These observations highlight that SCFAs promote macrophage elimination of K. pneumoniae via a LAMTOR2-dependent signal pathway and suggest that it is possible to intervene in K. pneumoniae pneumonia by targeting the gut microbiota.
KEYWORDS: K. pneumoniae, gut microbiota, immune responses, SCFAs, LAMTOR2, mechanisms
ABSRTACT
Klebsiella pneumoniae is a common cause of human-pneumonia-derived sepsis with high morbidity and mortality. The microbiota promotes and maintains host immune homeostasis. The mechanisms by which the gut microbiota affects the host defenses in the respiratory system systematically, however, remain poorly understood. Here, we show that gut microbiota depletion increases susceptibility to extracellular K. pneumoniae infections in terms of increased bacterial burdens in lung and decreased survival rates. Oral supplementation with gut microbiota-derived short-chain fatty acids (SCFAs), subsequently activating G protein-coupled receptor 43 (GPCR43), enhances a macrophage’s capacity to phagocytose invading K. pneumoniae. Furthermore, SCFAs and GPR43 increase macrophage bacterial clearance by upregulating LAMTOR2, which is further identified as an antibacterial effector and elucidated to facilitate phagosome-lysosome fusion and extracellular signal-regulated kinase (ERK) phosphorylation. Lastly, conditional ablation of Lamtor2 in macrophages decreases their antimicrobial activity, even though mice were pretreated with exogenous SCFA supplementation.
IMPORTANCE These observations highlight that SCFAs promote macrophage elimination of K. pneumoniae via a LAMTOR2-dependent signal pathway and suggest that it is possible to intervene in K. pneumoniae pneumonia by targeting the gut microbiota.
INTRODUCTION
Klebsiella pneumoniae is a severe multidrug-resistant (MDR) pathogen associated with high morbidity and mortality, which accounts for about one-third of all Gram-negative infections overall, owing to the limited availability of treatment options (1, 2). Being common natural inhabitants of our microbiome, the risk of worldwide spread of these MDR pathogens has become a recognized global threat (2). Therefore, there is an urgent need to expand our understanding of how host defenses limit the pathogenesis and dissemination of K. pneumoniae, and novel therapeutic strategies should be explored.
The past 2 decades witness the implications of gut microbiota in modulating the inflammatory responses both locally and systemically. Several studies demonstrate that alterations in the microbiome and the microbial metabolome are associated with a wide array of diseases (3–5), and concepts of the gut-liver axis, gut-lung axis, and gut-brain axis thus have been proposed, subsequently appreciated and accepted by the academic community. Indeed, the gut-lung axis has already been reported to be associated with respiratory diseases (6–8). Systemic roles of the gut microbiota are attributed mainly to microbiota-derived metabolites, including popularly studied short-chain fatty acids (SCFAs) (9–11). These bacterially derived metabolites are essential elements in the activation of G protein-coupled receptors (GPRs), such as GPR43, GPR41, and OLFR78 (12–15), and in the inhibition of histone deacetylases (HDACs) (16–18). Moreover, SCFAs, especially butyrate, have been demonstrated to promote regulatory T cell homeostasis in the colon (17, 18). Associations between SCFA metabolisms and the development of inflammatory disorders have thus been identified and accepted (19–21). Nevertheless, mechanisms by which SCFAs exert protective effects on distal organs remain controversial, especially in the respiratory system (22, 23).
Numerous pro- or anti-inflammatory mediators have been reported to participate in the elimination of extracellular invading pathogens by macrophages (24–26). LAMTOR2, the late endosomal/lysosomal adaptor mitogen-activated protein kinase (MAPK) and mammalian target of rapamycin (mTOR) activator/regulator complex 2, has been demonstrated to regulate dendritic cell (DC) homeostasis as well as endosomal biogenesis (27–29). In addition, the LAMTOR2 complex is of great significance in correct spatiotemporal extracellular signal-regulated kinase (ERK) phosphorylation and activation (30, 31). Combining multiomic analysis and in vitro or in vivo assays, we hypothesize that Lamtor2 deficiency may compromise the ability of macrophages to eliminate pathogenic bacteria.
In this study, by combining mouse models and multiomics analyses, we elucidate explicitly the mechanisms by which the gut microbiota promotes immune responses systematically in the lung during infection by the major human pathogen K. pneumoniae. Our work highlights the physiological functions of both SCFAs and LAMTOR2, demonstrates a heretofore-unrecognized microbiota-SCFA/GPR43-LAMTOR2-pERK-inducible nitric oxide synthase (iNOS) signaling pathway involved in host innate immune responses, and suggests that this pathway may be a novel therapeutic strategy to intervene in K. pneumoniae pneumonia by targeting the gut microbiota.
RESULTS
Gut microbiota depletion exacerbates K. pneumoniae-induced lung injury.
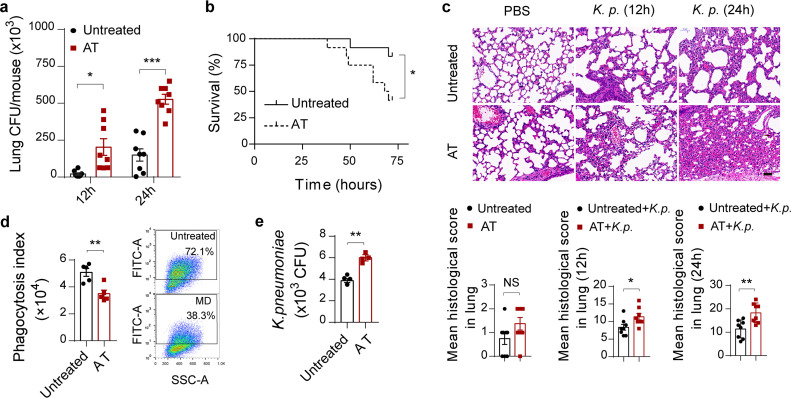
To determine the roles of the gut microbiota in regulating antibacterial immunity outside the intestinal lumen, a model of bacterial-infection-induced lung injury was applied. Briefly, we initially treated wild-type mice with broad-spectrum antibiotics in drinking water to deplete gut microbiota, as previously described (8), and then infected them intranasally with 1 × 105 CFU of K. pneumoniae. As with other studies (32, 33), in antibiotic-treated mice, sharp increases in the burdens of K. pneumoniae in the lung (Fig. 1a) and blood (see Fig. S1a in the supplemental material) were observed compared to levels in untreated controls. Pulmonary inflammatory cytokines, such as tumor necrosis factor alpha (TNF-α), interleukin 6 (IL-6), and IL-1β, and chemokines, such as chemokine C-X-C motif ligand 1 (CXCL1) and monocyte chemoattractant protein 1 (MCP-1), were all downregulated in antibiotic-treated mice (Fig. S1b). Bacterial-infection-induced mortality was remarkably elevated in antibiotic-treated mice (Fig. 1b). Taken together, these observations suggested that gut microbiota depletion resulted in compromised defenses against K. pneumoniae, as reflected by increased bacterial burdens in the lung and higher mortality.
FIG 1.
Gut microbiota depletion exacerbates K. pneumoniae-induced lung injury. (a) Pulmonary bacterial burdens in antibiotic-treated and untreated controls 12 h and 24 h after intranasal challenge with 1 × 105 CFU of K. pneumoniae. (b) Survival rates of antibiotic-treated and untreated controls infected by K. pneumoniae. (c) Representative hematoxylin and eosin (H&E) staining and quantification of pathological scores of lung sections derived from antibiotic-treated and untreated controls after K. pneumoniae infection. Scale bars, 50 μm. (d) Phagocytic capacity of isolated alveolar macrophages from antibiotic-treated and untreated controls after K. pneumoniae infection evaluated by flow cytometry (MOI = 100). (e) Bacterial loads of isolated alveolar macrophages from antibiotic-treated and untreated controls. Data are from three independent experiments (a, b, d, e) or one experiment representative of three independent experiments (c) (means ± SEM). The group size was 8 to 12 mice. The P values were determined using two-tailed Student t tests (a, c to e) or the log rank (Mantel-Cox) test (b). *, P < 0.05; **, P < 0.01; ***, P < 0.001. FITC-A, fluorescein isothiocyanate-area; NS, not significant. AT, antibiotic-treated mice.
Protective role of the gut microbiota during K. pneumoniae-induced sepsis. (a) Blood bacterial burdens in antibiotic-treated and untreated controls 12 h and 24 h after K. pneumoniae infection. (b) Pulmonary production of cytokines (TNF-α, 1L-1β, and IL-6) and chemokines (CXCL1 and MCP-1) in antibiotic-treated and untreated controls 24 h after K. pneumoniae infection. (c) Representative H&E staining and quantification of pathological scores of liver and kidney sections from antibiotic-treated and untreated controls 24 h after K. pneumoniae infection. Scale bars, 50 μm. The group size was 8 to 12 mice. Data are from three independent experiments (a, b) or one experiment representative of three independent experiments (c) (means ± SEM). The P values were determined using two-tailed Student t tests (a to c). *, P < 0.05; **, P < 0.01; ***, P < 0.001. NS, not significant; AT, antibiotic-treated mice. Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To further investigate whether the role of the gut microbiota is protective in the host’s respiratory defense against organ damage triggered by extracellular invading bacteria, we semiquantitatively determined pathology scores in lung, liver, and kidney between antibiotic-treated mice and untreated controls at various time points after K. pneumoniae challenge. Not surprisingly, all infected mice shared histological evidence of severe pneumonia. We found that antibiotic-treated mice displayed earlier and more severe inflammation in the lung (Fig. 1c), liver, and kidney (Fig. S1c). Collectively, these results indicated that the gut microbiota protects against lung injury during K. pneumoniae-induced sepsis.
Gut microbiota depletion impedes alveolar macrophage function.
The innate immune system acts as the first weapons of invading microorganisms, and residing macrophages in the lung are the key regulators of host innate immunity during bacterial infection and pneumonia (7, 34). Considering the protective effect of the gut microbiota on the host immune defense, we investigated whether microbiota depletion would affect the phagocytosis and clearance of K. pneumoniae by alveolar macrophages. We observed that primary alveolar macrophages originated from antibiotic-treated mice indeed had a markedly diminished capacity to phagocytize and eliminate K. pneumoniae compared to that of controls (Fig. 1f and g). With consideration of the results of other studies (32, 33), we demonstrated that the gut microbiota played protective roles during K. pneumoniae infections.
Gut microbiota depletion alters profiles of cecum metabolomics and gut microbiomes.
