Abstract
Dendritic cells (DCs) can orchestrate either immunogenic or tolerogenic responses to relay information on the functional state. Emerging studies indicate that circular RNAs (circRNAs) are involved in immunity; however, it remains unclear whether they govern DC development and function at the transcriptional level. In this study, we identified a central role for a novel circRNA, circSnx5, in modulating DC-driven immunity and tolerance. Ectopic circSnx5 suppresses DC activation and promotes the development of tolerogenic functions of DCs, while circSnx5 knockdown promotes their activation and inflammatory phenotype. Mechanistically, circSnx5 can act as a miR-544 sponge to attenuate miRNA-mediated target depression on suppressor of cytokine signaling 1 (SOCS1) and inhibit nuclear translocation of PU.1, regulating DC activation and function. Furthermore, the main splicing factors (SFs) were identified in DCs, of which heterogeneous nuclear ribonucleoprotein (hnRNP) C was essential for circSnx5 generation. Moreover, our data demonstrated that vaccination with circSnx5-conditioned DCs prolonged cardiac allograft survival in mice and alleviated experimental autoimmune myocarditis. Taken together, our results revealed circSnx5 as a key modulator to fine-tune DC function, suggesting that circSnx5 may serve as a potential therapeutic avenue for immune-related diseases.
Keywords: dendritic cell, circular RNA, immunity, circSnx5
Graphical Abstract
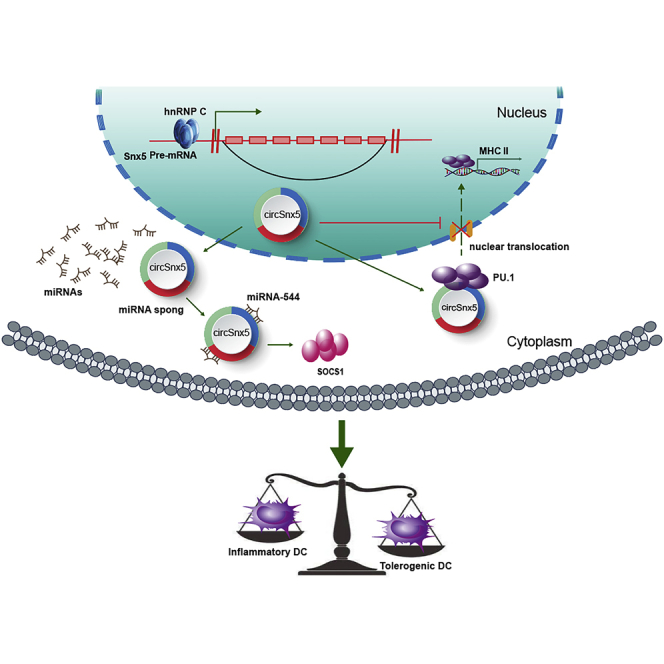
It remains unclear whether circRNAs govern DC development and function at the transcriptional level. Zhang and colleagues identified a novel circRNA, circSnx5, in modulating DC-driven immunity and tolerance by regulating the miR-544/SOCS1 axis and PU.1 nuclear translocation, suggesting that circSnx5 may serve as a potential therapeutic avenue for immune-related diseases.
Introduction
Dendritic cells (DCs) are potent antigen-presenting cells that play an essential role in immunity.1,2 The differentiation stage of DCs contributes to their various functions, promoting either an immunogenic or tolerogenic state.3, 4, 5, 6 Therefore, studying the orchestration of DC activation and function will improve our understanding of the immune response. Numerous studies have shown that the diverse functional fate of DCs is associated with various immune disorders.7,8 Moreover, a more comprehensive understanding of DCs as a cell therapy tool has provided a new perspective to treat immune-related diseases.9, 10, 11, 12 Also, note that DCs have been proven as therapeutic targets in autoimmune disease and transplantation.13, 14, 15 Therefore, identification and application of a novel regulator of DC functional properties in clinical settings for therapeutic purposes in immune disorders, such as autoimmune diseases and transplantation, have gained wide attention.
Circular RNAs (circRNAs), a novel subclass of non-coding RNAs (ncRNAs), originate from pre-mRNAs and are generated by a process called backsplicing.16, 17, 18, 19, 20 Their characteristics, including high abundance, stability, and evolutionary conservation, demonstrate diverse functions, such as harboring microRNA (miRNA) binding sites to serve as a sponge,21, 22, 23 regulating gene expression by acting as protein decoys,24 forming protein complexes, and encoding functional peptides.25, 26, 27, 28 In addition, circRNAs have exhibited tissue-specific expression and expression in pathological conditions, which are relevant to their effects in human diseases. Numerous studies suggest that circRNAs play important roles in the immune system,20,29 indicating that differential expression of circRNAs under pathological conditions is tightly linked to immune-related diseases. Interestingly, circMalat1 was found to be dysregulated in different functional DCs.30 However, study on this circRNA has only occurred at the expression level; therefore, an in-depth analysis about the function of circMalat1 or other circRNAs in DCs is needed. In short, the expression profiles and functions of newly identified circRNAs related to DC viability and functions still require further investigation.
In this study, we identified a central role for a novel circRNA, circSnx5, in governing DC-driven immunity and tolerance. In in vitro gain-of-function experiments, we demonstrated that circSnx5 could induce tolerogenic DCs and suppress the DC-based T cell response. In contrast, loss-of-function experiments suggest that circSnx5 knockdown promoted the inflammatory phenotype of DCs and active T cells. With regard to a mechanism, we demonstrated that circSnx5 could both act as a miR-544 sponge to attenuate miRNA-mediated target depression on the suppressor of cytokine signaling 1 (SOCS1) and inhibit nuclear translocation of PU.1 to regulate DC activation and function. Furthermore, we found that the splicing factor (SF) heterogeneous nuclear ribonucleoprotein (hnRNP) C was essential for circSnx5 generation. In in vivo studies, transfusion with circSnx5-overexpressing DCs mitigated the inflammatory response in an experimental autoimmune model of myocarditis and prolonged cardiac allograft survival in a heart transplantation mouse model.
Collectively, our findings show that circSnx5 serves as a critical rheostat in DCs to favor the development of tolerogenic functions and to maintain immune homeostasis, which provides a promising immunotherapeutic approach for interventions in immune-related diseases.
Results
Characterization of circSnx5 in DCs
According to our previous microarray analyses, differentially expressed circRNAs were identified in the rejecting cardiac allografts (Figures S1A and S1B). Since DCs are closely involved in allograft immunity, the top nine differentially expressed circRNAs were screened from bone marrow (BM)-derived DCs. qRT-PCR analyses showed that these circRNAs were dysregulated in lipopolysaccharide (LPS)-stimulated DCs (LPS-DCs) (Figure S1C). Furthermore, total RNAs from DCs were treated with RNase R (Figure S1C). Additionally, qRT-PCR analyses showed that all nine circRNAs were enriched in LPS-DCs (>0.1% of β-actin levels; Figure S1D). Among these, circRNA-015445 derived from the snx5 gene was selected for further study. According to circBase database annotation, circSnx5 was spliced from snx5 gene on chr2:144080461–144084914 and had a final-form length of 651 nt (Figure S2A). The circular expression level was quantified by qRT-PCR, followed by Sanger sequencing, and the result was consistent with the circBase data (Figure S2B). Divergent primers amplified circSnx5 in complementary DNA (cDNA) but not genomic DNA (gDNA), indicating that this RNA species was circular in form (Figure S2C). Furthermore, in RNase R digestion experiments, the circular nature of circSnx5 was confirmed by showing resistance to RNase R digestion (Figures S2D and S2E). LPS is known to induce the maturation of DCs, and it transforms DCs into an immunogenic state. To determine whether circSnx5 is functionally associated with DC functional status, circSnx5 expression was examined in DCs stimulated with or without LPS (1 μg/mL) by PCR analysis. circSnx5 expression decreased after a slight increase in LPS-DCs. There was no obvious change in its linear counterparts, snx5 gene expression, in DCs (Figure S2F). Fluorescence in situ hybridization (FISH) analysis further confirmed circSnx5 expression in DCs (Figure S2G), and subcellular localization determination via qRT-PCR demonstrated circRNA expression in the nucleus but mainly in the cytoplasm of DCs (Figure S2H). Collectively, we identified a DC-enriched circRNA (circSnx5) and showed that its expression was potentially associated with DC functional status.
circSnx5 Expression Induces a DC Tolerogenic Phenotype
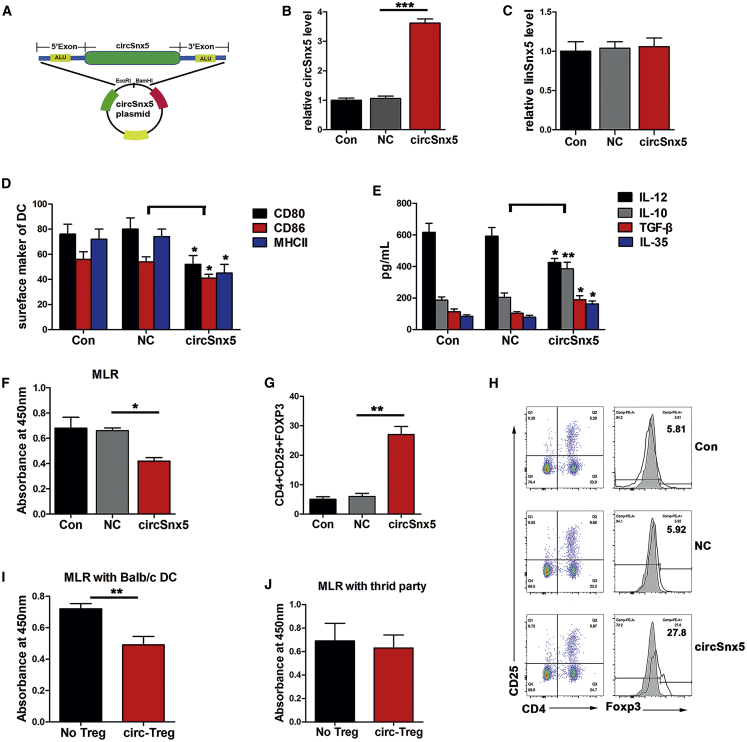
To evaluate the possible role of circSnx5 in DCs, a circular transcript expression plasmid was constructed (Figure 1A). The PCR assay showed that the circSnx5 plasmid significantly elevated circSnx5 levels, but not its linear counterpart (snx5 gene), in DCs (Figures 1B and 1C). Subsequently, the effects of circSnx5 on the phenotype and function of DCs were examined. After circSnx5 vector transduction into LPS-DCs for 6 h, upregulation of circSnx5 significantly inhibited the expression of co-stimulatory molecules CD80, CD86, and major histocompatibility complex class II (MHC class II) in LPS-DCs (Figure 1D). Furthermore, circSnx5 overexpression led to an increase in anti-inflammatory cytokines interleukin (IL)-10, IL-35, and transforming growth factor β (TGF-β) and a decrease in inflammatory cytokine IL-12 (Figure 1E), indicating that circSnx5 overexpression can block DC maturation. To further confirm the function of circSnx5 in DC-based T cell responses, an allogeneic mixed lymphocyte reaction (MLR) was performed in a CD4+ T cells-DCs coculture assay. Specifically, circSnx5 upregulation increased regulatory T cell (Treg) production from 5.81% to 27.8% (Figures 1G and 1H) and suppressed DC-induced T cell proliferation (Figure 1F). Moreover, DCs with circSnx5 induced more Treg expansion and induced Tregs showing antigen-specific immunosuppressive effects, which depressed T cell proliferation in the presence of BALB/c DCs as a stimulator but not DCs from third-party mice (KM mice) (Figures 1I and 1J). Taken together, these results established that circSnx5 suppressed DC activation and DC-based T cell responses.
