Abstract
Context
Germline mutations in the succinate dehydrogenase genes (SDHA/B/C/D, SDHAF2—collectively, “SDHx”) have been implicated in paraganglioma (PGL), renal cell carcinoma (RCC), gastrointestinal stromal tumor (GIST), and pituitary adenoma (PA). Negative SDHB tumor staining is indicative of SDH-deficient tumors, usually reflecting an underlying germline SDHx mutation. However, approximately 20% of individuals with SDH-deficient tumors lack an identifiable germline SDHx mutation.
Methods
We performed whole-exome sequencing (WES) of germline and tumor DNA followed by Sanger sequencing validation, transcriptome analysis, metabolomic studies, and haplotype analysis in 2 Italian-Australian families with SDH-deficient PGLs and various neoplasms, including RCC, GIST, and PA.
Results
Germline WES revealed a novel SDHC intronic variant, which had been missed during previous routine testing, in 4 affected siblings of the index family. Transcriptome analysis demonstrated aberrant SDHC splicing, with the retained intronic segment introducing a premature stop codon. WES of available tumors in this family showed chromosome 1 deletion with loss of wild-type SDHC in a PGL and a somatic gain-of-function KIT mutation in a GIST. The SDHC intronic variant identified was subsequently detected in the second family, with haplotype analysis indicating a founder effect.
Conclusions
This is the deepest intronic variant to be reported among the SDHx genes. Intronic variants beyond the limits of standard gene sequencing analysis should be considered in patients with SDH-deficient tumors but negative genetic test results.
Keywords: paraganglioma, succinate dehydrogenase, SDHC, intronic mutation, whole-exome sequencing
Involved in both the Krebs cycle and mitochondrial respiratory chain, succinate dehydrogenase (SDH) is a heterotetramer protein complex encoded by the SDHA, SDHB, SDHC, and SDHD genes [1]. Together with SDHAF2, which allows flavination and functioning of the SDHA subunit [2], these genes are collectively referred to as the “SDHx” genes. Loss-of-function SDHx variants inactivate SDH, leading to reactive oxygen species and succinate accumulation; the combined effect is inhibition of prolyl hydroxylases, resulting in decreased hydroxylation (inactivation) of hypoxia-inducible factor α, angiogenesis, cellular proliferation, and eventual tumorigenesis [1, 3, 4].
Consistent with the tumor-suppressor gene model, SDHx tumor syndromes demonstrate autosomal dominant inheritance due to heterozygous germline mutations, and variable penetrance and expressivity related to the timing of the somatic second hit, which is most commonly loss of heterozygosity, followed by somatic mutations, and, rarely, epigenetic inactivation [1, 3, 5]. SDHB tumor immunohistochemistry (IHC) is used to identify SDH-deficient tumors, with loss of any component of the SDH complex resulting in negative SDHB staining [2, 6-8]. In addition, the inactivation of SDH produces an excess of succinate, which can be directly assessed in tumor specimens, with high succinate:fumarate ratios indicative of SDH deficiency [2, 9].
Pheochromocytomas and paragangliomas (collectively, PPGLs) are the archetypal SDH-deficient tumor. Paragangliomas (PGLs) may involve any of the parasympathetic or sympathetic ganglia [3], making surveillance onerous and emphasizing the value of identifying causative mutations in PPGL kindreds to restrict follow-up to proven mutation carriers. PPGLs are regarded as the most heritable tumors in humans [5], with germline mutations identified in more than 30% of all-comers and 13% to 24% of sporadic cases [10-14]. Given the high mutation probability for most patients, and the clinical utility for patients and their families [15], genetic testing should be offered to all patients with PPGL [10]. Single/staged gene sequencing by direct sequencing has been supplanted by next-generation sequencing (NGS), addressing the marked genetic heterogeneity in PPGL with more than 15 genes implicated to date [5]. The role of SDH deficiency in PPGL is underscored by SDHx mutations accounting for approximately half of all heritable PPGL syndromes [2, 10]. SDH deficiency has also been implicated in smaller proportions of gastrointestinal stromal tumor (GIST), renal cell carcinoma (RCC), and pituitary adenoma (PA) [3]. These tumor types may coexist within individuals; however, no individuals or families have been reported to exhibit all 4 SDH-related tumors. Syndromes have been named according to specific tumor combinations: Carney triad for PGL, GIST, and pulmonary chondroma [16]; Carney-Stratakis dyad for PGL and GIST [17]; and 3P association syndrome (3PAs) for pheochromocytoma, PGL, and PA [18].
1. Materials and Methods
Following negative results from comprehensive PPGL gene testing in standard commercial NGS facilities, we investigated 2 families with various neoplasms, including PGL, GIST, RCC, and PA. We hypothesized that their tumor predilection may be due to a mutation in a novel PPGL gene or in a location in a known PPGL gene leading to altered gene transcription or expression that is not captured by standard genetic testing.
All clinical data were collated and updated prior to manuscript preparation in July 2019. All genetic investigations were performed in a clinical setting by nationally accredited laboratory processes. Prospective written consent to testing was obtained from all living patients, and from next of kin in the case of deceased relatives, prior to the genetic investigations. Subsequently, written consent to publication was obtained from all living patients and from next of kin in the case of deceased relatives. The publication was ethically approved by the Royal Adelaide Hospital Human Research Ethics Committee in accordance with the National Health and Medical Research Council guidelines.
A. Case Descriptions
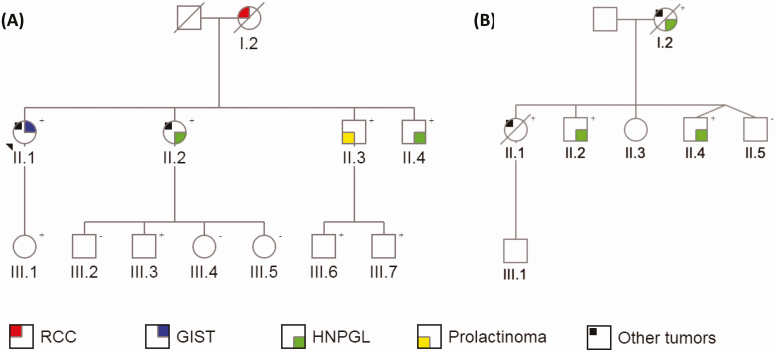
Pedigrees of the 2 Italian-Australian families are shown in Fig. 1 and clinical features are listed in Table 1. Genetic and IHC test results are summarized in Supplemental Table 1 [19].
Figure 1.