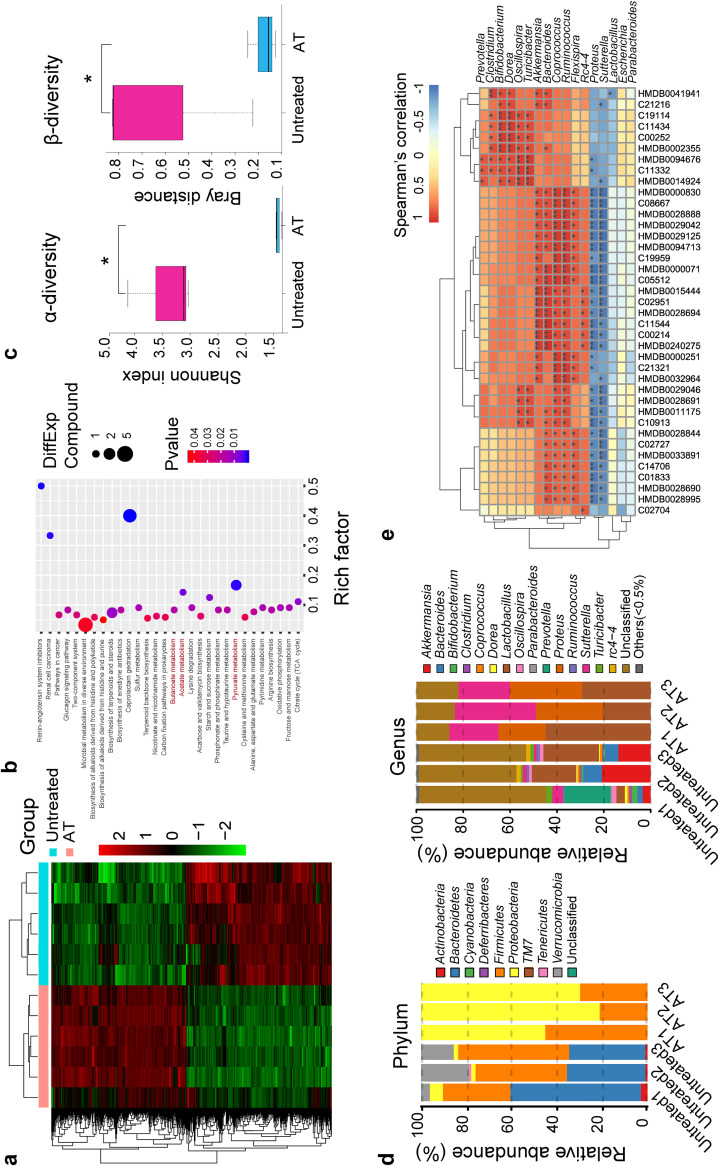
As previously described, the way that the microbiota modulates host immunity and susceptibility to infection depends largely on metabolites (35–38). To definitely determine the metabolic profiles of antibiotic-treated mice, we performed in-depth untargeted metabolomics sequencing of cecum samples by liquid chromatography-mass spectrometry (LC-MS). In total, 1,126 metabolites that differed significantly in abundance were identified and isolated in a comparison with controls based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Fig. 2a); these metabolites were associated primarily with carbohydrate and amino acid metabolism. Moreover, we carried out KEGG pathway enrichment analysis (Fig. 2b), which revealed that gut microbiota depletion elicited distinct shifts in metabolic pathways. Consistently with a previous work (39), we observed that SCFA metabolisms were dysregulated (Fig. 2b), which modulates host antimicrobial activity (14, 39, 40). Furthermore, we also analyzed metabolomics profiles of antibiotic-treated and K. pneumoniae-infected mice versus K. pneumoniae-infected controls to explore the influences of the gut microbiota on invading extracellular pathogens (Fig. S2a). Discriminations of SCFA metabolisms were also captured (Fig. S2b). Taken together, these results indicated that gut microbiota depletion may give rise to pronounced changes in metabolomics profiles whether specimens are infected with K. pneumoniae or not in a comparison with counterpart control mice.
FIG 2.
Gut microbiota depletion alters profiles of cecum metabolomics and associated gut microbiomes. (a) Hierarchical clustering heatmap of metabolites identified in cecum contents from antibiotic-treated and untreated controls. Red denotes increased expression; green denotes decreased expression. (b) Selected examples of KEGG pathway enrichment. Disturbances in SCFA metabolism are highlighted in red. (c) α-Diversity (Shannon index) and β-diversity (Bray-Curtis similarity index) of 16S rRNA genes from antibiotic-treated and untreated controls. (d) Discriminative operational taxonomic unit (OTU) abundances of taxonomic distributions at the phylum and genus levels in antibiotic-treated compared to untreated controls. (e) Spearman’s rank correlation between cecum metabolites and associated gut microbiota in antibiotic-treated and untreated controls. Red connections indicate a positive correlation, while blue connections are negative. The P values were determined using the hypergeometric test and the Benjamini-Hochberg false-discovery rate (FDR) correction (b) or the two-tailed Wilcoxon rank sum test (e). *, P < 0.05; **, P < 0.01; ***, P < 0.001. AT, antibiotic-treated mice.
Differential metabolites and KEGG pathway enrichment. (a) Hierarchical clustering heatmap of metabolites identified in cecum contents from K. pneumoniae-infected and antibiotic-treated mice versus K. pneumoniae-infected controls. Red denotes increased expression; green denotes decreased expression. (b) Selected examples of KEGG pathway enrichment. Disturbances in propionate and butyrate metabolism are highlighted in red. The P values were determined using the hypergeometric test and Benjamini-Hochberg FDR correction (b). AT, antibiotic-treated mice. Download FIG S2, TIF file, 1.3 MB (1.4MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To identify whether the metabolism dysfunction was associated with alterations in the gut microbiomes, we carried out 16S rRNA sequencing on the basis of V4 variable regions. We obtained a total of 736,749 high-quality gene sequences, which were then clustered into operational taxonomic units (OTUs) at a 97% similarity level. As with the previous studies (33), phylogenetic diversity analysis showed remarkable differences in the richness and diversity in antibiotic-treated mice (Fig. 2c), and the compositions of bacterial communities were also altered (Fig. 2d). Notably, the abundances of Parabacteroides, Bifidobacterium, Clostridium, Coprococcus, and Prevotella, at the genus level, which produced SCFAs, were all decreased in the antibiotic-treated mice (Fig. 2d). Moreover, similar alterations in the bacterial compositions were observed when all mice were infected with K. pneumoniae (Fig. S3a to c). Based on these findings, we proposed that microbiota depletion, whether the host is infected with K. pneumoniae or not, causes a gut microbiome disturbance, with lower abundances of SCFA-producing bacteria. A Spearman correlation analysis revealed an interrelationship between cecum metabolites and the corresponding gut microbiota among gut antibiotic-treated mice and controls infected with K. pneumoniae or not infected (Fig. 2e; Fig. S3d). It was therefore concluded that alteration of metabolites, SCFAs included, may in part be attributed to changes in gut microbial composition and function, subsequently leading to a dysregulation of host innate immunity.
Association of cecum metabolites and associated gut microbiomes. (a) α-Diversity (Shannon index) and β-diversity (Bray-Curtis similarity index) of 16S rRNA genes from K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. (b, c) Discriminative OTU abundances of taxonomic distributions at the phylum (b) and genus (c) levels between K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. (d) Spearman’s rank correlation between cecum metabolites and associated gut microbiota in K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. Red connections indicate a positive correlation, while blue connections are negative. The P values were determined using a two-tailed Wilcoxon rank sum test (d). *, P < 0.05; **, P < 0.01; ***, P < 0.001. AT, antibiotic-treated mice. Download FIG S3, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SCFAs enhance antimicrobial activity in macrophages.
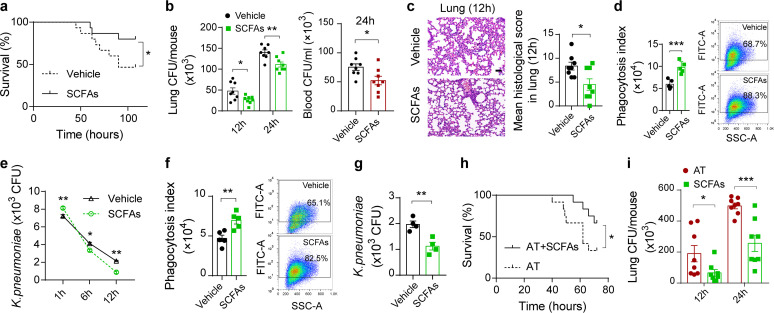
Recent studies have proposed that the vital link between the microbiota and host immunity is due to the production of SCFAs through bacterial metabolism (41). To assess influences of SCFAs on sepsis triggered by K. pneumoniae, mixtures of SCFAs were administered in drinking water before K. pneumoniae challenge (42). Consistently with the previous studies (14, 39), bacterial-infection-induced mortality was reduced in SCFA-treated mice (Fig. 3a). Furthermore, supplementation with SCFAs significantly reduced the bacterial burdens in the lung and blood (Fig. 3b). Pulmonary pathological scores were improved in SCFA-treated mice (Fig. 3c). Then, we validated whether SCFAs modulated alveolar macrophage physiological function with regard to the phagocytosis and clearance of K. pneumoniae. Comparative analysis showed that continuous exposure to SCFAs contributed to significantly increased capacities of macrophages to internalize and eliminate K. pneumoniae (Fig. 3d and e). Abilities to internalize and clear K. pneumoniae were also enhanced in SCFA-treated differentiated THP-1 cells (Fig. 3f and g). Additionally, we examined whether SCFA supplementation in antibiotic-treated mice could ameliorate immune dysfunction during K. pneumoniae-induced sepsis. As a result, supplementation of antibiotic-treated mice with SCFAs led to reduced mortality and decreased bacterial burdens in the lung (Fig. 3h and i). Therefore, we propose that SCFAs enhance host immunity in macrophages during K. pneumoniae infection.
FIG 3.
SCFAs promote inflammatory responses during K. pneumoniae-induced sepsis. (a) Survival rates of SCFA-treated mice and untreated controls postinfection with 1 × 105 CFU of K. pneumoniae. (b) Lung and blood bacterial burdens in SCFA-treated mice and untreated controls 12 h and 24 h after K. pneumoniae infection. (c) Representative H&E staining and quantification of pathological scores of lung sections from SCFA-treated mice and untreated controls 12 h after K. pneumoniae infection. Scale bars, 50 μm. (d) Phagocytic capacity of isolated alveolar macrophages from SCFA-treated mice and untreated controls. (e) Bacterial loads of isolated alveolar macrophages from SCFA-treated mice and untreated controls. (f, g) Phagocytosis (f) and clearance (g) of SCFA-treated differentiated THP-1 cells and untreated controls after K. pneumoniae infection. (h) Survival rates of antibiotic-treated mice treated with SCFAs or vehicle. (i) Pulmonary bacterial burdens in antibiotic-treated mice treated with SCFAs or vehicle. The group size was 8 to 12 mice. Data are from three independent experiments (b, d to g, i) or one experiment representative of three independent experiments (a, c, h) (means ± SEM). The P values were determined using a log rank (Mantel-Cox) test (a, h) or two-tailed Student t tests (b to g, i). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Gut microbiota depletion alters transcriptome profiles.
