Abstract
The proton pump inhibitor lansoprazole (LPZ) inhibits the growth of several cancer cell lines, including A549 and CAL 27. We previously reported that macrolide antibiotics such as azithromycin (AZM) and clarithromycin (CAM) potently inhibit autophagic flux and that combining AZM or CAM with the epidermal growth factor receptor inhibitors enhanced their antitumor effect against various cancer cells. In the present study, we conducted the combination treatment with LPZ and macrolide antibiotics against A549 and CAL 27 cells and evaluated cytotoxicity and morphological changes using cell proliferation and viability assays, flow cytometric analysis, immunoblotting, and morphological assessment. Combination therapy with LPZ and AZM greatly enhanced LPZ-induced cell death, whereas treatment with AZM alone exhibited negligible cytotoxicity. The observed cytotoxic effect was not mediated through apoptosis or necroptosis. Transmission electron microscopy of A549 cells treated with the LPZ + AZM combination revealed morphological changes associated with necrosis and accumulated autolysosomes with undigested contents. Furthermore, the A549 cell line with ATG5 knockout exhibited complete inhibition of autophagosome formation, which did not affect LPZ + AZM treatment-induced cytotoxicity, thus excluding the involvement of autophagy-dependent cell death in LPZ + AZM treatment-induced cell death. A549 cells treated with LPZ + AZM combination therapy retained the endosomal Alexa-dextran for extended duration as compared to untreated control cells, thus indicating impairment of lysosomal digestion. Notably, lysosomal galectin-3 puncta expression induced due to lysosomal membrane permeabilization was increased in cells treated with LPZ + AZM combination as compared to the treatment by either agent alone. Collectively, the present results revealed AZM-induced autolysosome accumulation, potentiated LPZ-mediated necrosis, and lysosomal membrane permeabilization, thus suggesting the potential clinical application of LPZ + AZM combination therapy for cancer treatment.
Keywords: lansoprazole, azithromycin, proton pump inhibitor, macrolide antibiotics, apoptosis, lysosome, autophagy, cancer
Introduction
Genetic mutations in cancer cells promote preferential clonal proliferation and survival over normal cells. In addition to genetic mutations which alter the inherent characteristics of cancer cells, the physical and biological properties of their microenvironment promote their growth (1,2). The constitutive activation of glycolysis in cancer cells leads to increased lactic acid production and tumor microenvironment acidification by extracellular proton (H+) transport through vacuolar V-ATPases. The functional expression of these (V)-ATPases on the cancer cell plasma membrane facilitates the maintenance of neutral intracellular and acidic extracellular pH (2). One of the key mechanisms for chemoresistance is the acidification of the tumor microenvironment (3,4).
Previous studies have revealed the antitumor effects and drug resistance reversal characteristics of proton pump inhibitors (PPIs) in numerous cancer types (4-10). It has been suggested that agents clinically used in peptic diseases to suppress gastric acid production, such as lansoprazole (LPZ), esomeprazole (EPZ), pantoprazole (PPZ), and omeprazole (OPZ), act by inhibiting H+/K+-ATPases to suppress V-ATPase-mediated H+ transport in tumor cells (2,7). This inhibition reverses extra-cellular acidification and suppresses formation of intracellular acidic vesicles, including lysosomes, to enhance tumor cell sensitivity to anticancer agents (4,5). PPIs have been revealed to induce apoptosis and antimigratory effects (4). PPI treatment synergized with doxorubicin in breast cancer cell lines (5), and PPZ was revealed to induce mitochondrial apoptosis and attenuate the NF-κB signaling pathway in glioma cells (8). In addition, PPIs reversed epithelial ovarian cancer paclitaxel resistance by alkalinizing the acidic tumor microenvironment created by V-ATPase D1 and inhibiting the multidrug resistance Yes-associated protein (9). In another study, the PPZ-mediated increased lysosomal pH caused inhibition of acid phosphatase activity and sensitized the chemo-resistant oral epidermoid carcinoma cells to vincristine (VCR) by reducing VCR lysosomal sequestration (10). These studies suggest combining PPIs with anticancer drugs as a promising approach to further enhance chemotherapy efficacy (3,4).
Autophagy recycles the cellular components and may facilitate cell survival after chemotherapy (11). PPZ has been revealed to inhibit autophagy in a time- and dose-dependent manner and sensitize cancer cells to anticancer drugs (12). PPZ was revealed to inhibit docetaxel-induced autophagy and reverse docetaxel resistance to potentiate its in vitro toxicity. This effect was confirmed in vivo in tumor sections with increased γH2AX foci and cleaved caspase-3 expression and decreased Ki67 expression (13). These results confirmed the involvement of autophagy as the underlying mechanism of docetaxel chemotherapy resistance. In contrast, EPZ has been reported to induce autophagy as a survival response to oxidative stress in human melanoma cells (14). Therefore, the role of PPIs in autophagic flux is still controversial, and their precise underlying molecular mechanisms are yet to be elucidated.
