Abstract
Long non-coding RNAs (lncRNAs) participate in the development and progression of prostate cancer (PCa). We aimd to identify a novel lncRNA, named lncRNA activated in metastatic PCa (lncAMPC), and investigate its mechanisms and clinical significance in PCa. First, the biological capacity of lncAMPC in PCa was demonstrated both in vitro and in vivo. The lncAMPC was overexpressed in tumor tissue and urine of metastatic PCa patients and promoted PCa tumorigenesis and metastasis. Then, a mechanism study was conducted to determine how the lncAMPC-activated pathway contributed to PCa metastasis and immunosuppression. In the cytoplasm, lncAMPC upregulated LIF expression by sponging miR-637 and inhibiting its activity. In the nucleus, lncAMPC enhanced LIFR transcription by decoying histone H1.2 away from the upstream sequence of the LIFR gene. The lncAMPC-activated LIF/LIFR expressions stimulated the Jak1-STAT3 pathway to simultaneously maintain programmed death-ligand 1 (PD-L1) protein stability and promote metastasis-associated gene expression. Finally, the prognostic value of the expression of lncAMPC and its downstream genes in PCa patients was evaluated. High LIF/LIFR levels indicated shorter biochemical recurrence-free survival among patients who underwent radical prostatectomy. Therefore, the lncAMPC/LIF/LIFR axis plays a critical role in PCa metastasis and immunosuppression and may serve as a prognostic biomarker and potential therapeutic target.
Keywords: prostate cancer, long non-coding RNA, competing endogenous RNA, histone H1.2, LIF/LIFR/Jak1/STAT3 pathway
Graphical Abstract
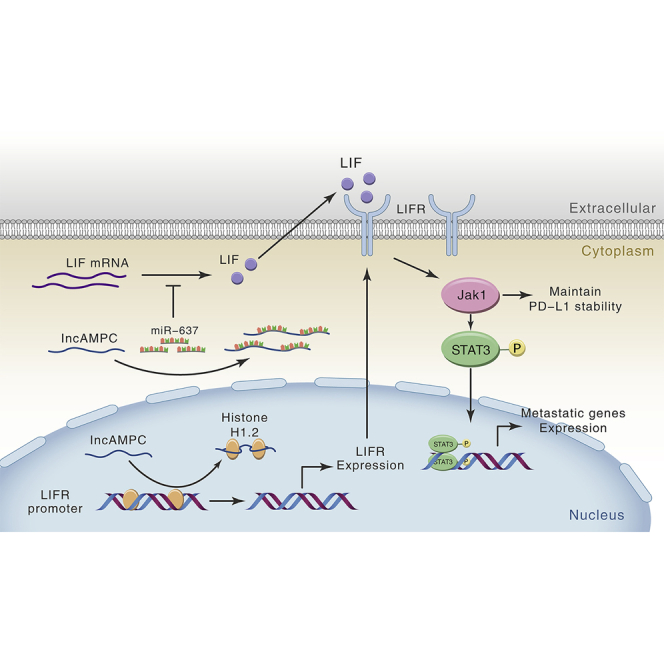
Ren and colleagues have identified lncAMPC, a novel lncRNA that is upregulated in metastatic PCa, and lncAMPC-activated LIF/LIFR expressions that are associated with poor prognosis. lncAMPC/LIF/LIFR plays a critical role in PCa metastasis and immunosuppression, as it could simultaneously maintain PD-L1 protein stability and promote metastatic genes expression in PCa.
Introduction
Prostate cancer (PCa) is the most common malignancy in men and accounts for 13% of all cancer-related deaths,1 with distant metastasis as the primary cause of death. The most frequent sites for metastatic PCa are the bone (84.0%) and regional lymph nodes (10.6%), while the incidence of visceral metastasis, including liver (10.2%), thorax (9.1%), and brain (3.1%) metastases, is relatively low.2 Overall, 18.4% of PCa patients have multiple metastatic sites. Distant metastasis, particularly visceral involvement, negatively impacts survival outcomes in PCa patients.3 Thus, prognostic biomarkers and comprehensive therapies are needed for patients with visceral metastasis. Metastasis is a multistep and complex process that includes the acquisition of invasive properties of metastatic cells from the primary tumor, dissemination to distant organs, and initiation of secondary lesions.4 Identifying molecules that play important roles in the tumorigenesis of PCa and understanding the molecular mechanisms underlying progression and metastasis are imperative for developing treatments for advanced PCa.
In PCa, long non-coding RNAs (lncRNAs) can promote cell proliferation, invasion, metastasis, and castration resistance.5, 6, 7 Large-scale RNA-sequencing (RNA-seq) technologies have drastically promoted the discovery of novel cancer-associated lncRNAs. Understanding the molecular mechanisms of these lncRNAs and how they regulate cancer could be crucial in determining their prognostic and therapeutic values. lncRNAs are involved in the biological process through diverse mechanisms, including recruiting chromatin-modifying complexes or transcriptional co-regulators at the transcription level8 and interacting with RNAs or proteins at the post-transcription level.9
In this study, we identified a novel lncRNA NR_046357.1, named lncRNA activated in metastatic PCa (lncAMPC), and investigated its contributions to metastasis and immunosuppression of PCa as well as its clinical significance for patients.
Results
lncAMPC Is a Novel lncRNA that Is Upregulated in Metastatic PCa
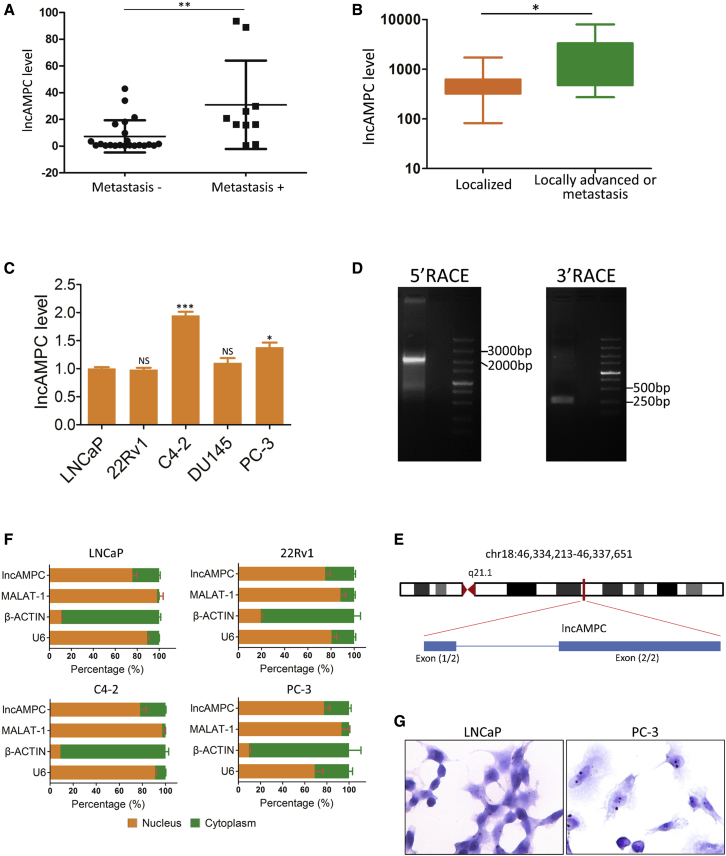
First, the quantitative real-time polymerase chain reaction (PCR) analysis of an independent set of PCa tissues validated that the lncAMPC expression in PCa tumor was significantly higher than that in its adjacent normal prostate tissue among 14/32 cases (Figure S1B). Interestingly, the lncAMPC was preferentially upregulated in metastatic PCa compared with localized PCa (Figure 1A). Moreover, another independent set of urine samples from PCa patients was analyzed. The expression of lncAMPC in locally advanced or metastatic PCa was consistently significantly higher than that in localized PCa (Figure 1B). Lastly, lncAMPC expression was detected in a panel of PCa cells (LNCaP, 22Rv1, C4-2, DU145, and PC-3), and we found that lncAMPC is highly expressed in aggressive PCa cells such as C4-2, DU145, and PC-3 (Figure 1C). The expression of lncAMPC was also evaluated in bladder cancer cell lines (J82 and T24) and renal carcinoma cell lines (786-O and ACHN). The lncAMPC levels in these non-PCa cell lines were significantly lower than those in PCa cell lines (Figure S1C). Located on chromosome 18 in humans, lncAMPC is composed of 2 exons with a poly(A) tail and has a full length of 1,887 nt, as revealed via a rapid amplification of cDNA ends assay (Figures 1D and 1E). Furthermore, the lncAMPC gene is moderately to highly conserved among placental mammals compared with the highly conserved coding genes (Figure S1D). A nuclear and cytoplasmic distribution assay (Figure 1F) and RNA in situ hybridization (Figure 1G) demonstrated that lncAMPC was expressed in both the nucleus and cytoplasm, with expression in the nucleus accounting for approximately 75% of its overall expression. These data demonstrate that lncAMPC is a highly expressed novel lncRNA in metastatic PCa.
