Abstract
Background
Mitochondrial fission regulator 2 (MTFR2) has been reported to promote proliferation, migration and invasion in tumors; however, little is known about its function in breast cancer. Thus, we investigated the effect of MTFR2 expression on prognosis of breast cancer.
Methods
The expression of MTFR2 in breast cancer tissues was detected by immunohistochemistry, and overall survival (OS) and recurrence free survival (RFS) were evaluated by the Log rank test and Cox model.
Results
We found that MTFR2 expression was significantly associated with clinical stage (P<0.001), T classification (P=0.005), N classification (P=0.001), M classification (P=0.041), HER2 expression (P= 0.001), and molecular subtypes (P=0.002), respectively. Compared with low MTFR2 expression, the patients with higher expression of MTFR2 exhibited significantly shorter OS and RFS (All P < 0.001). Both univariate and multivariate analyses showed that MTFR2 was an independent prognostic factor for OS (HR, 2.8, 95% CI 1.1–6.8, P = 0.023) and RFS (HR, 2.8, 95% CI 1.2–6.4, P = 0.015) in breast cancer patients. Moreover, in HER2 positive and TNBC subtype, the associations between high MTFR2 expression and poor OS and RFS were more pronounced.
Conclusion
Taken together, our results demonstrated that high MTFR2 expression was associated with poor prognosis of breast cancer patients, and such an association was more pronounced in the patients with aggressive tumors. Therefore, MTFR2 expression might be a potentially important prognostic biomarker and clinical target for patients with breast cancer.
Keywords: MTFR2, breast neoplasms, prognosis, biomarker, survival analysis
Introduction
Breast cancer has become the second cancer with high incidence and mortality globally, accounting for approximately 11.6% of all cancer deaths.1 It is a heterogeneous disease, which can be divided into 25 subtypes according to different histology and molecular profiles.2 More recently, despite targeted therapy (such as anti-estrogen and anti-HER2) has been widely used and improved prognosis, treatment outcomes for breast cancer remain relatively poor. Therefore, it is of great clinical significance to find new biomarkers that can effectively distinguish the patients with good prognosis between those with poor prognosis, and develop a new treatment scheme for patients with breast cancer.3,4
MTFR2 is also called family with sequence similarity 54, member A (FAM54A) and DUF729 domain containing 1(DUFD1, a 2 kb mRNA). It is located on chromosome 6q23.3 and plays a key role in mitochondria, promoting mitochondrial division and aerobic respiration in eukaryotic cells.5 Wang et al found that MTFR2 is one of the genes which are mostly correlated to dual specificity protein kinase TTK (TTK). It may regulate the expression of TTK by activating the transcription of TTK promoter, thus affecting the occurrence, treatment tolerance and recurrence of glioblastoma.6
TTK is located on chromosome 6q13-6q21.7 It enhances the activity of auroral kinase B through direct phosphorylation in the centromere, which affects cell proliferation and is necessary for chromosome alignment.8–10 Since mRNA and protein of TTK were overexpressed in breast cancer, MTFR2 might affect prognosis of breast cancer. Thus, we would explore the effect of expression of MTFR2 in breast cancer on clinical outcome.
Patients and Methods
Study Patients
We collected 139 patient samples with breast cancer from our hospital between January 2010 and December 2011. The median age at surgery was 53 years old (range: 32–91 years). All patients were followed up for 4 to 82 months with a median of follow up of 72 months. We ended our follow-up on July 30, 2017. All patients did not receive radiotherapy or chemotherapy before surgery. We collected clinical data, including age at operation, tumor size, lymph node status, breast grade, molecular subtype status, treatment method, and the time of recurrence and death in breast cancer patients. Stage of tumor was based on the American Joint Committee on Cancer (AJCC), the 8th edition. Written informed consents were given to all patients and this study was approved by the IRB committee of the Affiliated Yantai Yuhuangding Hospital of Qingdao University.