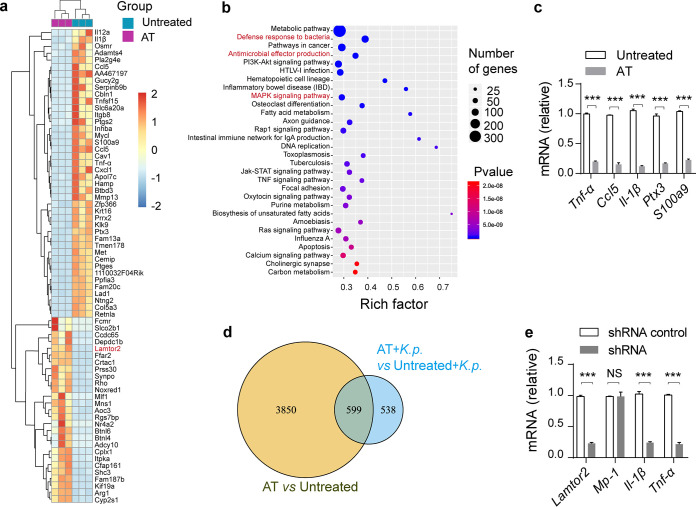
To refine our limited understanding of how the gut microbiota exerted its protective effects against invasion by extracellular pathogens, we performed genome-wide transcriptional sequencing of isolated alveolar macrophages and then analyzed gene expression profiles. Consistently with findings from previous literature (8), gut microbiota depletion had a remarkable influence on the alveolar macrophage transcriptome (Fig. 4a). Gene annotation revealed that substantially enriched KEGG pathways were involved in immunity, such as the defense response to bacteria and MAPK/reactive oxygen species (ROS) signaling cascades (Fig. 4b). To validate the RNA sequencing results, quantitative PCR with reverse transcription (RT-qPCR) was conducted. In line with sequencing results, levels of TNF-α, 1L-1β, CCL5, CXCL1, and S100A9 production were all decreased in antibiotic-treated mice (Fig. 4c). Moreover, we also analyzed differences of genome-wide transcriptional signatures between K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls (Fig. S4a, b). As with the above experiments, we conducted RT-qPCR assays to confirm the sequencing data (Fig. S4c). To search for commonly different expression genes among the four groups above, we performed a Venn diagram analysis (Fig. 4d). We observed a total of 599 genes, which consisted of 322 downregulated genes and 277 upregulated genes. Then, we tried to screen novel antimicrobial effectors during K. pneumoniae infection of RAW264.7 cells via short hairpin RNA (shRNA)-mediated RNA interference (RNAi). Consistently with the previous study (8), we observed that LAMTOR2 was dysregulated in antibiotic-treated mice, which might be attributed to the gut microbiome. In addition, LAMTOR2 was reported to limit Salmonella replications in vivo (31). Based on these data, we supposed that LAMTOR2, probably regulated by the gut microbiome, played a significant role in eliminating K. pneumoniae in macrophages. Fortunately, the immune responses were impaired with regard to downregulated proinflammatory effectors when Lamtor2 expression was knocked down (Fig. S4d and e and Fig. 4e). Taken together, these results suggested that gut microbiota depletion resulted in profound changes in the transcriptional profiles of alveolar macrophages whether K. pneumoniae infection was present or not in a comparison with counterpart controls.
FIG 4.
Gut microbiota depletion alters alveolar macrophage transcriptome profiles. (a) Hierarchical clustering heatmap of selected significant differentially expressed genes (DEGs) in alveolar macrophages from antibiotic-treated and untreated controls. Red denotes increased expression; green denotes decreased expression. (b) Selected examples of KEGG pathway enrichment. The defense response to bacteria, antimicrobial effector production, and the MAPK/ROS signaling pathway are highlighted in red. (c) Validation of Tnf-α, Ccl5, Il-1β, Cxcl1, and S100a9 mRNA expression using RT-qPCR. (d) Venn diagram of differentially expressed genes among four groups. (e) Responsiveness of shRNA-mediated Lamtor2 knockdown RAW264.7 cells and controls in terms of cytokine and chemokine production against K. pneumoniae infection. The group size was 8 to 12 mice. Data are from three independent experiments (c, e). The P values were determined using the hypergeometric test and Benjamini-Hochberg FDR correction (b) or two-tailed Student t tests (c, e). ***, P < 0.001. NS, not significant; AT, antibiotic-treated mice.
Gut microbiota depletion alters transcriptional profiles in infected mice. (a) Hierarchical clustering heatmap of selected significant DEGs between K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. Red denotes increased expression; green denotes decreased expression. (b) Selected examples of KEGG pathway enrichment. (c) Expression levels of Tnf-α, Il-10, Il-1β, Mcp-1, and Cxcl1 mRNA in K. pneumoniae-infected, antibiotic-treated mice compared to those of K. pneumoniae-infected controls. (d) Immunoblot analysis of LAMTOR2 in shRNA-mediated Lamtor2 knockdown RAW264.7 cells. (e) Immunoblot confirmation of the LAMTOR2 protein in Lamtor2−/− RAW264.7 macrophages. (f) Sequencing of targeted Lamtor2 mutations and alignment with those from wild-type RAW264.7 cells. Data are from three independent experiments (c) or one experiment representative of three independent experiments (d to f). The P values were determined using a hypergeometric test and Benjamini-Hochberg FDR correction (b) or a two-tailed Student t test (c). **, P < 0.01; ***, P < 0.001. AT, antibiotic-treated mice. Download FIG S4, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LAMTOR2 increases antimicrobial activity in macrophages.
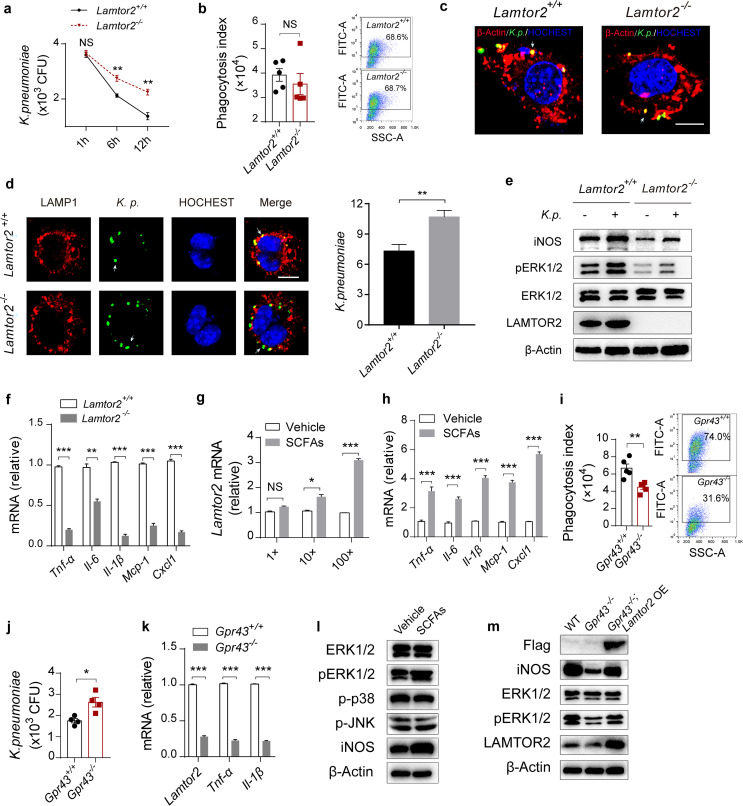
To further investigate the role of LAMTOR2 in K. pneumoniae-induced sepsis, we first investigated whether bacterial loads were altered in Lamtor2−/− RAW264.7 macrophages (Fig. S4f). As a result, bacterial loads were greater than those from normal controls (Fig. 5a), but there was no difference in the levels of uptake of K. pneumoniae (Fig. 5b and c). Next, we explored mechanisms involved in the LAMTOR2-mediated antibacterial ability of RAW264.7 cells. To start, the absence of Lamtor2 contributed to a sharp decrease in the count of phagosomes localized with lysosome (LAMP1) (Fig. 5d), which indicated an inappropriate phagosome-lysosome fusion. Additionally, it has been reported that ERK signaling triggers the production of several proinflammatory mediators, thus facilitating the host immune response (43). Consistently with a previous study (31), protein analysis showed noticeable decreases in ERK phosphorylation (pERK) and iNOS expression in Lamtor2−/− RAW264.7 macrophages (Fig. 5e), which impeded antimicrobial activity against intracellular pathogens. Cytokine and chemokine production was impaired in Lamtor2−/− RAW264.7 cells when they were infected with K. pneumoniae (Fig. 5f). Taken together, these results suggested that LAMTOR2 was necessary for phagosome-lysosome fusion and ERK activation, which collaboratively contributed to bacterial clearance.
FIG 5.
SCFAs facilitate ERK activation by upregulating LAMTOR2. (a, b) Bacterial loads (a) and phagocytic capacity (b) of Lamtor2−/− RAW264.7 cells and controls after K. pneumoniae infection. (c) Immunofluorescence analysis of the uptake of K. pneumoniae in Lamtor2−/− RAW264.7 cells and controls. Arrows indicate K. pneumoniae. Scale bars, 10 μm. (d, left) Immunofluorescence analysis of colocalization between phagosomes and lysosome. (Right) Count of phagosomes localized with LAMP1-stained lysosome. Arrows indicate K. pneumoniae. Scale bars, 10 μm. (e) Immunoblot analysis of phosphorylated (p-) ERK and iNOS expression levels in Lamtor2−/− RAW264.7 cells and controls 6 h after K. pneumoniae infection (MOI = 10). (f) RT-qPCR analysis of Tnf-α, Il-6, Il-1β, Mcp-1, and Cxcl1 mRNA expression in Lamtor2−/− RAW264.7 cells and controls 6 h after K. pneumoniae infection (MOI = 10). (g) RT-qPCR analysis of Lamtor2 mRNA expression in SCFA-treated RAW264.7 cells after K. pneumoniae infection (MOI = 10). (h) Responsiveness of isolated alveolar macrophages from SCFA-treated mice and untreated controls to K. pneumoniae in terms of cytokine and chemokine production in vitro (MOI = 10). (i, j) Phagocytosis (i) and clearance (j) of SCFA-treated Gpr43−/− RAW264.7 cells and controls. (k) RT-qPCR analysis of Lamtor2, Tnf-α, and Il-1β mRNA expression in Gpr43−/− RAW264.7 cells and controls, both of which were incubated with SCFAs. (l) Immunoblot analysis of pERK, p-JNK, p-p38, and iNOS expression levels in RAW264.7 cells treated with SCFAs and controls after 6 h of K. pneumoniae infection. (m) Immunoblot analysis of pERK and iNOS in Gpr43−/− and Gpr43−/− Lamtor2 overexpression (OE) RAW264.7 cells incubated with SCFAs 6 h after K. pneumoniae infection. The group size was 8 to 12 mice. Data are from three independent experiments (a, b, f to k) or one experiment representative of three independent experiments (c, d, e, l, m) (means ± SEM). The P values were determined using two-tailed Student t tests (a, b, d, f to k). *, P < 0.05; **, P < 0.01; ***, P < 0.001. NS, not significant; WT, wild type.