Figure 1.
lncAMPC Is Upregulated in Metastatic PCa
(A) Quantitative real-time PCR analysis of lncAMPC in an independent set of primary tumor tissues from patients with localized (n = 22) or metastatic (n = 10) PCa. (B) Quantitative real-time PCR analysis of lncAMPC in an independent set of urine samples from patients with localized (n = 113), locally advanced (n = 22), or metastatic (n = 22) PCa. (C) Quantitative real-time PCR analysis of lncAMPC expression in a panel of PCa cell lines (n = 3). (D) DNA gel electrophoresis of the 5′ RACE and 3′ RACE products of lncAMPC. (E) Schematic annotation of lncAMPC genomic locus in humans. Blue rectangles represent exons. (F) Quantitative real-time PCR analysis of lncAMPC in the subcellular fractions of PCa cell lines (n = 3). U6 and β-actin acted as nuclear and cytoplasmic markers, respectively. (G) RNA in situ hybridization of lncAMPC in LNCaP cells. The green fluorescent signal is from the fluorescein isothiocyanate (FITC)-lncAMPC RNA probe, and the blue fluorescent signal is nuclear DNA counterstained with DAPI. Results are presented as mean ± SD; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
lncAMPC Promotes Proliferation, Viability, Migration, and Invasion Capacities of PCa Cells
We designed three small interfering RNAs (siRNAs) targeting lncAMPC and found that all of them could reduce lncAMPC expression at different levels (Figure S1E). Two siRNAs—namely, silncAMPC-1 and silncAMPC-2—were used in subsequent analyses. We also successfully overexpressed lncAMPC in PCa cells via transfection of pcDNA3.1-lncAMPC (Figure S1F).
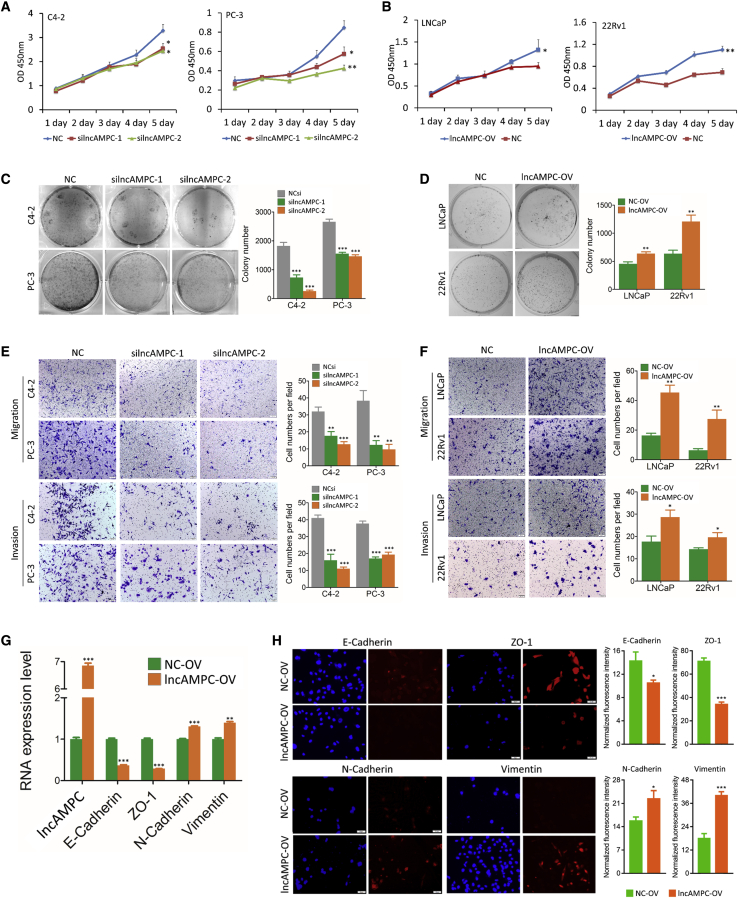
In the cell proliferation assay, suppression of lncAMPC by siRNAs reduced cell proliferation in C4-2 and PC-3 cells (Figure 2A), while proliferation was enhanced after lncAMPC overexpression in LNCaP and 22Rv1 cells (Figure 2B). In the colony assay, there were significantly fewer colonies in the silncAMPC groups than in the negative control siRNA (NCsi) group (Figure 2C). Meanwhile, there were significantly more colonies formed in the lncAMPC-overexpression (OV) group than in the overexpression control (NC-OV) group (Figure 2D). We also analyzed the effect of lncAMPC on cell migration and invasion. Compared with control groups, lncAMPC knockdown and overexpression significantly decreased (Figure 2E) and increased (Figure 2F) the proportion of migrated and invasive PCa cells, respectively.
Figure 2.
Effect of lncAMPC Knockdown or Overexpression on Oncogenic Activities In Vitro
(A) MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) assay testing the cell proliferation of C4-2 and PC-3 transfected with NCsi, silncAMPC-1, or silncAMPC-2 (n = 3). (B) MTS assay testing the cell proliferation of LNCaP and 22Rv1 transfected with lncAMPC-OV (lncAMPC overexpression) or NC-OV (overexpression control) (n = 3). (C) Colony assay testing the cell viability of C4-2 and PC-3 transfected with NCsi, silncAMPC-1, or silncAMPC-2 (n = 3). (D) Colony assay testing the cell viability of LNCaP and 22Rv1 transfected with lncAMPC-OV or NC-OV (n = 3). (E) Transwell assay testing the cell migration and invasion of C4-2 and PC-3 transfected with NCsi, silncAMPC-1, or silncAMPC-2 (n = 3). (F) Transwell assay testing the cell migration and invasion of LNCaP and 22Rv1 transfected with lncAMPC-OV or NC-OV (n = 3). (G) The mRNA expression levels of EMT markers in PC-3 transfected with lncAMPC-OV or NC-OV (n = 3). (H) Cell immunofluorescence analysis of the localization and expression of EMT markers in PC-3 transfected with lncAMPC-OV or NC-OV. Results are presented as means ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
We investigated whether lncAMPC influenced epithelial-to-mesenchymal transition (EMT) in PCa via quantitative real-time PCR and cell immunofluorescence. The quantitative real-time PCR analysis revealed that lncAMPC overexpression reduced the epithelial markers E-cadherin and ZO-1 and increased the mesenchymal markers N-cadherin and vimentin (Figure 2G). Cell immunofluorescence staining also revealed that lncAMPC overexpression induced a mesenchymal phenotype, downregulated E-cadherin and ZO-1, and upregulated N-cadherin and vimentin (Figure 2H). These results suggested that lncAMPC could promote PCa cell proliferation, invasion, and EMT.
