ABSTRACT
Chronic infection with human immunodeficiency virus (HIV) can cause progressive loss of immune cell function, or exhaustion, which impairs control of virus replication. However, little is known about the development and maintenance, as well as heterogeneity of immune cell exhaustion. Here, we investigated the effects of HIV infection on immune cell exhaustion at the transcriptomic level by analyzing single-cell RNA sequencing of peripheral blood mononuclear cells from four healthy subjects (37,847 cells) and six HIV-infected donors (28,610 cells). We identified nine immune cell clusters and eight T cell subclusters, and three of these (exhausted CD4+ and CD8+ T cells and interferon-responsive CD8+ T cells) were detected only in samples from HIV-infected donors. An inhibitory receptor KLRG1 was identified in a HIV-1 specific exhausted CD8+ T cell population expressing KLRG1, TIGIT, and T-betdimEomeshi markers. Ex-vivo antibody blockade of KLRG1 restored the function of HIV-specific exhausted CD8+ T cells demonstrating the contribution of KLRG1+ population to T cell exhaustion and providing an immunotherapy target to treat HIV chronic infection. These data provide a comprehensive analysis of gene signatures associated with immune cell exhaustion during HIV infection, which could be useful in understanding exhaustion mechanisms and developing new cure therapies.
KEYWORDS: HIV-1, single-cell RNA-seq, T cell dysfunction, immune exhaustion, KLRG1, NK cell impairment
GRAPHICAL ABSTRACT
Introduction
More than 76 million people have been infected with human immunodeficiency virus (HIV) since the epidemic was first recognized in the 1980s. Approximately 37 million people worldwide are currently living with HIV infection, of whom only ∼21 million have access to antiretroviral therapy (UNAIDS.org; http://www.unaids.org/en/resources/fact-sheet). The development of potent combination antiretroviral therapy (cART) has allowed HIV viremia to be controlled and significantly reduced the mortality of HIV-infected individuals; however, withdrawal of treatment leads to a rapid rebound of viremia, indicating that the host immune system remains unable to control viral replication [1]. HIV-induced dysfunction of the host immune system is one of the major causes of this recrudescence. Persistent exposure to viral antigens leads to chronic activation of immune cells, progressive loss of effector function to a state of exhaustion [2–6]. For example, exhausted CD8+ T cells lose their cytotoxic effector function and the ability to eradicate HIV-infected cells [7]. In addition, CD8+ T cell exhaustion prevents the differentiation of effector cells into memory cells with the ability to undergo rapid reactivation upon encounter with antigen [8]. Chronically activated CD4+ T cells also lose their effector functions, including the production of cytokines, such as IL-2 and IL-21, that sustain HIV-specific CD8+ T cells [9,10]. Consequently, HIV-induced CD4+ T cell depletion and exhaustion also results in dysfunction of HIV-specific CD8+ T cells, resulting in disease progression [8]. Therefore, understanding the mechanisms by which HIV infection leads to immune exhaustion is critical for the development of vaccines and therapies for HIV and, possibly, for other viral infections.
Sustained expression of high levels of inhibitory receptors such as PD-1, CTLA-4, CD160, TIGIT, and TIM-3 is a hallmark of T cell exhaustion. In HIV-infected individuals, expression of multiple inhibitory receptors has been demonstrated to correlate positively with plasma viral load and disease progression [4,11–16]. Notably, neutralizing antibody-mediated blockade of these receptors reverses T cell exhaustion by increasing the production of effector molecules and the proliferation of HIV-specific CD4+ and CD8+ T cells [4,11–14]. Blockade of PD-1 in simian immunodeficiency virus (SIV)-infected macaques results in expansion of SIV-specific CD8+ T cells, proliferation of memory B cells, decreased viremia, and prolonged survival [17,18]. Intriguingly, a recent report described that nivolumab (anti-PD-1) treatment of an HIV-infected patient with non-small-cell lung cancer restored the function of HIV-specific CD8+ T cells and decreased the HIV reservoir [19]. A clinical trial is currently ongoing to evaluate the safety and efficacy of PD-1 blockade in HIV-infected individuals with a low CD4+ T cell count (NCT03367754).
Despite the great promise of reversal of T cell exhaustion as a curative strategy for HIV, little is known about the mechanisms involved in immune cell exhaustion in HIV-infected individuals at the transcriptomic level. In addition, chronic HIV infection is associated with exhaustion not only of T cells but also of B and NK cells [20–22]. Therefore, it is possible that these three exhausted cell populations could cooperate in contributing to host immune system dysfunction, allowing unchecked HIV replication and disease progression.
Single-cell RNA sequencing (scRNA-seq) is a powerful technological advance to analyze the transcriptomic profiles of individual cells, thus enabling the heterogeneity of cells affecting biological processes to be analyzed [23]. scRNA-seq can reveal cellular identity as well as spatial organization and clonal distribution in complex heterogeneous immune populations [24,25]. The complexity of the immune system is a major obstacle to understanding the host immune response to HIV infection. The diversity and dynamic states of individual immune cells cannot be precisely revealed by traditional bulk expression profiles, which provide averaged values from highly heterogeneous populations [26]. In addition, there are limitations to identifying, isolating, and analyzing special or new cell populations, especially for the rare cell subtypes [27]. scRNA-seq has successfully identified new types of human blood dendritic cells (DCs), monocytes, and progenitors, resulting in a revised taxonomy of blood cells (Villani et al., 2017). Inspired by these studies, we reasoned that scRNA-seq could make it possible to analyze the effects of HIV infection on rare cell populations.
Recently, scRNA-seq technology has been applied to investigate viral diversity, heterogeneity of infection states, latency and reactivation, and virus–host interactions [28–30]. For example, Cohn et al. identified unique signature genes in reactivated latent CD4+ T cells, isolated from HIV infected individuals, which allow cell division without trigging the cell death pathways induced by HIV-1 replication [28]. In another report on HIV latency, scRNA-seq identified transcriptional heterogeneity in latent and reactivated CD4+ cells [29]. Other scRNA studies provided transcriptional dynamics in Zika and dengue viral infections [31] and during cytomegalovirus latency [32], and the heterogeneity of influenza virus infection [33]. These reports represent proof-of-concept that scRNA technology can provide unique insights into the virus–host interaction.
In this study, we performed scRNA-seq on PBMCs from HIV-infected and healthy donors to determine how HIV infection shapes the landscape of exhausted immune cells. We found that HIV infection induced the appearance of several cell clusters with novel gene signatures, including subsets of exhausted CD8+ and CD4+ T cells and CD8+ T cells that are highly responsive to interferon (IFN). One of the exhausted T cell signature genes was the inhibitory receptor KLRG1, which was co-expressed with TIGIT in exhausted T cells. Blockade of KLRG1 efficiently restored HIV-specific immune responses providing a promising novel target to develop immunotherapy for HIV infection. Finally, we performed integrated analysis of PBMCs and showed that HIV infection also induces B and NK cell dysfunction, which, together with T cell exhaustion, may contribute to HIV-related immunodeficiency.
Materials and methods
Ethical statement
Written informed consent was obtained from all donors or a parent under a study protocol approved by the Human Research Protection Program at UCSD.
Human subjects and PBMC isolation
Blood from healthy donors and PBMCs from HIV-infected donors were obtained from the UCSD AntiViral Research Center. Blood samples were isolated by Ficoll density centrifugation (Invitrogen) and PBMCs were removed, washed, and frozen until analyzed. scRNA-seq was performed on PBMCs from six HIV-infected donors, three each with high or low viral loads, and one healthy donor. In addition, we analyzed scRNA-seq datasets of PBMCs from three healthy donors acquired by using 10X genomics platform: one from 10x Genomics (8381 PBMCs) and two from a published study [34].
scRNA-seq library construction
PBMCs from the seven donors were thawed, and dead cells were removed using magnetic beads (AMSBIO, CB002) to ensure cell viability >80%. scRNA-seq libraries were prepared on the 10X Genomics platform using Chromium™ Single Cell 3’ v2 Reagent Kits and a Chromium instrument according to the manufacturer’s protocol. Libraries were sequenced on a HiSeq2500 platform to obtain 100-bp and 32-bp paired end reads using the following read length: read 1, 26 bp; read 2, 98 bp; and i7 index, 8 bp. The libraries were sequenced to reach ∼50,000 reads per cell.
scRNA-seq mapping
The scRNA-seq dataset was mapped against the human hg19 reference genome using CellRanger (v2.1; 10x Genomics). GRCh38-1.2.0 was used as a gene model for the alignment and was provided as part of the CellRanger pipeline as a compatible transcriptome reference. The Cell Ranger count function from the CellRanger pipeline was used to generate scgene counts for a single library. This pipeline was also used for alignment, filtering, and UMI counting.