Hormone receptor positivity was confirmed if allred score was above or equal to 3. HER2-positive expression was defined as score 3+, and negative expression with a score of 0 or 1+. To confirm gene amplification, the fluorescent in situ hybridization (FISH) was performed on tumors with a score of 2+. Our definition of molecular subtypes was as following: 1) Luminal/HER2 negative referred to estrogen receptor (ER) positive and/or progesterone receptor (PR) positive but negative for HER2; 2) HER2 positive represented HER2 positive rather than related to ER or PR status; 3) Triple-negative breast cancer (TNBC) was that ER, PR and, HER2 status were all negative.11
Immunohistochemistry (IHC)
IHC analysis was used to detect MTFR2 expression in 139 cases of breast cancer tissue samples. The procedures were carried out in a similar manner previously described (12). In a brief step, we cut the paraffin-embedded sample into 4 μm slices, bake at 60 °C for 2 hours, then dewaxed with xylene and rehydrated. The slices were soaked in EDTA antigen repair buffer and microwave for antigen repair. We treated the slices with 3% hydrogen peroxide in methanol, and then incubated it with 1% rabbit serum albumin. The slices were incubated overnight with anti-MTFR2 rabbit polyclonal antibodies at 4 °C. After washing, the tissue sections were stained with DAB and treated with biotin-labeled anti-rabbit secondary antibody. The nucleus was stained by hematoxylin.
According to the proportion of positively stained cancer cells, the samples were divided into 1 to 4 grades, which were <10%, 10–50%, 50–75%, and >75% positive cancer cells, respectively. The intensity of staining was marked with different depth of color: light yellow for weak staining, yellowish brown for moderate staining, and brown for strong staining, recorded as 1 to 3 grades, respectively. The difference of staining index was resolved by consensus. According to the measure of heterogeneity from the Log rank test statistics with respect to overall survival (OS), the cutoff values of high and low expression of MTFR2 was defined. A staining index score greater than or equal to 6 was defined as the high expression of MTFR2, while a score less than 6 was considered the low expression of MTFR2.
Statistical Analysis
All statistical analyses were performed using the SPSS 23 software, and the categorical variables (eg, MTFR2 expression) were analyzed using the Chi-square test or Fisher’s exact test. Recurrence free survival (RFS) was defined the time from first therapeutic operation until any recurrence or last follow-up or death from any cause. Overall survival (OS) was defined as the time from first therapeutic operation until death due to any cause or last follow-up. The survival analyses were conducted by the Kaplan–Meier method with the Log rank test and the Cox multivariable proportional hazard model. The stratified analysis of survival was also performed by several potential prognostic confounders. All p-values were two-tailed and a P< 0.05 was considered statistically significant.
Results
MTFR2 Expression in Study Patients’ Tumors
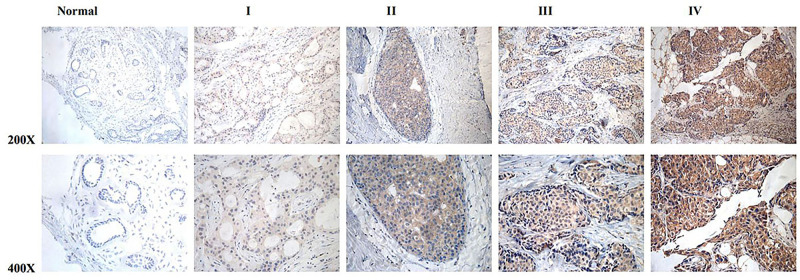
In this study, we performed the IHC in 139 patients. The clinical stages of these patients from I to IV were 24, 79, 32, and 4, respectively. The high MTFR2 expression was observed in 70 samples (50.4%) and the weak or no staining was detected in 69 cancer patients (49.6%) (Table 1). MTFR2 expression was found in the region containing cancer cells; while it was difficult to detect in normal breast or adjacent non-cancerous tissues. In subcellular localization, MTFR2 expression existed mainly in the cytoplasm as shown in Figure 1.
