Abstract
Enhancing the radioresponsiveness of colorectal cancer (CRC) is essential for local control and prognosis. However, consequent damage to surrounding healthy cells can lead to treatment failure. We hypothesized that short-chain fatty acids (SCFAs) could act as radiosensitizers for cancer cells, allowing the administration of a lower and safer dose of radiation. To test this hypothesis, the responses of three-dimensional-cultured organoids, derived from CRC patients, to radiotherapy, as well as the effects of combined radiotherapy with the SCFAs butyrate, propionate and acetate as candidate radiosensitizers, were evaluated via reverse transcription-quantitative polymerase chain reaction, immunohistochemistry and organoid viability assay. Of the three SCFAs tested, only butyrate suppressed the proliferation of the organoids. Moreover, butyrate significantly enhanced radiation-induced cell death and enhanced treatment effects compared with administration of radiation alone. The radiation-butyrate combination reduced the proportion of Ki-67 (proliferation marker)-positive cells and decreased the number of S phase cells via FOXO3A. Meanwhile, 3/8 CRC organoids were found to be non-responsive to butyrate with lower expression levels of FOXO3A compared with the responsive cases. Notably, butyrate did not increase radiation-induced cell death and improved regeneration capacity after irradiation in normal organoids. These results suggest that butyrate could enhance the efficacy of radiotherapy while protecting the normal mucosa, thus highlighting a potential strategy for minimizing the associated toxicity of radiotherapy.
Keywords: butyrate, colorectal cancer, FOXO3A, patient-derived organoid, radiosensitizer
Introduction
Colorectal cancer (CRC) was the third most commonly diagnosed malignancy and the second leading cause of cancer-related deaths worldwide in 2020 (1). As a primary treatment, enhancing the radio response of locally advanced rectal cancer is essential to achieving local control and improving long-term prognosis. Despite the effectiveness of radiotherapy in treating cancer, damage caused by radiation to the surrounding normal cells is often unavoidable (2). Therefore, it is necessary to design a strategy for effectively treating the tumor while minimizing these nontargeted side effects. One such approach includes the use of radiosensitizers, which can render cancer cells more sensitive to radiation, thereby enabling a lower and safer radiation dose to achieve the same therapeutic response (3). Over the past decade, several studies have addressed the efficacy of diverse radiosensitizers, including fluorouracil (5-FU), oxaliplatin, capecitabine and bevacizumab, in combination with radiotherapy for CRC. Specially, 5-FU achieved a complete pathological response rate of ~20-30% (4-6). However, in addition to their effects on cancer cells, these radiosensitizers are also reportedly toxic to healthy cells (7,8); therefore, while they may function to enhance the efficiency of radiation, they do not effectively minimize the adverse side effects.
Additional research has revealed the influence of intestinal microbes derived from diet on the development of CRC (9,10). As food flows through the colon, the fiber can be fermented by gut microbiota to produce short-chain fatty acids (SCFAs), such as propionate, acetate and butyrate, which play multifunctional roles in the intestinal epithelial cells (11). In particular, butyrate is used as an energy source in normal colonocytes. Under the Warburg effect, cancer cells rely on glucose as their primary energy source, resulting in nuclear accumulation of butyrate that functions as a histone deacetylase (HDAC) inhibitor (12-14). Considering that other HDAC inhibitors, including suberoylanilide hydroxamic acid (SAHA) and valproic acid, have been shown to enhance the radiosensitivity of cancer cells (15-17), we hypothesized that butyrate, along with other potential SCFAs, may serve as effective, cancer cell-specific and safe radiosensitizers.
To date, responses to radiotherapy have been investigated exclusively in animal models owing to the low success rate associated with the initiation of patient-derived cell cultures (18,19). However, several studies have demonstrated the value of 3D patient-derived organoid (PDO) culture systems as useful and physiologically relevant tools to study the treatment response (20,21). Organoids are self-organizing 3D structures grown from stem cells to mimic in vivo architecture and biology. Hence, organoids more closely mimic in vivo tissues than cells grown in two-dimensional tissue cultures, and are valuable tools for modeling human diseases, including cancers (22).
Therefore, in the present study, PDOs from CRC patients (CRC-PDO) were used as a model system for investigating the response to radiotherapy, and to evaluate the influence of combined radiotherapy with the SCFAs butyrate, propionate and acetate as candidate radiosensitizers through regulation of HDAC activity. Cumulatively, these results lay the foundation for development of a novel combined radiation/SCFA strategy for CRC and promote the utility of CRC-PDOs as a model system for investigating tumor evolution and modulating drug response.
Materials and methods
Human specimens
A total of 7 rectal cancer samples and 1 colon cancer tumor sample and the paired healthy tissue samples were obtained from 8 patients diagnosed with CRC between December 2017 and May 2018 at the Korea Cancer Center Hospital (Seoul, Korea). All tumors were staged according to the pathological tumor/node/metastasis classification (8th edition) of the American Joint Committee on Cancer (23). Surgically resected and endoscopic biopsy samples were verified by pathologists through hematoxylin and eosin staining as previously described (24). The 7 rectal samples were obtained during an endoscopic biopsy, and the 1 colon cancer sample was obtained by low anterior resection. The mean age of patients was 61.5 years (range, 53-85 years), and 4 patients were male and 4 patients were female. Clinical data, including patient information, such as age, sex and tumor locations, are provided in Table SI.
This study was approved by Ethics Committee of Korea Cancer Center Hospital (approval no. KIRAMS-2017-07-001). All research was performed in accordance with the approved guidelines and regulations of the institution. All samples were obtained from patients who had provided written informed consent for the use of their tissues for the purposes of research after the operation.
Organoid culture
Both tumor and adjacent normal tissues were collected, from which healthy crypts and tumor cells were isolated, as described previously (20,25), with minor modifications. Briefly, normal intestinal tissue fragments were incubated in 8 mM EDTA-phosphate buffered saline (PBS) for 20 min on ice and further incubated for 8 min at 37°C. Under these dissociation conditions, colonic crypts were mildly digested, thereby physically separating their bottom and top segments. Cancer tissues were incubated with collagenase type II (Sigma-Aldrich; Merck KGaA), dispase type II (Roche Applied Science) and Y-27632 (BioVision, Inc.) for 30 min at 37°C. Isolated crypts and cells were washed with PBS and centrifuged at 300 × g for 3 min at room temperature. The cells were then embedded in Matrigel on ice (growth factor reduced, phenol red free; Corning, Inc.) and seeded in 24-well plates, followed by addition of culture medium. The composition of the CRC-PDO culture medium was 1× B27 (Gibco; Thermo Fisher Scientific, Inc.), 1.25 mM N-acetyl cysteine (United States Pharmacopeia), 50 ng/ml human epidermal growth factor (BioVision, Inc.), 50 ng/ml human Noggin (PeproTech, Inc.), 10 nM gastrin (Sigma-Aldrich; Merck KGaA), 500 nM A83-01 (BioVision, Inc.) and 100 mg/ml primocin (InvivoGen). In experiments that prevented the Warburg effect, organoids were grown from high-glucose (3.151 g/l) to low-glucose (1 g/l) medium in same composition. Wnt signaling-related factors are essential for the growth of healthy colon organoids (normal-PDOs). However, in most cases, CRC is associated with mutations that aberrantly activate the Wnt signaling pathway (26). Therefore, for selection of tumor cells, CRC-PDOs were cultured without Wnt-related factors in the culture medium to ensure that normal tissue-derived organoids did not contaminate tumor tissues (27). The normal-PDOs were cultured in IntestiCult™ Organoid Growth Medium (Stemcell Technologies, Inc.) and CRC-PDO culture medium with CHIR99021 (Stemgent; ReproCELL) and R-spondin1 (R&D Systems, Inc.). To prevent anoikis, 10 µM Y-27632 was added to the culture medium for the first 2-3 days. Maintenance and freezing of organoids was carried out as described previously with slight modification (25). When organoids were >200 µm, they were passaged by pipetting using Gentle Cell Dissociation Reagent (Stemcell Technologies, Inc.) according to the manufacturer's instructions.