Single-Cell differential gene expression analysis
Secondary analysis on the raw counts generated from the Cell Ranger pipeline was performed using Seurat (v.2.2.0), an R package for single-cell genomics. The analysis was based on the pipeline presented in the Guided Clustering Tutorial by the Sajita Lab. First, the cells were filtered to exclude cells with a large number of genes or with a large amount of mitochondrial DNA. The data were then normalized using a global-scaling method known as “LogNormalize,” which normalizes gene expression measurements for each cell by total expression, multiplies the values by a scale factor, and log-transforms the results. The average expression for each gene was calculated to identify genes with highly variable expression and then scaled to remove background variation. Linear dimensional reduction principal component analysis was performed on the identified variable genes. Principal components were determined by the generated Jackstraw and Elbow plots and PC 10 was mostly used for our PBMC analysis. A graph-based clustering approach was used to cluster the cells by type. The resolution parameter was altered for each sample according to cell number. Typically, a resolution >0.8 was used to identify a large number of clusters so that even small cell populations could be identified. Clusters of the same cell type were recombined when the clusters were labelled. Non-linear dimension reduction (tSNE) was used to visualize the data, and the plots were color-coded by cell type.
Unsupervised clustering of PBMCs and T cells by specific markers with Seurat
Genes enriched in a specific cluster were identified by the mean expression of each gene across all cells in the cluster. Then the expression of each gene from the cluster was compared to the median expression of the same gene from cells in all other clusters. To identify cell clusters, Seurat Findclusters was utilized based on a graph-based clustering approach. Cells were clustered by a shared nearest neighbour (SNN) modularity optimization based clustering algorithm. Then clusters were further characterized based on known markers. The cluster-specific markers were based on the top 20 genes after finding differentially expressed genes for each cluster. We defined the DEG by average expression fold change (avg_log FC) and percentage of expressing cells (pct.). Generally, cluster-specific markers are with the mean expression avg_log FC > 1 and pct >0.5.
Identification of signature genes in exhausted T cells from HIV-Infected donors
T lymphocyte clusters were selected in each sample using Seurat vignettes. Additional rounds of calculating highly variable genes, scaling the data, and re-clustering the cells were performed on the T cell subsets. Up-regulated and down-regulated differentially expressed genes for each cluster were identified. T cell subsets were identified by the expression of cell type-specific markers in feature plots and heatmaps. Exhausted CD8+ T cell clusters were also identified using known exhaustion markers. Exhausted CD8+ T cell and effector memory CD8+ T cells from three donors with high HIV loads were separately analyzed. All pre-processing procedures and clustering were also performed on the distinct subpopulations. Analysis of differentially expressed genes in these two cell types enabled identification of signature genes from the heatmap and feature plots of these subpopulations.
Pseudotime analysis
Seurat data matrices were imported into monocle3 for pseudotime analysis [35,36]. The single-cell dataset was normalize and pre-processed using the DelayedArray packages in Bioconductor and the preprocessCDS function from monocle3, with a num_dim = 10. The dataset then went through one round of dimensionality reduction using UMAP to eliminate noise. The cells in the dataset were partitioned into supergroups for future distinct trajectory recognition. The supergroups were organized into trajectories using SimplePPT. The trajectory plots were visualized and coloured by cell types and pseudotime separately, with the start of the pseudotime set to be CD8 T effector memory cells.
Integrated analysis of PBMCs from healthy and HIV-Infected donors
Using the Seurat integration pipeline [37], the PBMC datasets from three donors with high HIV load and four healthy donors were combined by executing the following steps. Individual datasets were filtered, log-normalized, and scaled using the same strategies and parameters as above. After pre-processing the data and selecting genes, canonical correspondence analysis (CCA) analysis was performed to define the shared correlation space between the two healthy donor samples. Multi-CCA was performed on the three samples from high viral load donors. After non-overlapping subpopulations were identified, the CCA subspaces were aligned with dimensional reduction. Another round of clustering was then performed on the two Seurat objects. The two integrations were then combined using the same procedures as above. To detect conserved and differentially expressed genes markers across datasets, differential expression testing was performed on the integrated datasets from all five samples. By performing these two steps, further information was obtained about exhaustion markers, and dot plots were generated to display conserved and differentially expressed gene markers.
Flow cytometry
For phenotyping analysis of PBMC, cryopreserved PBMCs were rapidly thawed at 37°C and resuspended in complete RPMI 1640 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS), 1% penicillin–streptomycin (Hyclone), and 100 U/ml DNase I (StemCell). The cells were washed in complete medium and stained with the viability dye Zombie Aqua (BioLegend), AF700-conjugated anti-CD3 (OKT3), APC-Cy7-anti-CD8a (RPA-T8) from BioLegend and FITC-anti-TIGIT (MBSA43), PE-anti-KLRG1 (13F12F2) (from Thermo Fisher). Cells were washed with staining buffer (BioLegend) and permeabilized by intracellular staining permeabilization wash buffer (BioLegend). Intracellular factors, T-bet and Eomes, were stained by APC-anti-T-bet (4B10) and PE-eFluor 610-Eomes (WD1928) antibody. After all the staining, cells were washed with staining buffer for twice and fixed with FluoroFix buffer (BioLegend), and analyzed on a custom four-laser FACSCanto flow cytometer (BD Biosciences, San Jose, CA, USA). Fluorescence-minus-one (FMO) or isotype staining controls were prepared for gating, and UltraComp beads (Thermo Fisher) were individually stained with each antibody to allow compensation. Data were analyzed using FlowJo version 10 (Tree Star, Ashland, OR, USA).
KLRG1 functional assay
For the intracellular cytokine stimulation assay, cryopreserved PBMCs were rapidly thawed and rested in complete RPMI 1640 medium for 12 h. Cells were counted and seeded at a concentration of 1 million/ml. Pooled Gag and Nef peptides (National Institute of Health AIDS Reagent Program) were added into the cells to stimulate HIV-specific cells at a 2.5 ug/ml each peptide. Meanwhile, cells were blocked with isotype IgG (BioXcell), anti-KLRG1 (13F12F2) (Thermo Fisher) at 20 ug/ml. All of the cells were treated with Brefeldin A to inhibit the secretion of cytokines. After stimulation and blocking for 12 h, the cells were washed twice and then stained for viability with Aqua and CD8a, followed by permeabilization and intracellular staining of CD3, IFN-γ (EH12.2H7), and TNFα (MAb11) and acquisition on the flow cytometer as above. For the CFSE proliferation assay, cryopresevered PBMCs were washed with PBS and resuspened in PBS at a concentration of 1 million/ml. Then cells were labelled with 0.5 μM CFSE for 10 min at room temperature. The reaction was quenched by adding excess culture medium and further washed. The labelled cells were resuspended and stimulated with DMSO, pooled Gag and Nef peptides at a 2.5 ug/ml each peptide or T cell activator (StemCell, 10971) in the presence or absence of isotype IgG, anti-KLRG1 antibody. After stimulation for 7 days, cells were washed and then stained for viability with Aqua, anti-CD3 and CD8a antibodies. Cells were fixed and acquired on the flow cytometer.
Data availability
The RNA-seq data in this study is deposited at the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database with an accession number GSE157829.