Pedigrees of A, family 1 and B, family 2, highlighting succinate dehydrogenase (SDH)-related and other tumors in affected family members. Genetic status regarding the intronic SDHC variant c.20 + 74A >G is indicated in the top right-hand corner for all tested individuals. +, variant present; –, variant absent.
Table 1.
Tumor phenotype in affected relatives of family 1 and family 2
| Family | Individual (current age) | SDHC mutation status | Tumors (age at initial diagnosis, y) | SDHB tumor IHC | Succinate:fumarate ratio |
|---|---|---|---|---|---|
| 1 | I.2 (died 61) | N/T | RCC (60) | Positive | N/T |
| II.1 (59) | + | Desmoid tumor (50) | Positive | 10.600, 16.037, 11.681a | |
| HCC (50) | Positive | 29.458 | |||
| Gastric GIST (51) | Positive | 41.765, 6.815b | |||
| Solitary fibrous tumor of lung (53) | N/T | 13.895 | |||
| Adrenocortical adenoma (53) | N/T | N/T | |||
| Meningioma (59) | N/T | N/T | |||
| II.2 (57) | + | HNPGL (41) | Negative | 89.490 | |
| Ovarian serous cystadenoma and cellular fibroma (53) | N/T | 11.644 | |||
| Meningioma (54) | N/T | N/T | |||
| II.3 (54) | + | Prolactinoma (41) | N/T | N/T | |
| II.4 (52) | + | HNPGL (34) | Negative | 27.725 | |
| 2 | I.2 (died 61) | + | HNPGL (44) | Negative | N/T |
| Breast cancer (44) | N/T | N/T | |||
| Cholangiocarcinoma (61) | N/T | N/T | |||
| II.1 (died 38) | + | Diffuse gastric carcinoma (38) | Positive | N/T | |
| II.2 (38) | + | Suspected HNPGL (37) | N/T | N/T | |
| II.4 (34) | + | HNPGL (20) | Negative | N/T |
Abbreviations: +, mutation present; GIST, gastrointestinal stromal tumor; HCC, hepatocellular carcinoma; HNPGL, head and neck paraganglioma; IHC, immunohistochemistry; N/T, not tested; RCC, renal cell carcinoma.
aMultiple ratios determined from serial resections of recurrent desmoid tumor.
bMultiple ratios determined from multifocal gastric GIST resected simultaneously.
A-1. Family 1
The index family was of Italian ethnicity and consisted of 4 siblings with SDH-related tumors, with their mother having died from RCC. Preliminary genetic testing in the proband, II.2, was negative for SDHB, SDHC, SDHD, SDHAF2, VHL, RET, and TMEM127 mutations by direct gene sequencing, and for SDHB, SDHC, SDHD, and VHL copy number variants (CNVs) by multiplex ligation-dependent probe amplification (MLPA).
I.2, a lifelong nonsmoker, was diagnosed with RCC at age 60 years and underwent left nephrectomy, demonstrating an 8.5-cm RCC with focal sarcomatoid appearance, microscopic invasion of perinephric fat, and metastasis to peripelvic fat. SDHB tumor IHC was historically reported as positive, although the tissue slides are no longer available for review.
II.1 first presented with a multifocal mesenteric desmoid tumor at age 50. She underwent right hemicolectomy at diagnosis, followed by small bowel and mesenteric resection and medical therapy with ibuprofen, tamoxifen, celecoxib, and letrozole at age 54, and small bowel and sigmoid resection at age 55 for recurrent disease. Also at age 50, she was diagnosed with a well-differentiated hepatocellular carcinoma (HCC) for which she underwent resection at age 51 and radiofrequency ablation of a presumed second lesion at age 52. There was no evidence of HCC metastasis on computed tomography imaging or whole-body bone scan. At age 51, she underwent resection of 2 gastric GISTs, measuring 6 mm at the lesser gastric curve and 20 mm at the body of the stomach. At age 53, she was found to have an 11-mm nonfunctioning adrenocortical adenoma that is being monitored, and a solitary fibrous tumor of the lung that was resected. She was most recently diagnosed with a 4.2-cm left frontal meningioma at age 59 and is awaiting further management. SDHB IHC was performed on the desmoid tumor, HCC, and GIST, with all samples showing positive staining. The 2 GIST specimens shared a similar appearance, with predominant spindle cell morphology and positive IHC for c-Kit/CD117 and Discovered on GIST-1 (DOG1), all of which suggested a receptor tyrosine kinase–mediated tumor.
II.2 presented with a nonsecretory left cerebellopontine PGL at age 41 and underwent partial resection following embolization. SDH tumor IHC was negative for SDHB and positive for SDHA. She recently completed radiotherapy at age 57 for the PGL remnant, which reached a diameter of 4.2 cm and was encasing the left internal carotid artery and impinging on the brainstem. She was also diagnosed with an ovarian serous cystadenoma and ovarian cellular fibroma, both resected at age 53, and a parafalcine meningioma at age 54 that is under imaging surveillance.
II.3 was diagnosed with a 3.4-cm macroprolactinoma at age 41. He achieved a complete hormonal and tumor response with cabergoline.
II.4 was diagnosed with a nonsecretory, multifocal, skull base PGL at age 34 and underwent partial resection following embolization. The PGL remnant is stable on serial monitoring. SDH tumor IHC was negative for SDHB and positive for SDHA.
Because of the family history of PGL, all members of the second and third generations of family 1 apart from III.5 have been screened for PPGL via magnetic resonance imaging every 2 to 3 years and annual plasma metanephrines with no evidence of PPGL to date in any relatives other than II.2 and II.4.
A-2. Family 2
The second family was noteworthy for SDH-related and other neoplasia in 2 siblings and their mother. Preliminary genetic testing in the proband with PGL, II.4, was negative for SDHB, SDHC, SDHD, SDHAF2, VHL, RET, and TMEM127 mutations by direct gene sequencing, and for SDHB, SDHC, SDHD, and VHL CNVs by MLPA. All preliminary genetic tests in II.1, performed because of her history of gastric cancer, were negative, including CDH1, CTNNA1, MLH1, MSH2, MSH6, EPCAM, BRCA1, and BRCA2 by NGS; BRCA1, BRCA2, and PMS2 by direct gene sequencing; and CDH1, MLH1, MSH2, MSH6, EPCAM, and PMS2 by MLPA. In view of the negative genetic test results and shared Italian ethnicity, family 2 was selected for investigation for the mutation identified in family 1 during the course of this study.