lncAMPC Enhances Tumorigenesis and Metastasis of PCa In Vivo
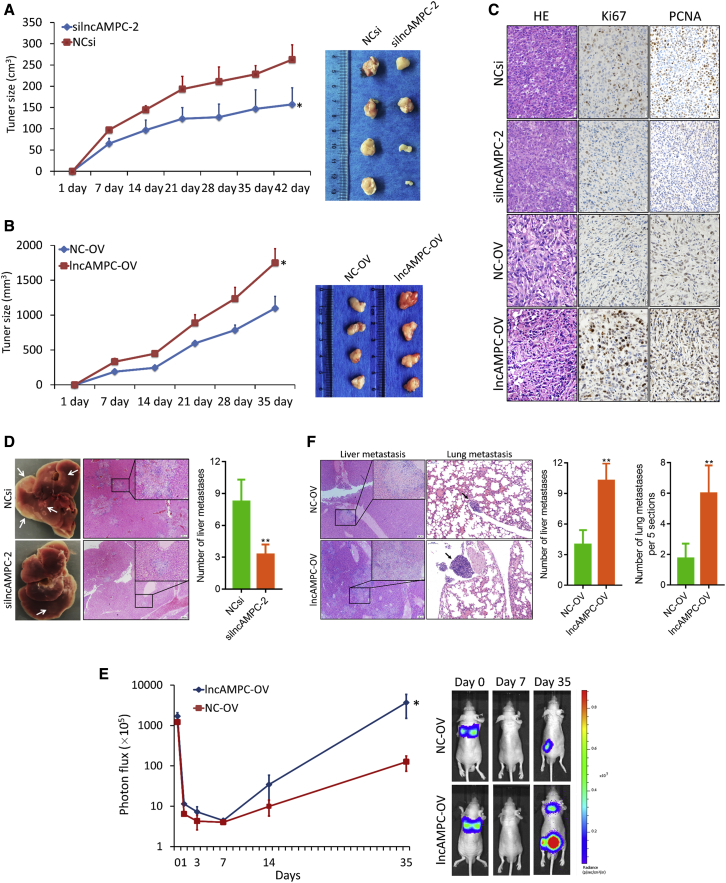
To determine whether lncAMPC mediates tumorigenesis in vivo, we conducted xenotransplantation experiments by subcutaneously injecting PC-3 cells with lncAMPC knockdown or overexpression. We found significantly lower growth curve and tumor volume in the silncAMPC group compared with the control group (Figure 3A). Meanwhile, the nude mice injected with cells overexpressing lncAMPC exhibited significantly increased tumor growth compared with the control group (Figure 3B). Furthermore, an immunohistochemistry (IHC) assay for Ki67 and proliferating cell nuclear antigen (PCNA) revealed that lncAMPC could enhance tumor cell proliferation (Figure 3C). Interestingly, there were fewer metastatic foci in the liver specimen harvested from the lncAMPC knockdown group than in the control group (Figure 3D).
Figure 3.
Effect of lncAMPC Knockdown or Overexpression on Xenograft Tumor Growth and Metastasis In Vivo
(A) The growth curve and harvested specimen of PC-3 subcutaneous xenograft tumors transfected with in vivo NCsi or silncAMPC-2 (n = 4). (B) The growth curve and harvested specimen of PC-3 subcutaneous xenograft tumors stably expressing lncAMPC-OV or NC-OV (n = 4). (C) The IHC analysis (Ki67 and PCNA) of tumors from subcutaneous xenograft models. (D) The liver specimen harvested from the subcutaneous xenograft models above and its H&E staining. (E) Luciferase signal intensities and representative images of mice over time after tail vein injection with PC-3 stably expressing lncAMPC-OV or NC-OV (n = 6). (F) The H&E staining of the liver and lung specimen harvested from the tail-vein-injection xenograft models above. Results are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
We further explored the role of lncAMPC in PCa metastasis in vivo by injecting PC-3 cells into the tail veins of nude mice. The PC-3 cells with stable overexpression of lncAMPC formed more metastatic foci in the mice than parental cells (Figure 3E). Hematoxylin and eosin (H&E) staining showed that the lncAMPC overexpression group had more large-sized metastatic tumor nests in the liver and lung than the control group (Figure 3F). These results suggested that lncAMPC could enhance tumorigenesis and metastasis of PCa in vivo.
lncAMPC Upregulates the Expression of LIF by Competitively Binding miR-637 and Then Inhibiting Its Activity in the Cytoplasm
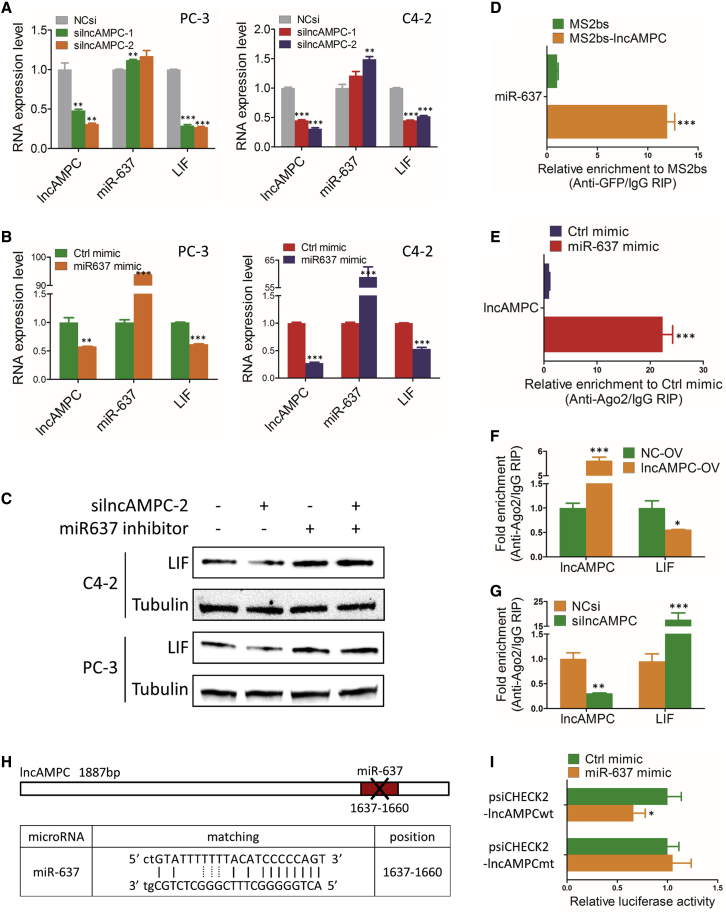
lncAMPC in the cytoplasm might act as a competing endogenous RNA (ceRNA) to competitively bind microRNAs (miRNAs) and cause the liberation of corresponding miRNA-targeted transcripts.10 First, we identified 6 putative lncAMPC-binding miRNAs using a miRDB prediction algorithm (Table S2). Evaluation of the expressions of these miRNAs and their corresponding targeted transcripts showed that the miR-637 level was higher in the lncAMPC knockdown group than in the control group. Importantly, lncAMPC knockdown significantly reduced the expression of miR-637-targeted LIF transcript (Figure 4A). As a member of cytokines, LIF was demonstrated as a direct target of miR-637, and its expression could be decreased by miR-637.11 Furthermore, we overexpressed miR-637 using miRNA mimics and confirmed that miR-637 mimics could simultaneously reduce lncAMPC and LIF transcript expressions (Figure 4B). This indicated that miR-637 bound to lncAMPC and induced the RNA degradation of lncAMPC. Western blot was further conducted to detect the protein level of LIF in PCa cells transfected with silncAMPC and/or miR-637 inhibitors. The protein level of LIF was downregulated by lncAMPC knockdown but upregulated by miR-637 inhibitors. Interestingly, the LIF level was also upregulated by co-transfection of silncAMPC and miR-637 inhibitors, indicating that silncAMPC-induced decrease in LIF could be rescued by miR-637 inhibition (Figure 4C).