Results
Atlas of PBMCs in healthy and HIV-Infected donors
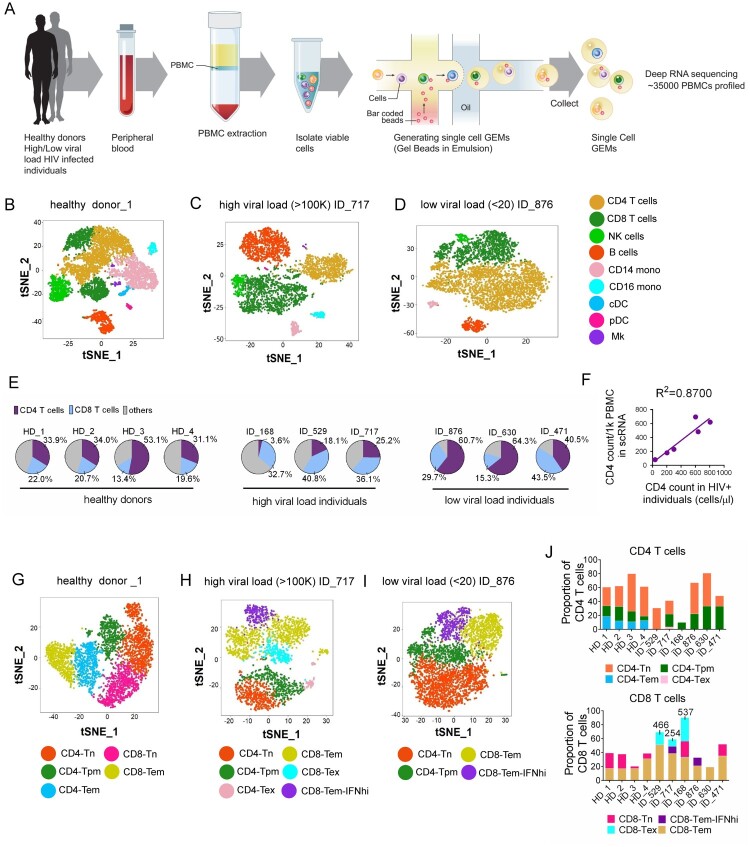
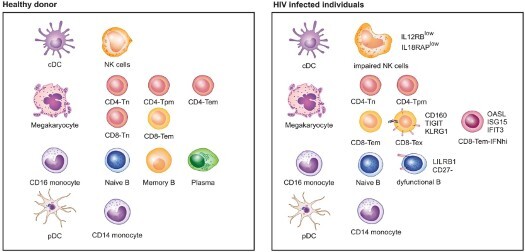
To examine the landscape of immune cell exhaustion induced by HIV infection, we isolated PBMCs from healthy and HIV-infected donors and performed scRNA-seq to identify cell clusters by expression cell type-specific gene signatures. Previous studies have suggested that expression of exhaustion genes such as PD-1, CTLA-4, CD160, TIGIT, and TIM-3 is associated with high HIV loads [11,12]. Therefore, we included three donors with a high viral load and three with a low viral load (>100,000 and <20 RNA copies/ml of plasma, respectively) in the analysis. A total of 12,852 and 15,758 PBMCs were sequenced from donors with high and low viral loads (hereafter referred to as HL-HIV-infected and LL-HIV-infected donors), respectively (4000–5500 PBMCs/donor). For comparison, we performed scRNA-seq of PBMCs from one healthy donor (ID HD_1) and obtained scRNA-seq data from other three healthy donors (ID HD_2: 10x Genomics; HD_3 and HD_4: two healthy controls, HC_1, HC_2, from a published study [34]). Donor characteristics are shown in Table 1. An overview of the approach is given in Figure 1A. Through unbiased analysis, we identified nine major cell clusters present in the healthy and HIV-infected donors based on expression of a unique gene signature: CD4+ T cells (CD3D+ CD8A− IL7Rhi), CD8+ T cells (CD3D+ CD8A+), natural killer cells (NK; CD3D− CD8A− IL7R− GNLYhi), B cells (MS4A1+), CD14+ monocytes (LYZhi CD14hi), CD16+ monocytes (LYZhi FCGR3Ahi), conventional dendritic cells (cDCs; LYZhi FCER1Ahi), plasmacytoid dendritic cells (pDCs; LYZlow IGJhi), and megakaryocytes (Mk; PPBP+) (Figure 1B–D, S1A-I). The healthy donor PBMC samples contained the expected proportions of major white blood cell classes: ∼50% T cells (CD4+:CD8+ T cells ∼2:1), 10–15% B cells, ∼10% NK cells, ∼20% monocytes, and ∼3% DCs (Figure S1I)[38].
Table 1.
Characteristics of HIV-infected individuals and healthy donors.
| PID | Age | Gender | Ethnicity | Duration of HIV infection, years |
Duration of viral suppression* |
ART regimen | Plasma HIV RNA (copies/mL) |
Stage of infection |
CD4+ count (cells/μL) |
Elite controllers |
|---|---|---|---|---|---|---|---|---|---|---|
| 529 | 59 | Male | Non-Hispanic | 0.3 | Intermittent suppression | GENVOYA | 585,100 | Chronic | 203 | No |
| 717 | 56 | Male | African American | 27 | Intermittent suppression | DESCOVY + TRUVADA + PREZISTA + PREZCOBIX + NORVIR |
185,072 | Chronic | 299 | No |
| 168 |
36 |
Male |
African American |
3.5 |
Never suppressed |
– |
259,111 |
Chronic |
37 |
No |
| 876 | 33 | Male | White | 7.6 | Fully suppressed | Triumeq | <20 | Chronic | 806 | No |
| 630 | 58 | Male | other race | 22 | Fully suppressed | ODEFSEY + TIVICAY | <20 | Chronic | 603 | No |
| 471 |
60 |
Male |
other race |
11 |
Fully suppressed |
JULUCA |
<20 |
Chronic |
638 |
No |
| HD1 | 22 | Female | Non-Hispanic | – | – | – | – | – | – | |
| HD2 | 23 | Male | White | – | – | – | – | – | – | |
| HD3 | 50–60 | Male | other race | – | – | – | – | – | – | |
| HD4 | 70–75 | Male | other race | – | – | – | – | – | – |
* Duration of viral suppression: fully suppressed (plasma HIV viral load < 50 copies/mL for >6 months), never suppressed (No ART treatment), intermittent suppression (ART treatment and plasma HIV viral load >50 copies/mL)
Figure 1.
Distinct cell clusters are identified by scRNA-seq of PBMCs from healthy and HIV-infected donors. (A) Overview of workflow. PBMCs were isolated from healthy donors and HIV-infected donors (three each with high and low viral loads [>100,000 and <20 RNA copies/ml plasma, respectively]). Single cells were captured by gel beads with primers and barcoded oligonucleotides and subjected to deep RNA-seq. (B–D) t-Distributed Stochastic Neighbor Embedding (t-SNE) projection of PBMCs from healthy donor HD_1 (B), high-load HIV-infected donor ID_717 (C), and low-load HIV-infected donor ID_876 (D), showing major cell clusters based on normalized expression of cell type-specific markers. NK, natural killer cells; CD14 mono, CD14+ monocytes; CD16 mono: CD16+ monocytes; cDC, conventional dendritic cell; pDC, plasmacytoid dendritic cell; Mk, megakaryocytes. (E) Pie charts showing the percentage CD4+ T cells, CD8+ T cells, and other PBMC subsets in the healthy and HIV-infected donors. (F) Linear regression analysis showing the correlation between CD4+ T cell counts calculated from scRNA analysis (cells/1000 PBMCs) vs flow cytometry (cells/μl) of PBMCs from HIV-infected donors. (G–I) tSNE projections for T cell subsets from healthy donor HD_1 (G), HL-HIV-infected donor ID_717 (H), and LL-HIV-infected donor ID_876 (I). Tn, naïve; Tpm, precursor memory; Tem, effector memory; Tex, exhausted; IFNhi, highly IFN-responsive. (J) Percentage of the indicated subclusters of CD4+ and CD8+ T cells from four healthy donor samples (HD_1, 2, 3, 4), three HL-HIV-infected donors (ID_529, _717, and _168), and LL-HIV-infected donors (ID_876, _630, and _471). See also Figure S1 and S2.
Consistent with the known effects of HIV infection, the absolute number and percentage CD4+ T cell counts were considerably lower in the three HL-HIV-infected donors (18.1%, 25.2%, 3.6%) compared with the four healthy donors (33.9%, 34.0%, 53.1%, 31.1%). In concert, the percentage (but not absolute cell numbers) of CD8+ T cells was increased in the HL-HIV-infected donors (40.8%, 36.1%, 32.7% versus 22.0%, 20.6%, 13.4%, 19.6%; Figures 1E and S1I). We also observed a high proportion of CD4+ T cells in the LL-HIV-infected donor samples (60.7%, 64.3%, 63.1%) (Figures 1E and S1I). Importantly, the absolute CD4+ T cell counts in the clinical blood samples and the numbers estimated from the scRNA-seq analysis showed a strong correlation (R2 = 0.87, Figure 1F), indicating that the scRNA-seq datasets accurately reflect the cell clusters present in the original blood samples.
