year | 2.5 (0.7–7.6) | 3 | 1550 | 89 | <0.001 |
Abbreviation: ICU, intensive care unit.
Greater than 50% is considered high heterogeneity, less than 50% is considered low heterogeneity. A low p‐value (<0.05) is consistent with high heterogeneity.
TABLE 4.
Meta‐analysis on laboratory features for confirmed patients with COVID‐19
| Normal range | COVID‐19 | Influenza type A | Influenza type B | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean (CI 95%) | Patient N | Study N | Mean (CI 95%) | Patient N | Study N | Mean (CI 95%) | Patient N | Study N | ||
| Leucocytes (WBCs) | 3.5–9.5 | 6.3 (× 10⁹ per L) (5.1–7.5) | 9268 | 62 | 6.4 (× 10⁹ per L) (6.4–6.5) | 16,962 | 48 | 7.4 (× 10⁹ per L) (6.2–606) | 940 | 9 |
| Increased | – | 13.3 (%) | – | – | 16 (%) | – | – | 10.5 (%) | – | – |
| Decreased | – | 26 (%) | – | – | 19 (%) | – | – | 23 (%) | – | – |
| Neutrophils | 1.8–6.3 | 4 (× 10⁹ per L) (3–8.5) | 8192 | 47 | 4.9 (4.6–5.2) | 8718 | 16 | 4.8 (4.5.5) | 561 | 5 |
| Increased | – | – | – | – | – | – | – | – | – | – |
| Decreased | – | – | – | – | 10 (%) | – | – | – | – | – |
| Lymphocytes | 1.1–3.2 | 1.13 (× 10⁹ per L) (0.9–1.2) | 10,185 | 61 | 1.3 (× 10⁹ per L) (1.1–1.4) | 8820 | 24 | 1.5 (× 10⁹ per L) (0.7–2.2) | 499 | 6 |
| Decreased | – | 62.5 (%) | – | – | 49 (%) | – | – | – | – | – |
| Platelets | 125–350 | 186.5 (× 10⁹ per L) (179–198) | 6356 | 39 | 192 (× 10⁹ per L) (187–199) | 10,792 | 29 | 179 (× 10⁹ per L) (159–199) | 350 | 4 |
| Decreased | – | 13 (%) | – | – | 24 (%) | – | – | 16 (%) | – | – |
| Increased | – | 28.5 (%) | – | – | 10 (%) | – | – | 3.5 (%) | – | – |
| CRP | 0–0.5 | 29.6 (mg/L) (16.7–42.5) | 1054 | 26 | 22.8 (mg/L) (22–35) | 5237 | 14 | 32.4 (mg/L) (28–35) | 287 | 3 |
| Increased | – | 81 (%) | – | – | 62 (%) | – | – | 43 (%) | – | – |
| Haemoglobin | 130–175 | 119 (g/L) (106–132) | 3062 | 37 | – | – | – | – | – | – |
| ESR | 0–15 | 28 (mm/h) (18–37) | 1149 | 11 | 21 (mm/h) (13–29) | 3209 | 8 | 15 (mm/h) (3–27) | 241 | 2 |
| Albumin | 40–55 | 36.8 (g/L) (24.5–46) | 1045 | 11 | – | – | – | – | – | – |
| Decreased | – | 80% | – | – | – | – | – | – | – | – |
| Interleukin‐6 | 0.0–7 | 7.9 (mg/ml) (6.8–8.6) | 99 | 2 | – | – | – | – | – | – |
| Increased | – | 52% | – | – | – | – | – | – | – | – |
| LDH | 120–250 | 280 (268–294) | 1783 | 9 | – | – | – | – | – | – |
| Increased | – | 70.3 (%) | – | – | – | – | – | – | – | – |
Note: Increased or decreased refers to values above or below the normal range.
Abbreviations: CRP, C‐reaction Protein; ESR, erythrocyte sedimentation rate; WBCs, white blood cells.