Organoid viability
For further dissociation into single cells, organoids were resuspended in TrypLE Express (Thermo Fisher Scientific, Inc.) via pipetting with a p200 pipette, and incubated at 37°C for 10 min. Cells were then pipetted multiple times to form a homogeneous resuspension and centrifuged at 600 × g for 5 min at 4°C, followed by discarding of the supernatant to obtain pelleted cells. The pellet was resuspended with Matrigel and divided into a 96-well plate (5,000 cells/10 µl Matrigel per well). After the Matrigel was polymerized, 100 µl culture media was added. Organoid viability was assessed as described previously (28) with slight modifications. Briefly, 3-[4,5-dimethylthiazol-2-yl]-2, 5 diphenyl tetrazolium bromide (MTT) solution was added to the organoid culture at a final concentration of 500 µg/ml. After incubation for 3 h, the medium was discarded, and 20 µl 2% sodium dodecyl sulfate solution was added to solubilize the Matrigel for 2 h. Next, 100 µl DMSO was added for 1 h to solubilize the reduced MTT, and the optical density was measured on a BioTek Eon microplate absorbance reader (BioTek Instruments, Inc.) at 562 nm. Matrigel without organoids (10 µl) was used as the control.
Treatment of organoids with compounds and radiation
To test radiosensitivity, the CRC-PDOs were irradiated with Matrigel in the plates after treatment for 6 h with 1 mM acetate (Sigma-Aldrich; Merck KGaA), butyrate (Sigma-Aldrich; Merck KGaA), propionate (Sigma-Aldrich; Merck KGaA) and/or 100 nM FOXO inhibitor, AS1842856 (EMD Millipore) at 37°C using a 137Cs γ-ray source (Atomic Energy of Canada Ltd.) at a dose rate of 3.81 Gy/min. The concentration of 1 mM SCFA was selected as described previously (29). The organoids were irradiated with three fractions of 5 Gy (one fraction per day), and their size was analyzed by ImageJ software 1.48 V (National Institutes of Health) on days 0 and 3. For analysis at the 2nd passage, organoids were treated with butyrate and/or ionizing radiation (IR). After 72 h, organoids were passaged by pipetting using Gentle Cell Dissociation Reagent with a 1:2-1:4 split ratio. After 72 h, organoids were evaluated using the EVOS FL Cell Imaging System (Thermo Fisher Scientific, Inc.).
Immunocytochemistry and immunohistochemistry
Intestinal stem cells can differentiate into all cell lineages of the intestinal epithelium, including goblet cells (mucin 2+; Muc2+), entero-endocrine cells (chromogranin A+; ChgA+) and enterocytes (VL1+); meanwhile all proliferating cells express Ki-67 (30). PDOs were fixed in 4% paraformaldehyde at room temperature for 24 h, embedded in paraffin and then dissected into sections (5-µm). Following treatment with Smartblock (CANDOR Bioscience) for 30 min at room temperature, the slides were incubated with primary antibodies against anti-Ki-67 (1:200; cat. no. ab16667; Abcam), anti-Muc2 (1:100; cat. no. ab90007; Abcam) and anti-ChgA (1:100; cat. no. 20086; ImmunoStar) at 4°C overnight. Slides were then incubated with Alexa Fluor 594 goat antibody to rabbit IgG (H+L) (1:200; cat. no. A11012; Thermo Fisher Scientific, Inc.) for 1 h at room temperature. Images were acquired using a EVOS FL Cell Imaging System (Thermo Fisher Scientific, Inc.).
The cellular efficacy of recognizing double-strand DNA breaks (DSB) and DNA damage response can be assessed by observing the foci of γ-H2AX (31). For the detection of γ-H2AX, organoids were seeded in 8-well glass chamber slides (LabTek Services, Ltd.). The cells were fixed with 4% paraformaldehyde for 30 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS. Unspecific epitopes on organoids were blocked with 5% bovine serum albumin (Gene Depot) in Tris-buffered saline for 1 h at room temperature and incubated with anti-γ-H2AX (1:100; cat. no. 05636; EMD Millipore) overnight at 4°C, followed by incubation with Alexa Fluor 488 goat antibody to mouse IgG (H+L) (1:200; cat. no. A11001; Thermo Fisher Scientific, Inc.) for 1 h at room temperature.
To characterize organoids and their tissue of origin, immunohistochemistry was performed with colorectal markers cytokeratin (CK)19, CK20 and CDX2, as described previously (32). Organoids and tissues were fixed in 4% paraformaldehyde at room temperature overnight and embedded in paraffin blocks. Subsequently, 3-µm sections were dewaxed in xylene and hydrated through a graded series of alcohol. Endogenous peroxidase activity was quenched by incubating the sections for 30 min in 0.3% H2O2 and incubated in Smartblock (CANDOR Bioscience) for 30 min at room temperature to reduced non-specific binding. Sections were incubated for 1 h at 37°C with anti-CDX2 (1:200; cat. no. 235R-16; Cell Marque; Sigma-Aldrich; Merck KGaA), CK20 (1:500; cat. no. 320M-16; Cell Marque; Sigma-Aldrich; Merck KGaA) and CK19 (1:400; cat. no. ab15463; Abcam). Detection was performed on an Envision/Horseradish Peroxidase system (Dako; Agilent Technologies, Inc.), and counterstained with hematoxylin for 10 min at room temperature. Finally, the sections were dehydrated through a graded series of alcohol, cleared in xylene and mounted. Images were acquired using the IX73 inverted microscope (Olympus Corporation; magnification, ×40).
EdU staining
EdU staining was performed with a Click-iT Plus EdU FACS kit (cat. no. C10424; Thermo Fisher Scientific, Inc.) and Click-iT® Plus EdU Imaging kits (cat. no. C10639; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Images were acquired using the EVOS FL Cell Imaging system fluorescence microscope (Thermo Fisher Scientific, Inc.; magnification, ×200).
Lactate assays
Lactate concentration was determined using a Lactate Assay kit (cat. no. K607-100; BioVision) according to the manufacturer's instructions. Optical density was measured on a BioTek Eon microplate absorbance reader (BioTek Instruments, Inc.) at 570 nm 30 min after the addition of the substrate. The mean values of lactate concentrations were calculated for each condition based on the data obtained from three independent experiments.
Reverse transcription-quantitative PCR (RT-qPCR)
Organoids were harvested from Matrigel by first washing them with cold PBS and incubating in cell recovery solution (Corning, Inc.) at 4°C for 1 h (33). Total RNA was isolated using a RNeasy mini kit (Qiagen GmbH). Subsequently, 2 µg RNA was used for cDNA synthesis using RevertAid first strand cDNA kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Gene expression levels were quantified using the SYBR Green PCR kit (Qiagen) and qPCRs were run on an ABI7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The qPCR thermocycling conditions were as follows: 94°C for 5 min, followed by 30 cycles at 94°C for 30 sec and 60°C for 30 sec, and a final extension step at 72°C for 10 min. The amount of target mRNA was normalized to that of GAPDH mRNA and analyzed as previously described (34). Primers are listed in Table SII. Cycle threshold (Cq) values were obtained using an auto baseline, which was applied to all amplicons of the same primer set by the corresponding PCR instrument software (Applied Biosystems; Thermo Fisher Scientific, Inc.). Triplicates with a Cq value difference of >0.5 were not considered and excluded from the analysis. Relative concentrations of cDNA for analyzing relative changes in gene expression were calculated using the 2−ΔΔCq method (35).