Figure 1.
Expressing circSnx5 Induced the Tolerogenic Phenotype of DCs
BM-derived DCs were cultured from BALB/c mice with the presence of LPS for 12 h and then respectively transfected with circSnx5 overexpressing plasmid or negative control (NC) for 6 h. (A) Schematic illustrating the major components of circSnx5 vector constructs. (B and C) The expressions of circSnx5 (B) and Snx5 mRNA (C) were determined with qRT-PCR in DCs transfected with circSnx5 vector or NC (n = 6 independent DC preparations). (D) DC maturation was detected by measuring the expression of MHC class II, CD80, and CD86 using flow cytometry (n = 4 independent DC preparations). (E) The supernatants were collected to assay the production of IL-12, IL-10, TGF-β, and IL-35 by ELISA (n = 4 independent DC preparations). (F–H) DCs were treated with mitomycin C (10 mg/mL, 2 h) and cocultured with allogeneic T cells from splenocytes of naive BALB/c mice for 48 h at a DC/T cell ratio of 1:20. (F) DC-triggered T cell proliferation was evaluated by BrdU-ELISA. (G and H) In the cocultured T cells, the numbers of Tregs (CD4+, CD25+, and Foxp3+) were assessed by flow cytometry (H) and the percentage of Tregs is shown in (G). n = 6 independent DC preparations. (I and J) Tregs (CD4+CD25+) isolated from T cells cocultured with circSnx5-DCs by magnetic-activated cell sorting (MACS) were added into coculture with DCs from BALB/c (I) or third-party mice (J) and T cells, with a Treg/T cell/DC ratio of 10:10:1. Then, the suppressive ability of the Tregs was assessed by T cell proliferation assays using BrdU-ELISA. NoTreg, without Treg; circ-Treg, Tregs induced by circSnx5-DCs (n = 5 independent DC preparations). Assays for each DC preparations were performed in triplicate. Con, DCs treated with 200 ng/mL LPS for 12 h; NC, negative control; circSnx5, LPS-DCs transfected with circSnx5 vector. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, by Student’s t test.
circSnx5 Knockdown Promotes a DC Inflammatory Phenotype
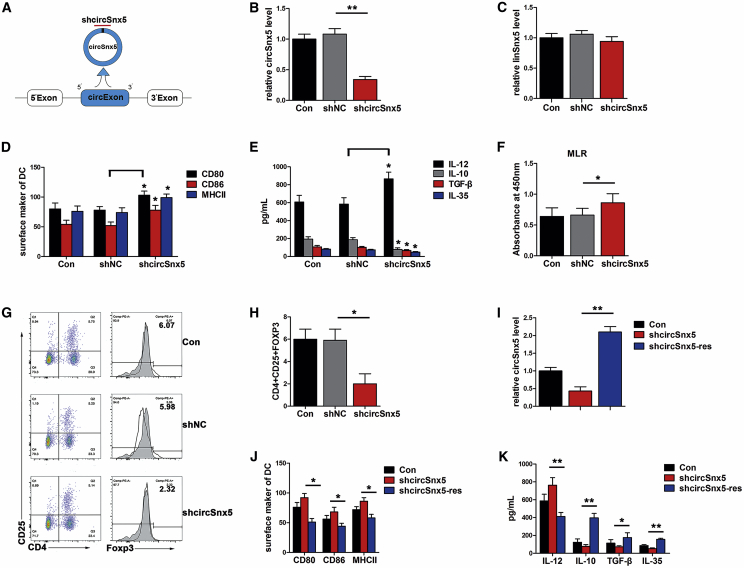
To further determine the function of circSnx5 in DCs, circSnx5 expression in DCs was disrupted by overexpressing small hairpin RNAs (shRNAs) that specifically target the circular junction of circSnx5 (Figure 2A). The qRT-PCR analysis indicated that circSnx5 shRNA (shcircSnx5) transfection significantly downregulated endogenous circSnx5 levels, but not its linear counterparts (Figures 2B and 2C). circSnx5 downregulation enhanced the expression of co-stimulators CD80, CD86, and MHC class II in LPS-DCs (Figure 2D). Furthermore, circSnx5 knockdown led to a decrease in anti-inflammatory cytokines IL-10, IL-35, and TGF-β and an increase in inflammatory cytokine IL-12 (Figure 2E), and it promoted T cell priming activity and reduced the number of Tregs (Figures 2F–2H), indicating that the circSnx5 knockdown promoted the inflammatory phenotype of DCs.
Figure 2.
circSnx5 Knockdown Promoted the Inflammatory Phenotype of DCs
DCs were transfected with a circSnx5 shRNA (shcircSnx5) or shRNA negative control (shNC) for 6 h before LPS treatment. (A) Schematic model of the shRNAs. shcircSnx5 targets the back-splice junction of circSnx5. (B and C) qRT-PCR for (B) circSnx5 and (C) Snx5 mRNA in DCs treated with shRNA as described above (n = 6 independent DC preparations). (D) DC subset treated with shRNA as described above was stained with the directly conjugated antibodies indicated and analyzed by flow cytometry (n = 4 independent DC preparations). (E) The supernatant was then harvested and analyzed for IL-12, IL-10, TGF-β, and IL-35 by ELISA (n = 4 independent DC preparations). (F–H) DCs were transfected with shcircSnx5 or NC for 6 h and then treated with 200 ng/mL LPS (12 h) and 10 μg/mL mitomycin C (2 h). (F) T cell proliferation was assessed by BrdU ELISA. (G and H) For Treg analysis, the mixed cells were stained with anti-CD4, anti-CD25, and then Foxp3 staining buffer and analyzed by flow cytometry (G) and the percentage of Tregs is shown in (H). n = 6 independent DC preparations. (I) qRT-PCR analysis for circSnx5 between shcircSnx5-transduced DCs and cells rescued by overexpression of circSnx5 (shcircSnx5-res). n = 6 independent DC preparations. (J) Co-stimulators (CD80, CD86, and MHC class II) were detected by flow cytometry in shcircSnx5-transduced or shcircSnx5-res DCs (n = 4 independent DC preparations). (K) Cytokine levels (IL-12, IL-10, TGF-β, and IL-35) were analyzed by ELISA in shcircSnx5-transduced or shcircSnx5-res DC supernatants (n = 4 independent DC preparations). Con, DCs treated with 200 ng/mL LPS for 12 h; NC, negative control; shcircSnx5, LPS-DCs were transfected with shcircSnx5. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, by Student’s t test.
To eliminate the possibility of off-target effects by the shcircSnx5 constructs, circSnx5 was re-expressed in DCs using shcircSnx5. After confirming the rescue of circSnx5 expression in DCs by qRT-PCR (Figure 2I), flow cytometry and an enzyme-linked immunosorbent assay (ELISA) were performed to measure expression levels of co-stimulators, that is, anti-inflammatory cytokines and inflammatory cytokines, which were both rescued (Figures 2J and 2K). Taken together, these results indicate that circSnx5 may serve as a critical regulator of the immunogenic and tolerogenic properties of DCs.