I.2 was diagnosed with a right jugular PGL at age 44 and underwent resection following embolization. Histopathology confirmed PGL with lymph node metastases. SDHB IHC was negative (SDHA IHC not performed). She was concurrently diagnosed with breast cancer, treated with mastectomy, axillary clearance, and adjuvant chemoradiotherapy. She remained in remission of her PGL and breast cancer at age 61, when she died of newly diagnosed metastatic cholangiocarcinoma.
II.1 presented with acute kidney injury and venous thromboembolism 2 months postpartum at age 38. She died from a presumed systemic inflammatory illness with multiple osteolytic lesions 3 weeks later. Postmortem examination revealed metastatic diffuse gastric carcinoma. Tumor IHC was positive for SDHB and SDHA.
II.4 underwent resection for a right jugulotympanic PGL at age 20. SDH tumor IHC was negative for SDHB and positive for SDHA.
The surviving second-generation members of family 2 recently underwent PPGL screening, revealing a likely head and neck PGL (HNPGL) recurrence in II.4 and a new diagnosis of likely HNPGL in II.2. PPGL screening was negative in II.3 and II.5.
B. DNA Extraction
Fresh blood samples were obtained from II.1 to 4 of family 1 and II.4 of family 2 for extraction of germline DNA from peripheral blood leukocytes. Among the deceased individuals, no DNA was available from I.2 of family 1, stored DNA was obtained from II.1 of family 2, and only tumor DNA was available from I.2 of family 2. Tumor DNA extraction was performed using formalin-fixed, paraffin-embedded (FFPE) tissue specimens of the 6-mm GIST in II.1 and the PGL in II.4 in family 1, and the PGL and breast cancer in I.2 in family 2. Other tumor specimens were not available for sequencing. Germline and tumor DNA were extracted using commercially available kits (Qiagen) according to manufacturer protocols.
C. Whole-Exome Sequencing
Whole-exome sequencing (WES) was performed in family 1 using the available germline and tumor DNA samples, the Roche NimbleGen SeqCap EZ MedExome Target Enrichment Kit, and the Illumina NextSeq 500 sequencing platform. The average of mean depth of coverage among all samples was 97×, and 94% of target bases were covered at 20× or greater. Bioinformatic analysis was performed at ACRF in Adelaide, Australia, using Genome Analysis Toolkit (GATK) HaplotypeCaller to detect small variants (typically < 50 base pairs, bp) and in-house scripts and Sequenza to analyze CNVs. Raw WES data were filtered for variants that were rare (< 1% population prevalence), possibly damaging (by snpEFF impact, splicing/binding predictions, or Genomic Evolutionary Rate Profiling [GERP] or Combined Annotation Dependent Depletion scores), and of high quality (by GATK internal filters). Germline variants were considered further if they were heterozygous in the germline DNA of all 4 siblings in family 1 with a GATK genotype quality score greater than 50 and depth of coverage greater than 30×. Variants in low-complexity regions or duplicated segments were discarded. Candidate genes were prioritized based on existing literature. In silico splice site assessment was performed using Alamut Visual, incorporating SpliceSiteFinder-like, MaxEntScan, NNSPLICE, and GeneSplicer prediction models.
The Roche NimbleGen SeqCap EZ MedExome Target Enrichment Kit and the Illumina NextSeq 500 sequencing platform were also employed in the preliminary testing of gastric cancer predisposition genes in II.1 of Family 2.
D. Sanger Sequencing
The leading germline variant of interest in family 1 was assessed using germline DNA from II.1 to 4 of family 1 and II.1 and II.4 of family 2, and tumor DNA from I.2 from family 2. Bidirectional genomic DNA sequencing was performed using primers designed via Primer3Plus and raw data were visualized using MutationSurveyor version 2.51 (SoftGenetics LLC).
Sanger sequencing for the leading germline variant of interest was later performed to facilitate predictive cascade testing in other relatives of family 1 and family 2.
E. Haplotype Analysis
Haplotype analysis was performed by considering rare variants (SNPs > 20× coverage, ExAC and UK10K allele frequencies < 0.01) in the WES data of II.2 of family 1 and II.1 of family 2 and mapping those loci that overlapped between the 2 individuals. For any rare variant identified in either individual, an unrelated individual would overlap at less than 1% of loci with random distribution throughout the genome. Conversely, relatedness due to identity by descent would be identified by a chain of shared rare variants that are nonrandomly distributed throughout the genome.
F. Transcriptome Analysis
Whole blood was obtained from II.2 of family 1 for transcriptome analysis to further investigate the leading germline variant of interest in family 1. RNA sequencing (RNA-Seq) was performed via the Illumina TruSeq LT platform using 150 bp reads and poly(A) selected RNA to deplete ribosomal RNA. Messenger RNA–enriched RNA-Seq libraries from the patient were sequenced on the Illumina NextSeq 500 platform using the stranded, paired-end protocol with a read length of 150 bp. Raw reads were adapter-trimmed and filtered for short sequences using Cutadapt v1.16, setting the minimum length option to 18, overlap 5 and error rate 0.2. The resulting FASTQ file containing 26.6 million read pairs was analyzed and quality-checked using the FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were mapped against the human reference genome (hg19) using the STAR spliced alignment algorithm (v2.5.3a with default parameters and –chimSegmentMin 20, –quantMode GeneCounts), returning an average unique alignment rate of 88.0%. Read alignments, including spliced reads, were visualized, interrogated, and graphically represented using the Integrative Genomics Viewer v2.3.80.
G. Krebs Cycle Metabolomic Studies
Available FFPE tumor specimens from the index family were tested in duplicate by mass spectrometry to measure succinate and fumarate levels and calculate succinate:fumarate ratios to aid the identification of SDH-deficient tumors as previously described [9]. Briefly, analysis of extracts was performed on a Prominence high-performance liquid chromatography system (Shimadzu) coupled to an API QTRAP 5500 mass spectrometer (SCIEX). Separation of target analytes from isobaric interferences was achieved using an Ascentis Express 100 Å~3.0 mm 2.7 μmRP Amide (Sigma-Aldrich) analytical column held at 40°C and isocratic elution using aqueous 0.4% formic acid with a flow rate of 0.5 mL/min. We routinely included certified reference materials for succinate and fumarate (Sigma-Aldrich). Multiple reaction monitoring with negative electrospray ionization was used for quantification. Each sample was run in duplicate, with an intra-assay coefficient of variation for succinate of 22% and for fumarate of 21%. Positive and negative controls were included in each run. We had previously determined a threshold for the succinate:fumarate ratio of 23.48 in SDH-deficient GIST FFPE specimens [9]. Recognizing that succinate:fumarate ratios are typically lower in HNPGL compared to sympathetic PGLs [20, 21], and in FFPE compared to fresh-frozen specimens [2, 21], we cautiously adopted a succinate:fumarate ratio of 65.00 that identifies most SDH-deficient PGLs [9].