Our group as well as other research groups have reported that macrolide antibiotics such as azithromycin (AZM) and clarithromycin (CAM) potently inhibit autophagic flux as an off-target effect (15-17). Combining AZM or CAM with the epidermal growth factor receptor inhibitors (e.g., gefitinib and erlotinib), which are potent inducers of autophagy, enhanced their antitumor effect against pancreatic and non-small cell lung cancer (NSCLC) cell lines (18,19). In addition, we revealed that concurrent inhibition of the ubiquitin-proteasome and autophagy-lysosome systems by bortezomib (proteasome inhibitor) and macrolides synergistically induced endoplasmic reticulum stress-mediated cytotoxicity in multiple myeloma and breast cancer cell lines (15,20). Since the combination of PPIs and macrolide antibiotics is a well-established clinical therapy for Helicobacter pylori infection in chronic gastritis (21), in the present study, it was investigated whether the LPZ + AZM drug combination could be repurposed for cancer treatment.
Materials and methods
Reagents
LPZ and OPZ were purchased from Wako Pure Chemical Industries and dissolved in dimethyl sulfoxide (DMSO) (Wako Pure Chemical Industries) to prepare 50 mM stock solutions. AZM and CAM were purchased from Tokyo Chemical Industry and dissolved in DMSO to prepare 10 mM stock solutions. Z-VAD-fmk, a pan-caspase inhibitor, was purchased from Peptide Institute, Inc. Necrostatin-1 (NEC-1), a specific inhibitor of receptor-interacting serine/threonine-protein kinase 1 (RIPK1), was purchased from Enzo Life Sciences. Thapsigargin was purchased from Nacalai Tesque, Inc. Staurosporine, TNF-α, and gefitinib were purchased from Wako Pure Chemical Industries. L-Leucyl-L-Leucine methyl ester (hydrochloride) (LLOMe) was purchased from Cayman Chemical Company. Cycloheximide was purchased from Calbiochem; Merck KGaA.
Cell lines and culture conditions
The human cancer cell lines, A549 (NSCLC), CAL 27 (oral squamous cell carcinoma), Detroit 562 (pharyngeal carcinoma), PANC-1 (pancreatic cancer), and HT-29 (colon adenocarcinoma) were obtained from the American Type Culture Collection. The A549 cell line was cultured in Roswell Park Memorial Institute-1640 medium, whereas all other cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM). Both media were supplemented with 10% fetal bovine serum (FBS) (Biosera) and 1% penicillin/streptomycin (Wako Pure Chemical Industries). Cell cultures were maintained at 37°C in a humidified incubator under 5% CO2 and 95% air. All cell line experiments were conducted within 10 passages after thawing. Mycoplasma contamination was tested routinely using the e-Myco™ Mycoplasma PCR Detection kit ver.2.0 (iNtRON Biotechnology, Inc.).
Cell viability and proliferation assays
The number of viable cells was assessed by the CellTiter Blue cell viability assay kit (Promega Corporation) according to the manufacturer's instructions. Briefly, cells were plated in a 96-well flat-bottom culture plate at a density of 3×103 cells/well and cultured for up to 72 h at 37°C in a CO2 incubator in the presence of LPZ or OPZ at various concentrations with/without either AZM or CAM at 50 µM. Fluorescence (560 nm excitation, 590 nm emission) was measured using fluorometer SpectraMax iD3 (Molecular Devises, LLC). For the positive control of RIPK1-dependent cell death, A549 cells were treated with 25 µM gefitinib in amino acid-free DMEM (cat. no. 048-33575; Wako Pure Chemical Industries) supplemented with 10% FBS and 1% penicillin/streptomycin as previously described (22). Cell confluency was used to monitor cell proliferation and was evaluated using the IncuCyte ZOOM 2016B software (Essen BioSciences).