Western blotting
For western blot analysis, organoids were washed with cold PBS and lysed in RIPA buffer (Thermo Fisher Scientific, Inc.). Proteins were quantified using the Bradford method and 20 µg protein/lane was resolved by 8-13.5% SDS-PAGE. The membranes were incubated with primary antibodies against cleaved poly-ADP-ribose polymerase (c-PARP; 1:1,000; cat. no. 5625; Cell Signaling Technology, Inc.), PARP (1:1,000; cat. no. 9542; Cell Signaling Technology, Inc.), cleaved caspase 3 (c-caspase 3; 1:1,000; cat. no. 9664; Cell Signaling Technology, Inc.), caspase 3 (1:1,000; cat. no. 9662; Cell Signaling Technology, Inc.), phosphorylated (p)-JNK (1:1,000; cat. no. 9251; Cell Signaling Technology, Inc.), JNK (1:1,000; cat. no. 9252; Cell Signaling Technology, Inc.), p-p38 (1:1,000; cat. no. 9211; Cell Signaling Technology, Inc.), p38 (1:1,000; cat. no. 9212; Cell Signaling Technology, Inc.), FOXO3A (1:1,000; cat. no. 2499; Cell Signaling Technology, Inc.), LaminB (1:1,000; cat. no. SC-6216; Santa Cruz Biotechnology, Inc.) and β-actin (1:5,000; cat. no. A5441; Sigma-Aldrich; Merck KGaA) overnight at 4°C, followed by incubation with corresponding horseradish peroxidase-conjugated secondary antibodies (1:2,000; cat. nos. SC-2357 and SC-516102; Santa Cruz Biotechnology, Inc.) for 1 h at room temperature. Proteins were visualized using enhanced chemiluminescence (Thermo Fisher Scientific, Inc.). Western blot images were analyzed using Bio-Rad ChemiDoc (Bio-Rad Laboratories, Inc.).
Targeted next-generation sequencing assay
To analyze the mutational status of tissues and organoids, they were harvested using cell recovery solution. DNA extraction and library construction were performed by Macrogen with Axen Cancer Panel 2 (Macrogen) comprising 170 cancer-related genes. The libraries were paired-end sequenced (2×150 bp) on a NextSeq 500 system (Illumina, Inc.) with high-throughput sequencing using synthesis technology to a depth coverage of ~2,000×.
Statistical analysis
The data obtained from a minimum of three independent experiments are expressed as the mean ± standard deviation. Unpaired two-tailed Student's t-tests were applied to determine significant differences between two groups. One-way ANOVA followed by Tukey's test was performed to compare the means between groups when performing multiple comparisons. P<0.05 was considered to indicate a statistically significant difference. Statistical analysis was performed using GraphPad Prism 7 (GraphPad Software, Inc.). Analysis of the mutation-annotated files was conducted using the maftools R package V 3.5.2, which included the generation of figure oncoplots (36).
Results
Butyrate exhibits anticancer effects against CRC-PDO and shows high specificity
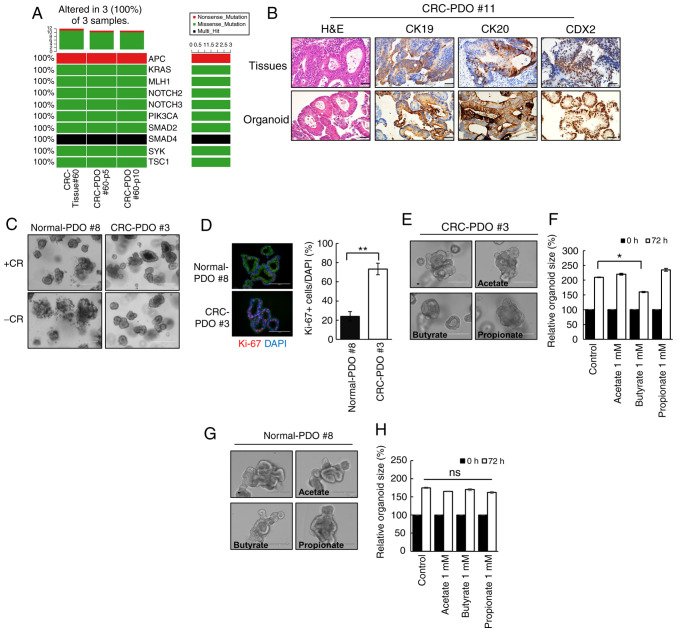
Immunocytochemistry confirmed that the established CRC-PDOs were positive for all markers of intestinal epithelium cell lineages (Muc2+, ChgA+, VL1+, Ki-67+; Fig. S1). In addition, the CRC-PDOs had the same genetic landscape as their matching tumor tissues, as demonstrated in the oncoplot analysis (Fig. 1A). Immunohistochemistry analysis revealed that CK19, CK20 and CDX2 were expressed in CRC-PDO #11 original tissues and organoids (Fig. 1B). Meanwhile other tissues and organoids were weakly positive for CK19, whereas strong expression of CK20 and CDX2 was detected in CRC-PDO #3 (Fig. S2). These features reflect those present in the patient tissue samples. The tumor organoids were selectively expanded by excluding Wnt-related factors from culture medium (Fig. 1C), and immunocytochemical analysis revealed that CRC-PDOs contained more proliferating cells compared with normal-PDOs (P<0.001; Fig. 1D), confirming that the organoid cultures reflect the characteristics of the primary tissue. Of the three SCFAs screened, only butyrate significantly decreased the organoid size (P=0.038), while propionate and acetate had no effects (Figs. 1E and F, and S3A and B). These observations were supported by the MTT assay results (Fig. S3C). By contrast, butyrate did not have any adverse effects on normal-PDOs (Fig. 1G and H). These findings suggest that butyrate may exhibit selective antitumor activity on CRC-PDO.
Figure 1.
Butyrate exhibits selective anticancer effects on CRC-PDOs. (A) Oncoplot of somatic cancer driver alterations in CRC-tissue #60 and CRC-PDO tissue #60 belonging to passage 5 and 10, including most of the known major cancer driver genes. (B) Immunohistochemical profile of FFPE sections of organoids and corresponding tissues for CK19, CK20 and CDX2 along with corresponding H&E staining. Magnification, ×40. Scale bar, 50 µm. (C) Images of normal-PDO #8 and CRC-PDO #3 morphology with CHIR99021 and R-spondin (CR+) or without (CR−). Scale bar, 400 µm. (D) Fluorescence microscopy images of normal-PDO #8 and CRC-PDO #3. Blue, DAPI; red, Ki-67; green, E-cadherin. Scale bar, 100 µm. (E) Morphology of CRC-PDO #3 treated with SCFA after 3 days. Scale bar, 200 µm. (F) Relative CRC-PDO #3 size 3 days after treatment with SCFA (n=3). Data are presented as mean ± SEM. *P<0.05, **P<0.01. (G) Morphology of normal-PDO #8 treated with SCFA after 3 days. Scale bar, 200 µm. (H) Relative normal-PDO #8 size 3 days after treatment with SCFA (n=3). Data are presented as mean ± standard error of the mean. *P<0.05, **P<0.01. ns, not significant; H&E, hematoxylin and eosin; SCFA, short-chain fatty acids; CRC, colorectal cancer; PDO, patient-derived organoid; CK, cytokeratin.