To identify which T cell subsets are selectively depleted or expanded in the HIV-infected donors, we performed unbiased clustering based on subset-specific marker genes. In the healthy donor PBMCs, three CD4+ T cell and two CD8+ T cell clusters (Figures 1G, S2A, and S2C-E) were identified based on the relative enrichment or depletion of signature genes as naïve CD4+ T cells (CD4-Tn: CD8A− CCR7+ IL7Rhi), effector memory CD4+ T cells (CD4-Tem: CD8A− IL7Rhi CCR7− GZMA+) [39], and a cluster that we defined as precursor memory cells (CD4-Tpm: CD8A− IL7Rhi CCR7low LTBhi). The putative CD4-Tpm cluster showed a similar signature to that of CD4-Tn cells (e.g. TCF7, FOXP1) but they did not express effector function-associated genes (e.g. GZMA, CCR5, NKG7), suggesting that they may represent a transitional state between naïve and effector memory status (Gattinoni et al., 2011; Youngblood et al., 2017). This cluster also harboured high expression of the TNF family molecule lymphotoxin β (LTB; Figure S2A), which is consistent with a recently described CD4-Tpm cytotoxic cell cluster [40]. In addition to the three CD4+ T clusters, PBMCs from the healthy donors also contained clusters consistent with naïve CD8+ T cells (CD8-Tn: CD8A+ CCR7hi) and effector memory CD8+ T cells (CD8-Tem: CD8A+ IL7R− CCR7− GZMA+ NKG7+; Figures 1G, S2A, and S2C-E).
Notably, the composition and proportion of T cell subtypes in PBMCs were markedly altered by HIV infection. Samples from the donors with high viral loads contained dramatically smaller populations of CD4-Tem and CD8-Tn, and three new cell clusters with unique gene signatures could be discerned; namely, exhausted memory CD8+ T cells (CD8-Tex), exhausted memory CD4+ T cells (CD4-Tex), and a population of CD8+ Tem cells with marked upregulation of IFN-response genes, which we termed CD8-Tem-IFNhi (Figures 1H, S2B and S2F-I). The Tex subpopulations were classified based on previous work showing that CD8+ T cells expressing high levels of the exhaustion markers CD160 and TIGIT were enriched in HIV-infected individuals [13,14]. In the three high-load HIV-infected donors, we found that 18.1%, 10.1%, and 33.9% of total CD8+ T cells carried the Tex gene signature. Similarly, CD4-Tex cells were characterized by expression of the exhaustion markers TIGIT and CTLA4. This is consistent with an earlier demonstration that CTLA4 is enriched in CD4-Tex cells during HIV chronic infection [12]. Finally, cells within the CD8-Tem-IFNhi cluster showed enrichment of IFN-stimulated genes such as OASL, ISG15, IFIT2, and IFIT3, consistent with expansion of the host antiviral immune response (Figures 1H, 1I, S2G). Another study also showed the interferon signatures in HIV-specific CD8+ T cells [41]. Interestingly, analysis of samples from the LL-HIV-infected donors showed a reduction in the CD4-Tem and CD8-Tn clusters and the appearance of a CD8-Tem-IFNhi cluster, similar to the observations in samples from HL-HIV-infected donors; however, there were no obvious CD4+ or CD8+ Tex cell populations in samples from these donors (Figures 1I, 1J, S2H, and S2I).
Collectively, these data identify the major PBMC and T cell subsets affected by HIV infection, including CD4+ and CD8+ Tex populations and highly IFN-responsive CD8+ Tem cells, each of which have unique signature profiles.
Identification of novel genes associated with T cell exhaustion
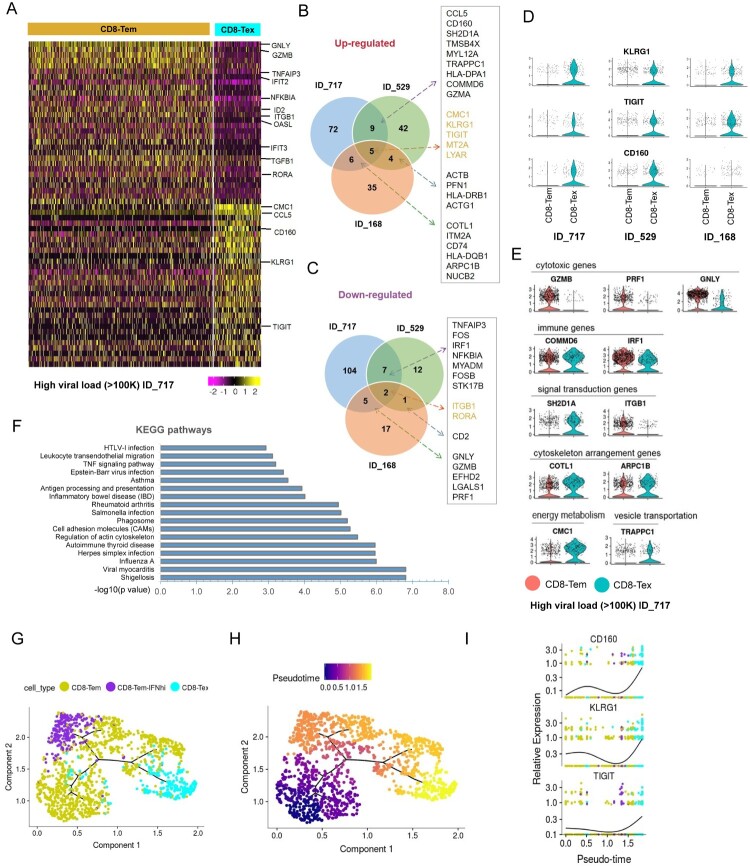
Since little is known about the gene signatures of Tex cells in HIV-infected donors, we further characterized the CD8+ Tex subsets by comparing their gene expression patterns with those of their unexhausted counterparts, which are typically Tem cells (Figure 2A and Table S1). From this analysis, we identified a total of 39 genes that were commonly altered in CD8-Tex cells from at least two of the three HL-HIV-infected donors (p < 0.01, roc test; Figure 2B and C). The 24 up-regulated genes in HL-HIV-infected donors included the known exhaustion markers TIGIT and CD160 (Figure 2D) [13,14]. These inhibitory receptors have also been identified as T cell exhaustion markers in pathogen-infected and tumor-bearing animals [13,14]. CD8-Tex cells from the HL-HIV-infected donors also showed up-regulated expression of another inhibitory receptor, killer cell lectin-like receptor subfamily G member 1 (KLRG1), which contains an immunoreceptor tyrosine-based inhibitory motif in the cytoplasmic domain (Figure 2D). KLRG1 is known to be a T cell differentiation marker [42]. Other genes specifically up-regulated in CD8-Tex cells from HL-HIV-infected donors included CMC1, SH2D1A, COMMD6, TRAPPC1, and COTL-1 (Figure 2B and E). Conversely, there were 15 genes commonly down-regulated in CD8-Tex cells from HL-HIV-infected donors, including IRF1, ITGB1, and genes encoding the cytotoxic T cell effector molecules granzyme B (GZMB), perforin 1 (PRF1), and granulysin (GNLY) (Figure 2C and E). These observations strongly suggest that CD8-Tex cells show less effector function phenotypes than normal CD8+ Tem cells.
Figure 2.