H. SDHC Promoter Methylation Analysis
Methylation status of the SDHC promoter region was determined in both GIST specimens from family 1 II.1. A Pyromark CpG assay was performed as previously described [22]. A total of 1500 ng of sample DNA was used for bisulfite conversion treatment in individual polymerase chain reaction (PCR) tubes, with 3 bisulfite conversion periods (60°C × 25 minutes, 60°C × 85 minutes, and 60°C × 175 minutes), separated by 3 DNA denaturation periods and followed by column purification as per the manufacturer’s instructions (Qiagen, EpiTect Bisulfite Kit, catalog No. 59104). Bisulfite-converted DNA was amplified using the PyroMark PCR Kit (catalog No. 978703) using primers targeting 4 CpG sites of the SDHC promoter (Chr1: 161 313 986; Chr1: 161 313 998; Chr1: 161 314 011, and Chr1: 161 314 022). Qiagen Epitec Control DNA, methylated (catalog No. 59655) and unmethylated (catalog No. 59665), were included as the assay quality controls. Following PCR, the amplicons were immobilized and single-strand templates produced via Streptavidin Sepharose High Performance beads (GE Healthcare, 17-5113-01), followed by annealing of the sequencing primer to the template. The samples were then analyzed on the PyroMark Q24 system, using PyroMark Gold Q24 Reagents Kit (catalog No. 970802) for quantitative measurement of methylation in 4 CpG sites of the SDHC promoter. Sequences surrounding the defined positions served as normalization and reference peaks for quantification and quality assessment of the analysis. Pyrograms were analyzed using Pyromark Q24 software (Qiagen), version 2.0.6, to calculate percentage methylation at each CpG, and mean methylation across all CpGs for each sample was calculated.
2. Results
A. Germline Genetic Analysis in Family 1
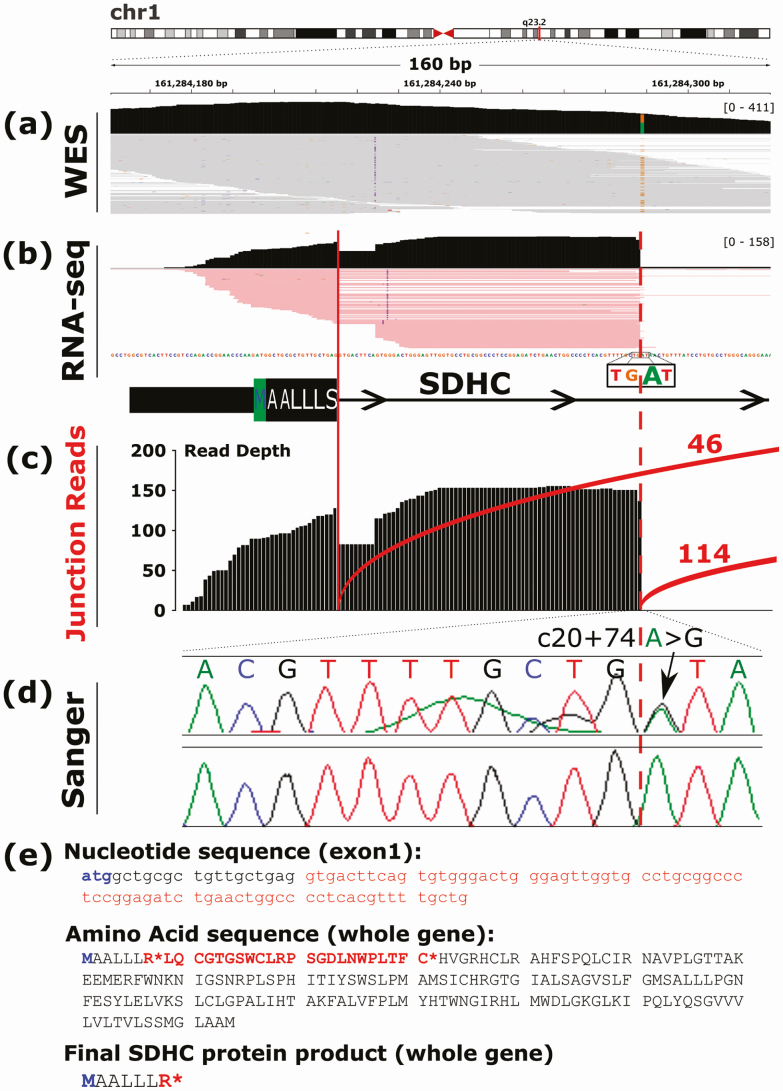
After filtration of raw data, WES of germline DNA revealed 19 581 rare variants with at least some evidence of pathogenicity, including 130 high-quality heterozygous variants in all 4 siblings of family 1. One variant was found in an intronic region of the known candidate gene, SDHC (GRCh37/hg19, Chr1:g.161284289A > G; NM_003001; c.20 + 74A>G; Fig. 2A), at greater than 20× coverage (21 wild-type [WT] reads, 32 mutant allele reads in II.1; 93,80 in II.2; 25,24 in II.3; and 35,30 in II.4). Sanger sequencing confirmed the variant in all 4 siblings (Fig. 2D). This SDHC intronic variant is situated in a conserved region (GERP 2.38) of intron 1 and has not been previously reported. It is absent in public genomic datasets, including: 1KGP; UK10K; ExAC; and gnomAD, containing 31 378 control alleles in the vicinity of this variant, including 106 alleles from patients of Southern European ethnicity. All 4 component splicing models of Alamut Visual predicted introduction of an alternate 5’ (donor) splice site at the location of the variant.
Figure 2.
DNA and RNA representations of the intronic SDHC variant c.20 + 74A > G in II.2 of family 1. A, Whole-exome sequencing result of germline DNA as depicted in Integrative Genomics Viewer. The heterozygous substitution of guanine (brown) for adenosine (green) is shown at genomic DNA position 161 284 289. B, RNA sequencing (RNA-Seq) result as depicted in Integrative Genomics Viewer showing alternative splicing of exon 1. The canonical splice site is indicated by the solid red line and the novel splice site by the dotted red line, coinciding with the A > G substitution. C, Junction counts of individual messenger RNA reads showing preferential expression of the aberrantly spliced transcript (n = 114) vs normal transcript (n = 46). D, Electrophoretogram confirming the germline DNA variant. E, Nucleotide, amino acid, and final protein product sequences produced by the 75-bp inclusion observed on RNA-Seq. The start codon is indicated in blue. The intronic inclusion in exon 1 created by the SDHC c.20 + 74A > G variant is indicated in red, and premature stop codons are indicated by the red asterisks.