4. DISCUSSION
Influenza and coronavirus are associated with respiratory diseases, which in most cases are asymptomatic, and the symptoms in patients can range from mild to severe disease and death. 32 After the influenza epidemic in 2017–2019, COVID‐19 reported as a contagious respiratory illness. The diagnosis of these viruses is essential. However, distinguishing these two viruses is challenging due to similar clinical signs and the same way of transmission. Our results show that fever and cough were the most common clinical symptoms in COVID‐19, influenza type A and influenza type B. In addition, runny nose was the third most common clinical finding among influenza A/B patients, while it was less common among COVID‐19. Among 75,164 patients with COVID‐19 infection, fatigue, sputum production and myalgia (muscle soreness) were the next most frequent clinical symptoms, while diarrhoea, rhinorrhoea, nausea and vomiting were less frequent. Within the 113,818 confirmed influenza type A patients, the next most frequent clinical manifestations were fatigue, sore throat, coryza, dyspnoea, rhinorrhoea, sputum production, chills, myalgia, pneumonia and headache.
Sore throat was less common in COVID‐19 patients (11.5%), in comparison to influenza type A (49%) and influenza type B (38%). Likewise, rhinorrhoea was less common in COVID‐19 patients (9.3%), in comparison to influenza type A (44.5%) and influenza type B (49%), as well as nausea and vomiting were less common in COVID‐19 (6%), in comparison to influenza type A (20%) and influenza type B (20%). On the other hand, fatigue was one of the most common clinical symptoms in influenza type A (60%), in comparison to COVID‐19 (27%) and influenza type B (21%). Dyspnoea was another less common clinical symptom in COVID‐19 (15%), in comparison to influenza type A (45.5%) and influenza type B (33%). Therefore, these clinical symptoms may help in first screening and distinguishing these respiratory viral infections from each other.
Our analysis indicated a history of recent travel or contact with endemic populations, contact history with another person with respiratory symptoms and being a healthcare worker were common risks amongst COVID‐19 confirmed cases. These data indicate, in coronavirus outbreaks, isolating infected individuals is one of the most important ways of controlling transmission.
ARDS (31.5%) was the most common amongst patients of influenza type A in comparison to COVID‐19 (26.6%) and influenza type B (0.8%). In addition, the most common comorbid chronic condition for COVID‐19 and influenza type A were hypertension (20%), and diabetes (19%)for influenza type B, as well as viral (24%) and fungi (20%) co‐infection occurred more frequently in influenza type B in comparison to two other virus infections, which indicate influenza type B may be pathogenic in people who have other infections.
We find that most of the patients with COVID‐19 had abnormal chest radiology (84%), in comparison to influenza type A (57%) and B (33%). GGO and consolidation in COVID‐19 patients being more frequent than in influenza type A and B patients. Radiologic findings and clinical symptoms such as sore throat indicated that the virus in COVID‐19 patients targets the lower respiratory system, while the upper respiratory system is more involved in influenza infections in comparison to COVID‐19.
Our analysis showed that the incubation period in COVID‐19 (6.4 days estimated from present literature to date) was more extended than influenza type A (3.4 days). Likewise, the duration of hospitalization in COVID‐19 patients (14 days) is longer than influenza type A (6.5 days) and influenza type B (6.7 days). These results suggest that the flu virus may show clinical signs earlier than the COVID‐19, and flu patients are discharged sooner than COVID‐19 patients from the hospital. Moreover, the hospital discharged rate of COVID‐19 (57.5%) is lower in comparison to influenza type A (82%) and influenza type B (87.5%) patients.
Our analysis showed that the mortality rates of COVID‐19, influenza types A and B are 6.5%, 6% and 3%, respectively. Based on WHO reports on 26 April 2020, out of 2804, 796 COVID‐19 confirmed cases and 193,710 cases died (6.9%) around the world, which is similar to our result. Among influenza type A, the mortality rates in subtypes H5N1 (42%) and H7N9 (30%) were higher than subtypes H1N1 (5.5%), H3N2 (1.7%) and non‐H1N1 (2%). The influenza mortality rate was associated with different age groups, in which a higher mortality rate is shown in people with ≥50‐year‐old ages (12%) in comparison to other age groups. These results indicated that older people are at risk of death from the flu. However, subtype H5N1 is fatal and life threatening for all age ranges.