Identification of novel signature genes in CD8-Tex cells from HIV-infected donors. (A) Heatmap showing differentially expressed genes in CD8-Tem and CD8-Tex cells from HL-HIV-infected donor ID_717. The signature genes are indicated to the right of the heatmap. The colour code below the map indicates the relative expression levels. (B and C) Venn graphs of conserved up-regulated (B) and down-regulated (C) genes in CD8-Tex cells from the three HL-HIV-infected donors. (D) Violin plots of the conserved up-regulated exhaustion-associated signature genes in CD8-Tex compared with CD8-Tem cells from the three HL-HIV-infected donors. Each dot represents a single cell and the shapes represent the expression distribution. The remaining cells are depicted on the x axis. (E) Violin plots of the up-regulated and down-regulated genes associated with the indicated functions in CD8-Tex compared with CD8-Tem cells from HIV-infected donor (ID_717). Each dot represents a single cell and the shapes represent the expression distribution. The remaining cells are depicted on the x axis. (F) GO analysis of the common genes in exhausted CD8+ memory T cells. (G and H) The trajectory plots of the pseudotime analysis shows two distinct trajectories of the CD8-Tem cells in HIV infected individuals. The start of the pseudotime was set to be CD8-Tem cells. The trajectory plots were visualized and coloured by cell types (G) and pseudotime (H) separately. (I) Three representative genes CD160, KLRG1, and TIGIT were identified by pseudotime analysis to be significantly enriched in the CD8-Tex cells.
Other immune response genes differentially expressed in HL-HIV-infected donor-derived Tex cells compared with Tem cells included COMMD6, an NF-κB-inhibiting protein, and the IFN-responsive transcription factor IRF1, which were up-regulated and down-regulated, respectively, in CD8-Tex cells, consistent with HIV-induced suppression of immune signalling (Figure 2E) [15,43]. In support of this, SH2D1A, which encodes the signalling inhibitor SLAM-associated protein (SAP), was also up-regulated in Tex compared with Tem cells [44]. SAP has important roles in signalling for T cell differentiation and the antiviral immune response [44]. Interestingly, Tex cells showed altered expression of genes involved in cytoskeletal function (TMSB4X, PFN1, ACTG1, COTL1, ARPC1B), energy metabolism (CMC1), and vesicle transportation (TRAPPC1) (Figure 2E). Gene Ontology analysis revealed that the differentially expressed genes common to Tex cells in HIV-infected donors were enriched in pathways involved in pathogen infection, actin cytoskeleton regulation, cell adhesion, antigen processing and presentation, and TNF signalling (Figure 2F).
The development of normal CD8 Tem cell into exhausted T cell was analyzed by “pseudotime” analysis as the cells differentiate asynchronously and heterogeneously. CD8 effector memory cells were ordered based on single cell transcriptomics by pseudotime analysis using monocle 3 [35,36]. Root state is defined as normal effector memory. CD8 effector memory cells branched into interferon high effector memory and exhausted cells, revealing a bifurcating trajectory of CD8 differentiation in HIV infected individuals. Exhausted cells are at the end of pseudotime, suggesting a terminal differentiation state (Figure 2G and H). Importantly, this exhaustion sub-branch was occupied by cells expressing high levels of exhaustion genes, CD160, KLRG1, and TIGIT (Figure 2I). Interferon high effector memory sub-branch showed high expression of IFIT3, ISG15 and OASL (Figure S2J). Taken together, these results identified novel signatures of KLRG1, CD160, and TIGIT characterizes exhausted CD8+ T cells in HIV-infected donors.
KLRG1 blockade effectively restores the function of HIV-specific CD8+ T cells
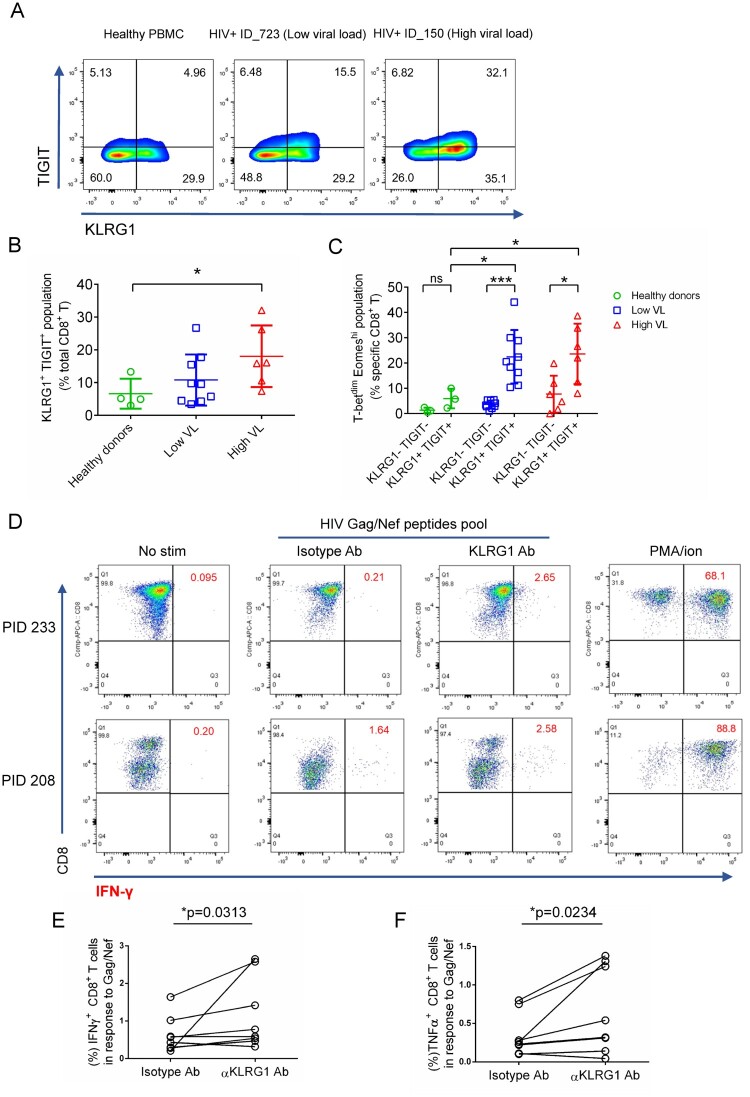
The observation that KLRG1 was strongly up-regulated and co-expressed with two known HIV induced exhaustion markers TIGIT, CD160 in Tex cell clusters from all three HL-HIV-infected donors raised the possibility that KLRG1 may be involved in T cell exhaustion. To test this hypothesis, we analyzed KLRG1 expression in PBMCs from healthy and HIV-infected donors by flow cytometry (Figure 3A and B). We found the frequency of KLRG1 and TIGIT expressing CD8+ T cells significantly increased in PBMCs from HIV-infected individuals (Low VL, 10.8%; High VL, 18.1%) compared with healthy donors (6.6%). Since PBMCs from higher viral load individuals showed severe exhaustion, we did see higher ratio of KLRG1+TIGIT+ populations in high viral load PBMCs. KLRG1 is widely used as a terminal differentiation marker in lymphocytes [45]. It is necessary to clarify the exact exhausted KLRG1+ subpopulation induced by HIV chronic infection. T-bet and Eomes had been proved to be highly associated with exhausted phenotype induced by HIV infection in a reciprocal pattern [46]. T-betdim Eomeshi bulk or HIV-specific CD8+ T cells showed poor functional characteristics [46]. Therefore, we further analyzed the percentages of T-betdim Eomeshi population in KLRG1+TIGIT+ CD8+ T cells. The result showed that the frequency was much higher in HIV-infected PBMCs (Low VL, 22.4%; High VL, 23.3%) compared with that from healthy donors (HD, 6.0%) (Figure 3C, S3A-C). In addition, in both high viral load and low viral load group, the percentage of T-betdim Eomeshi cells was much higher in KLRG1+TIGIT+ population (potential exhausted) compared with KLRG1-TIGIT- (non-exhausted) population (Figure 3C). This data suggests that KLRG1+TIGIT+ T-betdim Eomeshi population in CD8+ T cells represent a novel exhausted T cell population in HIV chronic infection.
Figure 3.