Table 1.
Clinicopathological Characteristics and MTFR2 Expression of Patients with Breast Cancer
| Variables | Number of Cases (%) |
|---|---|
| Age(years) | |
| ≤55 | 82(59.0%) |
| >55 | 57(41.0%) |
| Clinical stage | |
| I | 24(17.3%) |
| II | 79(56.8%) |
| III | 32(23.0%) |
| IV | 4(2.9%) |
| T classification | |
| T1 | 58(41.7%) |
| T2 | 68(48.9%) |
| T3 | 7(5.1%) |
| T4 | 6(4.3%) |
| N classification | |
| N0 | 63(45.3%) |
| N1 | 47(33.9%) |
| N2 | 15(10.8%) |
| N3 | 14(10.0%) |
| Grade | |
| I, II | 111(79.9%) |
| III | 28(20.1%) |
| Expression of ER | |
| Negative | 51(36.7%) |
| Positive | 88 (63.3%) |
| Expression of PR | |
| Negative | 61(43.9%) |
| Positive | 78(56.1%) |
| Expression of HER2 | |
| Negative | 105(75.5%) |
| Positive | 34(24.5%) |
| Expression of MTFR2 | |
| Low expression | 70(50.4%) |
| High expression | 69(49.6%) |
| Radiotherapy | |
| Not done | 96(69.1%) |
| Done | 43(30.9%) |
| Chemotherapy | |
| Not done | 41(29.5%) |
| Done | 98(70.5%) |
Figure 1.
MTFR2 protein overexpression in archived breast cancer tissues examined by immunohistochemistry. Representative IHC images of MTFR2 expression in normal human breast vs breast cancer tissues at different clinical stages.
Association of MTFR2 Expression with Clinicopathological Characteristics of Study Patients
As shown in Table 2, MTFR2 expression was significantly associated with clinical stage (P < 0.001), T classification (P = 0.003), N classification (P = 0.001), HER2 expression (P = 0.002), and molecular subtypes (P = 0.002), respectively and borderline significantly associated with M classification (P = 0.058). However, there were no significant associations with other variables including age, breast grade, ER status and PR status. Moreover, the Spearman correlation analysis showed that MTFR2 expression level was significantly correlated to clinical stages (r = 0.407, P < 0.001), T classification (r = 0.261, P = 0.002), N classification (r = 0.312, P < 0.001), M classification (r = 0.173, P = 0.041), and HER2 expression (r = 0.272, P = 0.001), respectively (Table 3). Thus, the IHC results revealed that increase of MTFR2 staining was positively correlated with advanced tumors, suggesting that high MTFR2 expression appeared to be associated with progression of breast cancer.
Table 2.
Associations Between Clinicopathological Characteristics and Expression of MTFR2 of Patients with Breast Cancer
| Characteristics | MTFR2 | χ2(P value) | F (P value) | |
|---|---|---|---|---|
| Low(n=70) n% | High(n=69) n% | |||
| Age(years) | 0.919 | 1.000 | ||
| ≤55 | 41(29.5%) | 41(29.5%) | ||
| >55 | 29(20.8%) | 28(20.2%) | ||
| Clinical stage | <0.001 | <0.001 | ||
| I | 18(12.9%) | 6(4.3%) | ||
| II | 46(33.1%) | 33(23.8%) | ||
