Abstract
Background
Early detection is essential to improve the survival and life quality of lung cancer (LC) patients. Changes of peripheral blood DNA methylation could be associated with malignancy but were mostly studied in Caucasians.
Methods
Here, in a Chinese population, we performed mass spectrometry assays to investigate the association between very early stage LC and methylation levels of RAPSN in the peripheral blood by a case–control cohort using of 221 LC patients (93.2% LC at stage I) and 285 unrelated cancer free control individuals.
Results
The odds ratios (ORs) of all CpG sites were evaluated for their risk to LC using inter-quartile analyses by logistic regression. In general, we observed an association between very early LC and decreased methylation of RAPSN_CpG_1.15 and RAPSN_CpG_3.4 (referring to Q4, OR range from 1.64 to 1.81, p<0.05). Stratified by gender, while hypomethylation of RAPSN_CpG_1.15, RAPSN_CpG_3.4 and RAPSN_CpG_7.14 were associated with LC in males (referring to Q4, ORs range from 1.94 to 2.31, p<0.05), RAPSN_CpG_2 and RAPSN_CpG_5 showed significantly lower methylation in female LC patients comparing to controls (referring to Q4, ORs range from 2.49 to 3.60, p<0.05). The risk of RAPSN hypomethylation to LC was enhanced by aging, and typically for people older than 55 years (referring to Q4, ORs range from 2.17 to 3.61 in six out of all 10 analyzed CpG groups, p<0.05).
Conclusion
Our study reveals an association between RAPSN hypomethylation in peripheral blood and LC and suggests the occurrence of altered blood-based methylation at the early stage of cancer.
Keywords: LC, early detection, DNA methylation, RAPSN, peripheral blood
Background
Lung cancer (LC) is malignancy-associated tumour occurring in the bronchial mucosal epithelium. The subgroup of LC mainly contains small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), whereas NSCLC accounts for 80–85% of all LC cases including adenocarcinoma, large cell carcinoma and squamous cell carcinoma.1–4 LC is the leading cause of cancer incidence and mortality worldwide, with increasing rates in recent decades.5 The prognosis of LC is associated with stage at initial diagnosis, with 5-year survival rates of stage I LC at 83%, stage II at 53%, stage III at 26% and stage IV at 6%.6 Taking advantage of the improved diagnostic methods, novel surgical techniques and chemotherapy drugs, the 5-year overall survival rate of LC patients is still only 16%, mainly due to the initial diagnosis at advanced stage which has already developed metastases or involved the lymph nodes.5 The key to improve the survival of LC patients is early diagnosis and following proper treatment.
As the principal targets of genetic changes and the principal drivers for distant metastases, epigenetic changes play an essential role in cancer and occur early in the progress of carcinogenesis.7 DNA methylation is one of the major signatures of epigenetic changes.8 Aberrant DNA methylation level is critical for carcinogenesis, for example, the hypomethylation of oncogenes and hypermethylation of tumor suppressor genes.9–11 DNA methylation is often associated with specific tissues and cell-types, recent studies also showed the associations between aberrant methylation in blood and cancers.12–14 Hypermethylation of DOK7, hypomethylation of HYAL2 and BRCA1 present the potentials as blood-based DNA methylation biomarkers for the screening of cancer.15–17 Hypermethylation of HOXA11 was reported for the diagnosis and prognosis of LC.18
Receptor-associated protein of the synapse (RAPSN) gene encodes 43 kDa receptor-associated protein of the synapse that is required for clustering of nicotinic acetylcholine receptors (nAChRs) at the neuromuscular junction (NMJ). RAPSN protein mainly distributes in cytoskeleton or postsynaptic membrane. RAPSN mutations could compromise neuromuscular transmission, and consequently cause congenital myasthenic syndrome (CMS).19 Tang et al have reported a connection between RAPSN hypomethylation in peripheral blood and breast cancer in Caucasian population.20 We hereby investigate the association between the RAPSN methylation level in blood and LC in Chinese population in a case–control study with 221 very early stage LC patients and 285 unrelated healthy control individuals using quantitative mass spectrometry.
Materials and Methods
Study Populations
This study was approved by the Ethics Committee of the Shanghai Chest Hospital and Jiangsu Provincial Hospital of Traditional Chinese Medicine in China with the approve ID of KS1407. This study was conducted in accordance with the Declaration of Helsinki. All the recruited people gave written informed consent. In the case cohort, peripheral blood from a total of 221 very early stage LC patients (93.2% at stage I, 4.5% at stage II, 2.3% at stage III) were collected at Shanghai Chest Hospital in the year of 2018. The diagnosis of LC was confirmed by thoracic surgery and pathology, and these blood samples were collected before surgery and any cancer related treatments. The cohort of LC patients has an average age of 55 years with 100 females (45.2%) and 121 males (54.8%). The clinical details of LC patients were shown in Table 1. Peripheral blood samples from unrelated self-reported healthy individuals were consecutively collected from physical examination center at the Jiangsu Provincial Hospital of Traditional Chinese Medicine in 2018. To match the LC cases, 143 females and 142 males were selected as controls with an average age of 55 years. No further conditions were used to match the cases and controls in this study. Due to the limitation of hospital based sample collection, environmental and life style factors, such as smoking and drinking habit, diet and nutrition, are not available in this study.
Table 1.
Clinical Characteristics of LC Patients
| Sample Characteristics | Type | N | % |
|---|---|---|---|
| Stage | I | 206 | 93.2% |
| II | 10 | 4.5% | |
| III | 5 | 2.3% | |
| Tumour Size | T1 | 192 | 86.9% |
| T2 | 24 | 10.9% | |
| T3 | 5 | 2.3% | |
| Lymph Node Involvement | pN0 | 214 | 96.8% |
| pN1andpN2 | 7 | 3.2% | |
| Tumour Length | ≤1 cm | 95 | 43.0% |
| >1 cm | 126 | 57.0% | |
| Age | <55 | 105 | 47.5% |
| ≥55 | 116 | 52.5% | |
| Gender | Male | 121 | 54.8% |
| Female | 100 | 45.2% |
Abbreviations: LC, lung cancer; N, number.
Sample Processing
All the peripheral blood samples of cases and controls were collected by Ethylene Diamine Tetraacetic Acid (EDTA) tubes. The blood samples were kept at 4 °C for less than 24 h and further stored at −80 °C till usage. The DNA was extracted from blood using the DNA Extraction Kit (TANTICA, Nanjing, China). The samples from LC cases and controls were processed in parallel.
Bisulfite Conversion
DNA extracted from each sample was bisulfite converted by EZ-96 DNA Methylation Gold Kit (Zymo Research, Orange, USA) according to the manufacturer’s instructions, and further amplified by bisulfite-specific primers (Table S1). After bisulfite treatment, all non-methylated cytosine (C) bases in CpG sites were converted to uracil (U), whereas all methylated C bases remained C. There are no single nucleotide polymorphisms (SNPs) or CpG sites in the primers and in the measured CpGs.