Butyrate enhances radiosensitivity in CRC-PDO
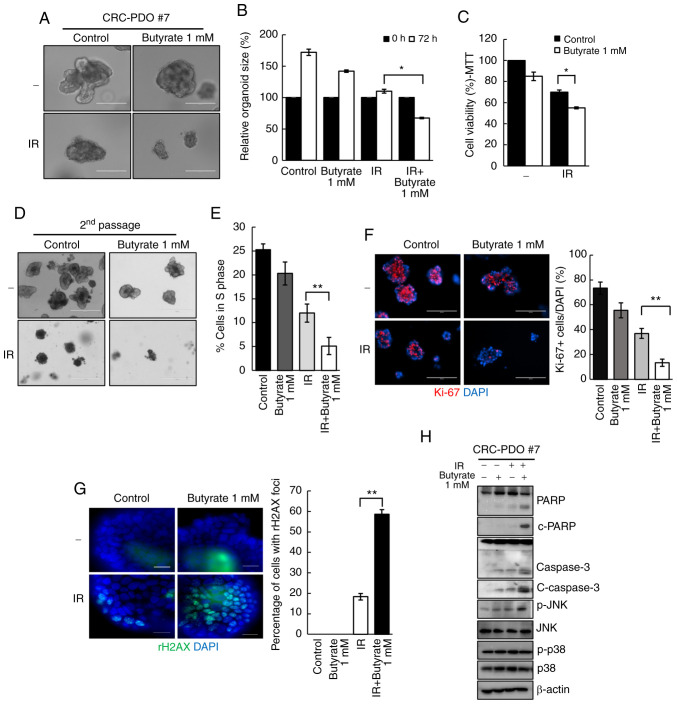
Organoids co-treated with butyrate and IR demonstrated a significant decrease in organoid size from 1.1-fold to 0.6-fold as compared to those treated only with IR (P=0.04; Fig. 2A and B). These observations were supported by the MTT assay results, wherein a significant reduction in organoid viability following butyrate and IR combination treatment was observed (Fig. 2C). As cancer consists of both tumorigenic and non-tumorigenic cancer cells, to further evaluate the antitumor effect of butyrate, the number of tumor organoids in the second passage were counted after PDO splitting. A total of 3 days after splitting, the number of formed tumor organoids significantly decreased after butyrate and IR treatment compared with that in the IR-only treatment group (Fig. 2D). These results suggested that butyrate enhances the effect of radiation on CRC organoids. To further elucidate if the loss of cell viability after butyrate and IR exposure was accompanied by changes in cell cycle arrest, the degree of EdU incorporation was measured by flow cytometry and it was identified that 12% of the cells were in the S phase after IR treatment, while only 5% were in the S phase after combination treatment (P=0.003; Fig. 2E).
Figure 2.
Butyrate enhances radiosensitivity in CRC-PDOs. (A) Morphology and (B) organoid size (n=3) of CRC-PDO #7 irradiated thrice with 5 Gy with or without butyrate. Scale bar, 200 µm. (C) MTT cell viability assay of organoids described in A (n=3). (D) Image of organoids after the second passage. Scale bar, 400 µm. (E) Percentage of CRC-PDO in S phase (n=3). (F) Left: Fluorescence microscopy images of organoids irradiated thrice with 5 Gy with or without butyrate. Blue, DAPI; red, Ki-67. Scale bar, 200 µm. Right: Statistical analysis representing Ki-67-positive cells per DAPI staining cells (n=3). (G) Fluorescence microscopy images showing γ-H2AX foci. Blue, DAPI; green, γ-H2AX. Right: Statistical analysis representing γ-H2AX foci (n=3). (H) Expression levels of PARP, c-PARP, caspase 3, c-caspase 3, p-JNK, JNK, p-p38 and p38 in the CRC-PDO. β-actin served as a loading control. Data are presented as mean ± standard error of the mean. *P<0.05, **P<0.01. CRC, colorectal cancer; PDO, patient-derived organoid; PARP, poly-ADP-ribose polymerase; c-, cleaved; p-, phosphorylated; IR, irradiation.
The effect of butyrate on the growth of CRC-PDOs was then evaluated. Organoids after treatment with butyrate and IR had a significantly lower number of Ki-67+ cells than those treated with IR alone (P=0.005; Figs. 2F and S4A). To investigate the effects of butyrate on radiation-induced DNA damage, γ-H2AX was analyzed by western blotting and it was identified that combination treatment with butyrate and radiation resulted in higher levels of γ-H2AX compared with radiation alone at 6 and 24 h post-RT (Fig. S4B). After irradiation, 18.3% of the cancer organoids were positive for γ-H2AX foci, whereas the combination treatment induced 58.6% of organoids to form γ-H2AX foci, indicating a significantly greater number of DSB (P<0.001; Fig. 2G).
Next, to evaluate the cellular response to radiation, the levels of apoptosis-related proteins were analyzed. The levels of c-PARP and c-caspase 3, which are considered hallmarks of apoptosis, were increased after IR and butyrate treatment relative to the respective levels after IR treatment alone. Moreover, p-JNK levels were increased after combination treatments. There were no changes in the expression levels of p-p38 (Fig. 2H). These findings suggest that butyrate acts as a radiosensitizer in CRC-PDO.
Butyrate does not act as a radiosensitizer in normal-PDO
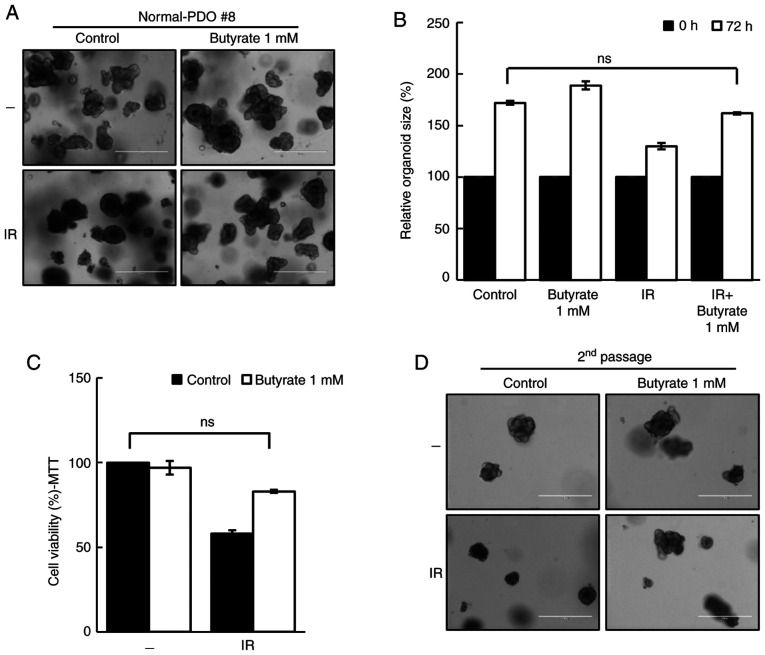
To evaluate the safety of butyrate, the response of butyrate in normal-PDOs was further investigated. Although the organoid size was reduced after irradiation, butyrate protected normal-PDOs from the damaging effects of IR (Fig. 3A and B). The MTT assay and second passage supported these results (Fig. 3C and D), thus suggesting that butyrate does not affect the radiosensitivity in normal-PDO.
Figure 3.