Figure 4.
lncAMPC Upregulates the Expression of LIF by Competitively Binding and Then Inhibiting the Activity of miR-637
(A) Quantitative real-time PCR analysis of miR-637 and LIF expressions in C4-2 and PC-3 transfected with NCsi, silncAMPC-1, or silncAMPC-2 (n = 3). (B) Quantitative real-time PCR analysis of lncAMPC and LIF expressions in C4-2 and PC-3 transfected with miR-637 mimics or control mimics (n = 3). (C) Western blot analysis of LIF expression in C4-2 and PC-3 transfected with silncAMPC-2 and/or miR-637 inhibitor (n = 3). (D) MS2-based RIP assay with anti-GFP antibody followed by miRNA quantitative real-time PCR to detect the binding ability of lncAMPC to miR-637 (n = 3). (E) Anti-Ago2 RIP assay followed by quantitative real-time PCR to detect the binding ability of Ago2 to lncAMPC (n = 3). (F and G) RIP assay of the enrichment of Ago2 on lncAMPC and LIF transcript relative to immunoglobulin G (IgG) in PC-3 cells transfected with lncAMPC-OV or NC-OV (F), and with NCsi or silncAMPC-2 (G) (n = 3). (H) Schematic outline of predicted binding site for miR-637 on lncAMPC; the red nucleotides (binding site) were mutated in the mutant constructs. (I) Luciferase activity of psiCHECK2-lncAMPCwt and psiCHECK2-lncAMPCmt upon transfection of miR-637 mimics in 293T cells (n = 3). Data are presented as the ratio of Renilla luciferase activity to Firefly luciferase activity. Results are presented as mean ± SD; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To further confirm the direct binding between lncAMPC and miR-637 at endogenous levels, we performed an MS2-based RNA immunoprecipitation (RIP) assay with anti-GFP antibody to pull down endogenous miRNAs associated with lncAMPC. The following quantitative real-time PCR analysis demonstrated that miR-637 was significantly enriched in products retrieved from MS2bs-lncAMPC compared to that from control MS2bs (Figure 4D). It is known that miRNAs bind to their targets and exert their translational repression or RNA degradation functions through the RNA-induced silencing complex (RISC). To investigate whether lncAMPC was regulated by miR-637 in such a manner, we performed a RIP assay with anti-Ago2 antibody, the core component of RISC, in PC-3 cells overexpressing miR-637. The following quantitative real-time PCR analysis showed that endogenous lncAMPC pull-down by Ago2 was significantly enriched in the miR-637 mimic group, compared with that in the control group (Figure 4E), confirming that miR-637 was the target of lncAMPC. Using an anti-Ago2 RIP assay in PC-3 cells with lncAMPC overexpression or knockdown, we then further evaluated whether lncAMPC would affect the enrichment of LIF transcript pull-down by Ago2. The succeeding quantitative real-time PCR analysis showed that lncAMPC overexpression simultaneously increased enrichment of Ago2 on lncAMPC and decreased enrichment on the LIF transcript (Figure 4F). Meanwhile, lncAMPC knockdown significantly increased the enrichment of Ago2 on the LIF transcript compared with the control group (Figure 4G). These results suggested that lncAMPC could compete with the LIF transcript for the Ago2-based RISC in PCa cells.
To further validate the binding efficiency of miR-637 to lncAMPC, we constructed luciferase reporters containing 1,887 nt of lncAMPC with wild-type (lncAMPCwt) or with mutated (lncAMPCmt) miR-637 binding sites (Figure 4H). The dual-luciferase reporter assay showed that transfection of miR-637 mimics significantly inhibited luciferase activity of wild-type reporter vector psiCHECK2-lncAMPCwt, whereas mutant reporter vector psiCHECK2-lncAMPCmt showed no response to miR-637 mimics (Figure 4I). These data suggested that lncAMPC contained a functional miR-637 binding site.
lncAMPC-Induced Migration and Invasion in PCa Cells Are Partially Rescued by miR-637 Inhibition
We designed a rescue experiment to evaluate the role of miR-637 in cell migration and invasion. The results showed that miR-637 inhibitors only partially rescued the silncAMPC-mediated reduction of migration or invasion effects (Figure S2A). Furthermore, transfection of lncAMPC-OV and miR-637 mimics simultaneously also partially rescued the enhanced migration or invasion effects via lncAMPC overexpression (Figure S2B). Collectively, these data suggest that, aside from miR-637 in cytoplasm, lncAMPC might interact with certain nuclear components to exert its functional effects in PCa.
lncAMPC Enhances the Transcription of LIFR by Decoying Histone H1.2 Away from the Upstream Sequence of LIFR Gene in the Nucleus
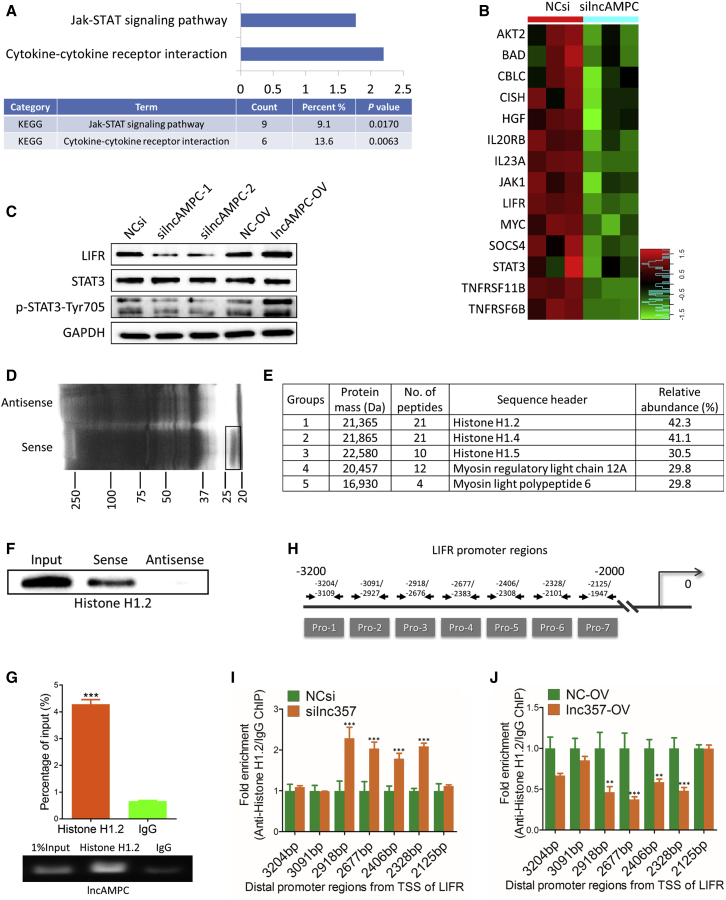
To identify the potential nuclear component that interacted with lncAMPC, we utilized a human gene-expression array to analyze global gene-expression change upon lncAMPC knockdown in PC-3 cells. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of changed genes revealed that lncAMPC knockdown markedly inhibited the activities of the Jak-STAT signaling and cytokine-cytokine receptor interaction pathways (Figure 5A). LIFR, an interleukin (IL)-6 receptor family member, is known as the major upstream receptor of the Jak1-STAT3 cascade.12 Based on the heatmap, 9.1%–13.6% of genes involved in the Jak-STAT signaling and cytokine-cytokine receptor interaction pathways, including LIFR, Jak1, and STAT3, were transcriptionally modulated by lncAMPC (Figure 5B). We further verified the microarray data via western blot and found that LIFR and p-STAT3 were suppressed by lncAMPC knockdown and activated by lncAMPC overexpression (Figure 5C).
Figure 5.
lncAMPC Enhances the Transcription of LIFR by Decoying Histone H1.2 Away from the Upstream Sequence of LIFR Gene
(A) KEGG pathway analysis of genes affected by lncAMPC knockdown in PC-3 from microarray data. (B) Heatmap showing the expression data of genes involved in cytokine-cytokine receptor interaction and Jak-STAT signaling pathway. (C) Western blot analysis of the indicated proteins in PC-3 transfected with NCsi, silncAMPC-1, silncAMPC-2, NC-OV, or lncAMPC-OV (n = 3). (D) RNA pull-down precipitates of lncAMPC-associated proteins were separated via SDS-PAGE and visualized using silver staining (n = 3). The black frame indicates the section of the gel cut out for mass spectrometry analysis. (E) Mass spectrometry analysis of the proteins specifically pulled down by lncAMPC. (F) Western blot analysis of histone H1.2 in the RNA pull-down precipitates retrieved with biotin-labeled lncAMPC or antisense RNA from the lysates of PC-3 cells (n = 3). (G) RIP assay of the enrichment of histone H1.2 with lncAMPC relative to IgG in the lysates of PC-3 cells (n = 3). (H) The 7 pairs of primers designed to cover the 3,200 bp–2,000 bp distal promoter regions from the transcription start site of LIFR. (I and J) Anti-histone H1.2 ChIP assay followed by quantitative real-time PCR to detect the binding ability of histone H1.2 to LIFR distal promoter regions in PC-3 cells transfected with NCsi or silncAMPC-2 (I) and NC-OV or lncAMPC-OV (J) (n = 3). Results are presented as mean ± SD; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
To elucidate the mechanisms underlying the upregulation of LIFR by lncAMPC, we performed an RNA pull-down assay to identify lncAMPC-interacting proteins (Figure 5D). The band specifically pulled down by lncAMPC was subjected to mass spectrometry analysis, and histone H1.2 was identified to potentially bind to lncAMPC (Figure 5E). In addition, western blot confirmed the presence of histone H1.2 in the RNA pull-down precipitates retrieved with biotin-labeled lncAMPC but not with antisense RNA (Figure 5F). A RIP assay was further performed to verify the specific interaction between lncAMPC and histone H1.2 (Figure 5G). These results confirmed that lncAMPC directly bound to histone H1.2 in the nucleus of PCa cells.