MALDI-TOF Mass Spectrometry
According to the benchmarking study of absolute DNA methylation techniques by Bock and colleagues,21 Mass spectrometric analysis of DNA methylation (EpiTyper assay) has comparable performance as pyrosequencing and bisulfite sequencing, and is a stable and mature locus-specific assay for the quantitative DNA methylation analysis and biomarker development. EpiTyper assay (Agena Bioscience, San Diego, USA) described by Yang et al was utilized to quantitatively measure the levels of methylation.15 In brief, the PCR amplified products of bisulfite-converted DNA were incubated with Shrimp Alkaline Phosphatase (SAP) and further transcribed to RNA by T7 transcriptase according to the standard protocol of Agena EpiTyper assay. The RNA was digested by RNase into small fragments and then by resin to get rid of ions. The final products were dispensed on a SpectroCHIP G384 by a Nanodispenser RS1000 apparatus (Agena, USA). DNA methylation levels were determined by comparing the intensities which contained methylated and non-methylated segments. We analyzed methylation levels on Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. The chips were read using a MassARRAY system (Agena Bioscience). The data were collected by SpectroACQUIRE v3.3.1.3 software and visualized by EpiTyper v1.3 software. The EpiTyper v1.3 software automatically calculate methylation levels quantitatively of each CpG site in the investigated amplicon. The MassArray generated measurable data for 14 CpG sites in the amplicon of RAPSN, and yielded 10 distinguishable peaks (Figure 1). On one hand, RAPSN_CpG_3.4 means the CpG 3 and CpG 4 located at the same fragment after the treatment of EpiTyper, and thus the methylation levels of CpG 3 and CpG 4 were presented as an average, which is applied to RAPSN_CpG_10.11 as well. On the other hand, a few fragments sharing the same mass, and to be brief we hereby showed the data as combined, such as RAPSN_CpG_1 and RAPSN_CpG_15, RAPSN_CpG_7 and RAPSN_CpG_14.
Figure 1.
Schematic diagram and sequence of RAPSN amplicon. (A) The locations of investigated 393 bp amplicon and the 14 measurable CpG sites; (B) Sequences of the RAPSN amplicon examined by EpiTyper assay (chr11:47,470,566–47,470,958, build 37/hg19, defined by the UCSC Genome Browser). The EpiTyper assay determined the methylation levels of 14 CpGs in this amplicon, and yielded 10 distinguishable peaks. The 14 measurable CpG sites are in light grey, and the undetectable CpG sites are in underline.
Statistical Analyses
All the statistical data were analyzed using SPSS Statistical 23 software. The differences between two or three groups were assessed by non-parametric tests or logistic regression models. The logistic regressions were adjusted for age, gender and experimental batches in this study. All statistical analyses were two-sided tested, and p values<0.05 were defined as significant difference.
Results
The Association Between RAPSN Methylation and Early Stage LC
To evaluate the association between the methylation levels of RAPSN in the peripheral blood DNA and LC, an 393 bp amplicon covering 10 distinguished CpG groups (CpG_1.15, CpG_2, CpG_3.4, CpG_5, CpG_6, CpG_7.14, CpG_8, CpG_9, CpG_10.11, CpG_12) were designed and analyzed in a case–control study with 221 very early stage LC cases and 285 age and gender matched healthy controls by Agena MALDI-TOF mass spectrometry. To highlight, 93.2% LC patients were collected at stage I before surgery and any cancer related treatments. Inter quartile ranges (IQR) were conducted to evaluate the ORs of all CpG sites in the RAPSN amplicon for the risk of LC by logistic regression adjusted for age, gender and experimental batches. The ORs of cases vs controls for CpG_1.15 and CpG_3.4 were 1.64 (95% CI=1.04–2.59, p=0.032) and 1.81 (95% CI=1.12–2.93, p=0.016) in Q3 compared to the highest quartile of methylation level (Q4), respectively (Table 2). Moreover, CpG_3.4 in the lowest quartile of methylation levels (Q1) showed significant association with increasing risk of LC comparing with the highest methylation quartile (Q4) (OR=1.81, 95% CI=1.12–2.93, p=0.036, Table 2). The rest CpG sites in the RAPSN amplicon showed no significant association with LC in any quartiles (Table 2).
Table 2.
The Methylation Levels of RAPSN in LC Cases Comparing to Controls by Quartile Analyses
| CpG Sites | Methylation | Control (N) | Case (N) | OR (95% CI) a | p-value |
|---|---|---|---|---|---|
| RAPSN_CpG_1.15 | Q4 (≥0.96) | 97 | 63 | 1.00 (reference) | - |
| Q3 (0.94–0.96) | 76 | 79 | 1.64 (1.04–2.59) | 0.032 | |
| Q2 (0.92–0.94) | 47 | 41 | 1.37 (0.80–2.33) | 0.249 | |
| Q1 (<0.92) | 62 | 38 | 0.93 (0.55–1.55) | 0.774 | |
| RAPSN_CpG_2 | Q4 (≥0.89) | 76 | 50 | 1.00 (reference) | - |
| Q3 (0.81–0.89) | 68 | 56 | 1.33 (0.80–2.20) | 0.278 | |
| Q2 (0.77–0.82) | 70 | 66 | 1.59 (0.96–2.65) | 0.074 | |
| Q1 (<0.77) | 68 | 49 | 1.20 (0.71–2.04) | 0.491 | |
| RAPSN_CpG_3.4 | Q4 (≥0.98) | 82 | 46 | 1.00 (reference) | - |
| Q3 (0.96–0.98) | 82 | 80 | 1.81 (1.12–2.93) | 0.016 | |
| Q2 (0.94–0.96) | 71 | 51 | 1.33 (0.79–2.22) | 0.285 | |
| Q1 (<0.94) | 47 | 44 | 1.81 (1.04–3.17) | 0.036 | |
| RAPSN_CpG_5 | Q4 (≥0.99) | 73 | 41 | 1.00 (reference) | - |
| Q3 (0.96–0.99) | 101 | 81 | 1.49 (0.91–2.42) | 0.110 | |
| Q2 (0.94–0.96) | 60 | 53 | 1.62 (0.95–2.79) | 0.078 | |
| Q1 (<0.94) | 48 | 46 | 1.71 (0.98–2.99) | 0.060 | |
| RAPSN_CpG_6 | Q4 (≥0.96) | 72 | 60 | 1.00 (reference) | - |
| Q3 (0.89–0.96) | 79 | 63 | 0.96 (0.59–1.54) | 0.853 | |
| Q2 (0.83–0.89) | 59 | 52 | 1.07 (0.64–1.77) | 0.807 | |
| Q1 (<0.83) | 71 | 46 | 0.73 (0.44–1.22) | 0.237 | |
| RAPSN_CpG_7.14 | Q4 (≥0.93) | 98 | 60 | 1.00 (reference) | - |
| Q3 (0.91–0.93) | 73 | 68 | 1.48 (0.93–2.37) | 0.098 | |
| Q2 (0.89–0.91) | 59 | 48 | 1.31 (0.79–2.17) | 0.292 | |
| Q1 (<0.89) | 52 | 45 | 1.39 (0.83–2.32) | 0.213 | |
| RAPSN_CpG_8 | Q4 (≥0.99) | 106 | 85 | 1.00 (reference) | - |
| Q3 (0.98–0.99) | 49 | 41 | 1.02 (0.61–1.71) | 0.928 | |
| Q2 (0.96–0.98) | 76 | 56 | 0.92 (0.59–1.45) | 0.735 | |
| Q1 (<0.96) | 51 | 39 | 0.94 (0.57–1.57) | 0.821 | |
| RAPSN_CpG_9 | Q4 (≥0.92) | 72 | 54 | 1.00 (reference) | – |
| Q3 (0.88–0.92) | 76 | 77 | 1.36 (0.84–2.20) | 0.213 | |
| Q2 (0.85–0.88) | 65 | 44 | 0.92 (0.54–1.56) | 0.762 | |
| Q1 (<0.85) | 69 | 46 | 0.89 (0.53–1.49) | 0.656 | |
| RAPSN_CpG_10.11 | Q4 (≥0.91) | 81 | 57 | 1.00 (reference) | – |
| Q3 (0.88–0.91) | 73 | 56 | 1.08 (0.66–1.76) | 0.761 | |
| Q2 (0.85–0.88) | 70 | 61 | 1.26 (0.77–2.06) | 0.353 | |
| Q1 (<0.85) | 58 | 47 | 1.14 (0.68–1.91) | 0.625 | |
| RAPSN_CpG_12 | Q4 (≥0.95) | 78 | 66 | 1.00 (reference) | – |
| Q3 (0.92–0.95) | 82 | 78 | 1.14 (0.72–1.79) | 0.577 | |
| Q2 (0.89–0.92) | 69 | 41 | 0.69 (0.41–1.15) | 0.157 | |
| Q1 (<0.89) | 53 | 36 | 0.79 (0.46–1.35) | 0.381 |
Note: alogistic regression adjusted for age, gender and experimental batches. p < 0.05 was considered as significant difference.