SCFAs facilitate ERK activation by upregulating LAMTOR2.
Next, we explored mechanisms by which SCFAs modulate innate immunity and investigated whether SCFAs collaborate with other inflammatory mediators to potentiate inflammatory and immune responses. Surprisingly, we observed that the expression level of Lamtor2 was upregulated in a concentration-dependent manner in SCFA-treated RAW264.7 cells 6 h after K. pneumoniae infection (Fig. 5g). As with the in vitro results, we found that the Lamtor2 expression level was elevated in vivo when mice were exposed to SCFAs after K. pneumoniae infection (Fig. S5a). Additionally, 6 h after the inoculation of K. pneumoniae in vitro, increased expression levels of cytokines (TNF-α, IL-1β, and IL-6) and chemokines (CXCL1 and MCP-1) were also observed compared to levels in controls without SCFA supplementation (Fig. 5h).
Determination of Lamtor2 mRNA expression in vivo and generation of Gpr43−/− RAW264.7 cells. (a) RT-qPCR analysis of Lamtor2 mRNA expression in isolated alveolar macrophages after K. pneumoniae infection. The P values were determined using two-tailed Student t tests. ***, P < 0.001. (b) Immunoblot validation of GPR43 in Gpr43−/− RAW264.7 cells. (c) Sequencing of targeted Gpr43 mutations in Gpr43−/− RAW264.7 cells and alignment with those from wild-type controls. Data are from three independent experiments (a) or one experiment representative of three independent experiments (b, c). Download FIG S5, TIF file, 0.8 MB (806.8KB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
It has been reported that GPR43/FFAR2 plays an important role in the gut-lung axis, serving as a sensor of host microbiota activity (14). Using Gpr43−/− RAW264.7 macrophages (Fig. S5b, c), we observed that the phagocytosis index was decreased compared to that of controls (Fig. 5i), and bacterial loads were tremendously increased (Fig. 5j). On the basis of these results, we proposed that SCFAs promoted K. pneumoniae uptake and clearance in a GPR43-dependent manner in macrophages. RT-qPCR assay showed that the expression level of Lamtor2 was decreased significantly in Gpr43−/− cells when they were treated with SCFAs and incubated with K. pneumoniae for 6 h, with the production of TNF-α and IL-1β significantly reduced (Fig. 5k). As previously depicted, SCFAs modulated MAPK signaling cascades in a GPR43-dependent manner in human renal cortical epithelial cells (HRCEs) (44). We further explored whether SCFAs/GPR43 had impacts on LAMTOR2 signaling in stimulating inflammatory responses. We investigated the protein levels of pERK, p-JNK, and p-p38 in alveolar macrophages when mice were exposed to SCFAs and then K. pneumoniae. The expression level of pERK was increased, with p-JNK and p-p38 exhibiting no observed alterations (Fig. 5l). We then overexpressed LAMTOR2 in normal and Gpr43−/− RAW264.7 macrophages (Fig. S6a). As expected, the production of inflammatory cytokines (TNF-α, IL-1β, and IL-6) and chemokines (CXCL1 and MCP-1) was elevated (Fig. S6b). Moreover, LAMTOR2 overexpression in Gpr43−/− RAW264.7 cells partly rescued ERK activation (Fig. 5m) and restored their ability to clear a K. pneumoniae infection (Fig. S6c). Therefore, we concluded that SCFA/GPR43 propagated phagocytosis in a LAMTOR2-independent manner, while promoting elimination of a K. pneumoniae infection in a LAMTOR2-dependent manner (Fig. 3d and Fig. 5b).
LAMTOR2 overexpression in RAW264.7 cells. (a) Immunoblot analysis of LAMTOR2 protein in LAMTOR2-overexpressed RAW264.7 cells. (b) Expression levels of Tnf-α, Il-6, Il-1β, Mcp-1, and Cxcl1 mRNA in LAMTOR2-overexpressed RAW264.7 cells after K. pneumoniae infection. (c) Bacterial loads of Gpr43−/− and Gpr43−/− Lamtor2 overexpression (OE) in RAW264.7 cells. Data are from three independent experiments (b, c) or one experiment representative of three independent experiments (a). The P values were determined using two-tailed Student t tests (b, c). **, P < 0.01; ***, P < 0.001. Download FIG S6, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lamtor2 conditional disruption attenuates antimicrobial activity in vivo.
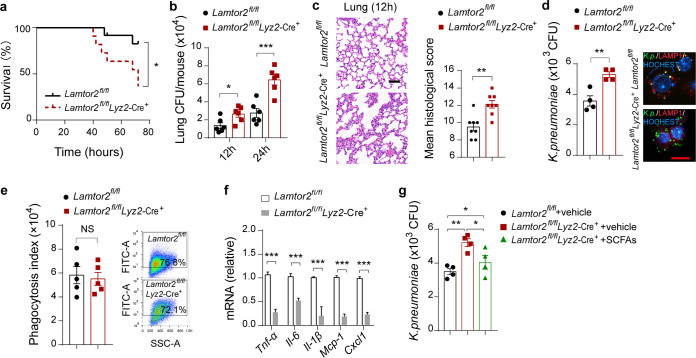
To investigate whether the functions of LAMTOR2 on macrophage activation are relevant in vivo, we constructed a Lamtor2 conditional knockout mouse using CRISPR/Cas9-mediated genome editing and crossed the mouse with Lyz2-Cre+ transgenic mice to generate Lamtor2fl/fl Lyz2-Cre+ mice (Fig. S7a to d). We observed that Lamtor2fl/fl Lyz2-Cre+ mice had an elevated mortality after K. pneumoniae infection compared to that of littermate Lamtor2fl/fl controls (Fig. 6a). In line with observations in vitro, bacterial burdens in the lungs and blood of Lamtor2fl/fl Lyz2-Cre+ mice were tremendously increased 12 h and 24 h after K. pneumoniae infection (Fig. 6b and Fig. S7e). In addition, we analyzed the pulmonary pathology of Lamtor2fl/fl Lyz2-Cre+ and littermate controls 12 h and 24 h after K. pneumoniae challenge. Consequently, an exacerbated lung pathology with enhanced interstitial inflammation, bronchitis, and large surfaces of confluent inflammation infiltration was observed (Fig. 6c and Fig. S7f). Moreover, bacterial loads were evaluated in isolated alveolar macrophages from Lamtor2fl/fl Lyz2-Cre+ mice and compared to those from Lamtor2fl/fl mice (Fig. 6d); the phagocytosis indexes were also evaluated and determined to have no alteration (Fig. 6e). In addition, the responsiveness of isolated primary alveolar macrophages derived from Lamtor2fl/fl Lyz2-Cre+ mice were decreased compared to those of Lamtor2fl/fl mice in response to invading K. pneumoniae (Fig. 6f) and lipopolysaccharide (LPS) (Fig. S7g), respectively.
FIG 6.
Lamtor2 loss of function leads to a decreased antimicrobial activity in vivo. (a) Survival rates of Lamtor2fl/fl Lyz2-Cre+ and littermate Lamtor2fl/fl mice intranasally infected with K. pneumoniae. (b) Pulmonary bacterial burdens of Lamtor2fl/fl Lyz2-Cre+ mice and controls 12 h and 24 h after K. pneumoniae infection. (c) Representative H&E staining and quantification of pathological scores of lung sections from Lamtor2fl/fl Lyz2-Cre+ mice and controls 12 h after K. pneumoniae infection. Scale bars, 50 μm. (d) Bacterial loads of isolated alveolar macrophages from Lamtor2fl/fl Lyz2-Cre+ mice and controls after K. pneumoniae infection. Scale bars, 20 μm. (e) Phagocytic capacity of isolated alveolar macrophages from Lamtor2fl/fl Lyz2-Cre+ mice and controls after K. pneumoniae infection. (f) Responsiveness of isolated alveolar macrophages from Lamtor2fl/flLyz2-Cre+ mice and controls to K. pneumoniae in terms of TNF-α, IL-6, 1L-1β, MCP-1, and CXCL1 production. (g) Bacterial loads of isolated alveolar macrophages from Lamtor2fl/fl Lyz2-Cre+ mice and controls treated with SCFAs after K. pneumoniae infection. The group size was 8 to 12 mice. Data are from three independent experiments (b, d to g) or one experiment representative of three independent experiments (a, c) (means ± SEM). The P values were determined using a log rank (Mantel-Cox) test (a), two-tailed Student t tests (b to f), or one-way ANOVA for multiple comparisons (g). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Generation of Lamtor2 conditional knockout (cKO) mice via CRISPR-Cas9 genome editing. (a) Schematic generation of Lamtor2 conditional knockout mice. Targeted exons 1 to 3 of the Lamtor2 gene were flanked by Loxp sites. Maps of the Lamtor2 wild-type allele, targeting vector, Lamtor2-targeted allele, and Lamtor2 knockout allele are shown. (b) Genotyping of wild-type (Lamtor2+/+), fl/+ (Lamtor2fl/+), and fl/fl (Lamtor2fl/fl) mice was performed by PCR. (c) RT-qPCR analysis of Lamtor2 mRNA expression in targeted alveolar macrophages. (d) Immunoblot analysis of the LAMTOR2 protein in targeted alveolar macrophages. (e) Blood bacterial burdens in Lamtor2fl/fl Lyz2-Cre+ and Lamtor2fl/fl mice 12 h and 24 h after K. pneumoniae infection. (f) Representative H&E staining and quantification of pathological scores of lung sections from Lamtor2fl/fl Lyz2-Cre+ mice and controls 24 h after K. pneumoniae infection. Scale bars, 50 μm. (g) Responsiveness of isolated alveolar macrophages from Lamtor2fl/fl Lyz2-Cre+ mice and controls against lipopolysaccharide (LPS) in terms of TNF-α, IL-6, 1L-1β, MCP-1, and CXCL1 production. Data are from three independent experiments (c, e, g) or one experiment representative of three independent experiments (b, d, f) (means ± SEM). The P values were determined using two-tailed Student t tests (c, e, f, g). **, P < 0.01; ***, P < 0.001. Download FIG S7, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we investigated whether isolated alveolar macrophages derived from Lamtor2fl/fl Lyz2-Cre+ mice treated with SCFAs exhibited an increased ability to eliminate extracellular invading bacteria compared to counterparts without SCFAs. Clearly, macrophages from Lamtor2 mutant mice treated with SCFAs harbored fewer K. pneumoniae organisms 6 h postinfection than untreated Lamtor2 mutant mice but more than treated Lamtor2fl/fl controls (Fig. 6g). Accordingly, these results indicated that SCFAs were associated with the LAMTOR2 signaling cascades in modulating antimicrobial activity in vivo.