Several limitations of this study exist. Publication bias and study heterogeneity are unavoidable in this type of study. Therefore, it should be considered when interpreting the outcomes of the reports and our final data‐set. Furthermore, this study likely overestimates disease severity due to a lack of screening of asymptomatic or mildly symptomatic individuals and subsequent publication bias related to these factors. Likely, many infected persons have not been detected, thus falsely elevating the rates of hospitalization and mortality compared to the milder symptomatic population. Whether this issue is the same for all viruses evaluated here is unknown. The lower quality analysis and reporting in some of the included publications is another limitation of the study. To prevent language bias, we included reports in languages other than English.
Additionally, we searched for a variety of sites and databases to prevent Internet platform bias. Using Egger's regression test, we did not find significant publication bias. Journal bias is an issue facing those who carry out a meta‐analysis, yet it does not usually affect the general conclusions. 33 However, we cannot reject the occurrence of other biases in this study, such as choice bias, since several journals are not indexed in Embase, Scopus, PubMed, Web of Science and the Cochrane library, and unpublished data from some regions of the world.
5. CONCLUSIONS
The results showed that despite the development of respiratory disease and similar transmission methods, COVID‐19 and influenza had many differences in terms of involvement and severity of the pulmonary injury, mortality rate, laboratory finding and clinical symptoms. Due to the high transmissibility and the lack of effective medication or vaccine for COVID‐19, timely detection of this viral infection and distinguishing from influenza are very important.
CONFLICT OF INTEREST
The authors have declared that no conflict of interests.
AUTHOR CONTRIBUTIONS
Conceived and designed the study: Ali Pormohammad, Juan‐Pablo Idrovo and Saied Ghorbani; comprehensive research: Saied Ghorbani, Alireza Khatami, Mohammad Hossein Razizadeh, Ehsan Alborzi and Ali Pormohammad; analysed the data: Ali Pormohammad; wrote and revised the paper: Ali Pormohammad, Saied Ghorbani, Juan‐Pablo Idrovo, Raymond J. Turner, Alireza Khatami, Mohammad Hossein Razizadeh, Ehsan Alborzi and Mohammad Zarei; participated in data analysis and manuscript editing: Ali Pormohammad, Saied Ghorbani, Juan‐Pablo Idrovo, Raymond J. Turner, Alireza Khatami, Mohammad Hossein Razizadeh, Ehsan Alborzi and Mohammad Zarei.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
Supporting information
Supporting Information 1
Supporting Information 2
Supporting Information 3
Supporting Information 4
Supporting Information 5
Supporting Information 6
Supporting Information 7
Supporting Information 8
Supporting Information 9
Contributor Information
Ali Pormohammad, Email: pormohammadali@yahoo.com.
Raymond J. Turner, Email: turnerr@ucalgary.ca.
REFERENCES
- 1. Kash JC, Taubenberger JK. The role of viral, host, and secondary bacterial factors in influenza pathogenesis. Am J Pathol. 2015;185(6):1528–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mertz D, Geraci J, Winkup J, Gessner BD, Ortiz JR, Loeb M. Pregnancy as a risk factor for severe outcomes from influenza virus infection: a systematic review and meta‐analysis of observational studies. Vaccine. 2017;35(4):521–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vaillant L, La Ruche G, Tarantola A, Barboza P. Epidemiology of fatal cases associated with pandemic H1N1 influenza 2009. Euro Surveill. 2009;14(33):19309. [DOI] [PubMed] [Google Scholar]
- 4. Ou Q, Lu Y, Huang Q, Cheng X. Clinical analysis of 150 cases with the novel influenza A (H1N1) virus infection in Shanghai, China. Biosci Trends. 2009;3(4):30–127. [PubMed] [Google Scholar]
- 5. Win MK, Chow A, Chen M, Lau YF, Ooi EE, Leo YS. Influenza B outbreak among influenza‐vaccinated welfare home residents in Singapore. Ann Acad Med Singap. 2010;39(6):448–452. [PubMed] [Google Scholar]
- 6. Hui DS, Rossi GA, Johnston SL. SARS, MERS and Other Viral Lung Infections. 2016. https://public.ebookcentral.proquest.com/choice/publicfullrecord.aspx?p=4625378. [Google Scholar]