circSnx5 Modulates the DC Functional State through the miR-544/SOCS1 Axis
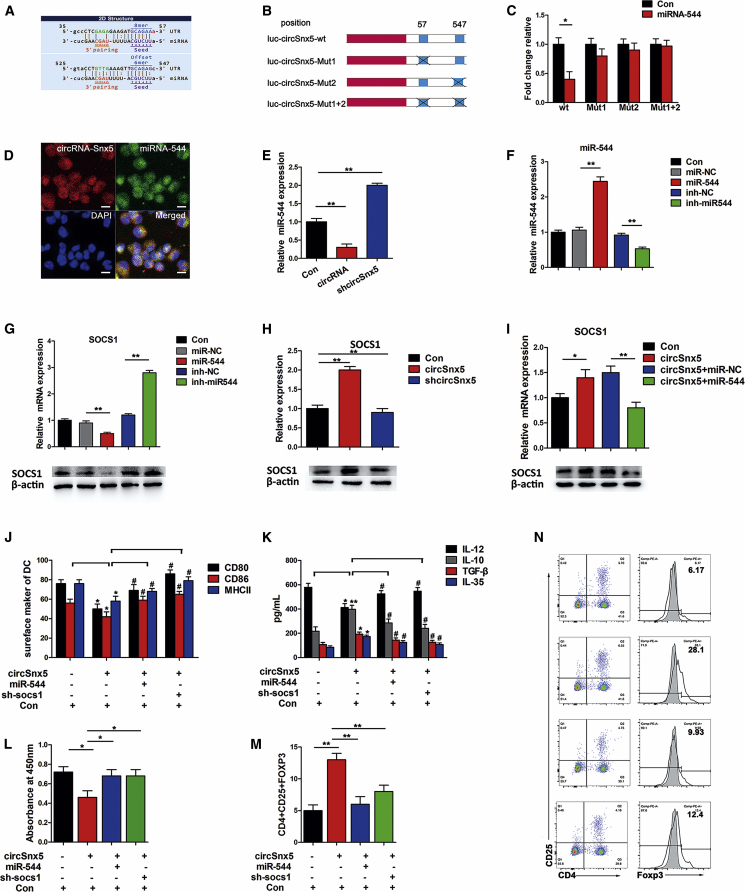
Given that circRNAs have been reported to function as sponges for miRNAs, we next investigated the ability of circSnx5 to bind to miRNAs. We used our previous comparative microarray analyses of circRNAs to build circSnx5-associated competing endogenous RNA (ceRNA) networks in which the selected miRNAs contain binding sites with circSnx5 (Figure S3A; Table S2). We identified six miRNAs having more than one targeting site for circSnx5. From these six miRNAs, we eliminated the miRNAs whose ID number was greater than 5,000, and thus three candidate miRNAs (miR-544, miR-22-5p, and miR-693-3p) were selected for the further study (Table S2). qRT-PCR analyses revealed that miR-544, miR-22-5p, and miR-693-3p expression levels were all increased after LPS stimulation for 12 h (Figure S3B). Additionally, the expression levels of these three miRNAs were decreased in DCs with high circSnx5 expression and increased after downregulation of circSnx5 (Figures S3C and S3D). The change in miR-544 expression was the most significant. These results suggest that circSnx5 may serve as a sponge for miR-544. Considering that SOCS1 is a pivotal regulator in DC function,31 the miR-544/SOCS1 axis was selected from the ceRNA networks. Bioinformatics analysis found two binding sites between miR-544 and circSnx5 (Figure 3A). A luciferase assay was performed by inserting either a wild-type (WT) circSnx5 sequence or mutated sequence at each binding site of miR-544 (Mut1, Mut2, or Mut1+2) into the luciferase construct (Figure 3B). Compared to control RNA, miR-544 reduced the luciferase reporter activity (Figure 3C), suggesting that miR-544 could interact with circSnx5 via a complementary seed region. In addition, the double confocal micrograph indicated the co-localization of circSnx5 and miR544 in DCs (Figure 3D). Subsequently, target miR-544 was downregulated upon circSnx5 overexpression in DCs (Figure 3E), suggesting that circSnx5 may function as a miR-544 sponge in DCs. To ascertain the relationship between miR-544 and SOCS1, we observed about 3-fold enrichment of SOCS1 with miR-544 downregulation compared with the negative control, whereas SOCS1 expression was reduced with miR-544 upregulation (Figure 3G) and in cells treated with shcircSnx5 (Figure 3H). More importantly, compared with the control cells, SOCS1 expression was inhibited by the circSnx5 plasmid, which was reversed by miR-544 mimics in DCs (Figure 3I). These data suggest that circSnx5 could function as a ceRNA for miR-544 to regulate SOCS1 expression.
Figure 3.
circSnx5 Modulated the Functional State of DCs through miR-544/SOCS1 Axis
(A) Binding sites of miR-544 in circSnx5. (B) Schematic of circSnx5 wild-type (WT) and mutant with deletions of miR-544 binding sites (Mut1, Mut2, or Mut1+2) luciferase reporter vectors. (C) Luciferase reporter assay for the luciferase activity of WT, Mut1, Mut2, or Mut1+2 in DCs co-transfected with miR-544 mimics. (D) Co-localization between miR-544 and circSnx5 was observed by RNA in situ hybridization in DCs after co-transfection with circSnx5 and miR-544 mimics. Nuclei were stained with DAPI. Images were taken by confocal microscopy. Scale bars, 50 μm. (E) qRT-PCR analysis of miR-544 level in DCs after transfection with circSnx5 vector shcircSnx5 or control RNA (Con). (F) LPS-DCs were transfected with miR-544 mimics, miR-negative control (miR-NC), miR-544 inhibitor (inh-miR544), or inhibitor-NC (inh-NC). miR-544 levels in DCs were analyzed by qRT-PCR. (G) Protein levels of SOCS1 in DCs transfected with miR-544 and miR-544 inhibitor using western blot assay. (H) Protein levels of SOCS1 in LPS-DCs transfected with circSnx5 vector or mock vector, respectively, using western blot assay. (I) Protein levels of SOCS1 using western blot assay in DCs transfected with shcircSnx5 or miR-544 as indicated. (J–N) DCs stimulated with LPS for 12 h co-transfected with circSnx5 vector, miR-544 mimics, or SOCS1 shRNA (sh-SOCS1). (J) Co-stimulators (CD80, CD86, and MHC class II) were detected by flow cytometry. (K) IL-10, IL-12, TGF-β, and IL-35 levels in supernatants were quantified by ELISA. (L) Allogeneic T cells were cocultured with these DCs and incubated with BrdU (10 mM, 24 h) to quantify T cell proliferation by BrdU-ELISA. (M and N) Flow cytometry assessment of the number of Tregs (CD4+, CD25+, and Foxp3+) in these cocultures, shown as percentages (N) and the percentage of Tregs is shown in (M). n = 6 independent DC preparations. Con, DCs treated with 200 ng/mL LPS for 12 h; NC, negative control; circSnx5, LPS-DCs transfected with circSnx5 vector; shcircSnx5, LPS-DCs transfected with shcircSnx5. Data are shown as mean ± SD. #p < 0.05 versus circSnx5 group; ∗p < 0.05, ∗∗p < 0.01, by Student’s t test.
As expected, circSnx5 could induce the tolerogenic phenotype of DCs with low co-stimulatory molecules and high anti-inflammatory cytokines, and this induction could be blocked by miR-544 overexpression or SOCS1 knockdown (Figures 3J and 3K). Furthermore, the MLR showed that circSnx5 induced T cell hyporesponsiveness and Treg expansion, which was also blocked by miR-544 overexpression or SOCS1 knockdown (Figures 3L–3N). Collectively, these findings demonstrate that circSnx5 inhibited DC maturation and accelerated Treg formation, at least partly through the miR-544/SOCS1 axis.
circSnx5 Represses PU.1 Nuclear Translocation and Downstream MHC Class II Expression in DCs
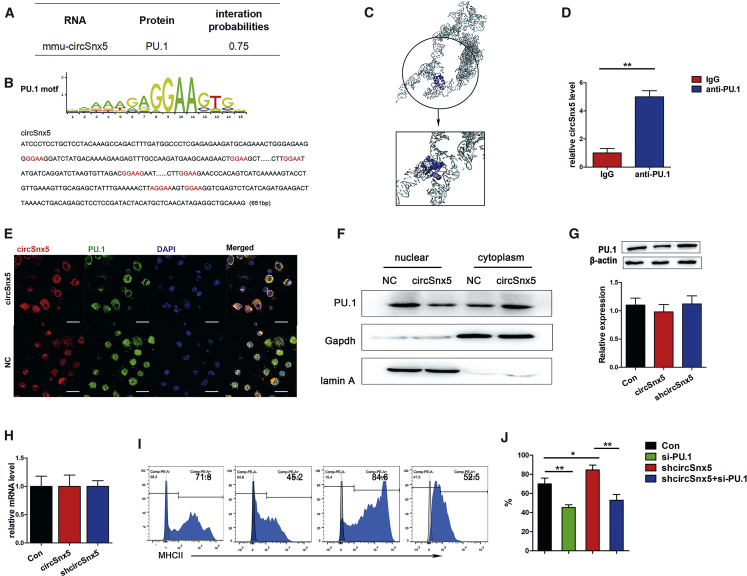
Using the RNA-protein interaction prediction (RPISeq) program, PU.1 and circSnx5 were likely to interact (Figure 4A), and several PU.1 motifs were observed in the circSnx5 sequence (Figure 4B). We then first used an online prediction tool to create a docking model to confirming the circSnx5 and PU.1 interaction (Figure 4C). Furthermore, RNA immunoprecipitation (RIP) assays were performed on DC extracts using antibodies against PU.1. As shown in Figure 4D, circSnx5 was obviously enriched in PU.1 immunoprecipitants relative to control immunoglobulin (Ig)G. To further verify the interactions between circSnx5 and proteins, we designed a circSnx5 RNA-specific pull-down probe and optimized comprehensive identification of RNA-binding proteins (RBPs) by mass spectrometry (ChIRP-MS) to identify circRNA-associated proteins in DCs (Figure S4A). The enriched proteins were analyzed with protein mass spectrometry, leading to the identification of 970 proteins in total. Consistent with the above-mentioned findings, PU.1 could be pulled down with the circSnx5, thus demonstrating that circSnx5 can directly bind to PU.1 (Figure S4B). Confocal microscopy clearly demonstrated the co-localization of circSnx5 and PU.1 in the cytoplasm of circSnx5-transfected DCs (Figure 4E), and western blotting showed that overexpression of circSnx5 increased the PU.1 expression level in the cytoplasm but not in the nucleus (Figure 4F; Figure S4C), indicating that circSnx5 partially inhibited nuclear translocation of PU.1 in DCs. Furthermore, circSnx5 overexpression or downregulation had no significant effect on PU.1 protein expression (Figure 4G) or PU.1 mRNA expression (Figure 4H) in DCs. To evaluate the downstream MHC class II expression, PU.1 siRNA and circSnx5 plasmids were transfected into DCs. Flow cytometric analysis showed that MHC class II expression was upregulated by circSnx5 siRNA introduction, which could be blocked by PU.1 siRNA (Figures 4I and 4J). Taken together, circSnx5 can bind to PU.1, thus inhibiting its transcriptional activity and downstream MHC class II expression in DCs.