Butyrate does not affect sensitivity of normal-PDOs. (A) Morphology and (B) organoid size (n=3) of normal-PDO #8 irradiated thrice with 5 Gy with or without butyrate. Scale bar, 400 µm. (C) MTT cell viability assay of organoids described in A (n=3). (D) Image of organoids after the second passage. Scale bar, 400 µm. Data are presented as mean ± standard error of the mean. ns, not significant; PDO, patient-derived organoid; IR, irradiation.
Butyrate enhances radiosensitivity via the Warburg effect
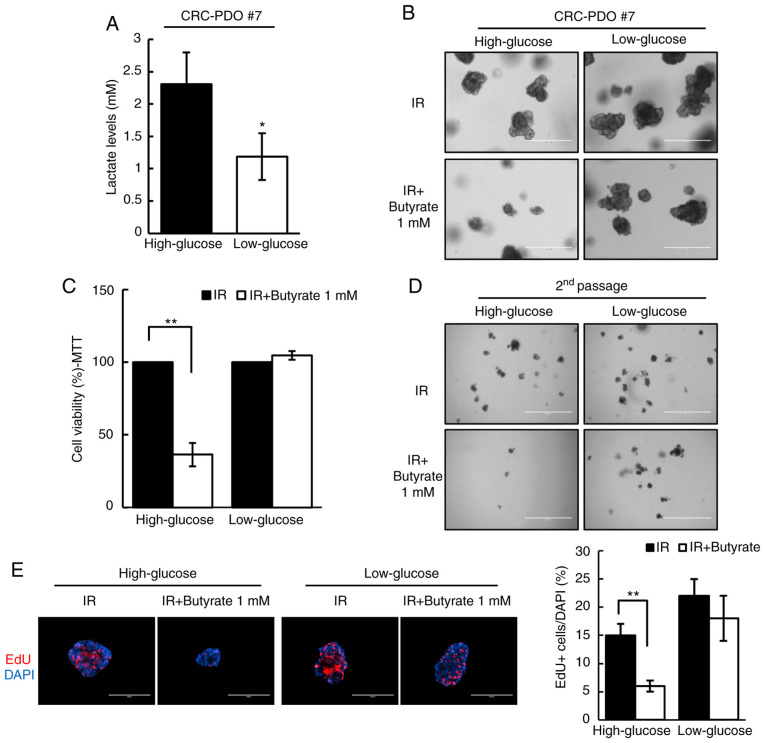
Butyrate has previously been shown to selectively suppress proliferation of cancer cell lines due to the difference in the Warburg effect between normal colon cells and colon cancer cells (37). Thus, to test whether the Warburg effect may account for the differential radiosensitizing effects of butyrate, the Warburg effect in CRC-PDOs was inhibited by changing the culture media from high-glucose (3.151 g/l) to low-glucose (1 g/l). Under these conditions, glucose levels were too low to support aerobic glycolysis, as evidenced by the detection of negligible levels of lactate, i.e., the end-product of glycolysis (P=0.01; Fig. 4A). Without the Warburg effect in the low-glucose condition, butyrate no longer affected the organoid size and viability after irradiation (Fig. 4B and C). In addition, the number of second passage organoids formed did not change after IR and butyrate treatment under the low-glucose condition (Fig. 4D). As shown by EdU staining, the proportion of S phase cells was reduced after IR and butyrate treatment in high-glucose medium (P<0.001), but not in low-glucose medium (Fig. 4E). These results demonstrate that butyrate increases radiosensitivity in CRC-PDOs in a Warburg effect-dependent manner.
Figure 4.
Radiosensitizing effects of butyrate are dependent on glucose concentration. (A) Lactate levels in CRC-PDO #7 grown in low- or high-glucose conditions (n=3). (B) CRC-PDO #7 grown in low- or high-glucose conditions irradiated thrice with 5 Gy with or without butyrate. Scale bar, 400 µm. (C) MTT cell viability assay of organoids described in (B) (n=3). (D) Image of organoids after the second passage. Scale bar, 1,000 µm. (E) Fluorescence microscopy images of EdU incorporation in CRC-PDO #7 cultured under low- or high-glucose conditions *P<0.05, **P<0.01. Scale bar, 200 µm. Blue, DAPI; red, EdU. CRC, colorectal cancer; PDO, patient-derived organoid; IR, irradiation.
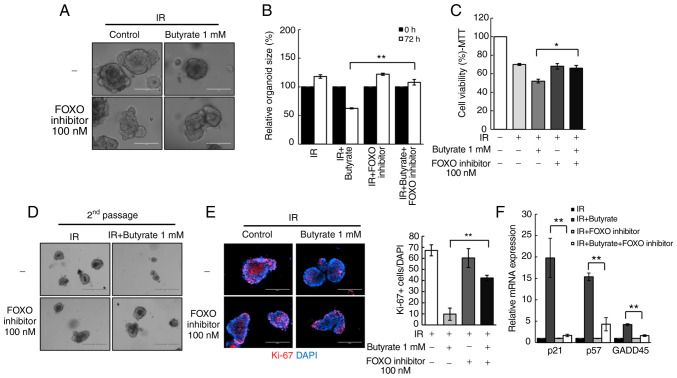
Butyrate regulates radio responses via FOXO3A in CRC-PDOs
To investigate the mechanism of the potential effects elicited by butyrate on radiosensitivity, the present study focused on transcription factors that regulate cell cycle genes. In particular, the influence of the transcription factor FOXO3A was examined, as a previous study suggested that butyrate regulates its transcriptional activity through HDAC inhibition (29). Treatment of CRC-PDOs with a FOXO inhibitor rescued the radiosensitizing effect of butyrate in CRC-PDOs (P<0.001; Fig. 5A and B). In addition, the MTT assay showed that the significant reduction in organoid viability following combination treatment, was also rescued by the FOXO inhibitor (P=0.03; Fig. 5C) along with the organoid-regenerating capacity analyzed by passaging (Fig. 5D). The FOXO inhibitor also reversed the proliferation inhibitory effects of butyrate after irradiation (P<0.001; Fig. 5E). RT-qPCR was conducted to examine the expression levels of cell cycle-related genes regulated by FOXO3A, namely GADD45, p21, and p57 (38,39). Results show that butyrate and IR increased the mRNA expression levels of GADD45 (P<0.001), p21 (P<0.001) and p57 (P<0.001) in CRC-PDOs, which were all decreased after treatment with the FOXO inhibitor (Fig. 5F). These findings suggest that butyrate increases radiosensitivity through induction of p21, p57, and GADD45 regulated by FOXO3A.
Figure 5.
Butyrate sensitizes CRC-PDOs through FOXO3A. (A) Morphology and (B) organoid size (n=3) of CRC-PDO #7 treated with IR and butyrate, with or without a FOXO inhibitor. Scale bar, 200 µm. (C) MTT cell viability assay of organoids described in (A) (n=3). (D) Image of organoids after the second passage. Scale bar, 400 µm. (E) Left: Fluorescence microscopy images of organoids treated with IR and butyrate with or without a FOXO inhibitor. Blue, DAPI; red, Ki-67. Right: Statistical analysis representing Ki-67-positive organoids (n=3). (F) Cell cycle-related gene expression levels in CRC-PDO #7 treated with IR and butyrate with or without a FOXO inhibitor analyzed by reverse transcription-quantitative PCR. The level of the target mRNA was normalized to that of GAPDH mRNA (n=3). Data are presented as mean ± standard error of the mean. *P<0.05, **P<0.01. CRC, colorectal cancer; PDO, patient-derived organoid; IR, irradiation.