Human ATG5 knockout by CRISPR/Cas9-mediated genome editing
The target sequences for CRISPER interference were as follows: Human ATG5 (exon 3), AAG AGT AAG TTA TTT GAC GT; non-targeting control, GTA GCG AAC GTG TCC GGC GT (23). Two complementary oligonucleotides with BpiI restriction sites for guide RNAs (gRNAs) were synthesized at Fasmac, Inc., and were cloned into the pSpCas9 (BB)-2A-Puro (pX459) V2.0 plasmid vector (gift from Dr Feng Zhang; plasmid cat. no. 48139; Addgene) (24) following an online protocol from the Feng Zhang laboratory (https://www.addgene.org/62988/). A549 cells (1×107 cells) were suspended in 100 µl of Opti-MEM I (cat. no. 31985-070, Thermo Fisher Scientific, Inc.) with 10 µg of pX459-gRNA and electroporated using the Super Electroporator NEPA 21 (NEPA GENE Co. Ltd.) with a 2-mm gap cuvette (cat.no. EC-002; NEPA GENE) under the following conditions: Poring pulse: Voltage, 120 V; pulse interval, 50 ms; pulse width, 10 ms; pulse number, 1 and attenuation rate 10%; transfer pulse: Voltage, 20 V; pulse interval, 50 ms; pulse width, 50 ms; pulse number, 5 and attenuation rate 40%. One day after electroporation, cells were incubated for 2 days with 2 µg/ml of puromycin dihydrochloride (Wako Pure Chemical Industries). The individual puromycin-resistant clones were isolated using a cloning ring. After successfully obtaining clones, A549/ATG5 knock out (KO) and A549/control cells were used for the following experiments.
Morphological assessments
Cells were seeded onto a 60-mm dish at 1×106 cells /dish and treated with various reagents for 48 h. After treatment with LPZ and/or AZM, the adherent cells were harvested by trypsinization. Cell spreads were prepared on glass slides using a Cytospin 4 centrifuge (Thermo Fisher Scientific, Inc.) [1,000 × g, for 5 min, at room temperature (RT)]. May-Grünwald-Giemsa staining was performed with May-Grünwald's stain solution (without dilution; cat. no. 15053; Muto Pure Chemicals) for 3 min at RT followed with Giemsa's stain solution (1 drop/1 ml H2O; cat. no. 15003; Muto Pure Chemicals) for 15 min at RT. Glass slides were examined under a digital light microscope (BZ-8100; Keyence Corporation) (objective magnification, ×100). Representative images were selected.
Transmission electron microscopy (TEM)
Cells were seeded onto a 60-mm dish at 1×106 cells/dish and treated with various reagents for 48 h. Then, the cells were fixed for 1 h at 4°C with 2.5% glutaraldehyde (TAAB Laboratories Equipment, Ltd.) in 0.1 M phosphate buffer (pH 7.3) (prepared with monobasic sodium phosphate and dibasic sodium phosphate; Wako Pure Chemical Industries), and fixed subsequently for 1 h at RT in 1% osmium tetroxide (Nisshin EM Co., Ltd.), dehydrated in graded ethanol (30-100%), and embedded in Quetol 812 epoxy resin (Nisshin EM Co., Ltd.) at 60°C for 2-4 days. ultrathin sections (60 nm) were obtained using an Ultracut J micro-tome (Reichert Jung), and the sections were stained with 4% lead nitrate (RT, 5 min) and saturated uranium acetate (RT, 10 min) and imaged using a transmission electron micro-scope JEM-1200EX II (JEOL, Ltd.) (magnification ranging from ×1,000 to ×10,000). All images were captured on films (Electron-microscopic film FG; Fujifilm).
Flow cytometry
To assess apoptosis, cells were suspended at 1×106 cells/ml in Annexin V binding buffer and stained using an Annexin V-FITC apoptosis detection kit (Nacalai Tesque, Inc.) according to the manufacturer's instructions to detect Annexin V and propidium iodide (PI) staining. Samples were analyzed by flow cytometry using the Attune Acoustic Focusing Cytometer (Life Technologies; Thermo Fisher Scientific, Inc.). Data analysis were performed with Attune Cytometric software v2.1 (Life Technologies; Thermo Fisher Scientific, Inc.).