Abbreviations: LC, lung cancer; N, number; OR, odd ratio; CI, confidence interval.
The Influence of Gender and Age on the Methylation Levels of RAPSN
Gender and age were reported to play essential roles in the pattern of DNA methylation.22,23 We further investigated the influence of gender and age to the methylation levels of RAPSN in LC cases and controls respectively. Interestingly, the methylation levels of RAPSN CpG sites showed significant differences between the two genders and between the age groups only in controls, but not in LC cases (Table S2). Thus, we further investigated the association between RAPSN methylation and LC stratified by gender and age groups.
The association between RAPSN methylation and LC in males and females were analyzed respectively by inter-quartile analysis using logistic regression adjusted for age and experimental batches. Interestingly, the LC associated RAPSN CpG sites variated by gender. In males, CpG_1.15, CpG_3.4 and CpG_7.14 showed significant differences between LC cases and controls but mostly in Q2 or Q3 comparing to the highest methylation quartile (Q4) with the ORs ranging from 1.94 to 2.31 (p < 0.05 for all, Table 3). In females, LC cases showed significant lower methylation at CpG_2 and CpG_5 than controls by comparing lower quartiles to the highest methylation (Q4) with ORs ranging from 2.49 to 3.60 (p < 0.05 for all, Table 4), but had no association at CpG_1.15, CpG_3.4 and CpG_7.14.
Table 3.
The Difference of RAPSN Methylation Levels Between Male LC Cases and Controls by Quartile Analyses
| CpG Sites | Methylation | Control (N) | Case (N) | OR (95% CI) a | p-value |
|---|---|---|---|---|---|
| RAPSN_CpG_1.15 | Q4 (≥0.96) | 48 | 30 | 1.00 (reference) | - |
| Q3 (0.94–0.96) | 35 | 48 | 2.31 (1.22–4.38) | 0.011 | |
| Q2 (0.92–0.94) | 26 | 26 | 1.67 (0.81–3.42) | 0.162 | |
| Q1 (<0.92) | 33 | 17 | 0.78 (0.37–1.64) | 0.507 | |
| RAPSN_CpG_2 | Q4 (≥0.89) | 38 | 36 | 1.00 (reference) | - |
| Q3 (0.81–0.89) | 37 | 31 | 0.95 (0.48–1.88) | 0.879 | |
| Q2 (0.77–0.81) | 32 | 31 | 1.15 (0.56–2.36) | 0.694 | |
| Q1 (<0.77) | 35 | 23 | 0.79 (0.37–1.67) | 0.535 | |
| RAPSN_CpG_3.4 | Q4 (≥0.98) | 43 | 25 | 1.00 (reference) | - |
| Q3 (0.96–0.98) | 43 | 46 | 1.94 (1.01–3.74) | 0.047 | |
| Q2 (0.95–0.96) | 21 | 20 | 1.71 (0.77–3.77) | 0.185 | |
| Q1 (<0.95) | 35 | 30 | 1.62 (0.80–3.29) | 0.183 | |
| RAPSN_CpG_5 | Q4 (≥0.98) | 44 | 38 | 1.00 (reference) | - |
| Q3 (0.96–0.98) | 39 | 23 | 0.70 (0.36–1.38) | 0.304 | |
| Q2 (0.94–0.96) | 19 | 15 | 0.94 (0.42–2.11) | 0.885 | |
| Q1 (<0.94) | 40 | 45 | 1.31 (0.71–2.41) | 0.393 | |
| RAPSN_CpG_6 | Q4 (≥0.95) | 37 | 36 | 1.00 (reference) | - |
| Q3 (0.88–0.95) | 36 | 38 | 1.07 (0.56–2.05) | 0.833 | |
| Q2 (0.82–0.88) | 38 | 22 | 0.56 (0.28–1.14) | 0.109 | |
| Q1 (<0.82) | 31 | 25 | 0.79 (0.39–1.60) | 0.513 | |
| RAPSN_CpG_7.14 | Q4 (≥0.93) | 44 | 27 | 1.00 (reference) | - |
| Q3 (0.91–0.93) | 44 | 39 | 1.43 (0.75–2.73) | 0.282 | |
| Q2 (0.89–0.91) | 24 | 31 | 2.07 (1.01–4.26) | 0.047 | |
| Q1 (<0.89) | 30 | 24 | 1.31 (0.64–2.70) | 0.462 | |
| RAPSN_CpG_8 | Q4 (≥0.99) | 49 | 46 | 1.00 (reference) | - |
| Q3 (0.98–0.99) | 29 | 27 | 1.00 (0.52–1.94) | 1.000 | |
| Q2 (0.96–0.98) | 38 | 26 | 0.75 (0.39–1.42) | 0.374 | |
| Q1 (<0.96) | 26 | 22 | 0.89 (0.44–1.79) | 0.743 | |
| RAPSN_CpG_9 | Q4 (≥0.91) | 44 | 40 | 1.00 (reference) | - |
| Q3 (0.88–0.91) | 31 | 36 | 1.30 (0.68–2.48) | 0.425 | |
| Q2 (0.85–0.88) | 36 | 18 | 0.56 (0.28–1.14) | 0.112 | |
| Q1 (<0.85) | 31 | 27 | 0.96 (0.49–1.89) | 0.914 | |
| RAPSN_CpG_10.11 | Q4 (≥0.90) | 43 | 34 | 1.00 (reference) | - |
| Q3 (0.88–0.90) | 28 | 23 | 1.09 (0.53–2.25) | 0.809 | |
| Q2 (0.85–0.88) | 36 | 37 | 1.48 (0.76–2.88) | 0.254 | |
| Q1 (<0.85) | 35 | 27 | 1.02 (0.52–2.00) | 0.964 | |
| RAPSN_CpG_12 | Q4 (≥0.95) | 37 | 36 | 1.00 (reference) | – |
| Q3 (0.92–0.95) | 41 | 41 | 1.07 (0.56–2.01) | 0.846 | |
| Q2 (0.89–0.92) | 37 | 25 | 0.68 (0.34–1.36) | 0.277 | |
| Q1 (<0.89) | 27 | 19 | 0.70 (0.33–1.48) | 0.348 |
Note: alogistic regression adjusted for age, gender and experimental batches; p < 0.05 was considered as significant difference.
Abbreviations: LC, lung cancer; N, number; OR, odd ratio; CI, confidence interval.
Table 4.
The Difference of RAPSN Methylation Levels Between Female LC Cases and Controls by Quartile Analyses
| CpG Sites | Methylation | Control (N) | Case (N) | OR (95% CI) a | p-value |
|---|---|---|---|---|---|
| RAPSN_CpG_1.15 | Q4 (≥0.97) | 36 | 19 | 1.00 (reference) | - |
| Q3 (0.94–0.97) | 54 | 45 | 1.86 (0.88–3.93) | 0.105 | |
| Q2 (0.92–0.94) | 21 | 15 | 1.61 (0.64–4.05) | 0.313 | |
| Q1 (<0.92) | 29 | 21 | 1.47 (0.66–3.28) | 0.346 | |
| RAPSN_CpG_2 | Q4 (≥0.89) | 38 | 14 | 1.00 (reference) | - |
| Q3 (0.83–0.89) | 27 | 16 | 1.86 (0.76–4.56) | 0.174 | |
| Q2 (0.77–0.83) | 42 | 44 | 3.60 (1.60–8.08) | 0.002 | |
| Q1 (<0.77) | 33 | 26 | 2.49 (1.09–5.72) | 0.031 | |
| RAPSN_CpG_3.4 | Q4 (≥0.98) | 39 | 21 | 1.00 (reference) | - |
| Q3 (0.96–0.98) | 39 | 34 | 1.69 (0.83–3.44) | 0.151 | |
| Q2 (0.94–0.96) | 33 | 21 | 1.26 (0.57–2.76) | 0.564 | |
| Q1 (<0.94) | 29 | 24 | 1.59 (0.74–3.40) | 0.237 | |
| RAPSN_CpG_5 | Q4 (≥0.99) | 39 | 15 | 1.00 (reference) | - |
| Q3 (0.96–0.99) | 52 | 46 | 2.55 (1.21–5.35) | 0.014 | |
| Q2 (0.95–0.96) | 16 | 14 | 2.61 (0.99–6.85) | 0.052 | |
| Q1 (<0.95) | 33 | 25 | 2.11 (0.94–4.70) | 0.069 | |
| RAPSN_CpG_6 | Q4 (≥0.96) | 39 | 28 | 1.00 (reference) | - |
| Q3 (0.90–0.96) | 35 | 25 | 0.99 (0.49–2.01) | 0.976 | |
| Q2 (0.86–0.90) | 30 | 26 | 1.18 (0.57–2.45) | 0.651 | |
| Q1 (<0.86) | 35 | 21 | 0.83 (0.40–1.72) | 0.614 | |
| RAPSN_CpG_7.14 | Q4 (≥0.94) | 37 | 25 | 1.00 (reference) | - |
| Q3 (0.91–0.94) | 46 | 37 | 1.23 (0.61–2.45) | 0.564 | |
| Q2 (0.89–0.91) | 35 | 17 | 0.74 (0.33–1.64) | 0.461 | |
| Q1 (<0.89) | 22 | 21 | 1.42 (0.64–3.15) | 0.390 | |
| RAPSN_CpG_8 | Q4 (≥0.99) | 57 | 39 | 1.00 (reference) | - |
| Q3 (0.98–0.99) | 20 | 14 | 1.10 (0.48–2.54) | 0.825 | |
| Q2 (0.96–0.98) | 38 | 30 | 1.24 (0.63–2.45) | 0.540 | |
| Q1 (<0.96) | 25 | 17 | 1.01 (0.48–2.11) | 0.986 | |
| RAPSN_CpG_9 | Q4 (≥0.92) | 38 | 24 | 1.00 (reference) | - |
| Q3 (0.88–0.92) | 35 | 31 | 1.60 (0.76–3.36) | 0.213 | |
| Q2 (0.84–0.88) | 34 | 32 | 1.68 (0.81–3.48) | 0.165 | |
| Q1 (<0.84) | 33 | 13 | 0.61 (0.27–1.40) | 0.246 | |
| RAPSN_CpG_10.11 | Q4 (≥0.92) | 41 | 24 | 1.00 (reference) | - |
| Q3 (0.89–0.92) | 29 | 26 | 1.53 (0.74–3.19) | 0.255 | |
| Q2 (0.86–0.89) | 37 | 24 | 1.11 (0.54–2.30) | 0.774 | |
| Q1 (<0.86) | 33 | 26 | 1.35 (0.65–2.77) | 0.421 | |
| RAPSN_CpG_12 | Q4 (≥0.95) | 41 | 30 | 1.00 (reference) | - |
| Q3 (0.92–0.95) | 41 | 37 | 1.30 (0.66–2.53) | 0.447 | |
| Q2 (0.90–0.92) | 24 | 15 | 0.90 (0.40–2.06) | 0.807 | |
| Q1 (<0.90) | 34 | 18 | 0.73 (0.35–1.53) | 0.405 |
Note: alogistic regression adjusted for age, gender and experimental batches; p< 0.05 was considered as significant difference.
Abbreviations: LC, lung cancer; N, number; OR, odd ratio; CI, confidence interval.
Next, we investigated the association between RAPSN methylation and LC in different age groups (stratified by the average age of 55 years old in this study) by inter-quartile analyses using logistic regression adjusted for gender and experimental batches. In the group of younger than 55 years, among the 10 distinguished CpG groups, only CpG_7.14 showed significantly lower methylation in LC cases than controls by comparing Q1 to the highest methylation quartile (Q4) with an OR of 2.26 (p=0.036, Table 5). In the group of older than or equal to 55 years old, LC cases showed significant lower methylation at six out of 10 measured CpG groups (CpG_1.15, CpG_2, CpG_5, CpG_7.14, CpG_8 and CpG_12) than controls with ORs for lower quartiles vs highest quartile (Q4) ranging from 2.17 to 3.61 (p < 0.05 for all, Table 6). These results suggested that decreased methylation levels in RAPSN may have higher risk to LC in elder people.
Table 5.
RAPSN Methylation Differences Between LC Cases and Controls (Age<55 Years Old)
| CpG Sites | Methylation | Control (N) | Case (N) | OR (95% CI) a | p-value |
|---|---|---|---|---|---|
| RAPSN_CpG_1.15 | Q4 (≥0.95) | 57 | 51 | 1.00 (reference) | - |
| Q3 (0.94–0.95) | 30 | 14 | 0.50 (0.24–1.06) | 0.072 | |
| Q2 (0.92–0.94) | 28 | 21 | 0.85 (0.42–1.68) | 0.633 | |
| Q1 (<0.92) | 33 | 19 | 0.55 (0.27–1.12) | 0.101 | |
| RAPSN_CpG_2 | Q4 (≥0.87) | 39 | 21 | 1.00 (reference) | - |
| Q3 (0.80–0.87) | 47 | 37 | 1.47 (0.73–2.94) | 0.279 | |
| Q2 (0.77–0.80) | 28 | 25 | 1.76 (0.81–3.84) | 0.152 | |
| Q1 (<0.77) | 34 | 22 | 1.25 (0.58–2.72) | 0.569 | |
| RAPSN_CpG_3.4 | Q4 (≥0.97) | 58 | 36 | 1.00 (reference) | - |
| Q3 (0.96–0.97) | 24 | 21 | 1.48 (0.71–3.05) | 0.294 | |
| Q2 (0.94–0.96) | 42 | 25 | 1.00 (0.52–1.92) | 0.997 | |
| Q1 (<0.94) | 24 | 23 | 1.57 (0.76–3.21) | 0.220 | |
| RAPSN_CpG_5 | Q4 (≥0.98) | 39 | 30 | 1.00 (reference) | - |
| Q3 (0.96–0.98) | 51 | 27 | 0.64 (0.32–1.26) | 0.194 | |
| Q2 (0.95–0.96) | 22 | 13 | 0.77 (0.33–1.80) | 0.552 | |
| Q1 (<0.95) | 36 | 35 | 1.30 (0.66–2.55) | 0.446 | |
| RAPSN_CpG_6 | Q4 (≥0.96) | 41 | 27 | 1.00 (reference) | - |
| Q3 (0.89–0.96) | 42 | 31 | 1.10 (0.56–2.17) | 0.783 | |
| Q2 (0.84–0.89) | 29 | 28 | 1.43 (0.70–2.93) | 0.330 | |
| Q1 (<0.84) | 36 | 19 | 0.80 (0.38–1.71) | 0.571 | |
| RAPSN_CpG_7.14 | Q4 (≥0.93) | 48 | 30 | 1.00 (reference) | - |
| Q3 (0.91–0.93) | 40 | 30 | 1.20 (0.62–2.34) | 0.585 | |
| Q2 (0.89–0.91) | 42 | 18 | 0.65 (0.31–1.36) | 0.251 | |
| Q1 (<0.89) | 18 | 27 | 2.26 (1.05–4.84) | 0.036 | |
| RAPSN_CpG_8 | Q4 (≥0.99) | 47 | 42 | 1.00 (reference) | - |
| Q3 (0.97–0.99) | 48 | 31 | 0.72 (0.38–1.35) | 0.303 | |
| Q2 (0.96–0.97) | 25 | 14 | 0.61 (0.28–1.35) | 0.226 | |
| Q1 (<0.96) | 28 | 18 | 0.73 (0.35–1.54) | 0.414 | |
| RAPSN_CpG_9 | Q4 (≥0.91) | 48 | 31 | 1.00 (reference) | - |
| Q3 (0.88–0.91) | 35 | 30 | 1.36 (0.69–2.68) | 0.368 | |
| Q2 (0.85–0.88) | 36 | 23 | 0.97 (0.48–1.97) | 0.938 | |
| Q1 (<0.85) | 29 | 21 | 1.11 (0.53–2.30) | 0.785 | |
| RAPSN_CpG_10.11 | Q4 (≥0.91) | 40 | 26 | 1.00 (reference) | - |
| Q3 (0.88–0.91) | 44 | 33 | 1.25 (0.63–2.49) | 0.521 | |
| Q2 (0.85–0.88) | 33 | 28 | 1.37 (0.67–2.81) | 0.387 | |
| Q1 (<0.85) | 31 | 18 | 0.97 (0.44–2.14) | 0.947 | |
| RAPSN_CpG_12 | Q4 (≥0.94) | 46 | 41 | 1.00 (reference) | - |
| Q3 (0.92–0.94) | 38 | 30 | 0.90 (0.47–1.72) | 0.752 | |
| Q2 (0.90–0.92) | 31 | 18 | 0.70 (0.34–1.44) | 0.329 | |
| Q1 (<0.90) | 33 | 16 | 0.54 (0.26–1.14) | 0.105 |
Note: alogistic regression adjusted for age, gender and experimental batches; p< 0.05 was considered as significant difference.
Abbreviations: LC, lung cancer; N, number; OR, odd ratio; CI, confidence interval.
Table 6.
RAPSN Methylation Differences Between LC Cases and Controls (Age≥55 Years Old)
| CpG Sites | Methylation | Control (N) | Case (N) | OR (95% CI) a | p-value |
|---|---|---|---|---|---|
| RAPSN_CpG_1.15 | Q4 (≥0.97) | 48 | 19 | 1.00 (reference) | - |
| Q3 (0.95–0.97) | 27 | 32 | 2.78 (1.30–5.95) | 0.008 | |
| Q2 (0.92–0.95) | 30 | 46 | 3.61 (1.74–7.52) | 0.001 | |
| Q1 (<0.92) | 29 | 19 | 1.56 (0.70–3.45) | 0.275 | |
| RAPSN_CpG_2 | Q4 (≥0.93) | 37 | 18 | 1.00 (reference) | - |
| Q3 (0.84–0.93) | 32 | 30 | 1.98 (0.92–4.26) | 0.082 | |
| Q2 (0.76–0.84) | 32 | 47 | 3.39 (1.56–7.37) | 0.002 | |
| Q1 (<0.76) | 33 | 21 | 1.63 (0.70–3.79) | 0.259 | |
| RAPSN_CpG_3.4 | Q4 (≥0.98) | 47 | 28 | 1.00 (reference) | - |
| Q3 (0.96–0.98) | 35 | 41 | 1.86 (0.96–3.61) | 0.067 | |
| Q2 (0.94–0.96) | 12 | 17 | 2.07 (0.85–5.07) | 0.109 | |
| Q1 (<0.94) | 40 | 30 | 1.34 (0.68–2.67) | 0.400 | |
| RAPSN_CpG_5 | Q4 (≥0.99) | 42 | 23 | 1.00 (reference) | - |
| Q3 (0.97–0.99) | 28 | 24 | 1.37 (0.64–2.93) | 0.421 | |
| Q2 (0.94–0.97) | 27 | 34 | 2.17 (1.05–4.51) | 0.037 | |
| Q1 (<0.94) | 37 | 35 | 1.49 (0.74–3.02) | 0.266 | |
| RAPSN_CpG_6 | Q4 (≥0.95) | 34 | 37 | 1.00 (reference) | - |
| Q3 (0.89–0.95) | 34 | 28 | 0.79 (0.39–1.58) | 0.504 | |
| Q2 (0.83–0.89) | 30 | 24 | 0.77 (0.38–1.59) | 0.486 | |
| Q1 (<0.83) | 35 | 27 | 0.66 (0.33–1.33) | 0.246 | |
| RAPSN_CpG_7.14 | Q4 (≥0.94) | 28 | 12 | 1.00 (reference) | - |
| Q3 (0.91–0.94) | 55 | 56 | 2.09 (0.95–4.58) | 0.067 | |
| Q2 (0.88–0.91) | 28 | 33 | 2.45 (1.03–5.81) | 0.043 | |
| Q1 (<0.88) | 23 | 15 | 1.4 (0.54–3.65) | 0.491 | |
| RAPSN_CpG_8 | Q4 (≥1.00) | 40 | 14 | 1.00 (reference) | - |
| Q3 (0.98–1.00) | 44 | 52 | 3.02 (1.44–6.33) | 0.003 | |
| Q2 (0.96–0.98) | 27 | 29 | 2.78 (1.23–6.32) | 0.014 | |
| Q1 (<0.96) | 23 | 21 | 2.68 (1.13–6.34) | 0.025 | |
| RAPSN_CpG_9 | Q4 (≥0.92) | 36 | 29 | 1.00 (reference) | - |
| Q3 (0.87–0.92) | 43 | 47 | 1.21 (0.63–2.34) | 0.564 | |
| Q2 (0.83–0.87) | 18 | 22 | 1.33 (0.59–3.01) | 0.488 | |
| Q1 (<0.83) | 37 | 18 | 0.60 (0.28–1.28) | 0.188 | |
| RAPSN_CpG_10.11 | Q4 (≥0.91) | 41 | 31 | 1.00 (reference) | - |
| Q3 (0.88–0.91) | 29 | 23 | 1.01 (0.49–2.11) | 0.970 | |
| Q2 (0.85–0.88) | 37 | 33 | 1.17 (0.58–2.33) | 0.661 | |
| Q1 (<0.85) | 27 | 29 | 1.41 (0.69–2.89) | 0.342 | |
| RAPSN_CpG_12 | Q4 (≥0.96) | 38 | 20 | 1.00 (reference) | - |
| Q3 (0.93–0.96) | 32 | 44 | 2.33 (1.13–4.79) | 0.022 | |
| Q2 (0.89–0.93) | 34 | 29 | 1.44 (0.68–3.05) | 0.336 | |
| Q1 (<0.89) | 30 | 23 | 1.34 (0.61–2.93) | 0.460 |
Note: alogistic regression adjusted for age, gender and experimental batches; p<0.05 was considered as significant difference.
Abbreviations: LC, lung cancer; N, number; OR, odd ratio; CI, confidence interval.
The Methylation Levels of RAPAN in LC Patients with Variant Clinical Characteristics
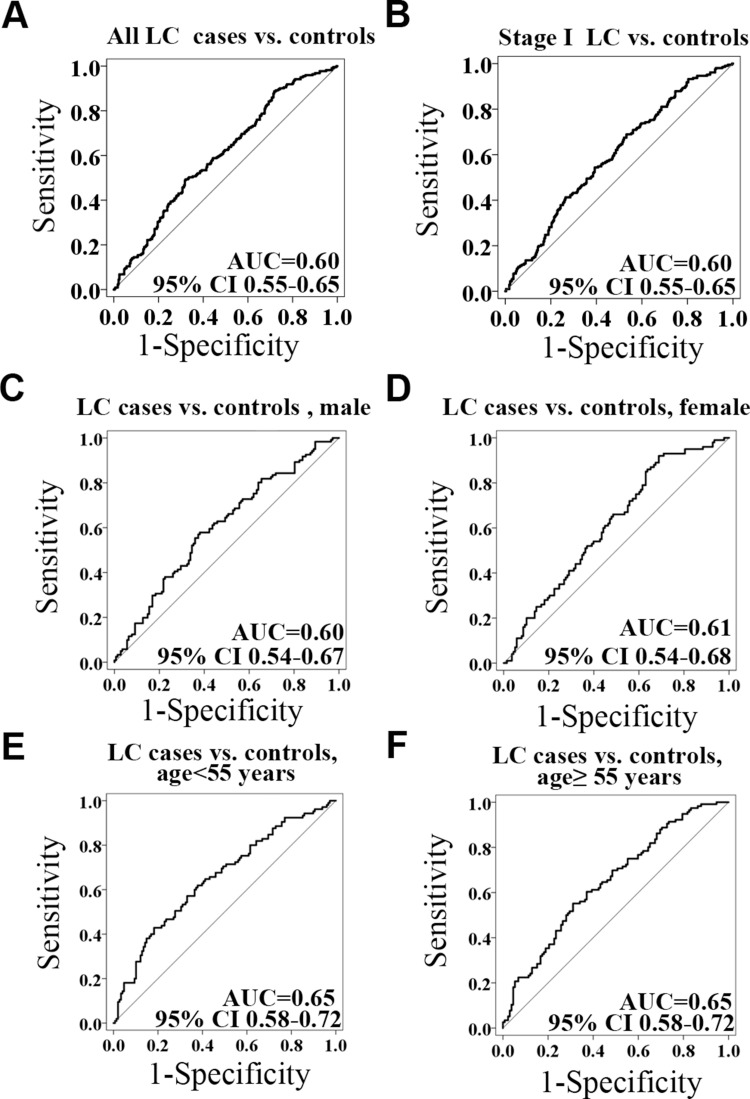
To understand the methylation patterns in the blood of LC, the methylation levels of RAPSN were further analyzed in LC patients stratified by different clinical characteristics. The methylation levels of CpG_7.14 of RAPSN was significantly lower in the LC patients with involved lymph nodes compared to the ones without (p=0.001, Table 7). However, due to the very limited sample size of pN1andpN2 patients, this result needs to be taken with caution. The methylation levels of other nine RAPSN CpG groups showed no significant difference among LC patients with variant tumour subtypes, stage, size, lymph node and tumour length. To estimate the potential clinical utility of RAPSN methylation as a marker for the detection of early LC, receiver operating characteristic (ROC) curve analyses were performed adjusted for possible confounding effects by logistic regression. When all measurable RAPSN CpG sites were considered, the methylation level of RAPSN showed a moderate discriminatory power for differentiating LC cases from cancer free controls (adjusted for age, gender and experimental batches, area under curve [AUC] =0.60, Figure 2A). To note, the discriminatory power of RAPSN methylation for the Stage I LC is comparable for the overall LC with an AUC of 0.61 (Figure 2B). The discrimination was not significantly improved when analyzed in separate genders (Figure 2C and D), but was influenced by age with increased AUC of 0.65 (Figure 2E and F).
Table 7.
RAPSN Methylation Levels in LC Patients with Different Clinical Characteristics
| Clinical | Group (N) | Methylation Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG _1.15 |
CpG _2 |
CpG _3.4 |
CpG _5 |
CpG _6 |
CpG _7.14 |
CpG _8 |
CpG _9 |
CpG _10.11 |
CpG _12 |
||
| Tumour Subtypes | adenocarcinoma in situ (47) | 0.94 | 0.83 | 0.96 | 0.96 | 0.88 | 0.91 | 0.98 | 0.88 | 0.88 | 0.93 |
| microinvasive adenocarcinoma (55) | 0.95 | 0.80 | 0.96 | 0.96 | 0.89 | 0.90 | 0.98 | 0.88 | 0.88 | 0.92 | |
| Invasive adenocarcinoma (119) | 0.94 | 0.81 | 0.96 | 0.96 | 0.90 | 0.91 | 0.98 | 0.89 | 0.87 | 0.93 | |
| p-value | 0.711 | 0.431 | 0.389 | 0.720 | 0.218 | 0.075 | 0.081 | 0.929 | 0.309 | 0.572 | |
| Tumour Size | T1 (192) | 0.94 | 0.81 | 0.96 | 0.96 | 0.90 | 0.91 | 0.98 | 0.88 | 0.88 | 0.93 |
| T2&T3 (29) | 0.94 | 0.82 | 0.96 | 0.96 | 0.91 | 0.91 | 0.98 | 0.90 | 0.87 | 0.94 | |
| p-value | 0.153 | 0.708 | 0.407 | 0.670 | 0.703 | 0.341 | 0.700 | 0.402 | 0.604 | 0.852 | |
| Tumour Length | ≤1cm (95) | 0.94 | 0.82 | 0.96 | 0.96 | 0.90 | 0.91 | 0.98 | 0.88 | 0.88 | 0.93 |
| >1cm (126) | 0.94 | 0.81 | 0.96 | 0.96 | 0.90 | 0.91 | 0.98 | 0.89 | 0.87 | 0.93 | |
| p-value | 0.878 | 0.484 | 0.932 | 0.262 | 0.704 | 0.273 | 0.125 | 0.565 | 0.191 | 0.320 | |
| Lymph Node | pN0 (214) | 0.94 | 0.81 | 0.96 | 0.96 | 0.9 | 0.91 | 0.98 | 0.89 | 0.88 | 0.93 |
| pN1andpN2 (7) | 0.94 | 0.87 | 0.97 | 0.96 | 0.85 | 0.86 | 0.98 | 0.87 | 0.86 | 0.94 | |
| p-value | 0.870 | 0.639 | 0.078 | 0.642 | 0.777 | 0.001 | 0.602 | 0.586 | 0.204 | 0.998 | |
| Tumour Stage | Stage I (206) | 0.94 | 0.81 | 0.96 | 0.96 | 0.90 | 0.91 | 0.98 | 0.88 | 0.88 | 0.93 |
| Stage II&III (15) | 0.94 | 0.86 | 0.96 | 0.95 | 0.90 | 0.90 | 0.98 | 0.89 | 0.87 | 0.94 | |
| p-value | 0.273 | 0.483 | 0.592 | 0.728 | 0.925 | 0.138 | 0.699 | 0.988 | 0.478 | 0.905 | |
Abbreviation: LC, lung cancer.
Figure 2.
The power of the RAPSN methylation to distinguish LC cases from controls. (A) The discrimination of LC cases from controls by RAPSN methylation. (B) The discrimination of stage I LC cases from controls by RAPSN methylation. (C–F) The methylation levels of all the measurable CpG sites in RAPSN distinguish LC cases from controls in females, males, in < 55 years old group and in ≥ 55 years old group, respectively. Logistic regression analysis was applied adjusted for age, gender and experimental batches.
Discussion
Lung cancer is a leading lethal cause of cancer globally and responsible for about 18.4% of all cancer deaths.24 Because of lacking effective methods for early detection, most LC patients were diagnosed at advanced stage or even had developed metastases. Therefore, the key way to improve the survival of LC is early detection. In our previous study, we reported an association between decreased RAPSN methylation level in peripheral blood and breast cancer.20 Here, we investigated the methylation levels of RAPSN in LC patients and healthy controls, and found a weak association between blood-based RAPSN hypomethylation and LC at very early stage.
RAPSN gene encoded rapsyn which is 43 kDa receptor-associated scaffold protein required for acetylcholine receptors (AChRs) clustering.25 Knock-in N88K in mice, a prevalent congenital myasthenic syndrome (CMS) mutation of Rapsyn, AChRs and other specific synaptic proteins fail to cluster on the muscle postsynaptic domain, and consequently mice die immediately after birth.26 In addition, rapsyn mutations also cause a decrease in AChR levels at the neuromuscular synapse of human, leading to severe CMS.27–29 Thus, RAPSN has a strong association with CMS at the muscle endplate. Until now, most researches mainly focused on the function of RAPSN in myasthenic syndrome, and the roles of RAPSN in cancer is barely reported except our previous study on breast cancer. Cyclin-dependent kinase 5 (CDK5) is one of the member in CDK family, which links to the interaction of rapsyn in the dispersal AChR cluster pathway.30 CDK5 overexpressed in many cancers including LC.31–34 Interestingly, the expression of CDK5 was also significantly related to gender with high expression in males.35 This might indirectly explain the reason that altered RAPSN methylation as a diagnostic biomarker for LC, especially in males. Many studies found that aberrant glycosylation plays a fundamental role in pathological processes of tumor development and progression.36 Adenylate kinase 9 (AK9) is a member of adenylate kinases family,37 and participates in N-glycosylation. Lam et al reported that defective of AK9 reduces N-glycosylation of AChR and rapsyn while a defective RAPSN impairs the clustering of AChR.38 Thus, we speculated RAPSN and the methylation of RAPSN connected with LC through CDK5 by cell cycle and AK9 by N-glycosylation pathway. Still, the specify molecular mechanism of RAPSN methylation and LC might be a topic for further studies as well. The expression of RAPSN could provide additional hint for the regulatory effect of DNA methylation. However, neither the fresh blood samples nor the RNA samples is available in this study. RAPSN is highly expressed in esophagus, heart, placenta and prostate, but has low base-line expression in lung and immune system.39 The hypomethylation of RAPSN may upregulate the function of RAPSN and subsequently accelerate downstream pathways.
In the males, three of 10 CpG groups in RAPSN amplicon were connected with LC, including CpG_1.15, CpG_3.4, CpG_7.14. While in the females, only the methylation levels of CpG_2 and CpG_5 were associated with LC. The different patterns of LC associated blood-based RAPSN methylation might be affected by sex differences and gender related life styles. Tobacco smoking has been considered as one of the major cause of LC.40,41 It has been reported that the methylation levels of certain genes are robustly connected with smoking exposure.42–45 Studies also showed that alcohol intake could induce epigenetic alterations, particularly in the pattern of DNA methylation which might be connected with alcohol-induced carcinogenesis.46 In China, the men have much higher exposure to tobacco smoking and alcohol than women, which may explain the reason that the males showed higher burden of LC related altered DNA methylation. However, the information of the smoking habit is not available in our hospital-based study, and hopefully could be considered and collected in the future investigations.
As known, the risk of LC increases with age. In this study, comparing the RAPSN methylation levels between LC cases and controls in different age groups, there were six CpG groups of significantly different (CpG_1.15, CpG_2, CpG_5, CpG_7.14, CpG_ 8 and CpG_12) in elder ages (age ≥ 55 years old) and only one CpG group (CpG_7.14) in people younger than 55 years old, which suggested the hypomethylation of RAPSN may link to higher cancer burden of older people. The mechanism of RAPSN and aging warrants further studies.
IN recent years, DNA methylation patterns have been linked with variant factors, such as diet and air pollutions.47 Especially, deficient diets may cause DNA hypomethylation and high-fat diet consumption may result in the changes of DNA methylation.43,48 As global hypomethylation and gene specific hypomethylation such as RAPSN hypomethylation were frequently reported, nutrition deficient and high-fat diet might be one of the reasons for higher cancer risk in elder people due to the dysfunction of metabolisms.47 Unfortunately, we are in lack of the information of diet and exposure to polluted air, which would better be considered in future studies.
Conclusions
We previously reported the hypomethylation of RAPSN in breast cancer in the Caucasian.20 This study disclosed an association between altered RAPSN methylation and very early stage LC in Chinese population, especially in male and older people. Here, we suggested that the different cancer types may share the same blood-based DNA methylation genes, but differ at the methylation levels and specific sites due to the modification of cancer types, or ethnic backgrounds, or different life styles. This observation may simplify and speed up the development of diagnostic markers for cancers.
Acknowledgments
We are thankful for the support of Ling Wang and Zheng Zhang with sample processing. This work was supported by the Nanjing Social Supporting Department and Social Supporting Ministry of Jiangsu Province granted from 20182020 and was supported by the Nanjing TANTICA Co. Ltd with grant number 2018LC01.1.
Disclosure
Feifei Di, Jun Wang, Yujie Wei, Yanman Zhang, and Rongxi Yang are employees of Nanjing TANTICA Biotechnology Co. Ltd. The authors report no other potential conflicts of interest for this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2018. 2018;68(1):7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 2.Barnett SA, Downey RJ, Zheng J, et al. Utility of routine PET imaging to predict response and survival after induction therapy for non-small cell lung cancer. Ann Thoracic Surgery. 2016;101(3):1052–1059. doi: 10.1016/j.athoracsur.2015.09.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moro-Sibilot D, Smit E, de Castro Carpeño J, et al. Non-small cell lung cancer patients with brain metastases treated with first-line platinum-doublet chemotherapy: analysis from the European FRAME study. Lung Cancer. 2015;90(3):427–432. doi: 10.1016/j.lungcan.2015.11.011 [DOI] [PubMed] [Google Scholar]
- 4.Brody H. Lung cancer. Nature. 2014;513(7517):S1. doi: 10.1038/513S1a [DOI] [PubMed] [Google Scholar]
- 5.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 6.Chansky K, Detterbeck FC, Nicholson AG, et al. The IASLC lung cancer staging project: external validation of the revision of the tnm stage groupings in the eighth edition of the TNM classification of lung cancer. J Thoracic Oncol. 2017;12(7):1109–1121. doi: 10.1016/j.jtho.2017.04.011 [DOI] [PubMed] [Google Scholar]
- 7.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3(4):253–266. doi: 10.1038/nrc1045 [DOI] [PubMed] [Google Scholar]
- 8.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–19. doi: 10.1038/35049533 [DOI] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341 [DOI] [PubMed] [Google Scholar]
- 11.Cairns P. Gene methylation and early detection of genitourinary cancer: the road ahead. Nat Rev Cancer. 2007;7(7):531–543. [DOI] [PubMed] [Google Scholar]
- 12.Tahara T, Maegawa S, Chung W, et al. Examination of whole blood DNA methylation as a potential risk marker for gastric cancer. Cancer Prevention Research. 2013;6(10):1093–1100. doi: 10.1158/1940-6207.CAPR-13-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marsit CJ, Koestler DC, Christensen BC, Karagas MR, Houseman EA, Kelsey KT. DNA methylation array analysis identifies profiles of blood-derived DNA methylation associated with bladder cancer. J Clinical Oncology. 2011;29(9):1133–1139. doi: 10.1200/jco.2010.31.3577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinclair KD, Allegrucci C, Singh R, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc Natl Acad Sci U S A. 2007;104(49):19351–19356. doi: 10.1073/pnas.0707258104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schott S, Yang R, Stöcker S, et al. HYAL2 methylation in peripheral blood as a potential marker for the detection of pancreatic cancer: a case control study. Oncotarget. 2017;8(40):67614–67625. doi: 10.18632/oncotarget.18757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyn H, Carmona FJ, Gomez A, et al. DNA methylation profiling in breast cancer discordant identical twins identifies DOK7 as novel epigenetic biomarker. Carcinogenesis. 2013;34(1):102–108. doi: 10.1093/carcin/bgs321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto T, Yamamoto N, Taguchi T, Tamaki Y, Noguchi S. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res Treat. 2011;129(1):69–77. doi: 10.1007/s10549-010-1188-1 [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Chen C, Ren X, Sun W. DNA methylation profiling identifies the HOXA11 gene as an early diagnostic and prognostic molecular marker in human lung adenocarcinoma. Oncotarget. 2017;8(20):33100–33109. doi: 10.18632/oncotarget.16528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cossins J, Burke G, Maxwell S, et al. Diverse molecular mechanisms involved in AChR deficiency due to rapsyn mutations. Brain. 2006;129(Pt 10):2773–2783. doi: 10.1093/brain/awl219 [DOI] [PubMed] [Google Scholar]
- 20.Tang Q, Holland-Letz T, Slynko A, et al. DNA methylation array analysis identifies breast cancer associated RPTOR, MGRN1 and RAPSN hypomethylation in peripheral blood DNA. Oncotarget. 2016;7(39):64191–64202. doi: 10.18632/oncotarget.11640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christoph B, Florian H, Francisco JC, et al. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nat Biotechnol. 2016;34(7):726–737. doi: 10.1038/nbt.3605 [DOI] [PubMed] [Google Scholar]
- 22.Horvath S, Zhang Y, Langfelder P, et al. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012;13(10):R97. doi: 10.1186/gb-2012-13-10-r97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davegårdh C, Hall Wedin E, Broholm C, et al. Sex influences DNA methylation and gene expression in human skeletal muscle myoblasts and myotubes. Stem Cell Res Ther. 2019;10(1):26. doi: 10.1186/s13287-018-1118-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 25.Froehner SC, Luetje CW, Scotland PB, Patrick J. The postsynaptic 43K protein clusters muscle nicotinic acetylcholine receptors in Xenopus oocytes. Neuron. 1990;5(4):403–410. doi: 10.1016/0896-6273(90)90079-u [DOI] [PubMed] [Google Scholar]
- 26.Xing G, Jing H, Zhang L, Cao Y, Mei L. A mechanism in Agrin signaling revealed by a prevalent Rapsyn mutation in congenital myasthenic syndrome. eLife Ences. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohno K, Engel AG, Shen XM, et al. Rapsyn mutations in humans cause endplate acetylcholine-receptor deficiency and myasthenic syndrome. Am J Hum Genet. 2002;70(4):875–885. doi: 10.1086/339465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ricardo AM, Vanessa D, Samuel IP, et al. Rapsyn mutations in myasthenic syndrome due to impaired receptor clustering. Muscle Nerve. 2003;28(3):293–301. doi: 10.1002/mus.10433 [DOI] [PubMed] [Google Scholar]
- 29.Banwell BL, Ohno K, Sieb JP, Engel AG. Novel truncating RAPSN mutations causing congenital myasthenic syndrome responsive to 3,4-diaminopyridine. NMD. 2004;14(3):202–207. doi: 10.1016/j.nmd.2003.11.004 [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez Cruz PM, Palace J, Beeson D. The neuromuscular junction and wide heterogeneity of congenital myasthenic syndromes. Int J Mol Sci 2018;19(6). doi: 10.3390/ijms19061677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang Q, Li L, Zhang J, et al. CDK5 is essential for TGF-β1-induced epithelial-mesenchymal transition and breast cancer progression. Sci Rep. 2013;3:2932. doi: 10.1038/srep02932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggers JP, Grandgenett PM, Collisson EC, et al. Cyclin-dependent kinase 5 is amplified and overexpressed in pancreatic cancer and activated by mutant K-Ras. Clinical Cancer Research. 2011;17(19):6140–6150. doi: 10.1158/1078-0432.ccr-10-2288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levacque Z, Rosales JL, Lee KY. Level of cdk5 expression predicts the survival of relapsed multiple myeloma patients. Cell Cycle. 2012;11(21):4093–4095. doi: 10.4161/cc.21886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demelash A, Rudrabhatla P, Pant HC, et al. Achaete-scute homologue-1 (ASH1) stimulates migration of lung cancer cells through Cdk5/p35 pathway. Mol Biol Cell. 2012;23(15):2856–2866. doi: 10.1091/mbc.E10-12-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun YQ, Xie JW, Chen PC, et al. Low expression of CDK5 and p27 are associated with poor prognosis in patients with gastric cancer. J Cancer. 2016;7(9):1049–1056. doi: 10.7150/jca.14778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7(23):35478–35489. 10.18632/oncotarget.8155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panayiotou C, Solaroli N, Karlsson A. The many isoforms of human adenylate kinases. Int J Biochem Cell Biol. 2014;49:75–83. doi: 10.1016/j.biocel.2014.01.014 [DOI] [PubMed] [Google Scholar]
- 38.Lam CW, Wong KS, Leung HW, Law CY. Limb girdle myasthenia with digenic RAPSN and a novel disease gene AK9 mutations. EJHG. 2017;25(2):192–199. doi: 10.1038/ejhg.2016.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duff MO, Olson S, Wei X, et al. Genome-wide identification of zero nucleotide recursive splicing in Drosophila. Nature. 2015;521(7552):376–379. doi: 10.1038/nature14475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asavasupreechar T, Chan MSM, Saito R, Miki Y, Boonyaratanakornkit V, Sasano H. Sex steroid metabolism and actions in non-small cell lung carcinoma. J Steroid Biochem Mol Biol. 2019;193:105440. doi: 10.1016/j.jsbmb.2019.105440 [DOI] [PubMed] [Google Scholar]
- 41.Smith CJ, Perfetti TA, King JA. Perspectives on pulmonary inflammation and lung cancer risk in cigarette smokers. Inhal Toxicol. 2006;18(9):667–677. doi: 10.1080/08958370600742821 [DOI] [PubMed] [Google Scholar]
- 42.Bergens MA, Pittman GS, Thompson IJB, et al. Smoking-associated AHRR demethylation in cord blood DNA: impact of CD235a+ nucleated red blood cells. Clin Epigenetics 2019;11(1):87. doi: 10.1186/s13148-019-0686-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Schöttker B, Florath I, et al. Smoking-associated dna methylation biomarkers and their predictive value for all-cause and cardiovascular mortality. Environ Health Perspect. 2016;124(1):67–74. doi: 10.1289/ehp.1409020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeilinger S, Kühnel B, Klopp N, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS One. 2013;8(5):e63812. doi: 10.1371/journal.pone.0063812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the european prospective investigation into cancer and nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22(5):843–851. doi: 10.1093/hmg/dds488 [DOI] [PubMed] [Google Scholar]
- 46.Varela-Rey M, Woodhoo A, Martinez-Chantar ML, Mato JM, Lu SC. Alcohol, DNA methylation, and cancer. Alcohol Research. 2013;35(1):25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kadayifci FZ, Zheng S, Pan YX. Molecular mechanisms underlying the link between diet and DNA Methylation. Int J Mol Sci 2018;19(12). doi: 10.3390/ijms19124055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu HL, Dong S, Gao LF, et al. Global DNA methylation was changed by a maternal high-lipid, high-energy diet during gestation and lactation in male adult mice liver. Br J Nutr. 2015;113(7):1032–1039. doi: 10.1017/s0007114515000252 [DOI] [PubMed] [Google Scholar]