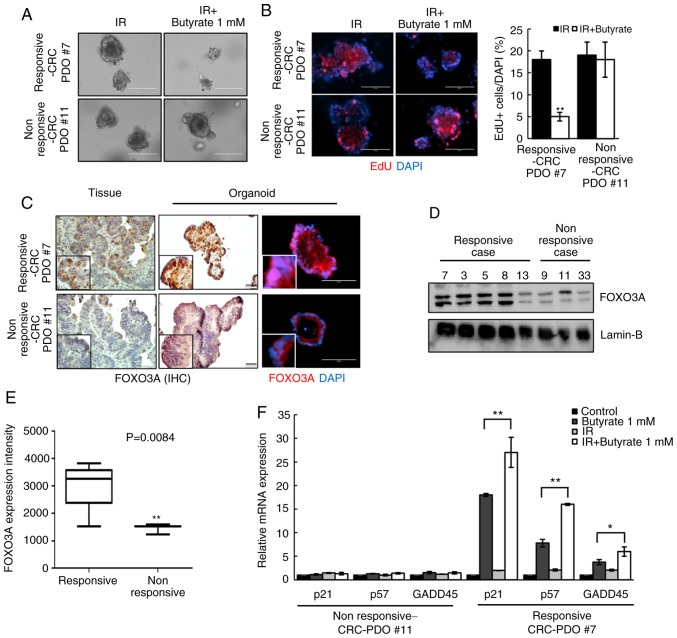
Radiosensitizing effect of butyrate is dependent on FOXO3A expression in CRC-PDOs
Although there was an overall reduction in organoid size following combined butyrate and IR treatment, no difference was observed in some cases (here-after referred to as non-responsive CRC-PDOs; Fig. 6A). EdU staining was, therefore, performed to compare the proliferative capacity of responsive and non-responsive CRC-PDOs. After treatment with butyrate and IR, the non-responsive CRC-PDO group had more EdU-incorporated cells than the responsive CRC-PDO group (P<0.001; Fig. 6B). Moreover, responsive CRC-PDOs, and their original tissues, showed stronger nuclear staining for FOXO3A than non-responsive-CRC-PDOs (Fig. 6C). Western blot analysis further showed that all five responsive CRC-PDO cases exhibited increased expression of FOXO3A compared with the non-responsive CRC-PDOs (Fig. 6D), and the intensity of FOXO3A expression was significantly increased compared with that in the three non-responsive CRC-PDO (P=0.0084; Fig. 6E). RT-qPCR analysis further showed that butyrate and IR combination treatment did not affect the expression levels of p21 (P<0.001), p57 (P<0.001) or GADD45 (P=0.048) in non-responsive CRC-PDOs in contrast to the effects observed in responsive CRC-PDOs (Fig. 6F). These results suggest that the radiosensitizing effect of butyrate may be dependent on the level of FOXO3A expression in CRC-PDOs.
Figure 6.
Radiosensitizing effects of butyrate are dependent on FOXO3A expression level in CRC-PDOs. (A) Images of CRC-PDO #7 irradiated thrice with 5 Gy with or without butyrate. Scale bar, 200 µm. (B) Fluorescence microscopy images of EdU incorporation into responsive-CRC-PDO #7 and non-responsive-CRC-PDO #11 after irradiation with butyrate. Scale bar, 200 µm. Blue, DAPI; red, EdU. (C) IHC and fluorescence microscopy images of FOXO3a in tissues, responsive-CRC-PDO #7 and non-responsive-CRC-PDO #11. Immunohistochemistry scale bar, 50 µm; fluorescence microscopy scale bar, 100 µm. Blue, DAPI; red, FOXO3A. (D) Protein expression of FOXO3A in the nuclear fraction of CRC-PDO. Lamin-B served as a marker for the nuclear fraction. (E) Box-and-whisker plots of FOXO3A expression in five cases of responsive-CRC-PDO and three cases of non-responsive-CRC-PDO. (F) CRC-PDO #7 was harvested at 24 h and analyzed by reverse transcription-quantitative PCR for cell cycle-related genes. The expression level of the target mRNA was normalized to that of GAPDH mRNA (n=3). Data are presented as mean ± standard error of the mean. *P<0.05, **P<0.01. CRC, colorectal cancer; PDO, patient-derived organoid; IR, irradiation; IHC, immunohistochemistry.
Discussion
The present study demonstrated that butyrate has a potent radiosensitizing effect on CRC-PDOs by increasing FOXO3A transcriptional activity and inducing cell cycle arrest regulated by p21, p57 and GADD45. However, butyrate protects against IR-induced damage in normal colon PDOs. To the best of our knowledge, this is the first study to suggest a role for butyrate in regulating the radio response and cellular function in CRC-PDOs. Previously, the underlying mechanism responsible for the anticancer function of butyrate was reportedly associated with mediating HDAC-dependent transcription activation (40,41). In the normal colon, butyrate is used as an energy source. However, since cancer cells rely on glucose as an energy source, butyrate accumulates in the nucleus to inhibit proliferation, thus inducing apoptosis only in cancer cells (13,37,42,43). Hence, targeting tumor metabolism has been described as a potential strategy for clinical applications (43,44). This specificity makes butyrate particularly attractive as a novel candidate radiosensitizer, as it would have minimal adverse effects on normal cells.
The present study further demonstrated that certain CRC-PDOs found to be non-responsive to butyrate and IR combination treatment had relatively lower expression levels of FOXO3A compared with responsive CRC-PDOs. Although responsive CRC-PDOs exhibited higher expression of FOXO3A compared with those non-responsive, variable expression was observed in both groups, highlighting the heterogeneity of patient samples that causes natural variation attributable to unique patient characteristics. Nevertheless, since butyrate appears to enhance radiosensitivity through FOXO3A, its expression may serve as a potential biomarker to predict the effectiveness of butyrate on radiotherapy.
FOXO transcription factors are emerging as critical transcriptional integrators among pathways regulating differentiation, proliferation, survival and the cell cycle (45-47). A previous study suggested that AZD6244 enhances the expression of FOXO3A, and suppresses colon cancer cell proliferation (48), while others reported that FOXO3A enhances the radiosensitivity of cancer cells through regulation of apoptosis and AMP-activated kinase (AMPK) signaling (49,50). Thus, FOXO3A is a vital regulator enhancing radiosensitivity. The present study further found that the cell cycle-related genes regulated by FOXO3A (p21, p57 and GADD45) were involved in the mechanism through which butyrate inhibits cell growth, and cell cycling in CRC-PDOs. These genes have been shown to negatively regulate the cancer cell cycle (41-43). Moreover, GADD45α inhibits the nitric oxide-regulated cytoplasmic localization of APE1, thereby enhancing the radiosensitivity of cervical cancer cells (51); while the negative regulation of p57 by miRNA221/222 may also contribute to radioresistance (52). Therefore, butyrate enhanced radiosensitivity by inhibiting HDAC and the FOXO3A/p21, p57 and GADD45 axis.
It was identified that butyrate enhanced the radiosensitivity via the Warburg effect. However, only the lactate levels in CRC-PDO were examined. AMPK negatively regulates the Warburg effect in cancer cells and suppresses the growth of tumors in vivo (53). Hence, it is necessary to evaluate other signaling pathways, such as AMP-activated kinase activation. Additionally, FOXO3A was recently shown to be negatively correlated with the expression of a number of glycolysis-associated genes and to inhibit glucose metabolism and tumor cell proliferation in melanoma (54). Furthermore, transcriptional activation of FOXO3A inhibits the Warburg effect in glioblastoma cells (55). Together these studies suggest that FOXO3A is associated with the Warburg effect, which warrants further investigation.