KLRG1 blockade effectively restores the function of HIV-specific CD8+ T cells. (A) Flow cytometry of KLRG1- and TIGIT-expressing CD8+ T cells from the indicated healthy and HIV-infected donors. Numbers indicate the percentage of KLRG1+ TIGIT+, KLRG1− TIGIT−, KLRG1+ TIGIT− and KLRG1− TIGIT+, -expressing cells. (B) KLRG1+ TIGIT+ population is increased in HIV HL-individuals. The percentage of KLRG1+ TIGIT+ cells was analysis by flow in healthy donors (n = 4), HIV LL- (n = 9) and HL- (n = 6) individuals. Mean ± SD, *p < 0.05, student’s t test. (C) A novel KLRG1+ TIGIT+T-betdimEomeshi CD8+ T cell population is significant up-regulated in HIV infected individuals. KLRG1 and TIGIT double negative or double positive cells was extracted from B. Expression of T-bet and Eomes was analyzed. The percentage of KLRG1+ TIGIT+T-betdimEomeshi was shown. Mean ± SD, *p < 0.05, ***p < 0.001, ns, not significant, student’s t test. (D-F) Blocking KLRG1 restores the activation of T cells to HIV peptides stimuli. Ex vivo PBMCs from chronically HIV-infected individuals were stimulated with HIV Gag/Nef peptide pool in the presence of isotype, KLRG1 blocking antibodies. (D) Representative flow cytometry plots with gating for CD8 T cells, showing IFN-γ responses of PBMCs from two HIV-infected individuals (PID 233 and 208). No HIV-1 Gag stimulation with an isotype control is shown as a negative control. A positive control with PMA and ionomycin (1:500) treatment is shown. (E and F). The frequency (%) of IFN-γ (E) and TNFα (F) positive CD8+ T cells (n = 8) is shown. p values were calculated by Wilcoxon matched-pairs signed ranked test. See also Figure S3.
To delineate the relationship between up-regulated KLRG1 and T cell dysfunction, we evaluated whether KLRG1 blockade on HIV-specific CD8+ T cells could restore the antiviral immune responses. In the cytokine stimulation assay, PBMCs from HIV-infected individuals were stimulated with HIV Gag/Nef peptide pool and treated with KLRG1 blocking or isotype antibodies, and then IFN-γ and TNFα expressing cells were evaluated to determine the restoration of HIV-specific T cell function. Incubation with KLRG1 blocking antibody (20 µg/mL) significantly increased the percentage of IFN-γ expressing HIV-specific CD8+ T cells from 0.21% to 2.65% in the sample of PID 233, and from 1.64% to 2.58% in the sample of PID 208 (Figure 3D). Meanwhile, the percentage of TNFα expressing HIV-specific CD8+ T cells increased from 0.26% to 1.31% and from 0.8% to 1.38%, respectively (Figure S3D). The statistical analysis for all of the PBMCs isolated from 8 HIV-infected donors showed that KLRG1 blockade could significantly restore cytokine responses (Figure 3E and F). We also evaluated the restoration of dysfunctional HIV-specific T cells utilizing CFSE proliferation assay. Blockade of KLRG1 significantly enhanced HIV-specific CD8+ T cells proliferation in Gag/Nef peptides pool stimulation group while no improvement was found in no stimulation group (Figure S3E, F). The percentage of the proliferated (CFSEdim) cells increased from 4.48% to 5.67% and from 3.14% to 7.15%, respectively. The statistical analysis for the PBMCs from 6 HIV-infected donors suggested the proliferation could be recovered by KLRG1 blockade. Collectively, blocking KLRG1 restored the cytokine production and the proliferation of HIV-specific CD8+ T cell.
Integrated analysis of HIV-infected individuals revealed heterogeneity of exhausted CD8+ T cells and immune cell dysfunction induced by HIV infection
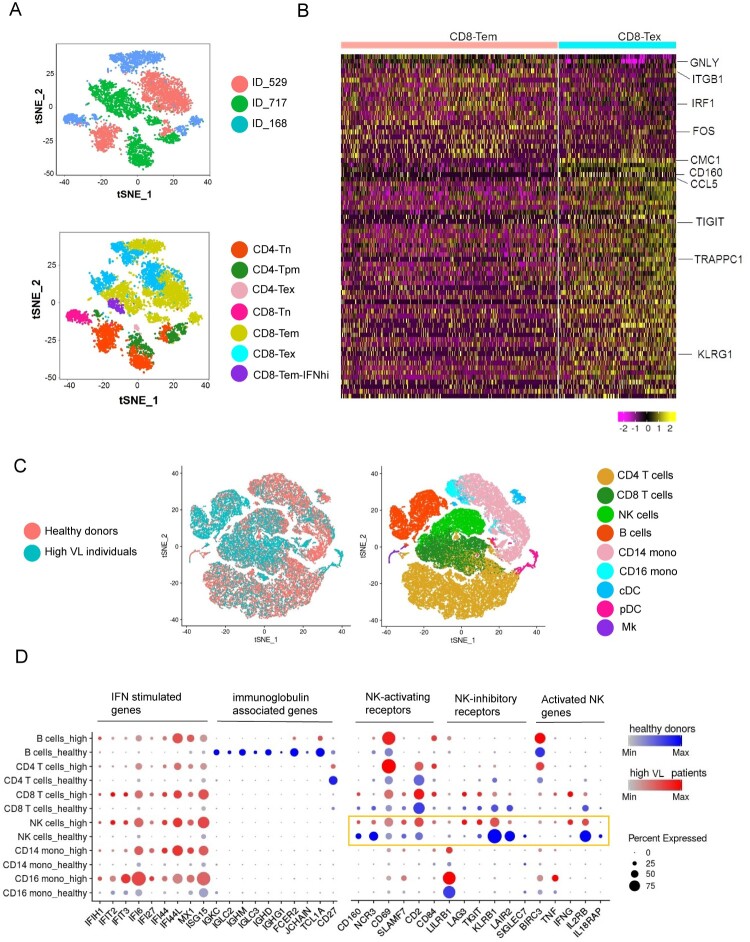
To characterize the heterogeneity of exhausted CD8+ T cells from different HIV-infected individuals, we performed integrated analysis for T cells. We observed heterogeneity in the HIV-infected donors (Figure 4A). This heterogeneity could reflect variations in the individual donor immune response to HIV infection and the different stages of HIV pathogenesis. However, the exhausted CD8+ T population from different HIV-infected individuals are close to each other (Figure 4A). Consistent with previous results, these exhausted population shared similar transcriptomics (Figure 4B). Therefore, exhausted CD8+ T clusters from different HIV-infected individuals are similar but exhibit heterogeneity.
Figure 4.
Integrated analysis of HIV-infected individuals revealed heterogeneity of exhausted CD8+ T cells and immune cell dysfunction induced by HIV infection. (A) tSNE plots of integrated datasets from three high viral load HIV-infected individuals. Tn, naïve; Tpm, precursor memory; Tem, effector memory; Tex, exhausted; IFNhi, highly IFN-responsive. (B) Heatmap of the scRNA-seq dataset from three high viral load HIV-infected individuals showing differentially expressed genes in in CD8-Tem and CD8-Tex cells. Colour bar below the map indicates the expression level. (C) tSNE plots of integrated datasets from the healthy and HIV-infected donors (left) and identification of nine major cell subpopulations (right). NK, natural killer cells; CD14 mono, CD14+ monocytes; CD16 mono: CD16+ monocytes; cDC, conventional dendritic cells; pDC, plasmacytoid dendritic cells; Mk, megakaryocytes. (D) Expression of “variable genes” in six of the cell clusters in healthy and HIV-infected donors. The colour intensity indicates the average expression level in a cluster and the circle size reflects the percentage of expressing cells within each cluster. See also Figure S4 and S5.
To extend our investigation to non-T cells, we performed an integrated analysis of the composition of PBMCs from the four healthy donors and three HL-HIV-infected donors. Whereas the cell clusters identified in PBMCs from the healthy donors were similar and overlapping, suggestive of a homogeneous cell composition (Figure S4A), we observed marked cluster heterogeneity in the HIV-infected donors (Figure S4B). Heterogeneity in these clusters could be the result of individual donor immune response to HIV or the stages of infection under various treatments. There are limited number of overlapping cells among the healthy donors and high viral load individuals. Nine clusters were identified with common marker genes in the integrated plots (Figure 4C and S4C). IFN-stimulated genes, such as IFIH1, IFIT2, IFI6, IFI27, IFI44, IFI44L, MX1 and ISG15, were significantly up-regulated in monocytes and CD4+, CD8+, B, and NK cells (Figure 4D). Sustained IFN signalling has been reported to be a key driver of immunosuppression, as demonstrated by CD4+ T cell depletion and CD8+ T cell expansion during HIV infection [47,48]. Blocking of the IFNα/β receptor in cART-suppressed HIV-infected humanized mice reversed HIV-induced type-I IFN and rescued the T cell response [2]. Therefore, suppression of IFN signalling is a potential therapeutic strategy in HIV-infected patients with chronic inflammation [2,5].