| III | 6(4.3%) | 26(18.7%) | ||
| IV | 0(0%) | 4(2.9%) | ||
| T classification | 0.005 | 0.003 | ||
| T1 | 36(25.9%) | 22(15.8%) | ||
| T2 | 33(23.8%) | 35(25.2%) | ||
| T3 | 1(0.7%) | 6(4.3%) | ||
| T4 | 0(0%) | 6(4.3%) | ||
| N classification | 0.001 | 0.001 | ||
| N0 | 40(28.8%) | 23(16.5%) | ||
| N1 | 24(17.3%) | 23(16.5%) | ||
| N2 | 5(3.6%) | 10(7.2%) | ||
| N3 | 1(0.7%) | 13(9.4%) | ||
| M classification | 0.041 | 0.058 | ||
| No | 70(50.4%) | 65(46.7%) | ||
| Yes | 0(0%) | 4(2.9%) | ||
| Grade | 0.704 | 0.833 | ||
| I, II | 55(39.6%) | 56(40.3%) | ||
| III | 15(10.8%) | 13(9.3%) | ||
| ER expression | 0.345 | 0.382 | ||
| Negative | 23(16.6%) | 28(20.1%) | ||
| Positive | 47(33.8%) | 41(29.5%) | ||
| PR expression | 0.806 | 0.865 | ||
| Negative | 30(21.6%) | 31(22.3%) | ||
| Positive | 40(28.8%) | 38(27.3%) | ||
| HER2expression | 0.001 | 0.002 | ||
| Negative | 61(43.9%) | 44(31.6%) | ||
| Positive | 9(6.5%) | 25(18.0%) | ||
| Subtype | 0.002 | 0.002 | ||
| Luminal/HER2(-) | 47(33.8%) | 39(28.0%) | ||
| HER2(+) | 9(6.5%) | 25(18.0%) | ||
| TNBC | 14(10.1%) | 5(3.6%) | ||
Table 3.
Spearman Correlation Between MTFR2 and Clinical Pathological Factors
| Variables | MTFR2 Expression | |
|---|---|---|
| Spearman Correlation | P value | |
| Clinical stage | 0.407 | <0.001 |
| T classification | 0.261 | 0.002 |
| N classification | 0.312 | <0.001 |
| M classification | 0.173 | 0.041 |
| Grade | −0.032 | 0.706 |
| ER expression | −0.080 | 0.348 |
| PR expression | −0.021 | 0.807 |
| HER2 expression | 0.272 | 0.001 |
| Subtype | 0.043 | 0.614 |
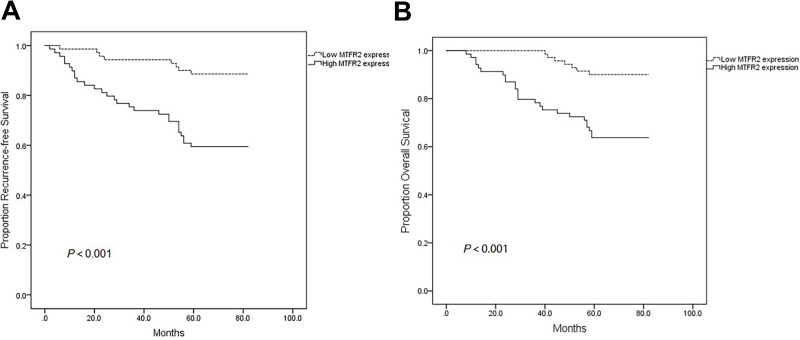
Association Between MTFR2 Expression and Survival
As shown in Figure 2A and B, the Kaplan–Meier analysis showed an negative association between MTFR2 expression and both of RFS and OS of patients with breast cancer (both P < 0.001). The cumulative rates of RFS and OS for patients with high MTFR2 expression were 59.4% and 63.8%, respectively, whereas the rates were 88.6% and 90.0% for patients with low MTFR2 expression, respectively. After both univariate and multivariate Cox proportional hazard regression analyses were performed, our results showed that N stage (aHR, 1.70, 95% CI, 1.24–2.33), MTFR2 expression (aHR, 3.85, 95% CI, 1.52–9.77) and HER2 expression (aHR, 4.81, 95% CI, 1.84–12.6) were independent prognostic factors for RFS. Furthermore, we found that N stage (aHR, 1.81, 95% CI, 1.30–2.52), MTFR2 expression (aHR, 3.30, 95% CI, 1.23–8.82) and HER2 expression (aHR, 5.02, 95% CI, 1.91–13.2) were also independent prognostic factors for OS (Table 4). However, we did not find significant associations between the treatment (eg, radiotherapy and chemotherapy) and both RFS and OS as shown in Table 4.