DISCUSSION
Pneumonia remains a leading cause of death and hospitalization worldwide, especially among children and the elderly (1, 2). Early and aggressive antibiotic treatments indeed exerted great control over these bacterial infections (45). Recent breakthroughs in our understanding of the protective role of the gut microbiota in our health have novel implications for respiratory and critical care medicine. In this study, we showed that the gut microbiota played a protective role in K. pneumoniae-induced lung impairment and highlighted that SCFAs enhanced macrophage phagocytosis of K. pneumoniae independently of LAMTOR2, while promoting host elimination of infected K. pneumoniae via a LAMTOR2-dependent signal pathway. We take a new step toward understanding the relationship between the gut microbiota and remote organs or immune cells with respect to the inflammatory response to extracellular invading pathogens.
It has become apparent that large communities of intestinal microbes not only fine-tune immune cell function locally in the vicinity of the mucosa but also exert a systemic influence on effector cells of the innate immune system at extraintestinal tissues or organs (46, 47). The mechanistic basis for these distal influences, however, has been incompletely characterized. In the lung, it has been suggested that resistance to multiple bacterial and viral pathogens is enhanced by the gut microbiota (8, 33). Trompette et al. once established the concept of the gut-lung axis in allergic airway disease by showing that gut microbiota-derived metabolites have an influence on the severity of allergic inflammation (48). Recently, Clarke demonstrated that gut microbiota depletion causes significant defects in the early innate response to lung infection by the major human pathogen K. pneumoniae and proposed that nod-like receptor ligands are required to facilitate early bacterial clearance from the lung (32). Consistently with these results, we show that the gut microbiota protects against lung injury during K. pneumoniae-induced sepsis, resulting in decreased bacterial burdens and higher survival rates.
Recent advances have identified SCFAs as an “indispensable linker” in host microbiota communication networks. Initially described as a fuel resource for epithelial cells (49), hepatocytes and peripheral tissues are now rapidly emerging as critical signals that directly influence immunity and cell function (42, 50). Prime mechanisms involved in SCFA immunomodulatory effects in neutrophils and macrophages are through binding and activating GPCRs. Indubitably, both Gpr41−/− and Gpr43−/− mice had reduced immune responses to Citrobacter rodentium infection (15). Work conducted by Galvão et al. suggested that the microbial metabolic sensor GPR43 modulated the lung’s innate immunity to bacterial pneumonia through binding ligand acetate (14). The cascades of SCFAs involved in regulating immune responses, including downstream targets, however, need to be further considered. In the present study, we show for the first time that SCFAs/GPR43 promote inflammatory responses against K. pneumoniae by activating the intracellular antibacterial effector LAMTOR2. LAMTOR2 initially was reported to combine with MP-1 to modulate MAPK signaling within a cell (27). Afterwards, LAMTOR2 was identified as a mediator in the immune response, as described for patients harboring a LAMTOR2 point mutation allele and, as a consequence, suffering from recurrent bronchopulmonary infections (51). Among the literature are observations that mice with LAMTOR2-deficient dendritic cells have a severe disturbance of the DC compartment (29). In the present report, we show that Lamtor2−/− RAW264.7 cells have increased bacterial loads, which were ascribed to inappropriate transport of phagosomes to lysosomes and impaired ERK activation. Furthermore, conditional ablation of Lamtor2 in vivo resulted in K. pneumoniae populating excessively where alveolar macrophage antimicrobial responses were not appropriately activated.
Our work highlights the concept that the antibacterial activity of alveolar macrophages in the distal lung is programmed and corrected systemically by signals derived from the gut microbiota. Nevertheless, whether there are other novel gut microbiota-derived factors and metabolites contributing to these effects remains to be further investigated. In addition, how these gut microbial metabolite SCFAs in combination with the host internal antibacterial effector LAMTOR2 regulate the host immune system needs to be further elucidated. More importantly, it remains to be determined whether the observed effects of gut microbiota depletion on K. pneumoniae infection also apply to infections with other important causative agents of pneumonia. On the other hand, characterizing microbial biomarkers has enormous potential for precision medicine and is a relatively simple way of translating microbiome research into clinical practice. In this report, we underlined for the first time the influence of gut microbiota depletion together with K. pneumonia challenge of the alveolar macrophage transcriptome and cecum metabolism, which were correlated with obviously observed phenotypes accompanied by a significantly diminished capacity to phagocytize and eliminate K. pneumoniae. Of clinical significance, thus, is how altered alveolar macrophage transcriptome and gut microbial metabolites are associated with K. pneumoniae pneumonia. Based on these data, therefore, it may be worthwhile exploring future new diagnostic biomarkers and novel therapeutics in bacterial pneumonia treatment targeting the gut microbiota.
MATERIALS AND METHODS
Mice.
Wild-type C57BL/6 mice were purchased from the Experimental Animal Center of Anhui Province (Hefei, China). Lamtor2fl/+ mice (C57BL/6J) were generated by CRISPR-Cas9-mediated genome-editing technology (Cyagen Biosciences, China) targeting exons 1 to 3. Lamtor2 conditional knockout mice were generated by crossing Lamtor2fl/fl mice with Lyz2-Cre+/− transgenic mice. Meanwhile, we obtained Lamtor2fl/fl littermate controls. All experiments involving mice were approved by the Institutional Animal Care and Use Committee of Anhui Medical University (Hefei, China).
Bacterial strains and cells.
K. pneumoniae (ATCC 43816) was cultured in Luria broth (LB) at 37°C overnight. HEK293T, RAW264.7, and THP-1 cell lines were all obtained from the American Type Culture Collection (ATCC) and cultured in endotoxin-free Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Thermo Fisher, USA). Moreover, THP-1 monocytes were incubated with 100 ng/ml phorbol myristate acetate (PMA) for at least 48 h to differentiate into macrophages.
Reagents and consumable material.
Main reagents and consumable material are listed in Table S1 in the supplemental material. Antibodies are listed in Table S2.
Reagents and materials. Download Table S1, PDF file, 0.02 MB (20.9KB, pdf) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibodies. Download Table S2, PDF file, 0.1 MB (74.1KB, pdf) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Models of microbiota depletion and K. pneumoniae infection.
Mice were treated with broad-spectrum antibiotics (ampicillin, 1 g · liter−1; neomycin sulfate, 1 g · liter−1; metronidazole, 1 g · liter−1; and vancomycin, 0.5 g · liter−1) administered in drinking water for 14 days (8, 33). Antibiotic treatment was stopped 3 days prior to infection. Then, antibiotic-treated mice and untreated controls were anesthetized with pelltobarbitalum natricum, inoculated intranasally with 1 × 105 CFU of K. pneumoniae in 50 μl of sterile phosphate-buffered saline (PBS), and sacrificed at different time points postinfection. Pathology scoring was conducted as described in reference 8; the total lung inflammation score was counted as the sum of the scores for each parameter, the maximum being 24, and the maximum score for liver and kidney sections was 12. To quantify bacterial burdens in lung, mice were sacrificed, and then organs were removed, homogenized in PBS, and cultivated on LB agar plates. For survival curves, mice were monitored 4 to 5 times daily until mice with characteristics of reduced movement, shivering, dyspnea, or circling were killed.
Isolation of alveolar macrophages.
Alveolar macrophages were isolated as previously described (52). Briefly, mice were sacrificed and immediately exsanguinated. Bronchoalveolar lavage fluid (BALF) was collected with 4 ml of 37°C sterile PBS containing 0.5 mM EDTA. Cells were pelleted and resuspended in RPMI 1640 supplemented with 5.0% (vol/vol) FBS and then were allowed to adhere to a tissue culture flask for 2 h (37°C, 5% CO2 [vol/vol]). In general, alveolar macrophage purity was more than 93% as analyzed by flow cytometry (FACS Celesta; BD Biosciences).
Phagocytosis and killing assays.
Bacterial phagocytosis and killing assays were performed essentially as described previously (8, 46). To begin, a suspension of bacteria was suspended in PBS (pH 9.0) and marked with carboxyfluorescein succinimidyl ester (CFSE; Invitrogen, the Netherlands), with stirring at 37°C for 30 min. Meanwhile, RAW264.7 cells, differentiated THP-1, or alveolar macrophages were incubated in complete medium without antibiotics for at least 2 h and then infected with K. pneumoniae (multiplicity of infection [MOI] = 100). After 1 h of incubation, cells were washed with complete medium without antibiotics but supplemented with 0.05% gentamicin (50 g · liter−1) and then were washed with cold PBS (pH 7.3). Subsequently, cells were analyzed by flow cytometry (FACS Celesta; BD Biosciences) for phagocytosis (the phagocytosis index is the geometric mean fluorescence times the percentage of positive cells) or incubated for the other times indicated (3 h, 6 h, and 12 h) and then lysed for killing assays. Briefly, cells were lysed with 0.3% (vol/vol) Triton X-100 for 5 min. Cell lysates were then serially diluted with PBS and inoculated on LB agar plates. Bacterial CFU were counted after incubation of the specimens at 37°C for 16 h.
Generation and validation of stable gene knockout cell lines.
To generate stable gene knockout cell lines, 5-μg plasmids encoding single guide RNAs (sgRNAs) were transfected using Lipofectamine 2000 (Invitrogen), with strict adherence to the manufacturer´s instructions. Puromycin selection was performed for 1 day at 1 mg · liter−1; cells were then split into a 96-well plate (Corning, USA), with 2 to 3 cells/well, and clones were picked 10 days later. Clones were expanded into 6-well dishes, and validation was performed by Western blotting and sequencing.
SCFA mixture treatment assay.