HIV infection is known to hyperactivate B cells but, paradoxically, it also suppresses the antibody response [47]. We found that PBMCs from HIV-infected donors contained an increased percentage of B cells compared with healthy donors (22.1%, 26.9%, 33.7% versus 8.7%, 15.1%, 4.0% and 7.5%; Figure S1I). However, compared with healthy donors, expression of immunoglobulin genes, including IGKC, IGLC2, IGHM, IGLC3, IGHD, IGHG1, FCER2, and JCHAIN, the naïve B cell marker TCL1A, and the activated B cell marker CD27 was abolished or dramatically down-regulated in HIV-infected donors (Figure 4D). In contrast, the B cell inhibitory receptor LILRB1 was up-regulated in PBMCs from HIV-infected donors (Figure 4D).
Growing evidence supports a role for NK cells in the antiviral response to HIV-1 [49]. NK cell cytokine secretion and cytotoxic functions are controlled by multiple activating and inhibitory receptors [50]. Of note, we found that HIV infection markedly altered the expression of several NK receptor genes (Figure 4D). Among the activating receptor genes, CD160 and NCR3 were down-regulated while CD69, SLAMF7, CD2, and CD84 were up-regulated. Conversely, the inhibitory receptor genes LILRB1, LAG3, and TIGIT were up-regulated and KLRB1, LAIR2, and SIGLEC7 were down-regulated. In addition, we observed increased expression of BIRC3, TNF, and IFNG, which have been proposed to be NK cell activation markers [22]. Despite the beneficial roles of NK cells in antiviral responses, their function is known to be impaired by chronic HIV infection [49]. Consistent with this, we found a reduction in the expression of genes involved in cytokine signalling proteins (e.g. IL12RB and IL18RAP). Costanzo et al. also observed a reduction in IL18RAP expression in NK cells in HIV-infected donors, and this was rescued in individuals inoculated with an HIV vaccine [22]. These data indicate that chronic HIV infection is associated with an impaired NK cell response. We also performed integration analysis of LL-HIV-infected individuals vs healthy donors and LL-HIV-infected vs HL-HIV-infected individuals and we found that transcriptomic changes were relatively small compared with integration analysis for HL-HIV-infected individuals vs healthy donors (Figure S5).
Discussion
Collectively, our results underscore the ability of scRNA-seq to overcome obstacles presented by the complexity and heterogeneity of the immune system to enable investigation of pathophysiological changes in very small cell populations [26]. In this study, we applied scRNA-seq to analyze how HIV infection perturbs the immune cell transcriptome, and we identified marked changes in the composition and function of CD4+, CD8+ T, and B cell subsets as well as the function of NK cells. Importantly, exhausted CD8+ T cell populations expressing CD160, TIGIT, and KLRG1 were identified here in three HL-HIV-infected individuals. KLRG1 co-expressing with TIGIT marked a new exhausted population and KLRG1 blocking antibody reversed the cytokines response of virus-specific CD8+ T cells. This validated the contribution of KLRG1+ population to exhaustion and suggested KLRG1 could be a novel target for immunotherapy against HIV infection.
Previous studies have described that repetitive and persistent antigen stimulation induced the up-regulation of KLRG1 in virus-specific CD8+ T cells [51]. KLRG1, as an inhibitory receptor, inhibits T cell and NK cell function when activated by its ligands [52,53]. In addition, another inhibitory receptor TIGIT which marks exhausted T cells in HIV infection co-expressed with KLRG1 (Figure 3A and B). Also, KLRG1+ CD8+ T cells showed decreased ability to secrete cytokines upon HIV stimulation compared with KLRG1- CD8+ T cells [54]. Therefore, it is reasonable that KLRG1 expression was induced by HIV chronic infection to impair the immune system. T-bet and Eomes are T-box transcription factors which regulate the expression of inhibitory receptors and the exhaustion of CD8+ T cells [55]. HIV chronic infection down-regulates T-bet and up-regulates Eomes which contributes to poor HIV-specific CD8+ T cell functionality [46]. The significant increase of T-betdim Eomeshi population in KLRG1+TIGIT+ CD8+ T cells in HIV-infected PBMCs compared with healthy PBMCs was consistent with these conclusions (Figure 3C). Further HIV-specific tetramer staining would help understand the expression pattern of these factors in HIV-specific CD8+ T cells. In addition, our KLRG1 blockade functional experiments further validate the contribution of KLRG1+ population to CD8+ T cell exhaustion (Figure 3D). Although, KLRG1 was a well-known differentiation maker for lymphocytes, it was dispensable for T cell and NK cell development and function after virus infection in vivo [56]. It suggests that KLRG1 is a promising and novel therapeutic target for HIV infection.
The immune cell exhaustion landscapes demonstrated by this and previous studies [57] provide a clearer illustration of HIV-induced immune deficiency. Chronic proinflammatory signalling induced by HIV infection and replication leads to activation of the immune system; however, the accompanying depletion of CD4+ T cells reduces their effector functions and, in turn, impairs the CD4+ T cell-dependent functions of CD8+ T cells and B cells. Thus, despite the fact that CD8+ T cell and B cell populations can be activated and expanded during HIV infection, the cells cannot differentiate into effector cells to counter the infection. Moreover, T and B cell exhaustion exacerbates the immune deficiency [47,48]. Taken together, our data support a vital role for chronic inflammation and immune cell exhaustion in the pathogenesis of HIV infection.
Supplementary Material
Acknowledgments
S.W. and Q.Z. designed and performed the experiments, analyzed the data, and wrote the first manuscript draft; H.H. and K.A. analyzed the data; M.A.Y.K. contributed to obtaining the PBMCs, and data analysis and interpretation; and T.M.R. contributed to the experimental design, data analysis and interpretation, and manuscript writing. All authors contributed to manuscript writing and approved the final version. We thank Drs. Douglas Richman, Jonathan Karn, and Mario Stevenson for helpful discussions, Kristen Jepsen at the IGM Genomics Center for help with scRNA-seq, Celsa Spina of UCSD CFAR for flow analysis and members of the Rana lab for helpful discussions and advice. We also thank Dr. Song Chen for advice and help in sample preparation and data analysis. This work was supported in part by grants from the National Institutes of Health (DA039562, DA046171, DA049524). Access to retrospectively collected blood samples from HIV-1-infected donors was made possible by the University of California, San Diego Center for AIDS Research, an NIH-funded program (P30 AI036214), which is supported by the following NIH Institutes and Centers: NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NIA, NIGMS, and NIDDK.
Funding Statement
This work was supported by National Institute on Drug Abuse: [Grant Number DA039562,DA046171,DA049524].
Declaration of interest statement
T.M.R. is a founder of ViRx Pharmaceuticals and has an equity interest in the company. The terms of this arrangement have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies.