Figure 4.
circSnx5 Repressed PU.1 Nuclear Translocation and Downstream MHC Class II Expression in DCs
(A) The predicted interaction between PU.1 and circSnx5 using the RNA-protein interaction prediction (RPISeq) program. Predictions with probabilities >0.5 were considered “positive,” indicating that the corresponding RNA and protein are likely to interact. (B) Enriched sequence motifs of PU.1 present in the circSnx5 sequence (red). (C) Graphical representation of 3D structures of the docking models of circSnx5 with PU.1 and zoomed-in images of the binding interface done by Hex docking software. (D) RIP experiments were performed using PU.1 or negative IgG antibody. Relative amounts of circSnx5 bound to IgG or PU.1 were detected by qRT-PCR. (E) RNA FISH showed co-localization of circSnx5 and PU.1 in DCs with or without circSnx5 overexpression. Scale bars, 100 μm. (F) Fractionation assays showed the expression of PU.1 in the nucleus and cytoplasm. (G) Western blotting analysis of PU.1 protein level in DCs after transfection with or without circSnx5 overexpression. (H) PU.1 mRNA expression level in DCs under different culture conditions detected by qRT-PCR assays. (H) DCs were transfected with or without shcircSnx5 or siPU.1 and then treated with 200 ng/mL LPS for 12 h. (I and J) MHC class II expression was detected by flow cytometry (I) and the percentage of Tregs is shown in (J). n = 5 independent DC preparations. Con, DCs treated with 200 ng/mL LPS for 12 h; NC, negative control. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, by Student’s t test.
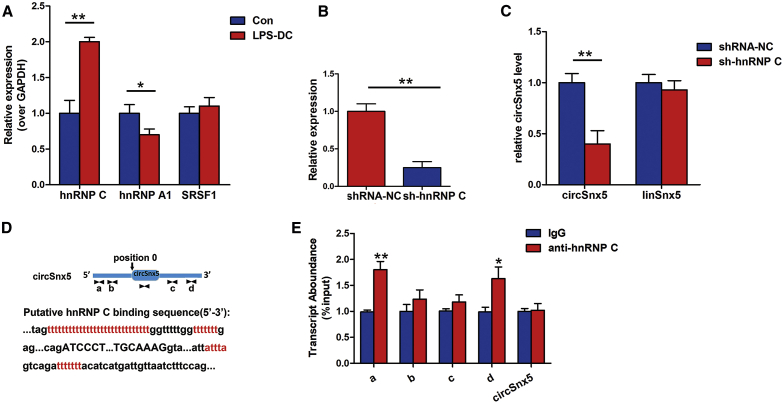
hnRNP C Promotes circSnx5 Generation in DCs
To ascertain the relationship between DC-associated SFs and circSnx5 generation, we first identified DC-abundant SFs. As shown in Figure 5A, LPS stimulation significantly increased hnRNP C expression, but it did not affect hnRNP A1 and SRSF1 expression. To further determine the roles of hnRNP C in circSnx5 generation, we applied siRNA to suppress the expression of hnRNP C (Figure 5B). qRT-PCR analyses showed that hnRNP C knockdown resulted in a significant reduction of circSnx5 expression in DCs, with no corresponding increase in the expression of the linear Snx5 isoform (Figure 5C). Given that SFs could directly bind to recognition motifs in the introns flanking circRNA-forming exons to promote circRNA generation, hnRNP C binding sequences were examined in circSnx5 and its flanking introns, identifying three putative hnRNP C binding sites (Figure 5D). RIP assays using anti-hnRNP C antibody was performed and showed that hnRNP C indeed bound to the putative binding sites on flanking introns of circSnx5 (Figure 5E). Taken together, these results suggested that hnRNP C is a critical element for circSnx5 production through binding to recognition sites in the flanking introns of circSnx5.
Figure 5.
hnRNP C Is Required for the Generation of circSnx5 in DCs
(A) qRT-PCR analysis of the SFs (hnRNP C, hnRNP A1, and SRSF1) in immature DCs (iDCs) and LPS-DCs. (B and C) qRT-PCR analysis of the expression of hnRNP C (B) and circSnx5 and linear Snx5 (C) in DCs transfected with hnRNP C siRNA. (D) Schematic illustrating the putative hnRNP C binding sites on the flanking introns in the circSnx5. The 5′ terminus of the circular exons of circSnx5 was defined as position 0. The putative hnRNP C-binding site is highlighted by the arrow located in the intron at the 5′ terminus and 3′ terminus of the circular exon (position −1000 to +1000). (E) The RIP assay was performed to verify hnRNP C binding with flanking introns of circSnx5. circSnx5 sequences were used as the negative control. qRT-PCR was used to detect the transcript abundance relative to input. n = 5 independent DC preparations. Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, by Student’s t test.
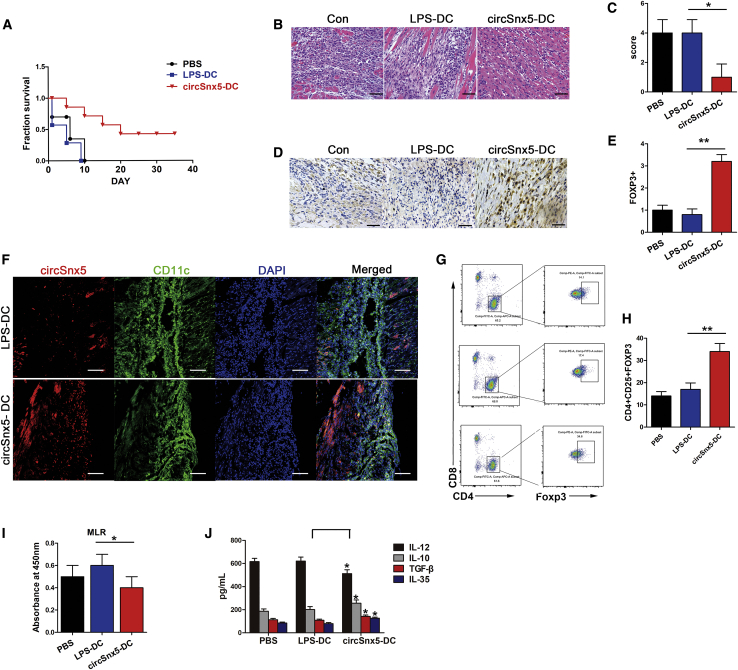
Transfer of circSnx5-Overexpressing DCs Protects against Acute Rejection after Cardiac Transplantation
Then, we aimed to demonstrate the feasibility and efficacy of circSnx5-conditioned DCs to induce tolerance in heart transplantation. In mice transfused with circSnx5-overexpressing DCs (circSnx5-DCs), the allograft survival was prolonged compared with LPS-DC and PBS transfusion (Figure 6A). Histological examination of cardiac allografts showed significantly reduced infiltration of inflammatory cells in recipient mice transfused with circSnx5-DC, whereas allografts from LPS-DC recipients showed severe myocyte damage (Figure 6B), with similar results obtained by grading (Figure 6C). Furthermore, immunohistochemical results showed that cardiac allografts from recipients transfused with circSnx5-DCs had greater numbers of Foxp3+ cells than did those transfused with LPS-DCs (Figures 6D and 6E). Moreover, we observed co-localization between circSnx5 and the DC marker CD11c in the cardiac allografts (Figure 6F). However, the number of Tregs was significantly elevated in the spleens of recipient mice injected with circSnx5-DCs (Figures 6G and 6H). In addition, an MLR assay showed reduced proliferation of splenocytes in the circSnx5-DC group compared to other groups (Figure 6I). Consistently, the inflammatory cytokine IL-12 in the serum was significantly decreased, although IL-10, IL-35, and TGF-β were significantly increased in circSnx5-DC-transfused mice (Figure 6J). Taken together, these results suggest that circSnx5-conditioned DCs can induce suppressive Tregs and prevent allograft rejection in vivo.
Figure 6.
Transfer of circSnx5-Overexpressing DCs Induced Treg Expansion and Protects Acute Rejection after Cardiac Transplantation
(A) Graft survival times of recipients is shown. The survival times in the groups were compared using Mann-Whitney U testing. (B and C) Histology typical of the three groups of mice. (B) Tissue sections were stained with H&E. The images are representative of several individuals from each group (n = 6 biological replicates). Scale bars, 200 μm. (C) Quantitative evaluation of myocardial inflammation. (D and E) Immunohistochemical analyses (D) were performed using anti-Foxp3 primary antibodies to detect Tregs. (E) The percentages of Foxp3+ cells were enumerated. The images are representative of several individuals from each group (n = 6 biological replicates). Scale bars, 50 μm. (F) Immunohistochemistry detection of the DC markers CD11c and circSnx5 in mouse transplanted tissues. The images are representative of several individuals from each group (n = 6 biological replicates). Scale bars, 20 μm. (G and H) Flow cytometry was performed to detect expression of Tregs (CD4+, CD25+, and Foxp3+) of spleens from recipients with circSnx5 overload compared with control mice (G), and are shown as percentages (H). (n = 6 biological replicates). (I) Splenic T cells were separated from recipient mice at day 7 post-transplantation. The proliferating activity of splenic T cells was assessed by BrdU-ELISA. (n = 6 biological replicates). (J) IL-10, IL-12, TGF-β, and IL-35 levels detected by ELISA in the plasma of recipient mice 14 days after heart transplantation. (n = 6 biological replicates). Data are shown as mean ± SD. ∗p < 0.05; ∗∗p < 0.01, by Student’s t test.