Subsequent RNA-Seq of whole blood from II.2 of family 1 showed aberrant splicing of SDHC with messenger RNA reads extending into intron 1 (Fig. 2B), due to conversion of a nonsplicing region (TG|AT) into a canonical splice site (TG|GT) because of the familial SDHC variant. There was evidence of preferential expression of the alternatively spliced transcript (n = 114) compared to the normal transcript (n = 46) (Fig. 2C). The alternatively spliced transcript is absent in publicly accessible databases (UCSC Genome Browser, Ensembl, GTEx, NCBI) as well as in-house RNA-Seq results from more than 700 samples.
The retained segment size is 75 bp due to the upstream inclusion of a common 2-bp SDHC insertion listed as benign by ClinVar (SDHC, NM_003001.3, c.20 + 11_20 + 12dupTG). Thus, frameshift does not occur. However, the retained intronic segment produces a premature stop codon immediately after exon 1. The final SDHC protein product is significantly shortened (Fig. 2E) and predicted to result in nonsense-mediated decay.
Overall, the familial SDHC variant fulfilled the American College of Medical Genetics and Genomics criteria for a pathogenic (class 5) variant (PVS1: null variant, PS3: functional evidence, PM2: absent from controls, PP1: cosegregation) [23].
Given the history of additional tumors in II.1 and II.2, germline data from these individuals was independently interrogated for variants in genes predisposing to GIST (KIT, PDGFRA), meningioma (NF2, SMARB1, SMARCE1, SUFU, LZTR1), and desmoid tumors (APC). Applying our basic filters, we found no germline variants in these genes.
B. Tumor Genetic Analysis in Family 1
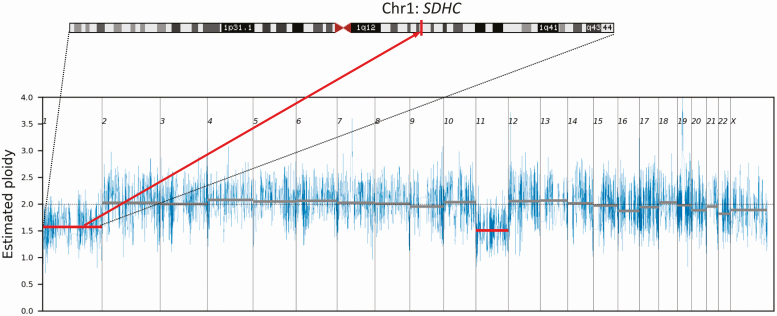
WES and copy number analysis of the PGL from II.4 of Family 1 revealed 0.4-0.5X ploidy loss of chromosome (Chr) 1 (Fig. 3). This may represent either loss of one copy of Chr 1 in 40% to 50% of tumor cells or loss of both copies of Chr 1 in 20% to 25% of tumor cells. Mutant allele frequency would be expected to be unchanged if both chromosomes were lost in a subset of cells or if there were unbiased loss of Chr 1 between different cells. By contrast, the SDHC intronic variant load on WES rose from 47% in II.4’s germline DNA to 60% in II.4’s tumor DNA. It was thus deduced that 40% of cells lost the SDHC WT allele (producing a 3:5 WT:mutant ratio in tumor DNA vs a 1:1 ratio in heterozygous germline DNA). This chromosomal loss was considered to be the second hit in the tumor-suppressor gene 2-hit model, thus supporting the germline SDHC intronic variant as the causative mutation. Whole chromosome loss of Chr 11 was also detected, as is commonly observed in PGL specimens [24].
Figure 3.
Chromosome 1 and 11 loss as demonstrated by tumor DNA whole-exome sequencing in the paraganglioma of II.4 from family 1. The position of the SDHC gene on chromosome 1 is indicated.
WES of the 6-mm GIST from II.1 of family 1 demonstrated a previously described [25] gain-of-function KIT mutation (GRCh37/hg19, Chr2:g.55593610T > G; ENST00000288135; p.Val559Gly/c.1676T > G). Other than the germline c.20 + 74A > G variant, no point mutations or copy number variants involving SDHC were found in this specimen. Each of the noncontiguous GIST specimens from II.1 of family 1 showed low SDHC promoter methylation: range 7% to 24%, average 15.3% in the 6-mm GIST; range 9% to 22%, average 14.8% in the 20-mm GIST. Further data are presented in Supplemental Table 2 and Supplemental Fig. 1 [19]. The methylation rate in these tumors fell in the bottom 10% of internal FFPE GIST control specimens (n = 15) [9], excluding SDHC promoter hypermethylation in the pathogenesis of this patient’s GISTs.
C. Genetic Linkage Between Family 1 and Family 2
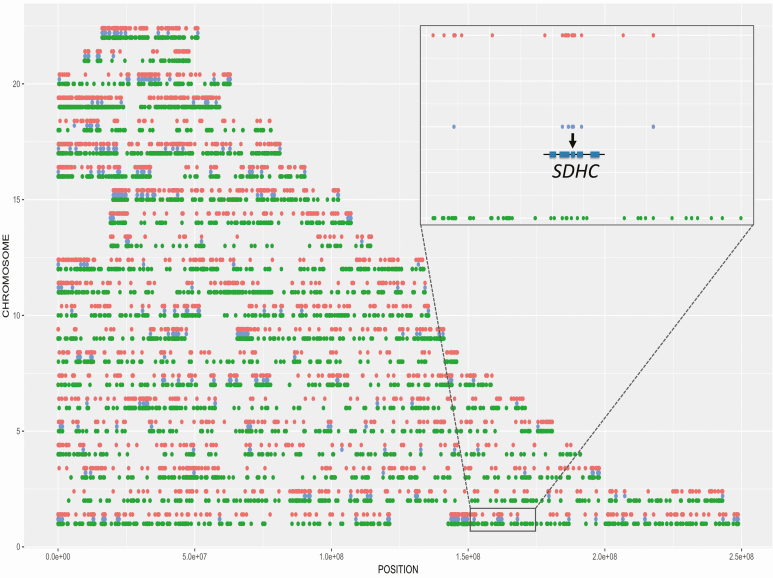
Sanger sequencing in family 2 using germline DNA from II.1 and II.4 and tumor DNA from the PGL and breast cancer of I.2 revealed the same SDHC intronic variant in these 3 family members with neoplasia. Because hotspot mutations have not been described in SDHC and because of the shared Italian ancestry of families 1 and 2, haplotype analysis was performed to investigate cryptic relatedness. This demonstrated multiple regions of identity by descent, including the SDHC gene, consistent with a shared common ancestor (Fig. 4). Expanded family history-taking revealed that the 2 families originated from the same small region in Italy.