References
- 1.Chun TW, Justement JS, Murray D, et al. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010 Nov 27;24(18):2803–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng L, Ma J, Li J, et al. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J Clin Invest. 2017 Jan 3;127(1):269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haas A, Zimmermann K, Oxenius A.. Antigen-dependent and -independent mechanisms of T and B cell hyperactivation during chronic HIV-1 infection. J Virol. 2011 Dec;85(23):12102–12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones RB, Ndhlovu LC, Barbour JD, et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J Exp Med. 2008 Nov 24;205(12):2763–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhen A, Rezek V, Youn C, et al. Targeting type I interferon-mediated activation restores immune function in chronic HIV infection. J Clin Invest. 2017 Jan 3;127(1):260–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bengsch B, Ohtani T, Khan O, et al. Epigenomic-Guided Mass cytometry Profiling Reveals disease-specific Features of exhausted CD8 T cells. Immunity. 2018 May 15;48(5):1029–1045.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wherry EJ. T cell exhaustion. Nat Immunol. 2011 Jun;12(6):492–499. [DOI] [PubMed] [Google Scholar]
- 8.Wherry EJ, Kurachi M.. Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol. 2015 Aug;15(8):486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porichis F, Kwon DS, Zupkosky J, et al. Responsiveness of HIV-specific CD4 T cells to PD-1 blockade. Blood. 2011 Jul 28;118(4):965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elsaesser H, Sauer K, Brooks DG.. IL-21 is required to control chronic viral infection. Science. 2009 Jun 19;324(5934):1569–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trautmann L, Janbazian L, Chomont N, et al. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat Med. 2006 Oct;12(10):1198–1202. [DOI] [PubMed] [Google Scholar]
- 12.Kaufmann DE, Kavanagh DG, Pereyra F, et al. Upregulation of CTLA-4 by HIV-specific CD4+ T cells correlates with disease progression and defines a reversible immune dysfunction. Nat Immunol. 2007 Nov;8(11):1246–1254. [DOI] [PubMed] [Google Scholar]
- 13.Peretz Y, He Z, Shi Y, et al. CD160 and PD-1 Co-expression on HIV-specific CD8 T cells Defines a subset with Advanced dysfunction. PLoS Pathog. 2012;8(8):e1002840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chew GM, Fujita T, Webb GM, et al. TIGIT marks exhausted T cells, Correlates with disease progression, and Serves as a target for immune restoration in HIV and SIV infection. PLoS Pathog. 2016 Jan;12(1):e1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Day CL, Kaufmann DE, Kiepiela P, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006 Sep 21;443(7109):350–354. [DOI] [PubMed] [Google Scholar]
- 16.Petrovas C, Casazza JP, Brenchley JM, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006 Oct 2;203(10):2281–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Velu V, Titanji K, Zhu B, et al. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature. 2009 Mar 12;458(7235):206–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shetty R D, Velu V, Titanji K, et al. PD-1 blockade during chronic SIV infection reduces hyperimmune activation and microbial translocation in rhesus macaques. J Clin Invest. 2012 May;122(5):1712–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guihot A, Marcelin AG, Massiani MA, et al. Drastic decrease of the HIV reservoir in a patient treated with nivolumab for lung cancer. Ann Oncol. 2018 Feb 1;29(2):517–518. [DOI] [PubMed] [Google Scholar]
- 20.Moir S, Ho J, Malaspina A, et al. Evidence for HIV-associated B cell exhaustion in a dysfunctional memory B cell compartment in HIV-infected viremic individuals. J Exp Med. 2008 Aug 4;205(8):1797–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavilio D, Lombardo G, Benjamin J, et al. Characterization of CD56-/CD16+ natural killer (NK) cells: a highly dysfunctional NK subset expanded in HIV-infected viremic individuals. Proc Natl Acad Sci U S A. 2005 Feb 22;102(8):2886–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costanzo MC, Kim D, Creegan M, et al. Transcriptomic signatures of NK cells suggest impaired responsiveness in HIV-1 infection and increased activity post-vaccination. Nat Commun. 2018 Mar 23;9(1):1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svensson V, Vento-Tormo R, Teichmann SA.. Exponential scaling of single-cell RNA-seq in the past decade. Nat Protoc. 2018 Apr;13(4):599–604. [DOI] [PubMed] [Google Scholar]
- 24.Giladi A, Amit I.. Single-Cell Genomics: A Stepping Stone for future Immunology Discoveries. Cell. 2018 Jan 11;172(1-2):14–21. [DOI] [PubMed] [Google Scholar]
- 25.Wagner A, Regev A, Yosef N.. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016 Nov 8;34(11):1145–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papalexi E, Satija R.. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018 Jan;18(1):35–45. [DOI] [PubMed] [Google Scholar]
- 27.Villani AC, Satija R, Reynolds G, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017 Apr 21;356(6335). doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohn LB, da Silva IT, Valieris R, et al. Clonal CD4(+) T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation. Nat Med. 2018 May;24(5):604–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Golumbeanu M, Cristinelli S, Rato S, et al. Single-Cell RNA-Seq Reveals transcriptional heterogeneity in latent and reactivated HIV-infected cells. Cell Rep. 2018 Apr 24;23(4):942–950. [DOI] [PubMed] [Google Scholar]
- 30.Rato S, Rausell A, Munoz M, et al. Single-cell analysis identifies cellular markers of the HIV permissive cell. PLoS Pathog. 2017 Oct;13(10):e1006678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanini F, Pu SY, Bekerman E, et al. Single-cell transcriptional dynamics of flavivirus infection. Elife. 2018 Feb 16;7, doi: 10.7554/eLife.32942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shnayder M, Nachshon A, Krishna B, et al. Defining the transcriptional landscape during cytomegalovirus latency with single-cell RNA sequencing. MBio. 2018 Mar 13;9(2). doi: 10.1128/mBio.00013-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russell AB, Trapnell C, Bloom JD.. Extreme heterogeneity of influenza virus infection in single cells. Elife. 2018 Feb 16;7, doi: 10.7554/eLife.32303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen W, Su W, Tang H, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu X, Hill A, Packer J, et al. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017 Mar;14(3):309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trapnell C, Cacchiarelli D, Grimsby J, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014 Apr;32(4):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butler A, Hoffman P, Smibert P, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018 Apr 2;36(5):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Cheung F, Shi R, et al. PBMC fixation and processing for Chromium single-cell RNA sequencing. J Transl Med. 2018 Jul 17;16(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gattinoni L, Lugli E, Ji Y, et al. A human memory T cell subset with stem cell-like properties. Nat Med. 2011 Sep 18;17(10):1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patil VS, Madrigal A, Schmiedel BJ, et al. Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Sci Immunol. 2018 Jan 19;3(19). doi: 10.1126/sciimmunol.aan8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buggert M, Nguyen S, de Oca G S-M, et al. Identification and characterization of HIV-specific resident memory CD8(+) T cells in human lymphoid tissue. Sci Immunol. 2018 Jun 1;3(24):eaar4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joshi NS, Cui W, Chandele A, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007 Aug;27(2):281–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maine GN, Mao X, Komarck CM, et al. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. EMBO J. 2007 Jan 24;26(2):436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu C, Nguyen KB, Pien GC, et al. SAP controls T cell responses to virus and terminal differentiation of TH2 cells. Nat Immunol. 2001 May;2(5):410–414. [DOI] [PubMed] [Google Scholar]
- 45.Pauken KE, Wherry EJ.. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015 Apr;36(4):265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buggert M, Tauriainen J, Yamamoto T, et al. T-bet and Eomes are differentially linked to the exhausted phenotype of CD8+ T cells in HIV infection. PLoS Pathog. 2014 Jul;10(7):e1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le Saout C, Hasley RB, Imamichi H, et al. Chronic exposure to type-I IFN under lymphopenic conditions alters CD4T cell homeostasis. PLoS Pathog. 2014 Mar;10(3):e1003976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Catalfamo M, Di Mascio M, Hu Z, et al. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc Natl Acad Sci U S A. 2008 Dec 16;105(50):19851–19856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scully E, Alter G.. NK cells in HIV disease. Curr HIV/AIDS Rep. 2016 Apr;13(2):85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vivier E, Tomasello E, Baratin M, et al. Functions of natural killer cells. Nat Immunol. 2008 May;9(5):503–510. [DOI] [PubMed] [Google Scholar]
- 51.Thimme R, Appay V, Koschella M, et al. Increased expression of the NK cell receptor KLRG1 by virus-specific CD8 T cells during persistent antigen stimulation. J Virol. 2005 Sep;79(18):12112–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ito M, Maruyama T, Saito N, et al. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J Exp Med. 2006 Feb 20;203(2):289–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grundemann C, Bauer M, Schweier O, et al. Cutting edge: identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J Immunol. 2006 Feb 1;176(3):1311–1315. [DOI] [PubMed] [Google Scholar]
- 54.Streeck H, Kwon DS, Pyo A, et al. Epithelial adhesion molecules can inhibit HIV-1-specific CD8(+) T-cell functions. Blood. 2011 May 12;117(19):5112–5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kao C, Oestreich KJ, Paley MA, et al. Transcription factor T-bet represses expression of the inhibitory receptor PD-1 and sustains virus-specific CD8+ T cell responses during chronic infection. Nat Immunol. 2011 May 29;12(7):663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grundemann C, Schwartzkopff S, Koschella M, et al. The NK receptor KLRG1 is dispensable for virus-induced NK and CD8+ T-cell differentiation and function in vivo. Eur J Immunol. 2010 May;40(5):1303–1314. [DOI] [PubMed] [Google Scholar]
- 57.Khaitan A, Unutmaz D.. Revisiting immune exhaustion during HIV infection. Curr HIV/AIDS Rep. 2011 Mar;8(1):4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data in this study is deposited at the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database with an accession number GSE157829.