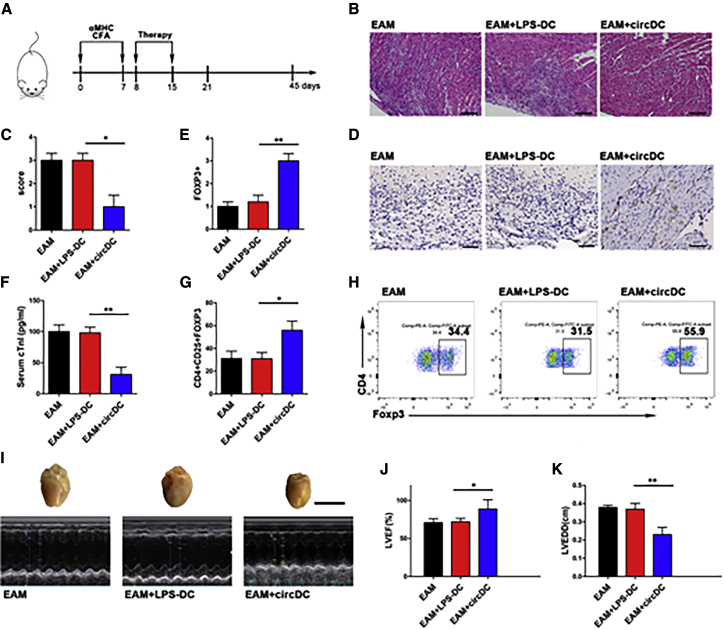
circSnx5-overexpressing DCs Reduce Cardiac Injury and Inflammation in Experimental Autoimmune Myocarditis (EAM)
CircSnx5-overexpressing DCs were transferred into EAM mice (Figure 7A). At 21 d after the first injection, the circDC mice were profoundly protected from EAM, developing minimal cardiac inflammation (Figure 7B) and exhibiting a lower inflammatory score (Figure 7C) and relatively normal serum troponin I levels (Figure 7F). In contrast, the hearts of LPS-DC mice developed considerable inflammation and intense inflammatory infiltration (Figure 7B), a high inflammatory score (Figure 7C), and elevated cardiac serum troponin I (Figure 7F). Subsequently, immunohistochemistry results showed that circSnx5 increased the number of Foxp3+-stained T cells (Figures 7D and 7E), which is consistent with flow cytometry analysis results showing that treatment with circDCs increased the percentage of CD4+CD25+Foxp3+ cells (Figures 7G and 7H). Moreover, treatment with circDCs increased the left ventricular ejection fraction and decreased the left ventricular end-diastolic dimension (LVEDD) in EAM mice at day 45 (Figures 7I–7K). In agreement, circDC treatment also significantly attenuated the heart size increase in mice (Figure 7I). Collectively, our results indicate that circSnx5 could modulate DC-driven immunity and tolerance by regulating the miR-544/SOCS1 axis and PU.1 nuclear translocation, suggesting that circSnx5 may serve as a potential therapeutic avenue for immune-related diseases (Figure 8).
Figure 7.
circSnx5-Overexpressing DCs Reduces Cardiac Injury and Inflammation in Experimental Autoimmune Myocarditis
(A) Schematic diagram of the experimental design. Experimental autoimmune myocarditis (EAM) was induced by injecting 100 μg of cardiac αMHC emulsified in CFA on days 0 and 7. circSnx5-DCs or LPS-DCs were injected into mice on days 8 and 15. Mice were killed at days 21 or 45 after the first immunization. (B and C) Representative (B) H&E staining of the hearts from EAM mice, LPS-DC mice, or circSnx5-DC mice at the peak of inflammation (day 21). Scale bars, 100 μm. (C) Inflammation was scored by percentage of myocardium infiltrated with mononuclear cells (n = 6 biological replicates). (D and E) Immunohistochemical labeling for Foxp3 (D) and counts of infiltrating Foxp3+ cells (E) in hearts harvested from different groups. Scale bars, 50 μm (n = 6 biological replicates). (F) Serum cardiac troponin-I (cTnI) levels in three different groups of mice were measured by ELISA on day 21 (n = 6 biological replicates). (G and H) Expression of CD4+CD25+Foxp3+ Tregs in splenic T cells was assessed by flow cytometry in three different groups on day 21 (H) and are shown as percentages (G). n = 6 biological replicates. (I) Images of hearts from three different mice groups at day 45. Scale bars, 1 cm. (J and K) Echocardiographic analysis images of left ventricular (LV) M-mode echocardiograms 45 days after injection. Left ventricular ejection fraction (LVEF) (J) and LV internal diastolic diameter (LVIDD) (K) are shown. circDCs, circSnx5-overexpression DCs (n = 6 biological replicates). Data are shown as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, by Student’s t test.
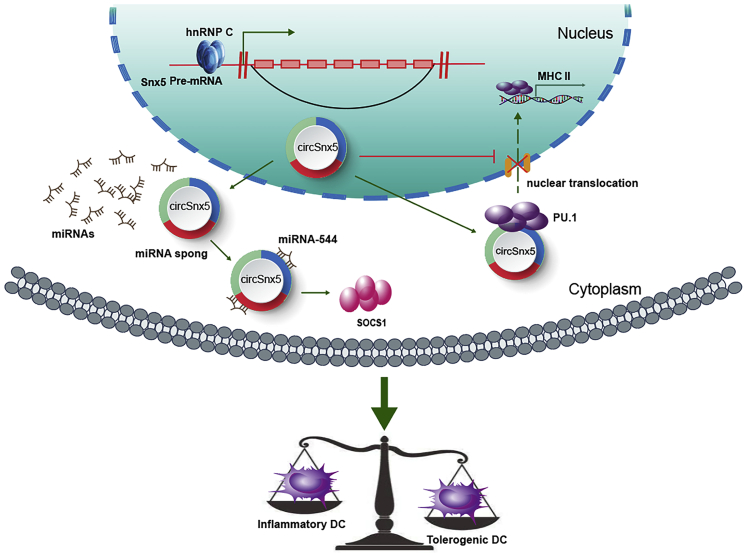
Figure 8.
Schematic of the Working Hypothesis
Schematic figure describing how circSnx5 can regulate DC-driven immunity and tolerance by regulating the miR-544/SOCS1 axis and PU.1 nuclear translocation.
Discussion
DCs play a dual role in linking and integrating innate and adaptive immune responses.1,2,4 DCs are front players in promoting unbalanced active immune responses, promoting the development of autoimmune diseases.32 A variety of regulatory factors are involved in the balance of maturity and immunological tolerance of DCs.33 Herein, we characterized a novel circRNA (circSnx5) that is stable and enriched expression in DCs. Ectopic circSnx5 expression suppressed DC activation and induced T cells hyporesponsiveness and antigen-specific Tregs, which predominantly mediate DC-based peripheral immune responses. Mechanistically, circSnx5 regulated the miR-544/SOCS1 axis and PU.1 nuclear translocation, thus greatly inhibiting the inflammatory differentiation of DCs and inducing immunological tolerance. Our results indicate that circSnx5 expression can induce immunological tolerance, which is supported by the findings that transfusion with circSnx5-DCs alleviated the inflammatory response in an EAM model and prolonged cardiac allograft survival. Overall, circSnx5 plays a central role in DC immune homeostasis and could be a potential target for novel immunotherapeutic approaches for specific treatment of immune-related diseases.
Previous studies have suggested that DCs are key regulatory cells for immune regulation, and their differentiation and function influence the outcome of innate and adaptive immune responses.3, 4, 5, 6 Altered DC distribution and/or disturbed key functions are emerging as a fundamental feature in both systemic and tissue-specific autoimmune diseases.34 During the past decades, numerous regulators that fine-tune DC function have been uncovered, including proteins, genes, and ncRNAs. For example, it has been reported that Lkb1 acts as a regulatory switch in DCs for controlling Treg homeostasis, immune response, and tolerance.35 Wnt1 leads to transcriptional silencing of CC/CXC chemokines in DCs, inducing adaptive immune resistance in human cancer cells.36 Cao and colleagues37 demonstrated that lnc-DC, a long ncRNA expressed exclusively in human DCs, highly expressed in human conventional DC (cDC) subsets could regulate DC differentiation by activating the transcription factor signal transducer and activator of transcription (STAT)3. circRNAs, novel, highly stable, and conserved ncRNAs, are involved in numerous pathologic and physiologic processes.38, 39, 40 However, the roles and mechanistic basis of circRNAs in DCs remain unclear. In this study, we identified a circRNA (circSnx5) derived from exon 4 to exon 10 of the Snx5 gene that is enriched in DCs. Furthermore, we demonstrated its functional association with the DC functional state. Functional experiments demonstrated that ectopic circSnx5 expression prevented the differentiation to the inflammatory phenotype of DCs, inducing T cells hyporesponsiveness and antigen-specific Tregs. Therefore, our findings suggest a critical role for circSnx5 in the immunological function of DCs, which uncovers a new role of circRNAs in immunity.
circRNAs are novel ncRNAs formed by covalently closed-loop RNA molecules composed of exon, intron, or even exon-intron sequences. They have been found to have a long half-life and to be evolutionally conserved and relatively stable in cells owing to their specific closed-loop structure.19,41 These features confer numerous potential functions to circRNAs, most of which act as sponges for miRNAs. In addition, crosslinking immunoprecipitation datasets suggest that circRNAs interact with many different RBPs to act as “protein sponges,” regulate protein function, act as scaffolds to mediate complex formation between specific enzymes and substrates, and recruit proteins to specific locations.42,43 Furthermore, only a subset of circRNAs, such as circ-ZNF609, circPINTexon2, and circ-SHPRH, have been shown to act as protein templates under specific conditions.26,44,45 A growing number of studies have revealed that circRNAs play potentially important roles in physiological and pathological conditions by distinct modes of action at the molecular level.43 Herein, we characterized a novel circRNA (circSnx5) that acts as a miR-544 sponge to attenuate miRNA-mediated target repression of SOCS1, thus modulating the cytokine microenvironment in DCs and reshaping the functional state of these cells. As circRNAs have also been shown to combine with RBPs to regulate their activities, mass spectrometry analysis was performed and confirmed that circSnx5 can directly bind to transcription factor PU.1 and inhibit MHC class II expression, modulating immunogenicity in DCs and reshaping the DC functional state.