As the most specific variant of linker histone H1, histone H1.2 is more abundant at repressed promoters and has the strongest correlation with low gene expression.13 Therefore, we hypothesized that lncAMPC might regulate LIFR transcription by binding and decoying histone H1.2 away from the promoter of the LIFR gene. To confirm this hypothesis, we performed a chromatin immunoprecipitation (ChIP) assay with the anti-histone H1.2 antibody and assessed the level of histone H1.2-bound LIFR promoter DNA. In total, 7 pairs of PCR primers were designed to amplify approximately 100- to 400-bp segments spanning the 3,200–2,000 bp upstream of the LIFR transcriptional start site (Figure 5H). ChIP-PCR assays in PC-3 cells with lncAMPC knockdown or overexpression confirmed that the enrichment of histone H1.2 in the distal promoter region of LIFR was significantly increased in lncAMPC-knockdown cells (Figure 5I), whereas it was decreased in lncAMPC-overexpressed cells (Figure 5J). These data confirmed our hypothesis that lncAMPC enhanced LIFR transcriptional activation by decoying histone H1.2 away from the upstream sequence of LIFR gene in PCa cell nucleus.
lncAMPC Activates the Jak1-STAT3 Signaling Pathway by Stimulating LIF/LIFR Expression
We further explored whether the LIF/LIFR-activated Jak1-STAT3 pathway was essential for lncAMPC oncogenic activity in PCa. The status of the LIF/LIFR/Jak1/STAT3 pathway was evaluated in subcutaneous xenograft tumors using IHC analysis (Figure S3A). Expressions of LIF, LIFR, and p-STAT3 were reduced in lncAMPC-knockdown tumors and increased in lncAMPC-overexpressed tumors, supporting the hypothesis that lncAMPC promoted PCa by activating the LIF/LIFR/Jak1/STAT3 pathway.
Next, we performed a rescue experiment to determine whether LIF/LIFR activation was functionally required for lncAMPC-induced invasion (Figure S3B). The results indicated that the number of invasive PCa cells increased due to lncAMPC overexpression but decreased by LIF or LIFR knockdown. Simultaneous inhibition of both LIF and LIFR using siRNAs could overcome the lncAMPC overexpression-mediated enhancement of invasion. These data confirmed that the LIF/LIFR-activated signaling pathway was essential for lncAMPC oncogenic activity in PCa.
PD-L1-Mediated Immunosuppression Partially Contributes to the Oncogenic Activity of lncAMPC-Activated LIF
In addition to changes in the expression of LIF, LIFR, and p-STAT3 in PC-3 subcutaneous xenograft models, IHC also showed that PD-L1 expression was positively correlated with the lncAMPC-activated LIF level (Figure S3A), which was further confirmed by The Cancer Genome Atlas (TCGA) public human PCa dataset (Figure S3C). In addition, the TCGA dataset indicated a positive correlation between RNF165 gene transcripts (lncAMPC is one of the transcripts produced from RNF165 gene) and PD-L1 expression (Figure S3C).
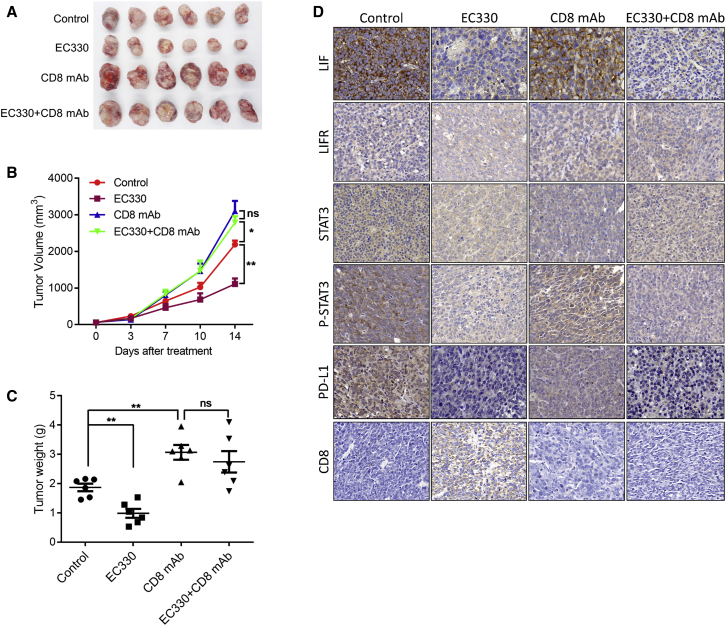
EC330 alone or combined with anti-CD8 monoclonal antibody (mAb) was applied in vivo to treat mouse RM-1 prostate tumors (Figures 6A–6C). Compared to the control group, the growth curve and tumor volume in the EC330-alone group were significantly decreased. However, the antitumor efficacy of EC330 was inhibited by anti-CD8 mAb. Furthermore, an IHC assay detecting the status of the LIF/LIFR/Jak1/STAT3 pathway, PD-L1, and CD8 revealed that the inhibition of LIF weakened the PD-L1-mediated immunosuppression in PCa (Figure 6D). These results indicated that PD-L1-mediated immunosuppression partially contributed to the oncogenic activity of lncAMPC-activated LIF in PCa.
Figure 6.
Inhibition of lncAMPC-Activated LIF Weakens the PD-L1-Mediated Immunosuppression in PCa
(A–C) The harvested specimen (A), growth curve (B), and tumor weight (C) of mouse RM-1 prostate tumors treated with EC330 alone or in combination with anti-CD8 mAb (n = 6). (D) IHC analysis evaluating the status of the LIF/LIFR/Jak1/STAT3 signaling pathway, PD-L1, and CD8 in the mouse RM-1 prostate tumors (n = 3). Results are presented as mean ± SD; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
LIF/LIFR Expression Is a Prognostic Factor for Outcomes of Radical Prostatectomy
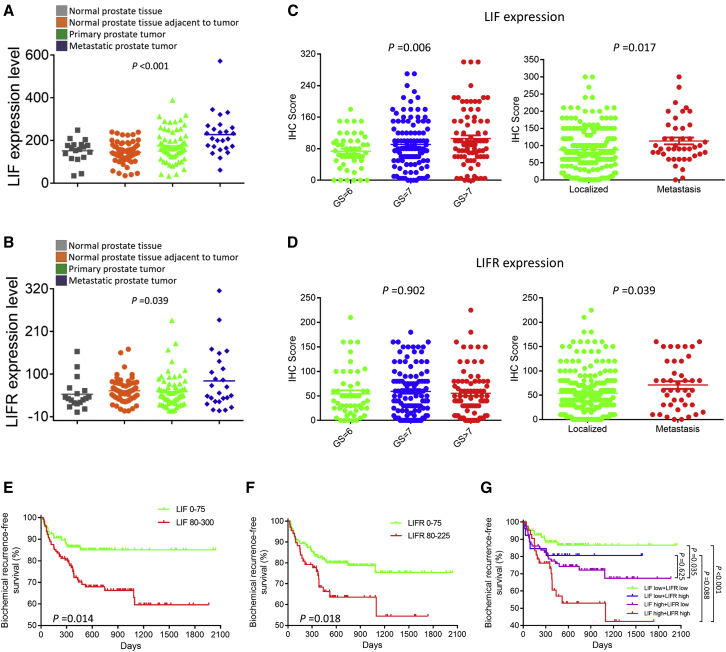
The public human PCa dataset was used to evaluate the expression levels of LIF and LIFR in tumors of different stages (Figures 7A and 7B). Among the 171 PCa patients from the Gene Expression Omnibus (GEO): GSE6919 dataset, LIF and LIFR expressions were the highest in metastatic PCa, followed by those in primary PCa. Meanwhile, LIF and LIFR expressions were the lowest in cases without PCa or normal prostate tissue adjacent to tumor.
Figure 7.