As conducted previously (14, 42), groups of 8 or 10 wild-type mice were pretreated with SCFA mixtures administered in drinking water (100 mM acetate, 25 mM propionate, and 25 mM butyrate) (Sigma-Aldrich) for 7 days (with fresh solutions three or four times a week). Then, mice were anesthetized with isoflurane and inoculated intranasally with 1 × 105 CFU of K. pneumoniae in 50 μl PBS. In addition, SCFA mixtures were added to RAW264.7 cells in vitro for 24 h at concentrations of 100× in the peripheral blood of healthy people (20 mM acetate, 0.5 mM propionate, and 0.5 mM butyrate). Then, RAW264.7 cells were infected with K. pneumoniae at an MOI of 1:100.
Immunofluorescence assay.
Quantification of bacterial killing and LAMTOR2-associated phagolysosome maturation were determined with a Zeiss LSM-800 confocal microscope (Carl Zeiss, Germany). Briefly, Lamtor2−/− RAW264.7 cells and wild-type controls (1 × 105 cells/well) were seeded in 4-well chamber slides (ThermoFisher Scientific, catalog [cat.] no. 155383) and then were infected with CSFE-K. pneumoniae (MOI = 100) for 1 h. Cells were next treated with gentamicin (100 g · liter−1) in incomplete medium to kill extracellular K. pneumoniae. Next, cells were immediately fixed on ice for 15 min with 4% paraformaldehyde after being washed twice with cold PBS containing gentamicin and then permeabilized with 0.5% TritonX-100 for 30 min at room temperature. Fixed and permeabilized cells were blocked with 5% bovine serum albumin in PBST (0.5% Tween 20 in PBS) for 30 min, incubated for 2 h with the primary antibody, and then washed three times with PBST and incubated for 60 min with the secondary antibody. Additionally, cells were stained with Hoechst 33258 (Sigma) to clarify the boundaries of the nucleus. Finally, images of macrophages were taken as z-stacks of multiple sections collected at 0.5-mm intervals at ×63 magnification with a Zeiss 800 inverted confocal microscope (Carl Zeiss, Germany). Data were processed with a Light Cycler 96 SW 1.1 (Roche, Germany).
DNA extraction and 16S rRNA sequencing.
Mouse fecal samples were collected before the mice were sacrificed, frozen immediately following collection, and stored at –80°C prior to analysis. Fecal samples were pulverized with a mortar and pestle in liquid nitrogen, and bacterial genomic DNA was then extracted with a QIAamp DNA stool minikit (Qiagen). V4 region amplicon sequencing (515F-GTGCCAGCMGCCGCGGTAA and 806R-GGACTACHVGGGTWTCTAAT) (53) of the 16S rRNA gene was performed on an Illumina HiSeq2500 sequencer at the Beijing Genomics Institute (BGI-Shenzhen, China). mothur (http://www.mothur.org/) (54) was used to obtain unique reads. Sequences of less than 200 bp and greater than 1,000 bp as well as sequences containing any primer mismatches, barcode mismatches, ambiguous bases, and homopolymer runs exceeding 6 bases were all excluded. All remaining sequences were assigned to operational taxonomic units (OTUs) with a 97% threshold of pairwise identity and then classified taxonomically using the RDP database (http://www.mothur.org/wiki/RDP_reference_files) (55). These taxonomies were used to construct summaries of the taxonomic distributions of OTUs, which can then be applied to calculate the relative abundances of microbiota at different levels.
RNA extraction, reverse transcription, quantitative real-time PCR, and RNA sequencing.
RNA was extracted from RAW264.7 cells or alveolar macrophages homogenates using an RNeasy Plus minikit (Qiagen, cat. no. 74134) by strictly following the manufacturer’s protocol. For reverse transcription, single-strand cDNA was synthesized using a PrimeScript 1st-strand cDNA synthesis kit (TaKaRa, cat. no. D6110A). Real-time PCR was performed using PrimeScript RT master mix (TaKaRa, cat. no. RR036A) by a three-step real-time PCR system (Light Cycler 96). The target gene expression levels were normalized to that of the housekeeping gene (Gapdh) mRNA, determined by the 2–ΔΔCT calculation method, where CT is the threshold cycle. Primers for real-time PCR are listed in Table S3. RNA sequencing was performed by the Beijing Genomics Institute (BGI-Shenzhen, China) using the BGISEQ-500 platform. The sequencing libraries were constructed as described in a previous study (56). Expression levels for each of the genes were normalized to the number of fragments per kilobase of the exon model per million mapped reads (FPKM) by RNA sequencing expectation maximization (RSEM). Differentially expressed genes (DEGs) were analyzed as indicated at http://david.abcc.ncifcrf.gov (57). Pathways enriched with DEGs were annotated in the KEGG database (Kyoto Encyclopedia of Genes and Genomes) (58).
Oligonucleotides. Download Table S3, PDF file, 0.1 MB (63.5KB, pdf) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cecal content metabolic sequencing.
Cecal contents were subsequently extracted at the death of the mice and then frozen immediately at –80°C. For metabolomics profiling, all cecum samples were thawed on ice, and a quality control (QC) sample, made by mixing and blending equal volumes (10 μl) of each cecum sample, was used to estimate a mean profile representing all the analytes encountered during analysis. We isolated and extracted metabolites (<1,500 Da) as follows. First, 100-μl cecum mixtures were precipitated with 200 μl methanol, and similarly, the QC sample was precipitated with methanol (1:2, vol/vol). All samples were subsequently centrifuged at 14,000 × g for 10 min at 4°C. The supernatants were subjected to metabolomics profiling by liquid chromatography mass spectrometry (LC-MS) at the Beijing Genomics Institute (BGI-Shenzhen, China). The acquired MS data pretreatments, including peak selection and grouping, retention time correction, a second peak grouping, and isotope and adduct annotation, were performed as previously described (59). LC-MS raw data files were converted into the mzXML format and then analyzed by the XCMS and CAMERA toolbox with the R statistical language (v3.4.1). The online KEGG database (http://www.genome.jp/kegg/) (58) and HMDB database (http://www.hmdb.ca) (60) were used to identify different metabolites. If a mass difference between observed and theoretical masses was less than 10 ppm, the metabolite name was reported and the molecular formulas of the matched metabolites were further validated by isotopic distribution measurements. Commercial reference standards were used to validate and confirm metabolites by comparison of their retention times and MS/MS spectra.
Flow cytometry.
For RAW246.7 cells or alveolar macrophages, flow cytometry was conducted as follows. Briefly, 500 μl of cold 50 mM EDTA in PBS was added to each well and incubated for at least 30 min at 37°C in 5% CO2 in a cell culture incubator. Subsequently, the cells were transferred to FACS tubes and centrifuged at 1,000 × g for 10 min. The supernatant was carefully removed, and the cell pellet was resuspended in 200 μl of freshly prepared staining solution. The samples were incubated in the dark for 25 min, and subsequently 200 μl of FACS buffer (2 mM EDTA in 10% PBS) was added until the sample was analyzed by a FACSCalibur flow cytometer (FACS Celesta, BD Biosciences). Electronic compensation was used to eliminate bleed-through fluorescence.
Statistical analysis.
All results are presented as means ± standard errors of the means (SEM). Statistical analysis was performed using unpaired Student t tests for two groups and one-way analysis of variance (ANOVA) or two-way ANOVA for multiple groups, with all data points showing a normal distribution. Mouse survival data were plotted as Kaplan-Meier curves and compared using the log rank (Mantel-Cox) test. Sample sizes were selected on the basis of preliminary results to ensure an adequate power. The study and experiments were not randomized or carried out in a blind manner. The results were considered statistically significant or very significant when P values were less than 0.05 or 0.01, respectively. All graphs were generated using Adobe Illustrator CC (2017 release) or GraphPad Prism 7.
Data availability.
Sequencing data and relevant files have been uploaded to public repositories. LC-MS data were deposited into the China National GenBank Database with the accession no. CNP0001290. The 16S sequencing data are available in GenBank with the accession no. MW011789 to MW012255. Transcriptome sequencing data can be found on the NCBI Sequence Read Archive (SRA) with the accession no. PRJNA662812.
ACKNOWLEDGMENTS
We sincerely thank Jiajia Li and Zhou Liu for technical support and Wanyin Tao (School of Life Sciences, University of Science and Technology of China) for critical discussions. We thank Jiang Li and Jiatao Liu for the suggestions on the layout of the drawings.
This work is supported by the National Natural Science Foundation of China (grants 81973983 and 81673242 to J.L.).
We declare no conflict of interest.
The animal study was reviewed and approved by the Animal Experimentation Ethics Committee of Anhui Medical University (approval LLSC20190253), and experiments were carried out in strict accordance with institutional guidelines for the care and use of laboratory animals.
Yanhu Lan, T.W., D.X., and J.L. conceived and designed the study. T.W., Yanhu Lan, C.S., F.X., G.Y., K.S., N.L., B.M., and Yanyan Liu performed the experiments. L.H. and M.X. helped with the confocal microscopy. T.W., Yanhu Lan, C.S., and F.X. analyzed the data. Yanhu Lan and T.W. wrote the manuscript. J.L., H.L., and D.X. revised the manuscript. H.W., Yan Liu, and Y.G. provided critical suggestion. All authors reviewed the manuscript.