Figure 4.
Haplotype analysis using exome data from each of the 22 autosomes showing regions unique to family 1 in red, regions unique to family 2 in green, and regions shared between the 2 families in blue. The multiple shared regions throughout the genome indicate identity by descent, and therefore a shared common ancestor between family 1 and family 2. The inset shows the shared region on chromosome 1 that includes SDHC.
D. Metabolomic Profiles
Succinate:fumarate ratios derived from the available tumors of family 1 are shown in Table 1. A high succinate:fumarate ratio consistent with SDH deficiency was documented in the PGL from II.2, in concordance with the negative SDHB IHC result for this tumor. The 6-mm GIST specimen from II.1 also exhibited a succinate:fumarate ratio consistent with SDH deficiency despite positive SDHB IHC, the WES finding of a known KIT mutation in this particular GIST specimen, and an SDH-sufficient ratio in the 20-mm GIST specimen from the same patient. The PGL from II.4 demonstrated an equivocal succinate:fumarate ratio, whereas SDHB IHC classified this tumor as SDH deficient. There is a lack of normative data with which to compare the ratios of the other tumor types; the ratios in Table 1 are provided for reference only. Overall, there was inconsistency between the metabolomic profiles and IHC results for the various neoplasms and the latter was prioritized as the gold standard by which to determine SDH deficiency.
E. Cascade Testing
The c.20 + 74 A > G SDHC variant was subsequently detected in III.1, III.3, III.6, and III.7 of family 1, and in II.2 of family 2. Apart from II.2 of family 2, who was recently diagnosed with a likely HNPGL, all of these mutation carriers appear to be unaffected to date.
The variant was absent in III.2, III.4, and III.5 of family 1, and in II.5 of family 2.
Cascade testing has not yet been performed in II.3 and III.1 of family 2.
3. Discussion
The SDHC intronic variant reported here is the deepest intronic variant to be reported among the SDHx genes, highlighting aberrant splicing as an important consideration in patients with unexplained familial PPGL syndromes and other SDH-deficient tumors. This was identified in 2 families with historical links to the same small region in Italy who were shown to be distantly related by haplotyping. Contrary to the fortuitous finding in this WES study, intronic variants beyond the first 10 bp of exon-intron junctions will usually be missed by WES as well as the NGS gene panels typically used in the clinical testing of PPGL patients. Whole-genome sequencing, though more costly, may be required in PPGL cases with negative routine genetic testing despite a characteristic phenotype. The extent to which this is necessary will depend on the frequency of intronic SDHx mutations in unexplained SDH-deficient tumor syndromes, noting that 18% to 19% of patients with SDH-deficient tumors by IHC lack identifiable SDHx mutations by current testing methodologies [26, 27].
SDH-related tumors have been described in the setting of a range of germline SDHx variant types, including start codon, missense, nonsense, and frameshift variants, and whole exon and gene deletions [1, 28]. Epigenetic variation has also been described with SDHC promoter hypermethylation now known to account for Carney triad [29] and the half of SDH-deficient GISTs that were previously considered unexplained because of a lack of germline SDHx mutations [2, 3]. Splice site mutations are another known mechanism of tumorigenesis, with germline SDHC splicing variants accounting for 15% of PGLs and 30% of GISTs [1]. However, such splicing variants are typically only 1 to 2 bp away from the intron-exon boundary. Of 557 publicly available SDHx variants reported in the Leiden Open Variant Database (http://www.lovd.nl/3.0/home) as anything but benign or likely benign, only 38 variants are intronic and suspected or proven to cause aberrant splicing according to the submitted classification. The deepest of such variants are only ±7 bp away from the exon-intron boundary. While the term deep intronic variant is reserved for variants more than 100 bp away from exon-intron boundaries [30], the c.20 + 74A > G SDHC variant is important to recognize because it falls outside the usual 10- to 20-bp region that is typically assessed in clinical genetic testing, explaining why the variant was undetected in the preceding sequential genetic testing that spanned 12 years in family 1.
The SDHC-related familial PGL syndrome (also referred to as hereditary PGL syndrome type 3; PGL3) is widely considered to be a less severe disorder than the more common familial syndromes associated with SDHD (PGL1) and SDHB (PGL4) mutations [3, 31]. PGL3 typically manifests as unifocal nonsecretory HNPGL with low malignant potential, reduced risk of RCC and PA, and overall low penetrance [3]. Consistent with this classical phenotype, our families developed nonsecretory HNPGLs with metastasis in only one case. All PGLs stained negative for SDHB and positive for SDHA, as expected. Furthermore, the one PGL available for WES exhibited Chr 1 loss consistent with loss of the WT SDHC allele.
Unusually for the SDHC gene in particular, family 1 also exhibited the other 3 tumor types that have been previously linked to SDH deficiency. However, none of these tumors was proven to be SDH deficient in this family. The RCC was deemed sufficient by SDHB IHC performed soon after surgery around the advent of SDH IHC, but contemporary IHC studies, metabolomic profiling, and WES were not possible in this specimen, which was later destroyed, or in the macroprolactinoma that was successfully treated with a dopamine agonist. The GIST stained positive for KIT, showed normal SDHB IHC, and had predominant spindle cell morphology, all consistent with the somatic gain-of-function KIT mutation found on WES. Metabolomic analysis conversely showed one GIST specimen to have a significantly increased succinate:fumarate ratio suggestive of SDH deficiency, although the limited evidence base of metabolomic profiling compared to SDHB IHC is recognized. Furthermore, we did not identify any SDHC somatic second hits on WES of the GIST and SDHC promoter hypermethylation studies were negative.
We observed other tumors that have been previously reported in patients with SDHx abnormalities—adrenocortical adenoma [16, 31, 32], meningioma [33], breast cancer [1, 33] and diffuse gastric cancer [34, 35]—as well as tumors that have not been previously linked with SDHx mutations—HCC, desmoid tumor, solitary fibrous tumor of the lung, ovarian serous cystadenoma, ovarian cellular fibroma and cholangiocarcinoma. Whether any of these tumors relate to the germline SDHC mutation remains to be elucidated because these tumors were either shown to be SDH sufficient by IHC or were unavailable for investigation. Succinate:fumarate ratios were calculated in available tumors, but the significance of these results is limited by the lack of comparative tumor-specific data.