SOCS1 is a pivotal regulator that participates in negative feedback of the Janus kinase (JAK)-STAT pathway, which plays a key role in the activation of DCs.46 Recent studies revealed the pivotal role of SOCS1 in regulating the phenotype and function of immune cells by negatively regulating the response of immune cells to cytokines. For example, SOCS1 and SOCS3 inhibit the activation of the JAK-signal transducer and limit the production of proinflammatory cytokines in DCs. Moreover, SOCS1 inhibits the nuclear factor κB (NF-κB) signaling pathway by stimulating the ubiquitylation and degradation of the Toll-IL-1 receptor (TIR) domain.46,47 In the present work, we show that SOCS1 is a target of circSnx5 and plays a vital role in the immune tolerance of DCs driven by circSnx5. However, whether other pathways play a role or interact with the miR-544/SOCS1 axis remains to be further studied.
circRNAs such as circPABPN1 and circANRIL function by interacting with a target protein. circPABPN1 suppresses the translation of nuclear poly(A) binding protein 1 mRNA by sequestering the RBP Hu antigen R (HUR) in human cervical carcinoma HeLa cells,48 and circANRIL impairs pre-rRNA processing and ribosome biogenesis by binding to pescadillo homolog 1.49 In addition, circRNAs also recruit proteins to specific cellular locations. circRNA FECR1 recruits TET1 to the promoter region of transcription factor FLI1, leading to the demethylation of CpG sites and active transcription.50 PU.1, a transcription factor, has been demonstrated to play key roles in DC function,51 and it can transactivate the CIITA pI promoter and regulate MHC class II expression.52 In this study, we confirmed that circSnx5 can directly bind to the transcription factor PU.1 and inhibit MHC class II expression, modulating immunogenicity in DCs and reshaping the DC functional state. During this regulation and control function, we speculate that circSnx5 can inhibit the transcriptional function of PU.1 by reducing its intracellular transport. However, the specific molecular mechanism of this function must be further elaborated in the future.
circRNA formation can be regulated by SFs.53,54 circRNA biogenesis is dependent on the canonical splicing machinery. Depletion of SFs suppresses the level of circRNAs, which play an important role in different biological process. For instance, SF ESRP1 promotes circBIRC6 biogenesis by binding to recognition sites in the introns flanking the circBIRC6-forming exons, indirectly regulating pluripotency in human embryonic stem cells (hESCs).55 In addition, circSMARCA5 increases expression of serine- and arginine-rich SF3 (SRSF3) RNA isoform containing exon 4, resulting in a non-productive non-sense-mediated decay (NMD) substrate.56 According to recent reports, hnRNP C, hnRNP A1, and SRSF1 are involved in the regulation of DC maturation.57, 58, 59 In our study, we examined altered SF expression in DCs, identifying hnRNP C as a potential mediator of circSnx5 generation in DCs. We also have identified and confirmed three putative hnRNP C binding sites in circSnx5-flanking introns using RIP assays. Our results suggest that hnRNP C is a critical element for circSnx5 production through binding to recognition sites in the flanking introns of circSnx5. However, additional studies are needed to further elucidate the underlying mechanisms of hnRNP C and circRNA formation.
In conclusion, circSnx5 was found to play a critical role in regulating DC-driven immunity and tolerance by regulating the miR-544/SOCS1 axis and PU.1 nuclear translocation, suggesting that it could be a promising therapeutic target in the treatment of immunological diseases.
Our study has certain limitations. First, although the effect of circSnx5 in animal experiments was clear, more detailed experiments in human DCs are needed to further verify their roles. In terms of functional verification, we mainly verified the function of circSnx5 on DCs, but whether circSnx5 can regulate other immune cells that play a role in immune regulation remains to be determined. Our study lays the foundation for the role of circRNAs in immune regulation, which provides a novel research direction for the specific treatment of immune-related diseases.
Materials and Methods
Animals
Male C57BL/6J and BALB/c mice (6–8 weeks old, 20–25 g) were purchased from Beijing Vital River Laboratory Animal Technology (Beijing, China). All animal care and handling procedures conformed to the Principles of Animal Care provided by the Guide for the Care and Use of Laboratory Animals (NIH publication 85-23, revised 2011, USA). All studies were conducted in accordance with the scheme approved by the Ethics of Animal Experiments of The Second Affiliated Hospital of Harbin Medical University (SYDW2019-147).
Generation and Transfection of BM-Derived DCs
DCs were derived from BM precursors of BALB/c mice, as described previously.60 Briefly, BM was flushed from tibias and femurs and passed through a 200-mm pore-size mesh to remove fibrous tissue. Furthermore, the red blood cells (Beyotime, China) were lysed, and the remaining cells were centrifuged, washed, and resuspended in Roswell Park Memorial Institute 1640 medium (HyClone, South Logan, UT, USA) containing 10% fetal bovine serum (ScienCell, Carlsbad, CA, USA), 1% penicillin/streptomycin (Beyotime, Shanghai, China), 10 ng/mL IL-4, and 20 ng/mL granulocyte-macrophage colony-stimulating factor (GM-CSF; PeproTech, Rocky Hill, NJ, USA). BM progenitors were added to six-well tissue culture plates (Corning Life Sciences, New York, NY, USA) and cultured at a concentration of 2 × 106 cells/mL in 2 mL of medium per well at 37°C, 5% CO2, followed by collection of the culture supernatant for further evaluation.
circSnx5 plasmids were synthesized by Hanbio (Shanghai, China). The endogenous 5′-flanking and 3′-flanking 1-kb introns cloned from the genome and fused to the corresponding ends of full-length circSnx5 were cloned into the specific vector; in particular, the exon-intron boundaries and canonical splicing sites at the 5′ and 3′ termini of circRNAs were preserved for correct splicing (Figure 1A). For shcircSnx5, the circular junction for circSnx5 was specifically targeted by shRNA oligonucleotides and individually cloned into the pGPH1/GFP/Neo vector between the BbsI and BamHI sites (Figure 2A). The targeting sequences of this shRNA oligonucleotides are listed in Table S1. shcircSnx5, miR-544 mimics, and RNA inhibitors, including SOCS1 shRNA, hnRNP C shRNA, and PU.1 siRNA, were synthesized by GenePharma (Shanghai, China). The sequences used are shown in Table S1. On the 6th day of culture, DCs were transfected with circSnx5 plasmid or shRNA, miRNA mimics or inhibitors, or plasmid DNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. At the 7th day of culture, that is, day 1 after transfection, the culture supernatant was collected for further evaluation after treatment with the Toll-like receptor (TLR) ligand LPS (200 ng/mL; Sigma-Aldrich, St. Louis, MO, USA) for 12 h. Transfected cells were harvested for indicated analyses 24 h after transfection.
RNase R Treatment
For RNase R treatment, 2 mg of total RNA was incubated for 20 min at 37°C with or without RNase R (Epicenter Technologies, Madison, WI, USA). RNA transcripts resistant to RNase R treatment were precipitated with ethanol and resuspended in RNase-free H2O for the indicated analyses.
RNA Preparation and qRT-PCR
RNA from different samples was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Nuclear and cytoplasmic RNAs were isolated using a cytoplasmic and nuclear RNA purification kit (Norgen Biotek, Thorold, ON, Canada), followed by reverse transcription using a Transcriptor first-strand cDNA synthesis kit (Roche, Basel, Switzerland) according to the manufacturers’ instructions.
To quantify the amount of mature miRNA, the Hairpin-it miRNA and U6 small nuclear RNA (snRNA) normalization RT-PCR quantitation kit (GenePharma, Shanghai, China) was used. Furthermore, incubation conditions for reverse transcription were conducted for 30 min at 25°C, 30 min at 42°C, and 5 min at 85°C. PCR was performed at the following settings: 40 cycles of 12 s at 95°C and 40 s at 62°C.
To quantify the amount of mRNA and circRNA, cDNA was amplified using a Transcriptor first-strand cDNA synthesis kit (Roche, Basel, Switzerland). Reverse transcription proceeded for 60 min at 50°C and 5 min at 85°C. PCR was conducted using Bestar SYBR Green qPCR master mix (DBI Bioscience, Germany) at the following settings: 40 cycles of 10 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Results were normalized to β-actin expression. The remaining primers are listed in Table S1.
RNA FISH
In situ hybridization was performed using a FISH kit (Ribobio, Guangzhou, China) according to the manufacturer’s instructions. The FISH assay was performed in DCs with or without co-transfection with circSnx5- and miR-544-expressing vectors. Probes specific to circSnx5 and miR-544 (Ribobio, Guangzhou, China) were used for hybridization. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). The images were acquired on a Leica TCS-SP5 LSM electron microscope (JEM-1220, JEOL, Japan).
Flow Cytometry Analysis
To analyze cell surface markers, single-cell suspensions of BM-derived DCs were prepared from spleen and lymph node (LN) samples for staining with fluorescein isothiocyanate (FITC)-CD11C (clone N418), phycoerythrin (PE)-CD80 (clone 16-10A1), PE-CD86 (clone GL1), or PE-MHC class II (clone M5/114.15.2) (eBioscience, San Diego, CA, USA). An appropriate isotype-matched control was included for each immunolabeling procedure. The indicated CD4+, CD25+, and Foxp3+ (Treg) populations were sorted using FITC-CD4 (clone RM4-5), allophycocyanin (APC)-CD25 (clone PC61.5), and PE-Foxp3 (clone FJK-16s) (eBioscience, San Diego, CA, USA). Cell surface staining was mostly performed at 4°C for 30 min. Intracellular staining was performed using an intracellular fixation and permeabilization buffer set (eBioscience). Flow cytometry data were acquired on a FACSDiva version 6.1.3 (BD Biosciences, San Jose, CA, USA) system and analyzed using FlowJo software (Tree Star).
ELISA
After transfection and LPS stimulation, cytokine secretion by DCs was measured. The levels of IL-12, IL-10, IL-35, and TGF-β (all purchased from Abcam, Cambridge, MA, USA) were determined by ELISA (ultra-sensitive mouse cardiac troponin-I [Tn-I]; Life Diagnostics, West Chester, PA, USA) according to the manufacturer’s instructions. Additionally, levels of these cytokines were determined in recipient mice 7 days after heart transplantation or at 21 days in EAM mice in the same manner.
MLR Assay
To determine the ability of differently treated DCs to induce Tregs, allogeneic splenic T cells (from C57BL/6 mice) were cultured with DCs at a DC/T cell ratio of 1:10 for 3 days in 10 μg/mL of mitomycin C-pretreated plates. Proliferation was assessed using an ELISA for 5-bromo-2′-deoxyuridine (BrdU) (BrdU-ELISA) according to the manufacturer’s instructions (Chemicon International, Temecula, CA, USA). The immunosuppressive function of isolated Tregs was also assessed by BrdU-ELISA. Tregs (CD4+CD25+) were cocultured with allogeneic effector T cells and mitomycin C-treated stimulators (splenic cells or DCs) in a Treg/T cell/stimulator ratio of 10:10:1. The proliferation of T cells was assessed by BrdU-ELISA. In several experiments, the levels of T cell proliferation in the splenic cells of recipient mice 7 days after transplantation or 21 days in EAM mice were also measured.