Cancer cell lines have long served as the primary work-horse model in radiobiology research. Indeed, some of the most well-known radiosensitizers such as 5-FU, curcumin and docetaxel (56-58) were discovered in cancer cell lines; however, these effects were not reproduced in patients. These inconsistent results are due to the highly heterogenous nature of tumors, which are composed of a mixture of sub-clones and various cell types, a characteristic that is not accurately reflected in cancer cell lines. Thus, our PDO culture system can be exploited for functional studies on radio responses within individual patients that cannot be achieved by cell lines or animal studies. Hence, we propose that PDOs will be a valuable tool for directly testing the radiosensitivity of a tumor in a personalized treatment approach.
Herein, it was demonstrated that 1 mM butyrate sensitizes CRC organoids to the cytotoxic effects of radiotherapy, while eliciting normal cell-protective effects. However, higher concentrations of butyrate have limited applications due to toxicity on normal intestinal cells (37). Hence, it is necessary to evaluate the optimal dose before butyrate is used in clinical settings as a radiosensitizer.
In conclusion, the results of the present study suggest that butyrate suppresses proliferation of three-dimensional CRC organoids and enhances radiation-induced cell death in CRC organoids through FOXO3A. However, butyrate does not increase radiation-induced cell death after irradiation in normal organoids. Thus, it may enhance the efficacy of radiotherapy while protecting the normal mucosa.
Supplementary Data
Acknowledgments
The biospecimens and data used in this study were provided by the Radiation Tissue Resources Bank of Korea Cancer Center Hospital (TB-2016-05).
Abbreviations
- CRC
colorectal cancer
- HDAC
histone deacetylase
- PDO
patient-derived organoid
- SAHA
suberoylanilide hydroxamic acid
- SCFAs
short-chain fatty acids
Funding
This study was supported by a grant of the Korea Institute of Radiological and Medical Sciences, funded by the Ministry of Science and ICT, Republic of Korea (grant no. 50542-2019).
Availability of data and materials
The datasets used or analyzed during this study are available from the corresponding author upon reasonable request.
Authors' contributions
MP, YH, and YK were involved in the conception and design of the study. MP, JK, and HJS performed the experiments. SMM, SBK and USS provided resources and analyzed the data. MP, YHH, and YK wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by Ethics Committee of Korea Cancer Center Hospital (approval no. KIRAMS-2017-07-001). All research was performed in accordance with the approved guidelines and regulations of the institution. All samples were obtained from patients who had provided written informed consent for the use of their tissues for the purposes of research after the operation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70:145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Guren MG, Dueland S, Skovlund E, Fossa SD, Poulsen JP, Tveit KM. Quality of life during radiotherapy for rectal cancer. Eur J Cancer. 2003;39:587–594. doi: 10.1016/S0959-8049(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Mu X, He H, Zhang XD. Cancer radiosensitizers. Trends Pharmacol Sci. 2018;39:24–48. doi: 10.1016/j.tips.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 4.Wolpin BM, Meyerhardt JA, Mamon HJ, Mayer RJ. Adjuvant treatment of colorectal cancer. CA Cancer J Clin. 2007;57:168–185. doi: 10.3322/canjclin.57.3.168. [DOI] [PubMed] [Google Scholar]
- 5.Willett CG, Duda DG, di Tomaso E, Boucher Y, Ancukiewicz M, Sahani DV, Lahdenranta J, Chung DC, Fischman AJ, Lauwers GY, et al. Efficacy, safety, and biomarkers of neoadjuvant bevacizumab, radiation therapy, and fluorouracil in rectal cancer: A multidisciplinary phase II study. J Clin Oncol. 2009;27:3020–3026. doi: 10.1200/JCO.2008.21.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crane CH, Eng C, Feig BW, Das P, Skibber JM, Chang GJ, Wolff RA, Krishnan S, Hamilton S, Janjan NA, et al. Phase II trial of neoadjuvant bevacizumab, capecitabine, and radiotherapy for locally advanced rectal cancer. Int J Radiat Oncol Biol Phys. 2010;76:824–830. doi: 10.1016/j.ijrobp.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 7.Tomiak A, Vincent M, Kocha W, Taylor M, Winquist E, Keith B, Sawyer M, Griffeth S, Whiston F, Stitt L. Standard dose (Mayo regimen) 5-fluorouracil and low dose folinic acid: Prohibitive toxicity? Am J Clin Oncol. 2000;23:94–98. doi: 10.1097/00000421-200002000-00025. [DOI] [PubMed] [Google Scholar]
- 8.Meta-Analysis Group In Cancer. Levy E, Piedbois P, Buyse M, Pignon JP, Rougier P, Ryan L, Hansen R, Zee B, Weinerman B, et al. Toxicity of fluorouracil in patients with advanced colorectal cancer: Effect of administration schedule and prognostic factors. J Clin Oncol. 1998;16:3537–3541. doi: 10.1200/JCO.1998.16.11.3537. [DOI] [PubMed] [Google Scholar]
- 9.Cho M, Carter J, Harari S, Pei Z. The interrelationships of the gut microbiome and inflammation in colorectal carcinogenesis. Clin Lab Med. 2014;34:699–710. doi: 10.1016/j.cll.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arthur JC, Jobin C. The struggle within: Microbial influences on colorectal cancer. Inflamm Bowel Dis. 2011;17:396–409. doi: 10.1002/ibd.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuwahara A. Contributions of colonic short-chain fatty acid receptors in energy homeostasis. Front Endocrinol (Lausanne) 2014;5:144. doi: 10.3389/fendo.2014.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bultman SJ. Molecular pathways: Gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin Cancer Res. 2014;20:799–803. doi: 10.1158/1078-0432.CCR-13-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Cao L, Tian Y, Zhang P, Ding C, Lu W, Jia C, Shao C, Liu W, Wang D, et al. Butyrate suppresses the proliferation of colorectal cancer cells via targeting pyruvate kinase M2 and metabolic reprogramming. Mol Cell Proteomics. 2018;17:1531–1545. doi: 10.1074/mcp.RA118.000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Folkvord S, Ree AH, Furre T, Halvorsen T, Flatmark K. Radiosensitization by SAHA in experimental colorectal carcinoma models-in vivo effects and relevance of histone acetylation status. Int J Radiat Oncol Biol Phys. 2009;74:546–552. doi: 10.1016/j.ijrobp.2009.01.068. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Wong P, Radany E, Wong JY. HDAC inhibitor, valproic acid, induces p53-dependent radiosensitization of colon cancer cells. Cancer Biother Radiopharm. 2009;24:689–699. doi: 10.1089/cbr.2009.0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flatmark K, Nome RV, Folkvord S, Bratland A, Rasmussen H, Ellefsen MS, Fodstad Ø, Ree AH. Radiosensitization of colorectal carcinoma cell lines by histone deacetylase inhibition. Radiat Oncol. 2006;1:25. doi: 10.1186/1748-717X-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Failli A, Consolini R, Legitimo A, Spisni R, Castagna M, Romanini A, Crimaldi G, Miccoli P. The challenge of culturing human colorectal tumor cells: Establishment of a cell culture model by the comparison of different methodological approaches. Tumori. 2009;95:343–347. doi: 10.1177/030089160909500312. [DOI] [PubMed] [Google Scholar]
- 19.Dangles-Marie V, Pocard M, Richon S, Weiswald LB, Assayag F, Saulnier P, Judde JG, Janneau JL, Auger N, Validire P, et al. Establishment of human colon cancer cell lines from fresh tumors versus xenografts: Comparison of success rate and cell line features. Cancer Res. 2007;67:398–407. doi: 10.1158/0008-5472.CAN-06-0594. [DOI] [PubMed] [Google Scholar]