LIF/LIFR Expression as a Prognostic Factor for Outcomes of Radical Prostatectomy
(A and B) The public human PCa dataset evaluating the expression levels of LIF (A) and LIFR (B) (n = 171). (C and D) IHC scores of LIF (C) and LIFR (D) expression in PCa patients having radical prostatectomy (n = 237). (E–G) Kaplan-Meier analyses of biochemical recurrence-free survival for PCa patients having radical prostatectomy using staining status of LIF (E), LIFR (F), and both of them (G) (n = 237). Results are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
The correlations between LIF/LIFR expressions and clinicopathological parameters, including Gleason score and metastatic status, were analyzed in our tissue microarray (TMA) cohort of 237 PCa patients who underwent radical prostatectomy (Figures 7C and 7D). LIF expression was the highest in patients with a Gleason score >7, followed by that in patients with Gleason scores of 7 and 6 (p < 0.01). Similarly, higher average expression of LIFR was observed in patients with higher Gleason scores, but the difference was not significant (p = 0.902). Compared to those in localized PCa subjects, LIF and LIFR levels were significantly elevated in tumor tissue samples from metastatic PCa (all p < 0.05). Kaplan-Meier analysis revealed that patients with higher LIF or LIFR expression had a lower biochemical recurrence-free survival rate (all p < 0.05) (Figures 7E and 7F). Notably, the subgroup of patients with both high LIF and LIFR expressions had the shortest biochemical recurrence-free survival (all p < 0.01) (Figure 7G). These data suggested that lncAMPC-activated LIF/LIFR expressions could serve as prognostic biomarkers for patients undergoing radical prostatectomy.
Discussion
There is currently no curative treatment modality for metastatic PCa, and metastatic PCa patients have a 5-year survival rate of only approximately 35%. Hence, understanding the mechanisms involved in PCa metastasis and identifying therapeutic targets and predictive biomarkers are crucial for improving patient prognosis.
The landscape of lncRNAs in tumors often changes during disease progression, and the expression of some lncRNAs may exhibit constant upregulation or downregulation.14 In our study, we found that lncAMPC was significantly upregulated in urine samples from locally advanced or metastatic PCa patients compared with localized PCa patients. In particular, urine biomarkers—including DNA, RNA, and protein—are, by far, the most developed biomarkers, because they are non-invasive.15 Several new urine lncRNA biomarkers, such as PCA3, MALAT-1, and FR0348383, have been developed in the past decade to detect PCa and monitor progression.16, 17, 18 Our data indicate that lncAMPC had potential value as a prognostic biomarker for patients undergoing radical prostatectomy.
As an uncharacterized lncRNA, lncAMPC was identified to be highly expressed in metastatic PCa patients, indicating that lncAMPC might be involved in distant metastasis. Many lncRNAs have been demonstrated to participate in the dysregulation of gene expression in PCa, which then contributes to cancer initiation, development, and progression.19 Gain- and loss-of-function experiments indicated that lncAMPC enhanced the biological capacities of cell growth and invasion in PCa. Furthermore, lncAMPC regulated EMT by inducing a mesenchymal phenotype. As an early event of tumor cell metastatic dissemination, EMT played a critical role in tumor metastasis by providing cells with a more motile, invasive potential.20 The aforementioned results were verified via in vivo experiments where lncAMPC enhanced both tumorigenesis and metastasis of PCa.
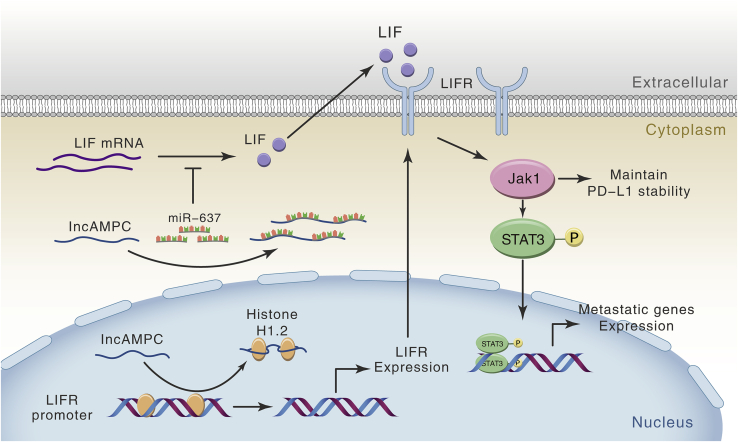
At a molecular level, lncRNAs have been reported to participate in several processes, ranging from chromatin remodeling and transcriptional regulation to protein translational control.21 Chromatin modification, epigenetic regulation, and alternative splicing by SCHLAP1, HOTAIR, and MALAT1 represent characterized examples of lncRNA-mediated control of cell invasiveness and metastasis.22, 23, 24 The subcellular localization of lncRNA in cells usually determines its possible specialized functions. Specifically, we found that lncAMPC was expressed in both the nucleus and the cytoplasm of PCa cells. This feature of discrete distribution in different cellular compartments is extensively shared by other lncRNAs,25,26 indicating the possibility of lncRNA shuttling between the nucleus and cytoplasm. Kino et al.27 showed that lncRNA Gas5 was localized both in the cytoplasm and the nucleus of HeLa cells, with its presence more predominant in the former compartment. They further demonstrated that Gas5 interacted with the ligand-activated glucocorticoid receptor in the cytoplasm and co-migrated with this glucocorticoid receptor into the nucleus. Miyagawa et al.28 found that lncRNA MALAT1 was actively retained in the nucleus through its nuclear retention elements, and altered MALAT1 was passively exported to the cytoplasm in the absence of these factors. How the lncAMPC shuttles between the cytoplasm and nucleus (using either active or passive localization mechanisms) is still unclear and remains to be answered. The discrete distribution of lncAMPC in PCa indicated its possible roles in diverse biological processes. The regulatory mechanisms of lncAMPC in the metastasis and immunosuppression of PCa are summarized in Figure 8. Yuan et al.29 demonstrated that lncRNA-activated by TGF-β (ATB) promoted the invasion-metastasis cascade of hepatocellular carcinoma cells via two distinct RNA-RNA interactions. Qu et al.30,31 also found that lncARSR not only acted as a ceRNA for miR-34 and miR-449 but also regulated Yes-associated protein (YAP) activity by blocking its phosphorylation in renal cancer.
Figure 8.
Schematic Diagram of lncAMPC-Based Regulatory Mechanism in PCa
miRNAs are usually regarded as negative regulators of gene expression by decreasing the stability of target RNAs or limiting their translation.32 Zhang et al.11 reported that miR-637, a primate-specific miRNA, negatively regulated STAT3 phosphorylation by suppressing autocrine LIF expression and exogenous LIF-triggered STAT3 in hepatocellular carcinoma. Accordingly, we hypothesized that lncAMPC upregulated the expression of LIF by competitively binding and then inhibiting the activity of miR-637 in PCa cell cytoplasm. This is consistent with the theory of ceRNAs proposed by Salmena et al.33 that cytoplasmic lncRNAs act as the “RNA sponges,” interacting with miRNAs and protecting target mRNAs from miRNA-mediated suppression.
Several recent studies have showed that many lncRNAs are involved in molecular regulatory networks through their interactions with proteins.8 As a key structural component of chromatin, histones have been reported to participate in the regulation of various gene expressions by lncRNAs in cancers.34 As archetypes of molecular functions, lncRNAs have recently been shown to execute, guide, decoy, and scaffold.35 If the lncRNA titrates transcription factors or other components away from chromatin, as is the case for lncAMPC, then the lncRNA develops into a molecular decoy.36 Based on our data, we hypothesize that lncAMPC enhances the transcription of LIFR by decoying histone H1.2 away from the upstream sequence of the LIFR gene in the PCa cell nucleus. As the key structural component of chromatin, histone H1 is not uniformly distributed along the genome. Notably, the abundance of histone H1.2 at the 3,200- to 2,000-bp distal promoter regions from the transcription start site is inversely proportional to gene expression.13 This provides a possible correlation between the abundance of histone H1.2 and the silencing of specific genes. Furthermore, additional studies suggested that histone H1 was not a global repressor of transcription but rather played a more dynamic and gene-targeted role, contributing to gene-specific transcriptional regulation.37 Hence, we hypothesize that histone H1.2 plays a critical role in the transcriptional regulation of LIFR by lncAMPC. However, the definite interaction mechanisms and binding sites for lncAMPC and histone H1.2 warrant further investigation.