Although most of the tumors in these families are individually rare and the combination of tumors would be exceedingly rare, we cannot exclude the incidental co-occurrence of sporadic tumors. Another possibility is multiple inherited neoplasia allele syndrome (known as MINAS) [36]; however, WES did not demonstrate suspicious germline variants in other relevant tumor predisposition genes. The apparently high burden of tumors in SDHx mutation carriers may also relate to close surveillance, especially in the absence of definitive evidence of SDH deficiency in the tumors.
The cryptic relatedness suspected in these families because of their shared ethnicity was confirmed by a contemporary form of haplotype analysis in which identity by descent was determined by comparing rare variants deduced from NGS and ExAC reference data. This methodology may be used to evaluate other suspected SDHx founder mutations. A partial SDHC gene deletion in apparently unrelated patients of Yemenite ethnicity has been suggested but unproven to be due to a founder mutation [31]. Previous genealogy work using demographic data traced a large cohort of French-Canadian patients with SDHC-related PGLs due to a truncating founder mutation [37]. Confirming cryptic relatedness is not only of biological interest, but also clinically significant as it guides cascade testing.
In conclusion, we report a novel SDHC pathogenic variant, c.20 + 74A > G, which represents the deepest intronic mutation in an SDHx gene. Although we showed the PGLs in these families to be SDH deficient, conclusive results were not reached in the other tumors that either showed positive SDHB IHC or were unable to be studied because of a lack of tumor specimens. Further research is required to assess the causative role of this SDHC mutation in the wide tumor spectrum described here. For now, SDHx intronic mutations should be considered in patients with SDH-related tumor types, especially in the approximately 20% of SDH-deficient tumors with no identifiable mutation on routine genetic testing [26, 27]. Large validation studies are required to determine the cost-benefit analysis of targeted testing for intronic mutations—for example, through RNA-Seq—in patients with PPGL and other seemingly inherited disorders [38].
Acknowledgments
We thank the families for their participation in this study; and Milena Babic, Rosalie Kenyon, Ming Lin, and Wendy Parker for technical support in DNA extraction and sequencing.
Financial Support: This work was supported by the Royal Adelaide Hospital A.R. Clarkson Scholarship and the Royal Australasian College of Physicians Servier Staff ‘Barry Young’ Research Establishment Fellowship (to S.M.C.D.) and a Royal Adelaide Hospital Health Services Charitable Gifts Board grant, and produced with the financial and other support of the Cancer Council SA’s Beat Cancer Project on behalf of its donors and the State Government of South Australia through the Department of Health.
Glossary
Abbreviations
- CNVs
copy number variants
- FFPE
formalin-fixed, paraffin-embedded
- GIST
gastrointestinal stromal tumor
- HCC
hepatocellular carcinoma
- HNPGL
head and neck paraganglioma
- IHC
immunohistochemistry
- MLPA
multiplex ligation-dependent probe amplification
- NGS
next-generation sequencing
- PA
pituitary adenoma
- PCR
polymerase chain reaction
- PGL
paraganglioma
- PPGLs
pheochromocytomas and paragangliomas
- RCC
renal cell carcinoma
- RNA-Seq
RNA sequencing
- SDH
succinate dehydrogenase
- SDHx
SDHA/B/C/D and SDHAF2 genes
- WES
whole-exome sequencing
- WT
wild-type
Additional Information
Disclosure Statement: The authors have nothing to disclose.
Data Availability
Restrictions apply to the availability of data generated and analyzed during this study to preserve patient confidentiality. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided.
References
- 1. Evenepoel L, Papathomas TG, Krol N, et al. Toward an improved definition of the genetic and tumor spectrum associated with SDH germ-line mutations. Genet Med. 2015;17(8):610-620. [DOI] [PubMed] [Google Scholar]
- 2. Gill AJ. Succinate dehydrogenase (SDH)-deficient neoplasia. Histopathology. 2018;72(1):106-116. [DOI] [PubMed] [Google Scholar]
- 3. Benn DE, Robinson BG, Clifton-Bligh RJ. 15 Years of paraganglioma: clinical manifestations of paraganglioma syndromes types 1-5. Endocr Relat Cancer. 2015;22(4):T91-T103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eijkelenkamp K, Osinga TE, Links TP, van der Horst-Schrivers ANA. Clinical implications of the oncometabolite succinate in SDHx-mutation carriers. Clin Genet. 2020;97(1):39-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toledo RA, Burnichon N, Cascon A, et al. Consensus statement on next-generation-sequencing-based diagnostic testing of hereditary phaeochromocytomas and paragangliomas. Nat Rev Endocrinol. 2017;13(4):233-247. [DOI] [PubMed] [Google Scholar]
- 6. van Nederveen FH, Gaal J, Favier J, et al. An immunohistochemical procedure to detect patients with paraganglioma and phaeochromocytoma with germline SDHB, SDHC, or SDHD gene mutations: a retrospective and prospective analysis. Lancet Oncol. 2009;10(8):764-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gill AJ, Benn DE, Chou A, et al. Immunohistochemistry for SDHB triages genetic testing of SDHB, SDHC, and SDHD in paraganglioma-pheochromocytoma syndromes. Hum Pathol. 2010;41(6):805-814. [DOI] [PubMed] [Google Scholar]
- 8. Gill AJ, Chou A, Vilain R, et al. Immunohistochemistry for SDHB divides gastrointestinal stromal tumors (GISTs) into 2 distinct types. Am J Surg Pathol. 2010;34(5):636-644. [DOI] [PubMed] [Google Scholar]
- 9. Kim E, Wright MJ, Sioson L, et al. Utility of the succinate:fumarate ratio for assessing SDH dysfunction in different tumor types. Mol Genet Metab Rep. 2017;10:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lenders JW, Duh QY, Eisenhofer G, et al. ; Endocrine Society Pheochromocytoma and paraganglioma: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2014;99(6):1915-1942. [DOI] [PubMed] [Google Scholar]
- 11. Neumann HP, Bausch B, McWhinney SR, et al. ; Freiburg-Warsaw-Columbus Pheochromocytoma Study Group Germ-line mutations in nonsyndromic pheochromocytoma. N Engl J Med. 2002;346(19):1459-1466. [DOI] [PubMed] [Google Scholar]