- 20.van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell. 2015;161:933–945. doi: 10.1016/j.cell.2015.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh AJ, Cook RS, Sanders ME, Aurisicchio L, Ciliberto G, Arteaga CL, Skala MC. Quantitative optical imaging of primary tumor organoid metabolism predicts drug response in breast cancer. Cancer Res. 2014;74:5184–5194. doi: 10.1158/0008-5472.CAN-14-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clevers H. Modeling development and disease with organoids. Cell. 2016;165:1586–1597. doi: 10.1016/j.cell.2016.05.082. [DOI] [PubMed] [Google Scholar]
- 23.Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR, Sullivan DC, et al., editors. AJCC Cancer Staging Manual. Springer International Publishing; 2017. [DOI] [Google Scholar]
- 24.Xie BY, Wu AW. Organoid culture of isolated cells from patient-derived tissues with colorectal cancer. Chin Med J (Engl) 2016;129:2469–2475. doi: 10.4103/0366-6999.191782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett's epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 26.Fujii M, Shimokawa M, Date S, Takano A, Matano M, Nanki K, Ohta Y, Toshimitsu K, Nakazato Y, Kawasaki K, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grabinger T, Luks L, Kostadinova F, Zimberlin C, Medema JP, Leist M, Brunner T. Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy. Cell Death Dis. 2014;5:e1228. doi: 10.1038/cddis.2014.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, Stappenbeck TS. The colonic crypt protects stem cells from microbiota-derived metabolites. Cell. 2016;167:1708–1720. doi: 10.1016/j.cell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36:5678–5694. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bozzi F, Mogavero A, Varinelli L, Belfiore A, Manenti G, Caccia C, Volpi CC, Beznoussenko GV, Milione M, Leoni V, et al. MIF/CD74 axis is a target for novel therapies in colon carcinomatosis. J Exp Clin Cancer Res. 2017;36:16. doi: 10.1186/s13046-016-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi H, Stappenbeck TS. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat Protoc. 2013;8:2471–2482. doi: 10.1038/nprot.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park M, Yoon HJ, Kang MC, Kwon J, Lee HW. MiR-338-5p enhances the radiosensitivity of esophageal squamous cell carcinoma by inducing apoptosis through targeting survivin. Sci Rep. 2017;7:10932. doi: 10.1038/s41598-017-10977-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28:1747–1756. doi: 10.1101/gr.239244.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang H, Tindall DJ. Dynamic FoxO transcription factors. J Cell Sci. 2007;120:2479–2487. doi: 10.1242/jcs.001222. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Tang N, Hadden TJ, Rishi AK. Akt, FoxO and regulation of apoptosis. Biochim Biophys Acta. 2011;1813:1978–1986. doi: 10.1016/j.bbamcr.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 40.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan HT, Tan S, Lin Q, Lim TK, Hew CL, Chung MC. Quantitative and temporal proteome analysis of butyrate-treated colorectal cancer cells. Mol Cell Proteomics. 2008;7:1174–1185. doi: 10.1074/mcp.M700483-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Pouillart PR. Role of butyric acid and its derivatives in the treatment of colorectal cancer and hemoglobinopathies. Life Sci. 1998;63:1739–1760. doi: 10.1016/S0024-3205(98)00279-3. [DOI] [PubMed] [Google Scholar]
- 43.Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nat Cell Biol. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vernieri C, Casola S, Foiani M, Pietrantonio F, de Braud F, Longo V. Targeting cancer metabolism: Dietary and pharmacologic interventions. Cancer Discov. 2016;6:1315–1333. doi: 10.1158/2159-8290.CD-16-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkenkamp KU, Coffer PJ. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem Soc Trans. 2003;31:292–297. doi: 10.1042/bst0310292. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt M, Fernandez de Mattos S, van der Horst A, Klompmaker R, Kops GJ, Lam EW, Burgering BM, Medema RH. Cell cycle inhibition by FoxO forkhead transcription factors involves down-regulation of cyclin D. Mol Cell Biol. 2002;22:7842–7852. doi: 10.1128/MCB.22.22.7842-7852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y, Zhou Y, Graves DT. FOXO transcription factors: Their clinical significance and regulation. Biomed Res Int. 2014;2014:925350. doi: 10.1155/2014/925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang JY, Chang CJ, Xia W, Wang Y, Wong KK, Engelman JA, Du Y, Andreeff M, Hortobagyi GN, Hung MC. Activation of FOXO3a is sufficient to reverse mitogen-activated protein/extra-cellular signal-regulated kinase kinase inhibitor chemoresistance in human cancer. Cancer Res. 2010;70:4709–4718. doi: 10.1158/0008-5472.CAN-09-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang JY, Xia W, Hu MC. Ionizing radiation activates expression of FOXO3a, Fas ligand, and Bim, and induces cell apoptosis. Int J Oncol. 2006;29:643–648. [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y, Pan W, Zhang S, Cao Y, Cheng H, Chen J, Sun X. Metformin can enhance the radiosensitivity of cholangiocarcinoma through AMPK-FOXO3a axis. Int J Clin Exp Med. 2016;9:13539–13550. [Google Scholar]
- 51.Li Q, Wei X, Zhou ZW, Wang SN, Jin H, Chen KJ, Luo J, Westover KD, Wang JM, Wang D, et al. GADD45α sensitizes cervical cancer cells to radiotherapy via increasing cytoplasmic APE1 level. Cell Death Dis. 2018;9:524. doi: 10.1038/s41419-018-0452-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milas L, Akimoto T, Hunter NR, Mason KA, Buchmiller L, Yamakawa M, Muramatsu H, Ang KK. Relationship between cyclin D1 expression and poor radioresponse of murine carcinomas. Int J Radiat Oncol Biol Phys. 2002;52:514–521. doi: 10.1016/S0360-3016(01)02693-1. [DOI] [PubMed] [Google Scholar]
- 53.Faubert B, Boily G, Izreig S, Griss T, Samborska B, Dong Z, Dupuy F, Chambers C, Fuerth BJ, Viollet B, et al. AMPK is a negative regulator of the warburg effect and suppresses tumor growth in vivo. Cell Metab. 2013;17:113–124. doi: 10.1016/j.cmet.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong Z, Yang J, Li L, Tan L, Shi P, Zhang J, Zhong X, Ge L, Wu Z, Cui H. FOXO3aSIRT6 axis suppresses aerobic glycolysis in melanoma. Int J Oncol. 2020;56:728–742. doi: 10.3892/ijo.2020.4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong Z, Zhong X, Lei Q, Chen F, Cui H. Transcriptional activation of SIRT6 via FKHRL1/FOXO3a inhibits the warburg effect in glioblastoma cells. Cell Signal. 2019;60:100–113. doi: 10.1016/j.cellsig.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence TS, Blackstock AW, McGinn C. The mechanism of action of radiosensitization of conventional chemotherapeutic agents. Semin Radiat Oncol. 2003;13:13–21. doi: 10.1053/srao.2003.50002. [DOI] [PubMed] [Google Scholar]
- 57.Chendil D, Ranga RS, Meigooni D, Sathishkumar S, Ahmed MM. Curcumin confers radiosensitizing effect in prostate cancer cell line PC-3. Oncogene. 2004;23:1599–1607. doi: 10.1038/sj.onc.1207284. [DOI] [PubMed] [Google Scholar]
- 58.Dunne AL, Mothersill C, Robson T, Wilson GD, Hirst DG. Radiosensitization of colon cancer cell lines by docetaxel: Mechanisms of action. Oncol Res. 2004;14:447–454. doi: 10.3727/0965040041791455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used or analyzed during this study are available from the corresponding author upon reasonable request.