REFERENCES
- 1.Bengoechea JA, Sa Pessoa J. 2019. Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev 43:123–144. doi: 10.1093/femsre/fuy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Navon-Venezia S, Kondratyeva K, Carattoli A. 2017. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev 41:252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 3.Blander JM, Longman RS, Iliev ID, Sonnenberg GF, Artis D. 2017. Regulation of inflammation by microbiota interactions with the host. Nat Immunol 18:851–860. doi: 10.1038/ni.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, Kurilshikov A, Bonder MJ, Jiang X, Tigchelaar EF, Dekens J, Peters V, Voskuil MD, Visschedijk MC, van Dullemen HM, Keszthelyi D, Swertz MA, Franke L, Alberts R, Festen EAM, Dijkstra G, Masclee AAM, Hofker MH, Xavier RJ, Alm EJ, Fu J, Wijmenga C, Jonkers D, Zhernakova A, Weersma RK. 2018. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med 10:eaap8914. doi: 10.1126/scitranslmed.aap8914. [DOI] [PubMed] [Google Scholar]
- 5.Blacher E, Bashiardes S, Shapiro H, Rothschild D, Mor U, Dori-Bachash M, Kleimeyer C, Moresi C, Harnik Y, Zur M, Zabari M, Brik RB, Kviatcovsky D, Zmora N, Cohen Y, Bar N, Levi I, Amar N, Mehlman T, Brandis A, Biton I, Kuperman Y, Tsoory M, Alfahel L, Harmelin A, Schwartz M, Israelson A, Arike L, Johansson MEV, Hansson GC, Gotkine M, Segal E, Elinav E. 2019. Potential roles of gut microbiome and metabolites in modulating ALS in mice. Nature 572:474–480. doi: 10.1038/s41586-019-1443-5. [DOI] [PubMed] [Google Scholar]
- 6.Ruane D, Chorny A, Lee H, Faith J, Pandey G, Shan M, Simchoni N, Rahman A, Garg A, Weinstein EG, Oropallo M, Gaylord M, Ungaro R, Cunningham-Rundles C, Alexandropoulos K, Mucida D, Merad M, Cerutti A, Mehandru S. 2016. Microbiota regulate the ability of lung dendritic cells to induce IgA class-switch recombination and generate protective gastrointestinal immune responses. J Exp Med 213:53–73. doi: 10.1084/jem.20150567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dockrell DH, Whyte MKB, Mitchell TJ. 2012. Pneumococcal pneumonia: mechanisms of infection and resolution. Chest 142:482–491. doi: 10.1378/chest.12-0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa e Melo F, Roelofs JJ, de Boer JD, Hoogendijk AJ, de Beer R, de Vos A, Belzer C, de Vos WM, van der Poll T, Wiersinga WJ. 2016. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 65:575–583. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang G, Huang S, Wang Y, Cai S, Yu H, Liu H, Zeng X, Zhang G, Qiao S. 2019. Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci 76:3917–3937. doi: 10.1007/s00018-019-03190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Illiano P, Brambilla R, Parolini C. 2020. The mutual interplay of gut microbiota, diet and human disease. FEBS J 287:833–855. doi: 10.1111/febs.15217. [DOI] [PubMed] [Google Scholar]
- 11.Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F. 2016. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 12.Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME, Brezillon S, Dupriez V, Vassart G, Van Damme J, Parmentier M, Detheux M. 2003. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem 278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- 13.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, Mackay CR. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- 14.Galvão I, Tavares LP, Correa RO, Fachi JL, Rocha VM, Rungue M, Garcia CC, Cassali G, Ferreira CM, Martins FS, Oliveira SC, Mackay CR, Teixeira MM, Vinolo MAR, Vieira AT. 2018. The metabolic sensor GPR43 receptor plays a role in the control of Klebsiella pneumoniae infection in the lung. Front Immunol 9:142. doi: 10.3389/fimmu.2018.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. 2013. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145:396–406.e1-10. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 16.Schulthess J, Pandey S, Capitani M, Rue-Albrecht KC, Arnold I, Franchini F, Chomka A, Ilott NE, Johnston DGW, Pires E, McCullagh J, Sansom SN, Arancibia-Carcamo CV, Uhlig HH, Powrie F. 2019. The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50:432–445.e7. doi: 10.1016/j.immuni.2018.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 18.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, Glickman JN, Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghorbani P, Santhakumar P, Hu Q, Djiadeu P, Wolever TM, Palaniyar N, Grasemann H. 2015. Short-chain fatty acids affect cystic fibrosis airway inflammation and bacterial growth. Eur Respir J 46:1033–1045. doi: 10.1183/09031936.00143614. [DOI] [PubMed] [Google Scholar]
- 20.Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, Vatanen T, Hall AB, Mallick H, McIver LJ, Sauk JS, Wilson RG, Stevens BW, Scott JM, Pierce K, Deik AA, Bullock K, Imhann F, Porter JA, Zhernakova A, Fu J, Weersma RK, Wijmenga C, Clish CB, Vlamakis H, Huttenhower C, Xavier RJ. 2019. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 4:293–305. doi: 10.1038/s41564-018-0306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duscha A, Gisevius B, Hirschberg S, Yissachar N, Stangl GI, Eilers E, Bader V, Haase S, Kaisler J, David C, Schneider R, Troisi R, Zent D, Hegelmaier T, Dokalis N, Gerstein S, Del Mare-Roumani S, Amidror S, Staszewski O, Poschmann G, Stuhler K, Hirche F, Balogh A, Kempa S, Trager P, Zaiss MM, Holm JB, Massa MG, Nielsen HB, Faissner A, Lukas C, Gatermann SG, Scholz M, Przuntek H, Prinz M, Forslund SK, Winklhofer KF, Muller DN, Linker RA, Gold R, Haghikia A. 2020. Propionic acid shapes the multiple sclerosis disease course by an immunomodulatory mechanism. Cell 180:1067–1080.e16. doi: 10.1016/j.cell.2020.02.035. [DOI] [PubMed] [Google Scholar]
- 22.Dalile B, Van Oudenhove L, Vervliet B, Verbeke K. 2019. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat Rev Gastroenterol Hepatol 16:461–478. doi: 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- 23.Dang AT, Marsland BJ. 2019. Microbes, metabolites, and the gut-lung axis. Mucosal Immunol 12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 24.Blander JM, Medzhitov R. 2006. On regulation of phagosome maturation and antigen presentation. Nat Immunol 7:1029–1035. doi: 10.1038/ni1006-1029. [DOI] [PubMed] [Google Scholar]
- 25.Berger SB, Romero X, Ma C, Wang G, Faubion WA, Liao G, Compeer E, Keszei M, Rameh L, Wang N, Boes M, Regueiro JR, Reinecker HC, Terhorst C. 2010. SLAM is a microbial sensor that regulates bacterial phagosome functions in macrophages. Nat Immunol 11:920–927. doi: 10.1038/ni.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Z, Cui P, Zhang K, Chen Q, Fang X. 2017. Transient receptor potential melastatin 2 regulates phagosome maturation and is required for bacterial clearance in Escherichia coli sepsis. Anesthesiology 126:128–139. doi: 10.1097/ALN.0000000000001430. [DOI] [PubMed] [Google Scholar]
- 27.Wunderlich W, Fialka I, Teis D, Alpi A, Pfeifer A, Parton RG, Lottspeich F, Huber LA. 2001. A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J Cell Biol 152:765–776. doi: 10.1083/jcb.152.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurzbauer R, Teis D, de Araujo ME, Maurer-Stroh S, Eisenhaber F, Bourenkov GP, Bartunik HD, Hekman M, Rapp UR, Huber LA, Clausen T. 2004. Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc Natl Acad Sci U S A 101:10984–10989. doi: 10.1073/pnas.0403435101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheffler JM, Sparber F, Tripp CH, Herrmann C, Humenberger A, Blitz J, Romani N, Stoitzner P, Huber LA. 2014. LAMTOR2 regulates dendritic cell homeostasis through FLT3-dependent mTOR signalling. Nat Commun 5:5138. doi: 10.1038/ncomms6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nada S, Hondo A, Kasai A, Koike M, Saito K, Uchiyama Y, Okada M. 2009. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J 28:477–489. doi: 10.1038/emboj.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taub N, Nairz M, Hilber D, Hess MW, Weiss G, Huber LA. 2012. The late endosomal adaptor p14 is a macrophage host-defense factor against Salmonella infection. J Cell Sci 125:2698–2708. doi: 10.1242/jcs.100073. [DOI] [PubMed] [Google Scholar]
- 32.Clarke TB. 2014. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun 82:4596–4606. doi: 10.1128/IAI.02212-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown RL, Sequeira RP, Clarke TB. 2017. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun 8:1512. doi: 10.1038/s41467-017-01803-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strieter RM, Belperio JA, Keane MP. 2002. Cytokines in innate host defense in the lung. J Clin Invest 109:699–705. doi: 10.1172/JCI15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meng X, Zhou HY, Shen HH, Lufumpa E, Li XM, Guo B, Li BZ. 2019. Microbe-metabolite-host axis, two-way action in the pathogenesis and treatment of human autoimmunity. Autoimmun Rev 18:455–475. doi: 10.1016/j.autrev.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Oliphant K, Allen-Vercoe E. 2019. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome 7:91. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavelle A, Sokol H. 2020. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 17:223–237. doi: 10.1038/s41575-019-0258-z. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D, Li S, Wang N, Tan H-Y, Zhang Z, Feng Y. 2020. The cross-talk between gut microbiota and lungs in common lung diseases. Front Microbiol 11:301. doi: 10.3389/fmicb.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sencio V, Barthelemy A, Tavares LP, Machado MG, Soulard D, Cuinat C, Queiroz-Junior CM, Noordine ML, Salome-Desnoulez S, Deryuter L, Foligne B, Wahl C, Frisch B, Vieira AT, Paget C, Milligan G, Ulven T, Wolowczuk I, Faveeuw C, Le Goffic R, Thomas M, Ferreira S, Teixeira MM, Trottein F. 2020. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep 30:2934–2947.e6. doi: 10.1016/j.celrep.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Antunes KH, Fachi JL, de Paula R, da Silva EF, Pral LP, dos Santos AÁ, Dias GBM, Vargas JE, Puga R, Mayer FQ, Maito F, Zárate-Bladés CR, Ajami NJ, Sant’Ana MR, Candreva T, Rodrigues HG, Schmiele M, Silva Clerici MTP, Proença-Modena JL, Vieira AT, Mackay CR, Mansur D, Caballero MT, Marzec J, Li J, Wang X, Bell D, Polack FP, Kleeberger SR, Stein RT, Vinolo MAR, de Souza APD. 2019. Microbiota-derived acetate protects against respiratory syncytial virus infection through a GPR43-type 1 interferon response. Nat Commun 10:3273. doi: 10.1038/s41467-019-11152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fachi JL, Felipe JS, Pral LP, da Silva BK, Correa RO, de Andrade MCP, da Fonseca DM, Basso PJ, Camara NOS, de Sales ESEL, Dos Santos Martins F, Guima SES, Thomas AM, Setubal JC, Magalhaes YT, Forti FL, Candreva T, Rodrigues HG, de Jesus MB, Consonni SR, Farias ADS, Varga-Weisz P, Vinolo MAR. 2019. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-dependent mechanism. Cell Rep 27:750–761.e7. doi: 10.1016/j.celrep.2019.03.054. [DOI] [PubMed] [Google Scholar]
- 42.Scott NA, Andrusaite A, Andersen P, Lawson M, Alcon-Giner C, Leclaire C, Caim S, Le Gall G, Shaw T, Connolly JPR, Roe AJ, Wessel H, Bravo-Blas A, Thomson CA, Kastele V, Wang P, Peterson DA, Bancroft A, Li X, Grencis R, Mowat AM, Hall LJ, Travis MA, Milling SWF, Mann ER. 2018. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci Transl Med 10:eaao4755. doi: 10.1126/scitranslmed.aao4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kasper CA, Sorg I, Schmutz C, Tschon T, Wischnewski H, Kim ML, Arrieumerlou C. 2010. Cell-cell propagation of NF-kappaB transcription factor and MAP kinase activation amplifies innate immunity against bacterial infection. Immunity 33:804–816. doi: 10.1016/j.immuni.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi M, Mikami D, Kimura H, Kamiyama K, Morikawa Y, Yokoi S, Kasuno K, Takahashi N, Taniguchi T, Iwano M. 2017. Short-chain fatty acids, GPR41 and GPR43 ligands, inhibit TNF-alpha-induced MCP-1 expression by modulating p38 and JNK signaling pathways in human renal cortical epithelial cells. Biochem Biophys Res Commun 486:499–505. doi: 10.1016/j.bbrc.2017.03.071. [DOI] [PubMed] [Google Scholar]
- 45.Giamarellou H. 2005. Multidrug resistance in Gram-negative bacteria that produce extended-spectrum beta-lactamases (ESBLs). Clin Microbiol Infect 11(Suppl 4):1–16. doi: 10.1111/j.1469-0691.2005.01160.x. [DOI] [PubMed] [Google Scholar]
- 46.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. 2010. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med 16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, Lienenklaus S, Weiss S, Staeheli P, Aichele P, Diefenbach A. 2012. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 48.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. 2014. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 49.Pomare EW, Branch WJ, Cummings JH. 1985. Carbohydrate fermentation in the human colon and its relation to acetate concentrations in venous blood. J Clin Invest 75:1448–1454. doi: 10.1172/JCI111847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Correa-Oliveira R, Fachi JL, Vieira A, Sato FT, Vinolo MA. 2016. Regulation of immune cell function by short-chain fatty acids. Clin Transl Immunology 5:e73. doi: 10.1038/cti.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bohn G, Allroth A, Brandes G, Thiel J, Glocker E, Schaffer AA, Rathinam C, Taub N, Teis D, Zeidler C, Dewey RA, Geffers R, Buer J, Huber LA, Welte K, Grimbacher B, Klein C. 2007. A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat Med 13:38–45. doi: 10.1038/nm1528. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Goncalves R, Mosser DM. 2008. The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14:Unit 14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R. 2012. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rooks MG, Garrett WS. 2016. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang J, Liang X, Xuan Y, Geng C, Li Y, Lu H, Qu S, Mei X, Chen H, Yu T, Sun N, Rao J, Wang J, Zhang W, Chen Y, Liao S, Jiang H, Liu X, Yang Z, Mu F, Gao S. 2017. A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience 6:1–9. doi: 10.1093/gigascience/gix024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang da W, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 58.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu R, Hong J, Xu X, Feng Q, Zhang D, Gu Y, Shi J, Zhao S, Liu W, Wang X, Xia H, Liu Z, Cui B, Liang P, Xi L, Jin J, Ying X, Wang X, Zhao X, Li W, Jia H, Lan Z, Li F, Wang R, Sun Y, Yang M, Shen Y, Jie Z, Li J, Chen X, Zhong H, Xie H, Zhang Y, Gu W, Deng X, Shen B, Xu X, Yang H, Xu G, Bi Y, Lai S, Wang J, Qi L, Madsen L, Wang J, Ning G, Kristiansen K, Wang W. 2017. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat Med 23:859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 60.Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, Bouatra S, Sinelnikov I, Arndt D, Xia J, Liu P, Yallou F, Bjorndahl T, Perez-Pineiro R, Eisner R, Allen F, Neveu V, Greiner R, Scalbert A. 2013. HMDB 3.0—the Human Metabolome Database in 2013. Nucleic Acids Res 41:D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Protective role of the gut microbiota during K. pneumoniae-induced sepsis. (a) Blood bacterial burdens in antibiotic-treated and untreated controls 12 h and 24 h after K. pneumoniae infection. (b) Pulmonary production of cytokines (TNF-α, 1L-1β, and IL-6) and chemokines (CXCL1 and MCP-1) in antibiotic-treated and untreated controls 24 h after K. pneumoniae infection. (c) Representative H&E staining and quantification of pathological scores of liver and kidney sections from antibiotic-treated and untreated controls 24 h after K. pneumoniae infection. Scale bars, 50 μm. The group size was 8 to 12 mice. Data are from three independent experiments (a, b) or one experiment representative of three independent experiments (c) (means ± SEM). The P values were determined using two-tailed Student t tests (a to c). *, P < 0.05; **, P < 0.01; ***, P < 0.001. NS, not significant; AT, antibiotic-treated mice. Download FIG S1, TIF file, 2.6 MB (2.6MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential metabolites and KEGG pathway enrichment. (a) Hierarchical clustering heatmap of metabolites identified in cecum contents from K. pneumoniae-infected and antibiotic-treated mice versus K. pneumoniae-infected controls. Red denotes increased expression; green denotes decreased expression. (b) Selected examples of KEGG pathway enrichment. Disturbances in propionate and butyrate metabolism are highlighted in red. The P values were determined using the hypergeometric test and Benjamini-Hochberg FDR correction (b). AT, antibiotic-treated mice. Download FIG S2, TIF file, 1.3 MB (1.4MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Association of cecum metabolites and associated gut microbiomes. (a) α-Diversity (Shannon index) and β-diversity (Bray-Curtis similarity index) of 16S rRNA genes from K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. (b, c) Discriminative OTU abundances of taxonomic distributions at the phylum (b) and genus (c) levels between K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. (d) Spearman’s rank correlation between cecum metabolites and associated gut microbiota in K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. Red connections indicate a positive correlation, while blue connections are negative. The P values were determined using a two-tailed Wilcoxon rank sum test (d). *, P < 0.05; **, P < 0.01; ***, P < 0.001. AT, antibiotic-treated mice. Download FIG S3, TIF file, 2.9 MB (3MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gut microbiota depletion alters transcriptional profiles in infected mice. (a) Hierarchical clustering heatmap of selected significant DEGs between K. pneumoniae-infected, antibiotic-treated mice and K. pneumoniae-infected controls. Red denotes increased expression; green denotes decreased expression. (b) Selected examples of KEGG pathway enrichment. (c) Expression levels of Tnf-α, Il-10, Il-1β, Mcp-1, and Cxcl1 mRNA in K. pneumoniae-infected, antibiotic-treated mice compared to those of K. pneumoniae-infected controls. (d) Immunoblot analysis of LAMTOR2 in shRNA-mediated Lamtor2 knockdown RAW264.7 cells. (e) Immunoblot confirmation of the LAMTOR2 protein in Lamtor2−/− RAW264.7 macrophages. (f) Sequencing of targeted Lamtor2 mutations and alignment with those from wild-type RAW264.7 cells. Data are from three independent experiments (c) or one experiment representative of three independent experiments (d to f). The P values were determined using a hypergeometric test and Benjamini-Hochberg FDR correction (b) or a two-tailed Student t test (c). **, P < 0.01; ***, P < 0.001. AT, antibiotic-treated mice. Download FIG S4, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Determination of Lamtor2 mRNA expression in vivo and generation of Gpr43−/− RAW264.7 cells. (a) RT-qPCR analysis of Lamtor2 mRNA expression in isolated alveolar macrophages after K. pneumoniae infection. The P values were determined using two-tailed Student t tests. ***, P < 0.001. (b) Immunoblot validation of GPR43 in Gpr43−/− RAW264.7 cells. (c) Sequencing of targeted Gpr43 mutations in Gpr43−/− RAW264.7 cells and alignment with those from wild-type controls. Data are from three independent experiments (a) or one experiment representative of three independent experiments (b, c). Download FIG S5, TIF file, 0.8 MB (806.8KB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LAMTOR2 overexpression in RAW264.7 cells. (a) Immunoblot analysis of LAMTOR2 protein in LAMTOR2-overexpressed RAW264.7 cells. (b) Expression levels of Tnf-α, Il-6, Il-1β, Mcp-1, and Cxcl1 mRNA in LAMTOR2-overexpressed RAW264.7 cells after K. pneumoniae infection. (c) Bacterial loads of Gpr43−/− and Gpr43−/− Lamtor2 overexpression (OE) in RAW264.7 cells. Data are from three independent experiments (b, c) or one experiment representative of three independent experiments (a). The P values were determined using two-tailed Student t tests (b, c). **, P < 0.01; ***, P < 0.001. Download FIG S6, TIF file, 1.0 MB (1MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Generation of Lamtor2 conditional knockout (cKO) mice via CRISPR-Cas9 genome editing. (a) Schematic generation of Lamtor2 conditional knockout mice. Targeted exons 1 to 3 of the Lamtor2 gene were flanked by Loxp sites. Maps of the Lamtor2 wild-type allele, targeting vector, Lamtor2-targeted allele, and Lamtor2 knockout allele are shown. (b) Genotyping of wild-type (Lamtor2+/+), fl/+ (Lamtor2fl/+), and fl/fl (Lamtor2fl/fl) mice was performed by PCR. (c) RT-qPCR analysis of Lamtor2 mRNA expression in targeted alveolar macrophages. (d) Immunoblot analysis of the LAMTOR2 protein in targeted alveolar macrophages. (e) Blood bacterial burdens in Lamtor2fl/fl Lyz2-Cre+ and Lamtor2fl/fl mice 12 h and 24 h after K. pneumoniae infection. (f) Representative H&E staining and quantification of pathological scores of lung sections from Lamtor2fl/fl Lyz2-Cre+ mice and controls 24 h after K. pneumoniae infection. Scale bars, 50 μm. (g) Responsiveness of isolated alveolar macrophages from Lamtor2fl/fl Lyz2-Cre+ mice and controls against lipopolysaccharide (LPS) in terms of TNF-α, IL-6, 1L-1β, MCP-1, and CXCL1 production. Data are from three independent experiments (c, e, g) or one experiment representative of three independent experiments (b, d, f) (means ± SEM). The P values were determined using two-tailed Student t tests (c, e, f, g). **, P < 0.01; ***, P < 0.001. Download FIG S7, TIF file, 1.6 MB (1.6MB, tif) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Reagents and materials. Download Table S1, PDF file, 0.02 MB (20.9KB, pdf) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Antibodies. Download Table S2, PDF file, 0.1 MB (74.1KB, pdf) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides. Download Table S3, PDF file, 0.1 MB (63.5KB, pdf) .
Copyright © 2020 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequencing data and relevant files have been uploaded to public repositories. LC-MS data were deposited into the China National GenBank Database with the accession no. CNP0001290. The 16S sequencing data are available in GenBank with the accession no. MW011789 to MW012255. Transcriptome sequencing data can be found on the NCBI Sequence Read Archive (SRA) with the accession no. PRJNA662812.