Protein and RNA Docking Program
To evaluate the predicted interaction between PU.1 and circSnx5, we first used an online prediction tool (http://rna.tbi.univie.ac.at/) to obtain the minimum RNA energy (−128.70 kcal/mol) secondary structure of circSnx5 and the predicted ring RNA tertiary structure. Protein structures of the transcription factor PU.1 were downloaded from the Protein Data Bank (PDB) (https://www.rcsb.org/). Hex docking software was used for RNA and protein docking, and 100 conformations were docked to select the conformation with the lowest energy. The docking model is shown in Figure 4C.
Luciferase Reporter Assay
To construct luciferase reporter vectors, plasmids, including circSnx5 WT (circSnx5-WT) and a mutant devoid of the miR-544 binding site (circSnx5-mut), were subcloned downstream of the coding region of the luciferase gene. psiCHECK2-circSnx5-WT1, -WT2, or -WT3 or psiCHECK2-circSnx5-mut 1, -mut2, or -mut3 was transfected with miR-544 mimics or negative control using Lipofectamine 2000 in 293T cells. The luciferase assays were performed using a luciferase assay kit (Promega, USA) according to the manufacturer’s instructions.
Western Blotting
The expression of SOCS1 and PU.1 was assessed by western blotting in DCs as previously described.9 Briefly, DC lysates were collected and blotted to determine SOCS1, PU.1, and β-actin expression using anti-PU.1, anti-SOCS1, and anti-β-actin antibodies (Abcam, USA). The protein levels were quantified by scanning densitometry (GS-710 imaging).
RIP Assay
A RIP assay was performed using anti-PU.1 or anti-hnRNP C antibody (Abcam, USA), and the coprecipitated RNAs were obtained using the Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, USA) according to the manufacturer’s instructions. IgG (Merck Millipore, Billerica, MA, USA) was used as a negative control (input group). Subsequently, these obtained RNAs were analyzed by qPCR with primers listed in Table S1. Total RNA (input control) and the isotype control were used to confirm the binding specificity of RNAs with PU.1, hnRNP C, or Ago.
RNA Pull-Down Assay
An RNA-protein pull-down kit (KT105-01, Gzscbio, Guangzhou, China) was used to detect interactions between circSnx5 and proteins. The biotin-labeled probe sequence is shown in Table S3. The biotin-labeled RNA was incubated with streptavidin magnetic beads (Invitrogen) at 37°C for 30 min, which were pre-washed twice with lysis buffer. The RNA-labeled beads were washed with 1 mL of wash buffer five or six times. DNA elution buffer (50 μL), RNase A (100 μg/mL), and RNase H (0.1 U/μL) were added, followed by incubation for 30 min, and then 10 μL of 5× loading buffer was added for elution at 100°C for 10 min. The eluted proteins were finally analyzed by mass spectrometry or western blot.
Heart Transplantation
At 24 h after injection with 0.3 mL of PBS only (negative control), 1.0 × 106 LPS-myeloid DCs (mDCs), or circSnx5-DCs from BALB/c mice caudal veins, the donor heart grafts were harvested from C57BL/6 mice and transplanted into the abdominal cavity of BALB/c recipient mice using microsurgical techniques. BM-derived DCs were cultured from donor mice and infected with circSnx5 vector on day 2. circSnx5-overexpressing DCs, control, or LPS-treated DCs were intravenously injected into recipient BALB/c mice 1 day prior to transplantation. Graft function was monitored by daily abdominal palpation postoperatively. Complete cessation of palpable heartbeats was defined as graft rejection, and direct visualization and histological examination of the graft were accessed for further verification.
Experimental Autoimmune-Induced EAM Mouse Model
Mice were subcutaneously injected with 100 μg of cardiac αMHC (MyHC-α614-629 sequence; Ontores Biotechnologies, Zhejiang, China) peptide emulsified in 1 mg/mL complete Freund’s adjuvant (CFA) (Sigma-Aldrich, St. Louis, MO, USA) on days 0 and 7. Additionally, EAM mice were injected with circSnx5-DCs (5 × 106 cells/mouse) PBS or LPS-DCs via the caudal vein twice a week following the first immunization with myosin (Figure 7A). Mice were evaluated for the development of inflammatory infiltration and dilated cardiomyopathy (DCM) onset on days 21 and 45, respectively.
Histological Analyses of Heart Tissue
Seven days after transplantation, all allografts from the recipients were harvested. Then, a part of each allograft was embedded in paraffin for hematoxylin and eosin staining. Additionally, the paraffin-embedded sections were stained with Foxp3 (Wanleibio, China). Hematoxylin and eosin staining was assessed as grades from 0 (none) to 4 (severe), according to the 2014 classification of the International Society for Heart and Lung Transplantation for Acute Cellular Rejection.61
For EAM mice, heart weight was measured at 21 days after myosin injection, and the hearts were stained with hematoxylin and eosin and assayed according to a semiquantitative scale as described previously:40 0, no inflammatory infiltrates; 1, small foci of inflammatory cells between myocytes; 2, larger foci of >100 inflammatory cells; 3, involvement of <10% of a cross-section; 4, involvement of >30% of a cross-section.
For immunohistochemical analysis, sections were dewaxed, subjected to antigen retrieval, and incubated with anti-Foxp3 antibody (Wanleibio, Shenyang, China) and secondary antibody. All histological evaluations were performed in a double-blinded manner by two professional staff members. Images were captured using a Leica TCS-SP5 LSM electron microscope (JEM-1220, JEOL, Japan).
Echocardiography
Following anesthesia with 1%–2% isoflurane, the mice heart rates were maintained at approximately 450 bpm. Transthoracic echocardiography was performed to measure the LVEDD and left ventricular end-systolic dimension (LVESD), LV fractional shortening, and LV ejection fraction in long axis M-mode analysis from the average of three independent cardiac cycles, using a Hitachi Vision 900 system (Hitachi Medical System, Tokyo, Japan) with a 13-MHz transducer.
Statistical Analysis
Statistical analyses were performed with SPSS 23.0 (SPSS, USA) and GraphPad Prism 6 (GraphPad, La Jolla, CA, USA). Data are expressed as means ± SD. Survival of cardiac allografts was compared using the Mann-Whitney U test. Variances between two groups were analyzed by Student’s t tests. For other comparisons, analysis of variance was calculated by one-way ANOVA. A p value <0.05 was considered statistically significant.
Author Contributions
M.Z. and M.E. conceived and designed this research. Q.C. performed experiments, analyzed the data, and wrote the manuscript. G.M. and J.W. performed the experiments and revised the article. P.S., T.L., H.Z., N.W., Z.T., W.W., and Y.Z. helped with the experiments and analyzed the data. J.W. and J.T. helped to revise the article. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81670459 to M.Z. and 81972143 to J.W.); the National Key R&D Program of China (grant 2016YFC1301100 to B.Y.); the Key Laboratory of Myocardial Ischemia, Harbin Medical University, Ministry of Education (KF201816 to Q.C. and KF201829 to G.M.); the Youth Innovation Science Research Foundation of the Second Affiliated Hospital of Harbin Medical University (kycx2018-01); and by the Harbin Medical University Graduate Research and Practice Innovation Project (YJSSJCX2018-66HYD).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.07.001.
Contributor Information
Mingyan E, Email: emingyan889@163.com.
Maomao Zhang, Email: maomaolp1983@163.com.