Figure 2.
Kaplan–Meier plots of RFS and OS according to MTFR2 expression level. (A) RFS of all patients with high MTFR2 expression vs low MTFR2 expression. (B) OS of all patients with high MTFR2 expression vs low MTFR2 expression.
Table 4.
Univariate and Multivariable Analysis on Survival of Patients with Breast Cancer
| Characteristics | RFS | OS | ||||||
|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||
| HR (95% CI) | P | aHR (95% CI) | P | HR (95% CI) | P | aHR (95% CI) | P | |
| Age | 0.098 | 0.117 | 0.226 | 0.154 | ||||
| ≤55 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| >55 | 1.74 (0.90–3.34) | 2.31 (0.98–4.96) | 1.53 (0.77–3.07) | 1.82 (0.80–4.13) | ||||
| T stage | <0.001 | 0.292 | <0.001 | 0.582 | ||||
| T1/T2 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| T3/T4 | 5.08 (2.38–10.9) | 2.01 (0.55–7.40) | 5.26 (2.43–11.4) | 1.43 (0.40–5.17) | ||||
| N stage | <0.001 | 0.001 | <0.001 | <0.001 | ||||
| N0/N1 | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| N2/N3 | 3.32 (1.70–6.45) | 1.70 (1.24–2.33) | 4.15 (2.07–8.32) | 1.81 (1.30–2.52) | ||||
| Clinical stage | <0.001 | 0.567 | <0.001 | 0.286 | ||||
| I/II | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| III/IV | 4.96 (2.56–9.61) | 1.62 (0.31–8.50) | 6.53 (3.18–13.4) | 2.44 (0.47–12.58) | ||||
| Grade | 0.290 | 0.381 | 0.927 | 0.797 | ||||
| I/II | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| III | 1.48 (0.72–3.08) | 1.47 (0.62–3.44) | 1.04 (0.45–2.41) | 0.88 (0.34–2.31) | ||||
| MTFR2 | <0.001 | 0.005 | 0.001 | 0.017 | ||||
| Low | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| High | 4.29 (1.96–9.43) | 3.85 (1.52–9.77) | 4.39 (1.90–10.1) | 3.30 (1.23–8.82) | ||||
| ER | 0.994 | 0.675 | 0.903 | 0.821 | ||||
| Negative | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Positive | 0.99 (0.51–1.97) | 1.37 (0.32–5.93) | 0.96 (0.47–1.96) | 1.18 (0.28–5.06) | ||||
| PR | 0.807 | 0.142 | 0.988 | 0.259 | ||||
| Negative | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Positive | 1.09 (0.56–2.11) | 3.42 (0.66–17.6) | 1.01 (0.50–2.02) | 2.54 (0.50–12.8) | ||||
| HER2 | 0.001 | 0.001 | <0.001 | 0.001 | ||||
| Negative | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Positive | 2.91 (1.51–5.62) | 4.81 (1.84–12.6) | 3.66 (1.83–7.31) | 5.02 (1.91–13.2) | ||||
| Group | 0.897 | 0.223 | 0.854 | 0.972 | ||||
| non-TNBC | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| TNBC | 1.06 (0.41–2.74) | 1.08 (0.73–3.86) | 0.91 (0.32–2.58) | 1.02 (0.40–2.59) | ||||
| Radiotherapy | 0.619 | 0.122 | 0.962 | 0.206 | ||||
| No | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Yes | 0.83 (0.40–1.72) | 0.52 (0.23–1.19) | 0.98 (0.47–2.07) | 0.57 (0.24–1.36) | ||||
| Chemotherapy | 0.548 | 0.501 | 0.692 | 0.767 | ||||
| No | 1.00 | 1.00 | 1.00 | 1.00 | ||||
| Yes | 0.81 (0.40–1.62) | 1.32 (0.58–3.00) | 0.86 (0.41–1.82) | 1.15 (0.47–2.82) | ||||
Abbreviation: aHR, adjusted with the variables listed in this table.