- 12. Sbardella E, Cranston T, Isidori AM, et al. Routine genetic screening with a multi-gene panel in patients with pheochromocytomas. Endocrine. 2018;59(1):175-182. [DOI] [PubMed] [Google Scholar]
- 13. Brito JP, Asi N, Bancos I, et al. Testing for germline mutations in sporadic pheochromocytoma/paraganglioma: a systematic review. Clin Endocrinol (Oxf). 2015;82(3):338-345. [DOI] [PubMed] [Google Scholar]
- 14. Currás-Freixes M, Piñeiro-Yañez E, Montero-Conde C, et al. PheoSeq: a targeted next-generation sequencing assay for pheochromocytoma and paraganglioma diagnostics. J Mol Diagn. 2017;19(4):575-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Buffet A, Ben Aim L, Leboulleux S, et al. ; French Group of Endocrine Tumors (GTE) and COMETE Network Positive impact of genetic test on the management and outcome of patients with paraganglioma and/or pheochromocytoma. J Clin Endocrinol Metab. 2019;104(4):1109-1118. [DOI] [PubMed] [Google Scholar]
- 16. Carney JA. Gastric stromal sarcoma, pulmonary chondroma, and extra-adrenal paraganglioma (Carney triad): natural history, adrenocortical component, and possible familial occurrence. Mayo Clin Proc. 1999;74(6):543-552. [DOI] [PubMed] [Google Scholar]
- 17. Carney JA, Stratakis CA. Familial paraganglioma and gastric stromal sarcoma: a new syndrome distinct from the Carney triad. Am J Med Genet. 2002;108(2):132-139. [DOI] [PubMed] [Google Scholar]
- 18. Xekouki P, Szarek E, Bullova P, et al. Pituitary adenoma with paraganglioma/pheochromocytoma (3PAs) and succinate dehydrogenase defects in humans and mice. J Clin Endocrinol Metab. 2015;100(5):E710-E719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Sousa S, Toubia J, Hardy TSE, et al. Aberrant splicing of SDHC in families with unexplained succinate dehydrogenase-deficient paragangliomas - supplementary material. Figshare. Deposited 4 September 2020. doi: 10.6084/m9.figshare.12917279.v1 [DOI] [PMC free article] [PubMed]
- 20. Richter S, Gieldon L, Pang Y, et al. Metabolome-guided genomics to identify pathogenic variants in isocitrate dehydrogenase, fumarate hydratase, and succinate dehydrogenase genes in pheochromocytoma and paraganglioma. Genet Med. 2019;21(3):705-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Richter S, Peitzsch M, Rapizzi E, et al. Krebs cycle metabolite profiling for identification and stratification of pheochromocytomas/paragangliomas due to succinate dehydrogenase deficiency. J Clin Endocrinol Metab. 2014;99(10):3903-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Flanagan S, Lee M, Li CC, Suter CM, Buckland ME. Promoter methylation analysis of IDH genes in human gliomas. Front Oncol. 2012;2:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dannenberg H, de Krijger RR, Zhao J, et al. Differential loss of chromosome 11q in familial and sporadic parasympathetic paragangliomas detected by comparative genomic hybridization. Am J Pathol. 2001;158(6):1937-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joensuu H, Rutkowski P, Nishida T, et al. KIT and PDGFRA mutations and the risk of GI stromal tumor recurrence. J Clin Oncol. 2015;33(6):634-642. [DOI] [PubMed] [Google Scholar]
- 26. Castelblanco E, Santacana M, Valls J, et al. Usefulness of negative and weak-diffuse pattern of SDHB immunostaining in assessment of SDH mutations in paragangliomas and pheochromocytomas. Endocr Pathol. 2013;24(4):199-205. [DOI] [PubMed] [Google Scholar]
- 27. Papathomas TG, Oudijk L, Persu A, et al. SDHB/SDHA immunohistochemistry in pheochromocytomas and paragangliomas: a multicenter interobserver variation analysis using virtual microscopy: a Multinational Study of the European Network for the Study of Adrenal Tumors (ENS@T). Mod Pathol. 2015;28(6):807-821. [DOI] [PubMed] [Google Scholar]
- 28. Andrews KA, Ascher DB, Pires DEV, et al. Tumour risks and genotype-phenotype correlations associated with germline variants in succinate dehydrogenase subunit genes SDHB, SDHC and SDHD. J Med Genet. 2018;55(6):384-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Haller F, Moskalev EA, Faucz FR, et al. Aberrant DNA hypermethylation of SDHC: a novel mechanism of tumor development in Carney triad. Endocr Relat Cancer. 2014;21(4):567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaz-Drago R, Custódio N, Carmo-Fonseca M. Deep intronic mutations and human disease. Hum Genet. 2017;136(9):1093-1111. [DOI] [PubMed] [Google Scholar]
- 31. Else T, Marvin ML, Everett JN, et al. The clinical phenotype of SDHC-associated hereditary paraganglioma syndrome (PGL3). J Clin Endocrinol Metab. 2014;99(8):E1482-E1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Richter S, Klink B, Nacke B, et al. Epigenetic mutation of the succinate dehydrogenase C promoter in a patient with two paragangliomas. J Clin Endocrinol Metab. 2016;101(2):359-363. [DOI] [PubMed] [Google Scholar]
- 33. Niemeijer ND, Papathomas TG, Korpershoek E, et al. Succinate dehydrogenase (SDH)-deficient pancreatic neuroendocrine tumor expands the SDH-related tumor spectrum. J Clin Endocrinol Metab. 2015;100(10):E1386-E1393. [DOI] [PubMed] [Google Scholar]
- 34. Hansford S, Kaurah P, Li-Chang H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1(1):23-32. [DOI] [PubMed] [Google Scholar]
- 35. Habano W, Sugai T, Nakamura S, et al. Reduced expression and loss of heterozygosity of the SDHD gene in colorectal and gastric cancer. Oncol Rep. 2003;10(5):1375-1380. [PubMed] [Google Scholar]
- 36. Whitworth J, Skytte AB, Sunde L, et al. Multilocus inherited neoplasia alleles syndrome: a case series and review. JAMA Oncol. 2016;2(3):373-379. [DOI] [PubMed] [Google Scholar]
- 37. Bourdeau I, Grunenwald S, Burnichon N, et al. A SDHC founder mutation causes paragangliomas (PGLs) in the French Canadians: new insights on the SDHC-related PGL. J Clin Endocrinol Metab. 2016;101(12):4710-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bagnall RD, Ingles J, Dinger ME, et al. Whole genome sequencing improves outcomes of genetic testing in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2018;72(4):419-429. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Restrictions apply to the availability of data generated and analyzed during this study to preserve patient confidentiality. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided.