Abstract
A linezolid-resistant E.faecalis strain harboring optrA and cfr resistance genes were isolated from a patient in china, which had no mutations in rplC, rplD, rplV, and 23S rRNA gene. Transformation indicated that optrA and cfr were located on two different plasmids and both could be transferred to recipient strain, resulting in the increase of MICs of linezolid and chloramphenicol. Cfr, carried by an 11,872-bp plasmid, was enclosed with an IS110 transposase in upstream and an IS3-like transposase in downstream, while optrA was on an 8357-bp plasmid. As far as we know, this is the first report of an E.faecalis clinical strain co-harboring optrA and cfr in China.
Keywords: E.faecalis, linezolid, cfr, optrA
Linezolid is considered as a last resort drug for the treatment of severe infections caused by multidrug-resistant gram-positive pathogens including vancomycin-resistant Enterococcus spp. (VRE), methicillin-resistant Staphylococcus spp. and Streptococcus pneumoniae.1 Although most of gram-positive cocci remain susceptible to linezolid, resistant isolates of Enterococcihave been reported worldwide. The main resistance mechanisms of Enterococcus spp to linezolid include the mutation of 23S rRNA gene,2 acquired resistance genes such as cfr, cfr(B), optrAor poxtA,3,4 and mutation of ribosomal proteins coding genes like rplC, rplD, and rplV.5 The cfr and cfr(B) genes encode a rRNA methyltransferase causing resistance of oxazolidinones, chloramphenicols, tetracycline, lincomycins, pleuromutilin, and streptogramin A and decreasing sensitivity of macrolide.3,4,6–9 Cfr or cfr(B) and optrA, poxtA along with optrA has been previously reported on the same plasmid in E. faecalis, Staphylococcus sciuri, or Enterococcus spp. from swine and farm environment.10–13 Here, we firstly reported the emergence of linezolid-resistant E. faecalis clinical strain with cfr and optrA in China.
E.faecalis strain EF02 was isolated from the midstream urine of a 72-year-old patient with diabetes during hospitalization in November, 2018. Prior to the isolation of E.faecalis strain EF02, isepamicin was used for the treatment of infection. Bacterial identification was conducted by MALDI-TOF (VITEK MS, BioMérieux). Antimicrobial susceptibility testing was performed by broth microdilution according to CLSI guideline.14 E.faecalis strain EF02 was resistant to linezolid (MIC =8 mg/L), nitrofurantoin (MIC =64 mg/L), tetracycline (MIC =64 mg/L), erythromycin (MIC =64 mg/L), chloramphenicol (MIC =64 mg/L) and levofloxacin (MIC =32 mg/L). However, this strain was susceptible to vancomycin (MIC =1 mg/L), teicoplanin (MIC =0.5 mg/L), and ampicillin (MIC =2 mg/L). PCR detection and sequencing revealed that E.faecalis strain EF02 was positive for cfr and optrA without mutation among rplC, rplD, rplVand 23S rRNA gene. The plasmids extracted from the donor strain E.faecalis EF02 were then guided into the recipient strain E.faecalis OG1RF by the electrotransformationmethod.3,4 The transformants were selected on brain heart infusion agar containing 3 mg/L linezolid and 10 mg/L chloramphenicol. Colonies that grew on these selective plates were further confirmed by antimicrobial susceptibility testing and PCR for the detection cfr and optrA genes. Transformants harboring the plasmid with optrA and cfr were named L13/optrA and L18/cfr, respectively. Comparing with the recipient strain, the linezolid and chloramphenicol MICs of transformants increased 4–8 fold (Table 1).
Table 1.