Prognostic of MTFR2 Expression in Aggressive Subtypes
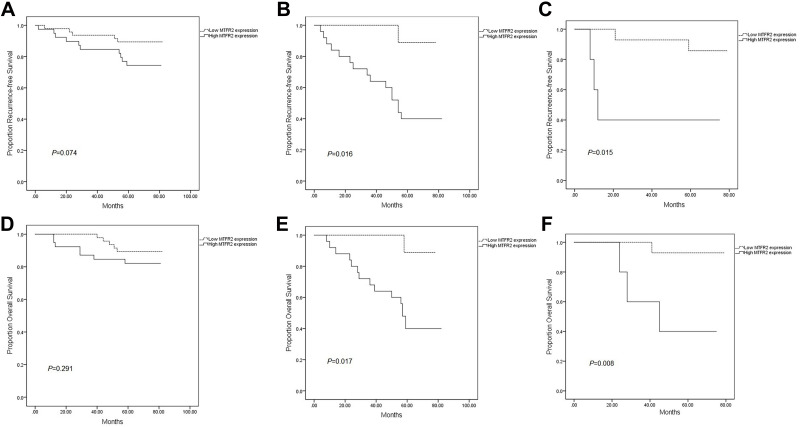
We assessed survival according to MTFR2 expression in each molecular subtype. The expression of MTFR2 had no significant effect on survival in the patients with negative expression of luminal/HER2 (Figure 3A; P=0.074), while the RFS differed significantly in both HER2 subtype (Figure 3B; P=0.016) and TNBC (Figure 3C; P=0.015), similarly, OS differed significantly in the HER2 subtype (Figure 3E; P=0.017) and TNBC (Figure 3F; P=0.008), but no significant difference in the patients with Luminal/HER2 negative expression (Figure 3D; P=0.291).
Figure 3.
Kaplan–Meier plots of RFS and OS according to MTFR2 expression level in each subtype. RFS differed significantly according to MTFR2 expression in the HER2 and TNBC subtypes (B and C), but it did not differ in the other subtypes (A). OS differed significantly according to MTFR2 expression in the HER2and TNBC subtypes (E and F), but it did not differ in luminal/HER2 negative (D). P values were calculated by Log rank tests.
Discussion
In the present study, MTFR2 expression levels were relatively higher in cancer lesions than those in normal tissues. MTFR2 was highly expressed in breast cancer tissues, and it was significantly correlated with clinical stage, T, N, classification and HER2 expression level of breast cancer. The expression of MTFR2 increased with the progression of breast cancer, indicating that high expression of MTFR2 could be related to the progression of breast cancer. Our findings revealed that MTFR2 was an independent prognostic factor for prognosis of breast cancer patients, and the patients with high MTFR2 expression had worse RFS and OS than those with corresponding low expression. Lu et al also found that MTFR2 can promote growth, migration, invasion and tumor progression in breast cancer cells.12 Thus, it appears that MTFR2 expression may have clinical significance as a novel predictor of prognosis and one of potential new targets for future targeted therapy of breast cancer.
Previous studies found that MTFR2 was highly expressed in mice testicular cells.5 MTFR2 encodes a protein in the mitochondria and promotes mitochondrial fission and anti-DNA oxidative damage.13,14 Mitochondria play a key role in induction of intrinsic apoptosis.15 Compared to those in normal cells, mitochondria in cancer cells express a higher level of reactive oxygen species (ROS) and reductant,16–18 whereas the excessive ROS damage lipids and DNA.19,20 In addition, mitochondrial fission can induce glycolysis reprogramming of cancer-related myofibroblasts, accelerating tumor growth and angiogenesis.21
Recently, Wang found that MTFR2 promoted the proliferation, migration, and invasion of oral squamous carcinoma.22 Wang et al also found that MTFR2-dependent regulation of TTK was involved in maintaining glioma stem-like cells (GSCs) in glioblastoma and could be a potential new druggable target for glioblastoma.6 It might regulate the expression of TTK by activating the transcription of TTK promoter for participation in up-regulation and expression of GSCs in glioblastoma. Besides the effect on cell proliferation, TTK also played important roles in centrosome duplication, DNA damage response, and organ development.8 Daniel et al demonstrated that reduced TTK levels could cause abnormal mitoses, induce apoptosis and decrease survival of breast cancer cells.23 Maire et al found that TTK depletion would seriously impair the viability and ability to form colonies of TNBC cell lines.24 Therefore, TTK could be an independent prognostic biomarker and it is biologically plausible that MTFR2 might activate regulation, promote expression of TTK in breast tumor cells, and have an important regulatory role in the occurrence and development of breast cancer. However, the exact mechanisms need further investigation.
Intriguingly, we also found that expression of MTFR2 was associated with aggressive subtypes, particularly for HER2-positive and TNBC subtypes, indicating that MTFR2 may have clinical significance as a new target for improved outcome and individualized treatment of patients with breast cancer. Furthermore, MTFR2 expression was significantly correlated with HER2 expression status, implying that MTFR2 may provide additional effective value for targeted therapy in patients with breast cancer. Therefore, MTFR2 might be a valuable biomarker for predicting prognosis and guiding future plan of follow up of breast cancer patients.
In this study, several limitations need to be addressed. First, the retrospective review with a small sample size is our main limitation, which may result in biased estimates of association. Moreover, since this is a relatively small study and all patients were recruited from a single hospital, the selection bias may exist. However, our preliminary findings from such a small sample size may help generate a hypothesis for testing or validation in future large prospective studies via consortia or multi-centers. Finally, the exact molecular mechanisms underlying the associations remain unclear, thus, more mechanistic investigation is warranted.
Conclusion
In conclusion, we found that high expression of MTFR2 is correlated with breast cancer progression, and MTFR2 expression was significantly associated with survival of breast cancer patients. Moreover, the prognostic effect of MTFR2 expression was even more pronounced in aggressive tumors. Our results may support that MTFR2 might serve as an independent prognostic biomarker and a potential therapeutic target for breast cancer.
Funding Statement
There is no funding to report.
Data Sharing Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of the Yantai Yuhuangding Hospital Affiliated Hospital of Qingdao University and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Disclosure
The authors declare that they have no competing interests. These authors contributed equally to the manuscript: Wenjie Lu, Rukun Zang and Yuncheng Li.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2.Hurvitz SA, Lalla D, Crosby RD. Use of the metastatic breast cancer progression (MBC-P) questionnaire to assess the value of progression-free survival for women with metastatic breast cancer. Breast Cancer Res Treat. 2013;142(3):603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang M, Ji S, Shao G, Zhang J, Zhao K, Wang Z. Effect of exosome biomarkers for diagnosis and prognosis of breast cancer patients. Clinical Translational Oncol. 2018;20(7):906–911. [DOI] [PubMed] [Google Scholar]
- 4.Fu S, Cheng J, Wei C, et al. Development of diagnostic SCAR markers for genomic DNA amplifications in breast carcinoma by DNA cloning of high-GC RAMP-PCR fragments. Oncotarget. 2017;8(27):43866–43877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monticone M, Panfoli I, Ravera S, et al. The nuclear genes Mtfr1 and Dufd1 regulate mitochondrial dynamic and cellular respiration. J Cell Physiol. 2010;225(3):767–776. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Xie Y, Bai X, et al. Targeting dual specificity protein kinase TTK attenuates tumorigenesis of glioblastoma. Oncotarget. 2018;9(3):3081–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Y, Wang A, Lin J, et al. Mps1/TTK: a novel target and biomarker for cancer. J Drug Target. 2017;25(2):112–118. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Winey M. The MPS1 family of protein kinases. Annu Rev Biochem. 2012;81:561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foijer F, Xie SZ, Simon JE, et al. Chromosome instability induced by Mps1 and p53 mutation generates aggressive lymphomas exhibiting aneuploidy-induced stress. Proc Natl Acad Sci U S A. 2014;111(37):13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuckle H, Benn P, Pergament E. Cell-free DNA screening for fetal aneuploidy as a clinical service. Clin Biochem. 2015;48(15):932–941. doi: 10.1016/j.clinbiochem.2015.02.011 [DOI] [PubMed] [Google Scholar]
- 11.Yoon CI, Ahn SG, Bae SJ, et al. High A20 expression negatively impacts survival in patients with breast cancer. PLoS One. 2019;14(8):e0221721. doi: 10.1371/journal.pone.0221721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu G, Lai Y, Wang T, et al. Mitochondrial fission regulator 2 (MTFR2) promotes growth, migration, invasion and tumour progression in breast cancer cells. Aging. 2019;11(22):10203–10219. doi: 10.18632/aging.102442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonachini L, Monticone M, Puri C, et al. Chondrocyte protein with a poly-proline region (CHPPR) is a novel mitochondrial protein and promotes mitochondrial fission. J Cell Physiol. 2004;201(3):470–482. doi: 10.1002/jcp.20126 [DOI] [PubMed] [Google Scholar]
- 14.Monticone M, Tonachini L, Tavella S, et al. Impaired expression of genes coding for reactive oxygen species scavenging enzymes in testes of Mtfr1/Chppr-deficient mice. Reproduction. 2007;134(3):483–492. doi: 10.1530/REP-07-0199 [DOI] [PubMed] [Google Scholar]
- 15.Giorgi C, Baldassari F, Bononi A, et al. Mitochondrial Ca2+ and apoptosis. Cell Calcium. 2012;52(1):36–43. doi: 10.1016/j.ceca.2012.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles‘ heel? Nat Rev Cancer. 2014;14(11):709–721. doi: 10.1038/nrc3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace DC. Mitochondrial genetic medicine. Nat Genet. 2018;50(12):1642–1649. doi: 10.1038/s41588-018-0264-z [DOI] [PubMed] [Google Scholar]
- 18.Baulies A, Montero J, Matias N, et al. The 2-oxoglutarate carrier promotes liver cancer by sustaining mitochondrial GSH despite cholesterol loading. Redox Biol. 2018;14:164–177. doi: 10.1016/j.redox.2017.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulda S, Galluzzi L, Kroemer G. Targeting mitochondria for cancer therapy. Nat Rev Drug Discov. 2010;9(6):447–464. [DOI] [PubMed] [Google Scholar]
- 20.Dixon SJ, Stockwell BR. The role of iron and reactive oxygen species in cell death. Nat Chem Biol. 2014;10(1):9–17. doi: 10.1038/nchembio.1416 [DOI] [PubMed] [Google Scholar]
- 21.Jelluma N, Brenkman AB, van den Broek NJF, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132(2):233–246. doi: 10.1016/j.cell.2007.11.046 [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Xiong M, Jiang L, Chen Z, Shao Y. MTFR2 Promotes the Proliferation, Migration, and Invasion of Oral Squamous Carcinoma by Switching OXPHOS to Glycolysis. Front Oncol. 2020;10:858. doi: 10.3389/fonc.2020.00858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniel J, Coulter J, Woo J-H, Wilsbach K, Gabrielson E. High levels of the Mps1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc Natl Acad Sci U S A. 2011;108(13):5384–5389. doi: 10.1073/pnas.1007645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maire V, Baldeyron C, Richardson M, et al. TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS One. 2013;8(5):e63712. [DOI] [PMC free article] [PubMed] [Google Scholar]