Supplemental Information
References
- 1.Qian C., Cao W. Dendritic cells in the regulation of immunity and inflammation. Semin. Immunol. 2018;35:3–11. doi: 10.1016/j.smim.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Iberg C.A., Jones A., Hawiger D. Dendritic cells as inducers of peripheral tolerance. Trends Immunol. 2017;38:793–804. doi: 10.1016/j.it.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mildner A., Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40:642–656. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 4.Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenbarth S.C. Dendritic cell subsets in T cell programming: location dictates function. Nat. Rev. Immunol. 2019;19:89–103. doi: 10.1038/s41577-018-0088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akkaya B., Oya Y., Akkaya M., Al Souz J., Holstein A.H., Kamenyeva O., Kabat J., Matsumura R., Dorward D.W., Glass D.D., Shevach E.M. Regulatory T cells mediate specific suppression by depleting peptide-MHC class II from dendritic cells. Nat. Immunol. 2019;20:218–231. doi: 10.1038/s41590-018-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horwitz D.A., Fahmy T.M., Piccirillo C.A., La Cava A. Rebalancing immune homeostasis to treat autoimmune diseases. Trends Immunol. 2019;40:888–908. doi: 10.1016/j.it.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J., Abram C.L., Hu Y., Lowell C.A. CARD9 mediates dendritic cell-induced development of Lyn deficiency-associated autoimmune and inflammatory diseases. Sci. Signal. 2019;12 doi: 10.1126/scisignal.aao3829. eaao3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei H.J., Gupta A., Kao W.M., Almudallal O., Letterio J.J., Pareek T.K. Nrf2-mediated metabolic reprogramming of tolerogenic dendritic cells is protective against aplastic anemia. J. Autoimmun. 2018;94:33–44. doi: 10.1016/j.jaut.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Moussion C., Mellman I. The dendritic cell strikes back. Immunity. 2019;50:533. doi: 10.1016/j.immuni.2019.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Wilgenhof S., Corthals J., Heirman C., van Baren N., Lucas S., Kvistborg P., Thielemans K., Neyns B. Phase II study of autologous monocyte-derived mRNA electroporated dendritic cells (TriMixDC-MEL) plus ipilimumab in patients with pretreated advanced melanoma. J. Clin. Oncol. 2016;34:1330–1338. doi: 10.1200/JCO.2015.63.4121. [DOI] [PubMed] [Google Scholar]
- 12.Yuan X., Qin X., Wang D., Zhang Z., Tang X., Gao X., Chen W., Sun L. Mesenchymal stem cell therapy induces FLT3L and CD1c+ dendritic cells in systemic lupus erythematosus patients. Nat. Commun. 2019;10:2498. doi: 10.1038/s41467-019-10491-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wadwa M., Klopfleisch R., Buer J., Westendorf A.M. Targeting antigens to Dec-205 on dendritic cells induces immune protection in experimental colitis in mice. Eur. J. Microbiol. Immunol. (Bp.) 2016;6:1–8. doi: 10.1556/1886.2015.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spiering R., Margry B., Keijzer C., Petzold C., Hoek A., Wagenaar-Hilbers J., van der Zee R., van Eden W., Kretschmer K., Broere F. DEC205+ dendritic cell-targeted tolerogenic vaccination promotes immune tolerance in experimental autoimmune arthritis. J. Immunol. 2015;194:4804–4813. doi: 10.4049/jimmunol.1400986. [DOI] [PubMed] [Google Scholar]
- 15.Ettinger M., Gratz I.K., Gruber C., Hauser-Kronberger C., Johnson T.S., Mahnke K., Thalhamer J., Hintner H., Peckl-Schmid D., Bauer J.W. Targeting of the hNC16A collagen domain to dendritic cells induces tolerance to human type XVII collagen. Exp. Dermatol. 2012;21:395–398. doi: 10.1111/j.1600-0625.2012.01474.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 18.Patop I.L., Wüst S., Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38:e100836. doi: 10.15252/embj.2018100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santer L., Bär C., Thum T. Circular RNAs: a novel class of functional RNA molecules with a therapeutic perspective. Mol. Ther. 2019;27:1350–1363. doi: 10.1016/j.ymthe.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu C.X., Li X., Nan F., Jiang S., Gao X., Guo S.K., Xue W., Cui Y., Dong K., Ding H. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177:865–880.e21. doi: 10.1016/j.cell.2019.03.046. [DOI] [PubMed] [Google Scholar]
- 21.Sun M., Zhao W., Chen Z., Li M., Li S., Wu B., Bu R. circ_0058063 regulates CDK6 to promote bladder cancer progression by sponging miR-145-5p. J. Cell. Physiol. 2019;234:4812–4824. doi: 10.1002/jcp.27280. [DOI] [PubMed] [Google Scholar]
- 22.Hall I.F., Climent M., Quintavalle M., Farina F.M., Schorn T., Zani S., Carullo P., Kunderfranco P., Civilini E., Condorelli G., Elia L. circ_Lrp6, a circular RNA enriched in vascular smooth muscle cells, acts as a sponge regulating miRNA-145 function. Circ. Res. 2019;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X., Wang S., Wang H., Cao J., Huang X., Chen Z., Xu P., Sun G., Xu J., Lv J., Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of circRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu Z., Li P., Fan L., Wu M. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front. Immunol. 2018;9:9. doi: 10.3389/fimmu.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Z., Li P., Fan L., Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy Front. Immunol. 2018;9:9. doi: 10.3389/fimmu.2018.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Zhang G., Liu Y., Chen R., Zhao D., McAlister V., Mele T., Liu K., Zheng X. GDF15 regulates Malat-1 circular RNA and inactivates NFκB signaling leading to immune tolerogenic DCs for preventing alloimmune rejection in heart transplantation. Front. Immunol. 2018;9:2407. doi: 10.3389/fimmu.2018.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kobayashi T., Yoshimura A. Keeping DCs awake by putting SOCS1 to sleep. Trends Immunol. 2005;26:177–179. doi: 10.1016/j.it.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Sozzani S., Del Prete A., Bosisio D. Dendritic cell recruitment and activation in autoimmunity. J. Autoimmun. 2017;85:126–140. doi: 10.1016/j.jaut.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Qian C., Cao X. Dendritic cells in the regulation of immunity and inflammation. Semin. Immunol. 2018;35:3–11. doi: 10.1016/j.smim.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Funes S.C., Manrique de Lara A., Altamirano-Lagos M.J., Mackern-Oberti J.P., Escobar-Vera J., Kalergis A.M. Immune checkpoints and the regulation of tolerogenicity in dendritic cells: implications for autoimmunity and immunotherapy. Autoimmun. Rev. 2019;18:359–368. doi: 10.1016/j.autrev.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Du X., Wei J., Long L., Tan H., Guy C., Dhungana Y., Qian C., Neale G., Fu Y.X. LKB1 orchestrates dendritic cell metabolic quiescence and anti-tumor immunity. Cell Res. 2019;29:391–405. doi: 10.1038/s41422-019-0157-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kerdidani D., Chouvardas P., Arjo A.R., Giopanou I., Ntaliarda G., Guo Y.A., Tsikitis M., Kazamias G., Potaris K., Stathopoulos G.T. Wnt1 silences chemokine genes in dendritic cells and induces adaptive immune resistance in lung adenocarcinoma. Nat. Commun. 2019;10:1405. doi: 10.1038/s41467-019-09370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y.G., Kim M.V., Chen X., Batista P.J., Aoyama S., Wilusz J.E., Iwasaki A., Chang H.Y. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 2017;67:228–238.e5. doi: 10.1016/j.molcel.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng W.L., Marinov G.K., Liau E.S., Lam Y.L., Lim Y.Y., Ea C.K. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861–871. doi: 10.1080/15476286.2016.1207036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., Liu T., Wang X., He A. Circles reshaping the RNA world: from waste to treasure. Mol. Cancer. 2017;16:58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xia P., Wang S., Ye B., Du Y., Li C., Xiong Z., Qu Y., Zusen Fan Z. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity. 2018;48:688–701.e7. doi: 10.1016/j.immuni.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 43.Huang A., Zheng H., Wu Z., Chen M., Huang Y. Circular RNA-protein interactions: functions, mechanisms, and identification. Theranostics. 2020;10:3503–3517. doi: 10.7150/thno.42174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang M., Zhao K., Xu X., Yang Y., Yan S., Wei P., Liu H., Xu J., Xiao F., Zhou H. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 2018;9:4475. doi: 10.1038/s41467-018-06862-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 46.Kobayashi T., Yoshimura A. Keeping DCs awake by putting SOCS1 to sleep. Trends Immunol. 2005;26:177–179. doi: 10.1016/j.it.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 47.Evel-Kabler K., Song X.T., Aldrich M., Huang X.F., Chen S.Y. SOCS1 restricts dendritic cells’ ability to break self tolerance and induce antitumor immunity by regulating IL-12 production and signaling. J. Clin. Invest. 2006;116:90–100. doi: 10.1172/JCI26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen N., Zhao G., Yan X., Lv Z., Yin H., Zhang S., Song W., Li X., Li L., Du Z. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018;19:218. doi: 10.1186/s13059-018-1594-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chopin M., Lun A.T., Zhan Y., Schreuder J., Coughlan H., D’Amico A., Mielke L.A., Almeida F.F., Kueh A.J., Dickins R.A. Transcription factor PU.1 promotes conventional dendritic cell identity and function via induction of transcriptional regulator DC-SCRIPT. Immunity. 2019;50:77–90.e5. doi: 10.1016/j.immuni.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Kitamura N., Yokoyama H., Yashiro T., Nakano N., Nishiyama M., Kanada S., Fukai T., Hara M., Ikeda S., Ogawa H. Role of PU.1 in MHC class II expression through transcriptional regulation of class II transactivator pI in dendritic cells. J. Allergy Clin. Immunol. 2012;129:814–824.e6. doi: 10.1016/j.jaci.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 53.Chen Y.G., Chen R., Ahmad S., Verma R., Kasturi S.P., Amaya L., Broughton J.P., Kim J., Cadena C., Pulendran B. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell. 2019;76:96–109.e9. doi: 10.1016/j.molcel.2019.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao W., Cui Y., Liu L., Qi X., Liu J., Ma S., Hu X., Zhang Z., Wang Y., Li H. Splicing factor derived circular RNA circUHRF1 accelerates oral squamous cell carcinoma tumorigenesis via feedback loop. Cell Death Differ. 2020;27:919–933. doi: 10.1038/s41418-019-0423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017;8:1149. doi: 10.1038/s41467-017-01216-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barbagallo D., Caponnetto A., Cirnigliaro M., Brex D., Barbagallo C., D’Angeli F., Morrone A., Caltabiano R., Barbagallo G.M., Ragusa M. circSMARCA5 inhibits migration of glioblastoma multiforme cells by regulating a molecular axis involving splicing factors SRSF1/SRSF3/PTB. Int. J. Mol. Sci. 2018;19:480. doi: 10.3390/ijms19020480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia R., Li X., Yu C., Fan M., Guo J. The splicing factor hnRNP C regulates expression of co-stimulatory molecules CD80 and CD40 in dendritic cells. Immunol. Lett. 2013;153:27–32. doi: 10.1016/j.imlet.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 58.da Glória V.G., Martins de Araújo M., Mafalda Santos A., Leal R., de Almeida S.F., Carmo A.M., Moreira A. T cell activation regulates CD6 alternative splicing by transcription dynamics and SRSF1. J. Immunol. 2014;193:391–399. doi: 10.4049/jimmunol.1400038. [DOI] [PubMed] [Google Scholar]
- 59.Chen C., Cao M., Wu D., Li N., Peng J., Song L., Qi P., Zhang M., Zhao J. KH-type splicing regulatory protein mediate inflammatory response in gastric epithelial cells induced by lipopolysaccharide. Cell Biol. Int. 2017;41:871–878. doi: 10.1002/cbin.10804. [DOI] [PubMed] [Google Scholar]
- 60.Son Y.-I., Shin-ichi Egawa S.-i., Tatsumi T., Redlinger R.E., Jr., Kalinski P., Kanto T. A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J. Immunol. Methods. 2002;262:145–157. doi: 10.1016/s0022-1759(02)00013-3. [DOI] [PubMed] [Google Scholar]
- 61.Szymańska S., Grajkowska W., Pronicki M. Pathologic diagnosis of antibody-mediated rejection in endomyocardial biopsy after heart transplantation based on renewed International Society for Heart and Lung Transplantation criteria. Pol. J. Pathol. 2014;65:176–181. doi: 10.5114/pjp.2014.45785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.