MICs for E. Faecalis EF02, Transformants, and the Recipient Strains (μg/ML)
| Antimicrobial Agents | E. faecalis EF02 | L13/optrA | L18/cfr | E. faecalis OG1RF |
|---|---|---|---|---|
| Linezolid | 8 | 8 | 8 | 1 |
| Tetracycline | >64 | 0.25 | 0.25 | 0.25 |
| Erythromycin | >64 | 4 | 8 | 2 |
| Chloramphenicol | >64 | 8 | 8 | 2 |
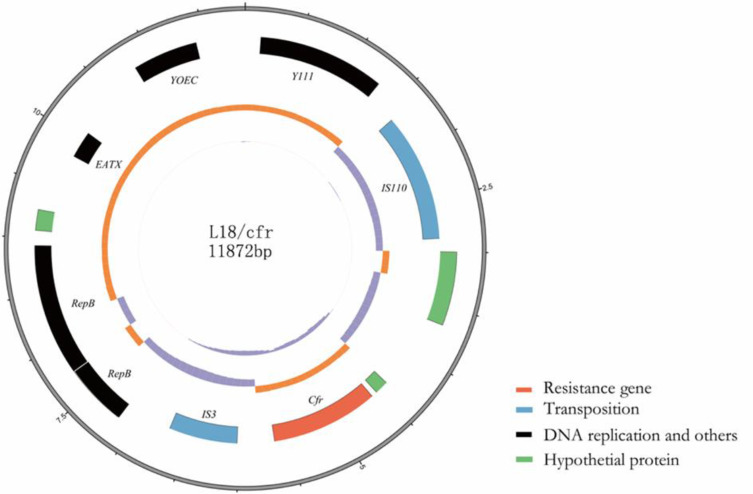
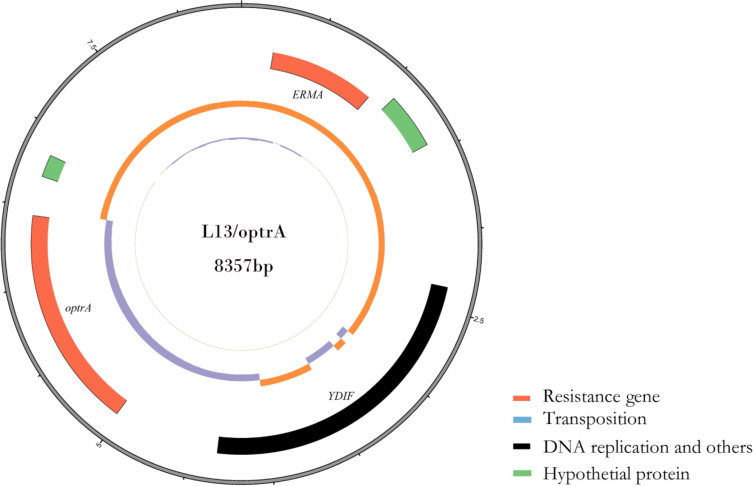
PCR mapping according to plasmid DNA sequencing of the L18/cfr plasmid suggested that an IS110 transposase was located upstream of cfr and an IS3-like transposase was located downstream of cfr. Mobile elements like these might lead to the transfer of resistance genes among plasmids. L18/cfr plasmid contained eight open reading frames encoding Y111, IS110, cfr, IS3, RepB, RepB, EATX, and YOEC (Figure 1). However, L13/optrA plasmid, which contained three open reading frames encoding ERMA, YDIF, and optrA (Figure 2). The result of multilocus sequence typing indicated that E.faecalis strain EF02 belonged to ST 330. The DNA sequencing of plasmid pEF-L18/cfr and pEF-L13/optrA has been submitted to the NCBI database (NCBI number: MT874923 and MT874924).
Figure 1.
Circular representation of the L18/cfr plasmid. Moving from inside to outside in the plasmid circular map, slots1–3 (slot 1, GC skew; slot 2, GC content; slot 3, open reading frames: Y-family DNA polymerase Y111, IS110 family transposase, cfr, IS3 family transposase, two replication protein RepB, antitoxin epsilon EATX, Probable integrase YOEC.).
Figure 2.
Circular representation of the L13/optrA plasmid. Moving from inside to outside in the plasmid circular map, slots1–3 (slot 1, GC skew; slot 2, GC content; slot 3, open reading frames: 23S rRNA adenine(2058)-N(6)-methyltransferase ERMA, Putative ATP-binding protein YDIF, optrA).
cfrand optrA genes have been reported in various gram-positive bacteria, they can be transferred to recipient bacteria by transformation experiments and cause an increase in the MIC values of linezolid and chloramphenicol.3,4,15 The cfr and optrA located on one same plasmid in a strain have been reported. Fan R and Morroni G et al reported Staphylococcus sciuri isolated from pig origin in Germany and E. faecium isolates in Italy carrying both cfr and optrA.9,16 However, no conjugatant or transformant was acquired to demonstrate the resistance mediated by cfr or optrA. Li et al also reported co-producing cfr and optrA of Staphylococcus sciuri, and got the transformant with optrA.11 Similarly, two E. faecium clinical isolates carrying cfr and optrA were collected in Italy, in which optrA was transferred from donor to recipient, whereas cfr was not transferrable.17
In our study, we found cfr and optrA genes carried by E. faecalis EF02 were located on two different plasmids, and both plasmids could be transferred from the donor strain to the recipient by transformation experiments, making an increase of MICs of linezolid and chloramphenicol. To our knowledge, this is the first report of linezolid-resistant E. faecalis co-producing cfr and optrA in a clinical isolate in China.
Funding Statement
This study was supported by the Science and Technology Project of Quanzhou (2019N030S) and China Antimicrobial Surveillance Network (WI207259). The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
Abbreviations
VRE, vancomycin-resistant Enterococcus spp.
Ethics Approval and Consent to Participate
The study was approved by the Ethics Board of Fujian Medical University.
Consent for Publication
Not applicable.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Sadowy E. Linezolid resistance genes and genetic elements enhancing their dissemination in enterococci and streptococci. Plasmid. 2018;99:89–98. [DOI] [PubMed] [Google Scholar]
- 2.Wong A, Reddy SP, Smyth DS, Aguero-Rosenfeld ME, Sakoulas G, Robinson DA. Polyphyletic emergence of linezolid-resistant staphylococci in the United States. Antimicrob Agents Chemother. 2010;54(2):742–748. doi: 10.1128/AAC.00621-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Wang Y, Schwarz S, et al. Transferable multiresistance plasmids carrying cfr in Enterococcus spp. from swine and farm environment. Antimicrob Agents Chemother. 2013;57:42–48. doi: 10.1128/AAC.01605-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y, Lv Y, Cai J, et al. A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother. 2015;70:2182–2190. doi: 10.1093/jac/dkv116 [DOI] [PubMed] [Google Scholar]
- 5.Mendes RE, Deshpande LM, Farrell DJ, Spanu T, Fadda G, Jones RN. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J Antimicrob Chemother. 2010;65:2329–2335. doi: 10.1093/jac/dkq331 [DOI] [PubMed] [Google Scholar]
- 6.Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother. 2000;44:2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vester B. The cfr and cfr-like multiple resistance genes. Res Microbiol. 2018;169:61–66. doi: 10.1016/j.resmic.2017.12.003 [DOI] [PubMed] [Google Scholar]
- 8.Antonelli A, D’Andrea MM, Brenciani A, et al. Characterization of poxtA, a novel phenicol-oxazolidinone-tetracycline resistance gene from an MRSA of clinical origin. J Antimicrob Chemother. 2018;73:1763–1769. doi: 10.1093/jac/dky088 [DOI] [PubMed] [Google Scholar]
- 9.Fan R, Li D, Fessler AT, Wu C, Schwarz S, Wang Y. Distribution of optrA and cfr in florfenicol-resistant Staphylococcus sciuri of pig origin. Vet Microbiol. 2017;210:43–48. doi: 10.1016/j.vetmic.2017.07.030 [DOI] [PubMed] [Google Scholar]
- 10.Kuroda M, Sekizuka T, Matsui H, et al. Complete genome sequence and characterization of linezolid-resistant enterococcus faecalis clinical isolate KUB3006 carrying a cfr(b)-transposon on its chromosome and optrA-Plasmid. Front Microbiol. 2018;9:2576. doi: 10.3389/fmicb.2018.02576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D, Wang Y, Schwarz S, Cai J, Shen J. Co-location of the oxazolidinone resistance genes optrA and cfr on a multiresistance plasmid from Staphylococcus sciuri. J Antimicrob Chemother. 2016;71:1474. doi: 10.1093/jac/dkw040 [DOI] [PubMed] [Google Scholar]
- 12.Lazaris A, Coleman DC, Kearns AM, et al. Novel multiresistancecfr plasmids in linezolid-resistant methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecium (VRE) from a hospital outbreak: co-location of cfr and optrA in VRE. J Antimicrob Chemother. 2017;72:3252–3257. [DOI] [PubMed] [Google Scholar]
- 13.Hao W, Shan X, Li D, et al. Analysis of a poxtA- and optrA-co-carrying conjugative multiresistance plasmid from Enterococcus faecalis. J Antimicrob Chemother. 2019;74:1771–1775. doi: 10.1093/jac/dkz109 [DOI] [PubMed] [Google Scholar]
- 14.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing., 30th ed Wayne, PA: CLSI supplement M100. Clinical and Laboratory Standards Institute; 2020. [Google Scholar]
- 15.Shen J, Wang Y, Schwarz S. Presence and dissemination of the multiresistance gene cfr in Gram-positive and Gram-negative bacteria. J Antimicrob Chemother. 2013;68:1697–1706. doi: 10.1093/jac/dkt092 [DOI] [PubMed] [Google Scholar]
- 16.Morroni G, Brenciani A, Antonelli A, et al. Characterization of a multiresistance plasmid carrying the optra and cfr resistance genes from an enterococcus faecium clinical isolate. Front Microbiol. 2018;9:2189. doi: 10.3389/fmicb.2018.02189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenciani A, Morroni G, Vincenzi C, et al. Detection in Italy of two clinical Enterococcus faecium isolates carrying both the oxazolidinone and phenicol resistance gene optrA and a silent multiresistance gene cfr. J Antimicrob Chemother. 2016;71:1118–1119. doi: 10.1093/jac/dkv438 [DOI] [PubMed] [Google Scholar]