LIF, a member of the IL-6 family cytokines, can bind to a heterodimer of the specific LIFR to activate downstream signaling through the Jak/STAT, mitogen-activated protein kinase (MAPK), and phosphatidylinositol 3-kinase (PI3K) signaling pathways.38 We found that the LIF/LIFR/Jak1/STAT3 signaling pathway mediated the metastasis and immunosuppression process activated by lncAMPC in PCa. The oncogenic activity of LIF in different solid tumors has been described; however, its role in tumor-induced immunosuppression remains largely unexplored. Notably, in our study, CD8+ T cells were recruited to the tumor tissue upon LIF blockade by EC330 or silncAMPC. Similar findings were observed in glioblastoma and ovarian cancer models.39 We demonstrated for the first time that lncAMPC-activated LIF positively regulated PD-L1 expression in PCa. Recent studies showed that activated Jak1/2 could upregulate PD-L1 expression.40 Chan et al.41 showed that IL-6-activated JAK1 maintained PD-L1 protein stability, possibly by phosphorylating PD-L1 (Tyr112) and enhancing its association with N-glycosyltransferase STT3A. Collectively, these data indicated that blocking lncAMPC-activated LIF signaling might eliminate PD-L1/PD-1-driven cancer immune escape, suggesting that the combination of targeted therapies and immune checkpoint inhibitors might be effective against PCa.
Our results have not only potential therapeutic implications by targeting lncAMPC-activated LIF in PCa but also prognostic implications by using the IHC scores of LIF/LIFR as prognostic biomarkers in patients undergoing radical prostatectomy. We found that higher expressions of LIF/LIFR were associated with higher Gleason scores and metastatic PCa. Similarly, Liu et al.42 reported that LIF levels were the highest in both the primary tumor and serum in metastatic PCa patients, followed by localized PCa patients and healthy donors. Furthermore, survival analysis indicated that PCa patients with higher LIF/LIFR expressions had shorter biochemical recurrence-free survival. Similar results were reported in non-small-cell lung cancer, breast cancer, colorectal cancer, and nasopharyngeal cancer patients.43, 44, 45
In summary, our results indicate that lncAMPC expression is significantly upregulated in metastatic PCa and is associated with poorer prognosis. Mechanistically, lncAMPC upregulates LIF expression by competitively binding and then inhibiting the activity of miR-637 in the cytoplasm and enhances LIFR transcription by decoying histone H1.2 away from the upstream sequence of LIFR gene in the nucleus. The stimulated LIF/LIFR expression activates the Jak1-STAT3 signaling pathway to simultaneously maintain PD-L1 protein stability and promote metastasis-associated gene expression. Therefore, the lncAMPC/LIF/LIFR axis may be a biomarker for unfavorable prognosis in PCa patients and serve as a therapeutic target.
Materials and Methods
Identification of lncAMPC
We performed RNA-seq of 65 pairs of PCa and matched adjacent normal tissues obtained via radical prostatectomy (Table S1) and identified 439 deregulated lncRNA transcripts (fold change ≥ 1.8).46 We stratified these patients by Gleason score and pathological stage and found that lncAMPC was associated with Gleason score and pathological tumor node metastasis (TNM) stage (Figure S1A).
Patients and Clinical Specimens
All tissue and urine samples were obtained from Changhai Hospital, Shanghai, China. In total, 32 primary PCa tissues from patients undergoing radical prostatectomy and 157 urine samples from patients with positive prostate biopsy were collected for evaluating the expression of lncAMPC. A TMA cohort of 237 PCa patients who underwent radical prostatectomy was recruited for evaluating the expressions of LIF and LIFR. All patients provided written informed consent, and this study was approved by the Clinical Research Ethics Committee of Changhai Hospital.
Xenograft Model
All experiments involving mice were approved by the Institutional Animal Care and Use Committee of Changhai Hospital. PC-3 cells stably transfected with LV-lncAMPC or LV-control for lncAMPC were used for in vivo experiments. In the subcutaneous model, the stably transfected PC-3 cells (3 × 106) were mixed with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) at a ratio of 1:1 and subcutaneously injected into 6-week-old male nude mice. Tumors were evaluated weekly for 5 weeks, and the harvested tumor weight was also measured. For the metastasis model, 2 × 106 single cells were injected into the tail vein of nude mice. Metastases were monitored using the IVIS Lumina II System (Caliper Life Sciences, Waltham, MA, USA) 10 min after intraperitoneal injection of 4.0 mg luciferin (Promega, Durham, NC, USA) diluted in 50 μL saline. Formalin-fixed, paraffin-embedded sections from the liver and lung with metastatic lesions were subjected to H&E staining.
For the subcutaneous model of lncAMPC knockdown, the in vivo-jetPEI DNA and siRNA Delivery Kit (Polyplus-Transfection, New York, NY, USA) was used to knock down lncAMPC in vivo according to the manufacturer’s instructions. After confirming the existence of the subcutaneous tumors, 20 μg siRNA and 2.4 μL in vivo-jetPEI reagent were dissolved in 12.5 μL 10% glucose solution, mixed, and diluted into 50 μL. After incubation at room temperature for 15 min, the complexes were injected into the tumors 3 times per week.
For the subcutaneous model of mouse RM-1 prostate cells, the RM-1 cells (2 × 106) were injected into the male C57BL/6 mice. After confirming the existence of subcutaneous tumors, mice were treated with oral EC330 (2.5 mg/kg, 7 times per week) alone or in combination with intraperitoneally injected anti-CD8 mAb (200 μg, 2 times per week). Tumors were evaluated every 3 days for 2 weeks, and the harvested tumor weight was also measured.
Statistical Analysis
All statistical analyses were performed using SPSS v.17.0 software (Abbott Laboratories, Chicago, IL, USA). Data are presented as the mean ± SD. Statistical differences between the indicated experimental groups were compared using a two-tailed Student’s t test or one-way ANOVA with Dunnett’s multiple comparisons test as appropriate. A p < 0.05 was considered statistically significant.
Detailed Experimental Materials and Methods
Detailed descriptions of the experimental procedures and statistical analyses are provided in the Supplemental Materials and Methods.
Author Contributions
Conceptualization and Methodology, S.R., Y.S., W.Z., X.S., and R.C.; Investigation, W.Z., X.S., R.C., and S.R.; Formal Analysis and Data Curation, W.Z., X.S., R.C., Y.Z., S.P., Y.C., and X.N.; Technical and Material Support, G.X., Z.F., Y.L., Z.C., L.Z., and G.L.; Writing – Original Draft, W.Z. and X.S.; Writing – Review and Editing, R.C., Y.S., and S.R.; Supervision, Y.S.; Funding Acquisition, S.R., Y.S., W.Z., and X.S.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81802581 to W.Z. and 81872105 to S.R.), Shanghai Sailing Program (18YF1422600 to W.Z. and 19YF1447300 to X.S.), National Key R&D Program of China (2017YFC0908002 to S.R.), and Shanghai Clinical Medical Center of Urological Diseases Program (2017ZZ01005 to Y.S.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.06.013.
Contributor Information
Yinghao Sun, Email: sunyhsmmu@126.com.
Shancheng Ren, Email: renshancheng@gmail.com.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2016. CA Cancer J. Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Gandaglia G., Abdollah F., Schiffmann J., Trudeau V., Shariat S.F., Kim S.P., Perrotte P., Montorsi F., Briganti A., Trinh Q.D. Distribution of metastatic sites in patients with prostate cancer: A population-based analysis. Prostate. 2014;74:210–216. doi: 10.1002/pros.22742. [DOI] [PubMed] [Google Scholar]
- 3.Gandaglia G., Karakiewicz P.I., Briganti A., Passoni N.M., Schiffmann J., Trudeau V., Graefen M., Montorsi F., Sun M. Impact of the Site of Metastases on Survival in Patients with Metastatic Prostate Cancer. Eur. Urol. 2015;68:325–334. doi: 10.1016/j.eururo.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 4.Sethi N., Kang Y. Unravelling the complexity of metastasis - molecular understanding and targeted therapies. Nat. Rev. Cancer. 2011;11:735–748. doi: 10.1038/nrc3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramnarine V.R., Kobelev M., Gibb E.A., Nouri M., Lin D., Wang Y., Buttyan R., Davicioni E., Zoubeidi A., Collins C.C. The evolution of long noncoding RNA acceptance in prostate cancer initiation, progression, and its clinical utility in disease management. Eur. Urol. 2019;76:546–559. doi: 10.1016/j.eururo.2019.07.040. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y., Pitchiaya S., Cieślik M., Niknafs Y.S., Tien J.C., Hosono Y., Iyer M.K., Yazdani S., Subramaniam S., Shukla S.K. Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat. Genet. 2018;50:814–824. doi: 10.1038/s41588-018-0120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao G., Yao J., Kong D., Ye C., Chen R., Li L., Zeng T., Wang L., Zhang W., Shi X. The Long Noncoding RNA TTTY15, Which Is Located on the Y Chromosome, Promotes Prostate Cancer Progression by Sponging let-7. Eur. Urol. 2019;76:315–326. doi: 10.1016/j.eururo.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Tsai M.C., Manor O., Wan Y., Mosammaparast N., Wang J.K., Lan F., Shi Y., Segal E., Chang H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 10.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J.F., He M.L., Fu W.M., Wang H., Chen L.Z., Zhu X., Chen Y., Xie D., Lai P., Chen G. Primate-specific microRNA-637 inhibits tumorigenesis in hepatocellular carcinoma by disrupting signal transducer and activator of transcription 3 signaling. Hepatology. 2011;54:2137–2148. doi: 10.1002/hep.24595. [DOI] [PubMed] [Google Scholar]
- 12.Heinrich P.C., Behrmann I., Haan S., Hermanns H.M., Müller-Newen G., Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem. J. 2003;374:1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Millán-Ariño L., Islam A.B., Izquierdo-Bouldstridge A., Mayor R., Terme J.M., Luque N., Sancho M., López-Bigas N., Jordan A. Mapping of six somatic linker histone H1 variants in human breast cancer cells uncovers specific features of H1.2. Nucleic Acids Res. 2014;42:4474–4493. doi: 10.1093/nar/gku079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhamija S., Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer. 2016;139:269–280. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 15.Roobol M.J., Haese A., Bjartell A. Tumour markers in prostate cancer III: biomarkers in urine. Acta Oncol. 2011;50(Suppl. 1):85–89. doi: 10.3109/0284186X.2010.524935. [DOI] [PubMed] [Google Scholar]
- 16.de Kok J.B., Verhaegh G.W., Roelofs R.W., Hessels D., Kiemeney L.A., Aalders T.W., Swinkels D.W., Schalken J.A. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- 17.Wang F., Ren S., Chen R., Lu J., Shi X., Zhu Y., Zhang W., Jing T., Zhang C., Shen J. Development and prospective multicenter evaluation of the long noncoding RNA MALAT-1 as a diagnostic urinary biomarker for prostate cancer. Oncotarget. 2014;5:11091–11102. doi: 10.18632/oncotarget.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang W., Ren S.C., Shi X.L., Liu Y.W., Zhu Y.S., Jing T.L., Wang F.B., Chen R., Xu C.L., Wang H.Q. A novel urinary long non-coding RNA transcript improves diagnostic accuracy in patients undergoing prostate biopsy. Prostate. 2015;75:653–661. doi: 10.1002/pros.22949. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 22.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat. Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinn J.L., Kertesz M., Wang J.K., Squazzo S.L., Xu X., Brugmann S.A., Goodnough L.H., Helms J.A., Farnham P.J., Segal E., Chang H.Y. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao H., Wang L., Zhan H., Dai J., Chang Y., Wu F., Liu T., Liu Z., Gao C., Li L., Song X. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 2019;15:e1008144. doi: 10.1371/journal.pgen.1008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015;16:20. doi: 10.1186/s13059-015-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kino T., Hurt D.E., Ichijo T., Nader N., Chrousos G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyagawa R., Tano K., Mizuno R., Nakamura Y., Ijiri K., Rakwal R., Shibato J., Masuo Y., Mayeda A., Hirose T., Akimitsu N. Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA. 2012;18:738–751. doi: 10.1261/rna.028639.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan J.H., Yang F., Wang F., Ma J.Z., Guo Y.J., Tao Q.F., Liu F., Pan W., Wang T.T., Zhou C.C. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 30.Qu L., Ding J., Chen C., Wu Z.J., Liu B., Gao Y., Chen W., Liu F., Sun W., Li X.F. Exosome-Transmitted lncARSR Promotes Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Qu L., Wu Z., Li Y., Xu Z., Liu B., Liu F., Bao Y., Wu D., Liu J., Wang A. A feed-forward loop between lncARSR and YAP activity promotes expansion of renal tumour-initiating cells. Nat. Commun. 2016;7:12692. doi: 10.1038/ncomms12692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 33.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang Y., Wang J., Lian Y., Fan C., Zhang P., Wu Y., Li X., Xiong F., Li X., Li G. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol. Cancer. 2017;16:42. doi: 10.1186/s12943-017-0612-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ulitsky I., Bartel D.P. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan Y., Nikitina T., Zhao J., Fleury T.J., Bhattacharyya R., Bouhassira E.E., Stein A., Woodcock C.L., Skoultchi A.I. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 38.Nicola N.A., Babon J.J. Leukemia inhibitory factor (LIF) Cytokine Growth Factor Rev. 2015;26:533–544. doi: 10.1016/j.cytogfr.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pascual-García M., Bonfill-Teixidor E., Planas-Rigol E., Rubio-Perez C., Iurlaro R., Arias A., Cuartas I., Sala-Hojman A., Escudero L., Martínez-Ricarte F. LIF regulates CXCL9 in tumor-associated macrophages and prevents CD8+ T cell tumor-infiltration impairing anti-PD1 therapy. Nat. Commun. 2019;10:2416. doi: 10.1038/s41467-019-10369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prestipino A., Emhardt A.J., Aumann K., O’Sullivan D., Gorantla S.P., Duquesne S., Melchinger W., Braun L., Vuckovic S., Boerries M. Oncogenic JAK2V617F causes PD-L1 expression, mediating immune escape in myeloproliferative neoplasms. Sci. Transl. Med. 2018;10:10. doi: 10.1126/scitranslmed.aam7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan L.C., Li C.W., Xia W., Hsu J.M., Lee H.H., Cha J.H., Wang H.L., Yang W.H., Yen E.Y., Chang W.C. IL-6/JAK1 pathway drives PD-L1 Y112 phosphorylation to promote cancer immune evasion. J. Clin. Invest. 2019;129:3324–3338. doi: 10.1172/JCI126022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y.N., Niu S., Chen W.Y., Zhang Q., Tao Y., Chen W.H., Jiang K.C., Chen X., Shi H., Liu A. Leukemia Inhibitory Factor Promotes Castration-resistant Prostate Cancer and Neuroendocrine Differentiation by Activated ZBTB46. Clin. Cancer Res. 2019;25:4128–4140. doi: 10.1158/1078-0432.CCR-18-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng H., Qu J., Jin N., Xu J., Lin C., Chen Y., Yang X., He X., Tang S., Lan X. Feedback Activation of Leukemia Inhibitory Factor Receptor Limits Response to Histone Deacetylase Inhibitors in Breast Cancer. Cancer Cell. 2016;30:459–473. doi: 10.1016/j.ccell.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Yu H., Yue X., Zhao Y., Li X., Wu L., Zhang C., Liu Z., Lin K., Xu-Monette Z.Y., Young K.H. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nat. Commun. 2014;5:5218. doi: 10.1038/ncomms6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu S.C., Tsang N.M., Chiang W.C., Chang K.P., Hsueh C., Liang Y., Juang J.L., Chow K.P., Chang Y.S. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J. Clin. Invest. 2013;123:5269–5283. doi: 10.1172/JCI63428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren S., Wei G.-H., Liu D., Wang L., Hou Y., Zhu S., Peng L., Zhang Q., Cheng Y., Su H. Whole-genome and Transcriptome Sequencing of Prostate Cancer Identify New Genetic Alterations Driving Disease Progression. Eur. Urol. 2018;73:322–339. doi: 10.1016/j.eururo.2017.08.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.