- 7. Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID‐19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Eur Surveill. 2020;25(10):2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID‐19. J Am Med Assoc. 2020;323(14):1406–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Riou J, Althaus CL. Pattern of early human‐to‐human transmission of Wuhan 2019 novel coronavirus (2019‐nCoV), December 2019 to January 2020. Euro Surveill. 2020;25(4):2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu JT, Leung K, Bushman M, et al. Estimating clinical severity of COVID‐19 from the transmission dynamics in Wuhan, China. Nat Med. 2020;4(8):506–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sanche S, Lin Y, Xu C, Romero‐Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(7):7–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Coburn BJ, Wagner BG, Blower S. Modeling influenza epidemics and pandemics: insights into the future of swine flu (H1N1). BMC Med. 2009;7(1):30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neuzil KM, Zhu Y, Griffin MR, et al. Burden of interpandemic influenza in children younger than 5 years: a 25‐year prospective study. J Infect Dis. 2002;185(2):147–152. [DOI] [PubMed] [Google Scholar]
- 14. Louie JK, Acosta M, Jamieson DJ, Honein MA. Severe 2009 H1N1 influenza in pregnant and postpartum women in California. N Engl J Med. 2010;362(1):27–35. [DOI] [PubMed] [Google Scholar]
- 15. McElhaney JE, Beran J, Devaster J‐M, et al. AS03‐adjuvanted versus non‐adjuvanted inactivated trivalent influenza vaccine against seasonal influenza in elderly people: a phase 3 randomised trial. Lancet Infect Dis. 2013;13(6):485–496. [DOI] [PubMed] [Google Scholar]
- 16. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID‐19 in Wuhan, China: a retrospective cohort study. Lancet. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Contini A. Virtual screening of an FDA Approved drugs database on two COVID‐19 coronavirus proteins; 2020.
- 18. Zhavoronkov A, Aladinskiy V, Zhebrak A, et al. Potential COVID‐2019 3C‐like Protease inhibitors designed using generative deep learning approaches; 2020.
- 19. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. Ann Int Med. 2009;151(4):264–269. [DOI] [PubMed] [Google Scholar]
- 20. Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 21. Bai S, Wang J, Zhou Y, et al. Analysis of the first cluster of cases in a family of novel coronavirus pneumonia in Gansu Province. Zhonghua Yu Fang Yi Xue Za Zhi. 2020;54:E005. [DOI] [PubMed] [Google Scholar]
- 22. Zhang M, Wang X, Chen Y, et al. Clinical features of 2019 novel coronavirus pneumonia in the early stage from a fever clinic in Beijing. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43(3):215–218. [DOI] [PubMed] [Google Scholar]
- 23. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. J Am Med Assoc. 2020;323(11):1061–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guan W‐J, Ni Z‐Y, Hu Y, et al. Clinical characteristics of 2019 novel coronavirus infection in China. MedRxiv. 2020. [Google Scholar]
- 27. Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected by SARS‐CoV‐2 in Wuhan, China. Allergy. 2020;75(7):1730–1741. [DOI] [PubMed] [Google Scholar]
- 28. Cheng Y, Luo R, Wang K, et al. Kidney impairment is associated with in‐hospital death of COVID‐19 patients. medRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl J Med. 2020;382(13):1199–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu X‐W, Wu X‐X, Jiang X‐G, et al. Clinical findings in a group of patients infected with the 2019 novel coronavirus (SARS‐Cov‐2) outside of Wuhan, China: retrospective case series. Br Med J. 2020;368(606):102–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song F, Shi N, Shan F, et al. Emerging coronavirus 2019‐nCoV pneumonia. Radiology. 2020;295(1):210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hui D, Rossi G, Johnston S. Respiratory Syncytial Virus‐‐SARS, MERS and Other Viral Lung Infections. Lausanne, Switzerland: European Respiratory Society; 2016. [PubMed] [Google Scholar]
- 33. Sutton AJ, Duval S, Tweedie R, Abrams KR, Jones DR. Empirical assessment of effect of publication bias on meta‐analyses. Br Med J. 2000;320(7249):1574–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information 1
Supporting Information 2
Supporting Information 3
Supporting Information 4
Supporting Information 5
Supporting Information 6
Supporting Information 7
Supporting Information 8
Supporting Information 9
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