Immunoblotting
Immunoblotting was performed as previously described (17). Briefly, the cells were seeded onto 60 mm dish at 1×106 cells/dish and treated with various reagents for optimal duration. Thereafter, the cells were lysed with RIPA lysis buffer (Nacalai Tesque, Inc.) supplemented with a protease and phosphatase inhibitor cocktail (Nacalai Tesque, Inc.). Protein concentrations were quantified by the Bradford assay (Thermo Fisher Scientific, Inc.). Proteins (15 µg) were loaded and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (7.5, 10, and 15% gels were used) and transferred to Immobilon-P membranes (EMD Millipore). The membranes were probed at 4°C for overnight with the following primary antibodies (Abs): Anti-caspase-3 Ab (1:1,000; product no. 9662S), anti-poly (ADP-ribose) polymerase (PARP) Ab (1:1,000; cat. no. 9542S), anti-RIPK1 Ab (1:1,000; product no. 4926S), and anti-phospho-RIPK1 (Ser166) mAb (1:1,000; cat. no. 65746S). All the aforementioned antibodies were purchased from Cell Signaling Technology, Inc. Anti-p62/SQSTM1 mAb (1:1,000; cat. no. sc-28359), anti-H+/K+ ATPase-β (C-4) mAb (1:1,000; cat. no. sc-374094), anti-GAPDH mAb (1:1,000; cat. no. sc-32233), and anti-β-actin mAb (1:1,000; cat. no. sc-47778) were obtained from Santa Cruz Biotechnology, Inc. and anti-LC3B Ab (1:4,000; cat. no. NB600-1384) was purchased from Novus Biologicals, LLC. Anti-mixed lineage kinase domain-like protein (MLKL) Ab (1:1,000; product code ab183770) and anti-phospho-MLKL (Ser358) Ab (1:1,000; product code ab187091) were purchased from Abcam. Immunoreactive proteins were detected at RT for 1 h with horseradish peroxidase-conjugated secondary Abs (anti-mouse: cat. no. 115-035-003, at 1:5,000 dilution; anti-rabbit: cat. no. 711-035-152, at 1:5,000 dilution; Jackson ImmunoResearch Laboratories, Inc.) and visualized with an enhanced chemiluminescence reagent (EMD Millipore). Protein bands were imaged and analyzed using the ChemiDoc XRS system (Bio-Rad Laboratories, Inc.). For positive control of necroptosis, HT-29 cells were pre-treated with Z-VAD-fmk (20 µM) for 30 min, and subsequently treated with cycloheximide (10 µg/ml) and TNF-α (20 ng/ml) for 8 h as previously described (22). For positive control of cleaved caspase-3 and cleaved PARP, CAL 27 cells were treated with 1 µM staurosporine for 4 h.
Fluorescent dextran uptake
Cells were cultured in CELLview 35-mm glass-bottom cell culture dishes with four compartments (cat. no. 627870; Greiner Bio-One). Two days after seeding (8×104 cells/dish), cells were incubated for 8 h with 50 µg/ml Alexa Fluor 488 dextran (molecular weight: 10,000; cat. no. D22910; Thermo Fisher Scientific, Inc.) in a CO2 incubator at 37°C. The medium containing fluorescent dextran was subsequently replaced with fresh medium containing LysoTracker Red (50 nM) (Life Technologies; Thermo Fisher Scientific, Inc.) and cultured further for up to 6 h with LPZ or AZM alone or in combination at 37°C, 5% CO2 under humidified conditions on a stage top incubator (Carl Zeiss AG). Fluorescence images were captured by confocal micros-copy (LSM 700; Carl Zeiss) (objective magnification, ×63) and analyzed using the ZEN 2.3 SP1 Black Edition software (Carl Zeiss AG). Alexa488 fluorescence intensity or number of particles of each cell at 0 and 6 h were analyzed by ImageJ (1.50i; National Institutes of Health), and the fold change of fluorescence intensity or number of particles were calculated.
Galectin puncta assay for lysosomal membrane permeabilization detection
The lysosomal membrane permeabilization was assessed by fluorescence immunocytochemistry. Cells were fixed with methanol at -20°C for 10 min, blocked with 10% normal goat serum in TBST, and incubated with anti-galectin-3 (1:100 at 4°C, overnight; cat. no. 87985; Cell Signaling Technology, Inc.) and anti-lysosome-associated membrane protein-2 (LAMP-2; 1:100 at 4°C, overnight; cat. no. sc-18822, Santa Cruz Biotechnology, Inc.); the latter is a lysosomal and late endosomal marker (25). Then, cells were incubated with anti-mouse IgG Alexa 488 and anti-Rabbit IgG Alexa 555 antibodies (cat. nos. A11029 and A21428; Molecular Probes; Thermo Fisher Scientific, Inc.) (1:250 at RT for 1 h). Confocal microscopic observation was performed using an LSM 700 confocal laser scanning microscope (Carl Zeiss AG) (objective magnification, ×63). As a positive control for lysosomal membrane permeabilization (LMP), cells were treated with 1 mM LLOMe for 3 h.
Statistical analysis
All the quantitative data were expressed as the mean ± standard deviation (SD). Statistical analysis except for the galectin-3 puncta assay and band intensity analysis of immunoblots were performed with two-way ANOVA, followed by Bonferroni's multiple comparison test. For the galectin-3 puncta assay and immunoblotting analysis, one-way ANOVA followed by Bonferroni's multiple comparison test was used. A P-value <0.05 was considered to indicate a statistically significant difference. All analyses were performed with GraphPad Prism 5 software (GraphPad Software, Inc.).
Results
Combination treatment with LPZ and AZM reveals enhanced cytotoxicity in cancer cells
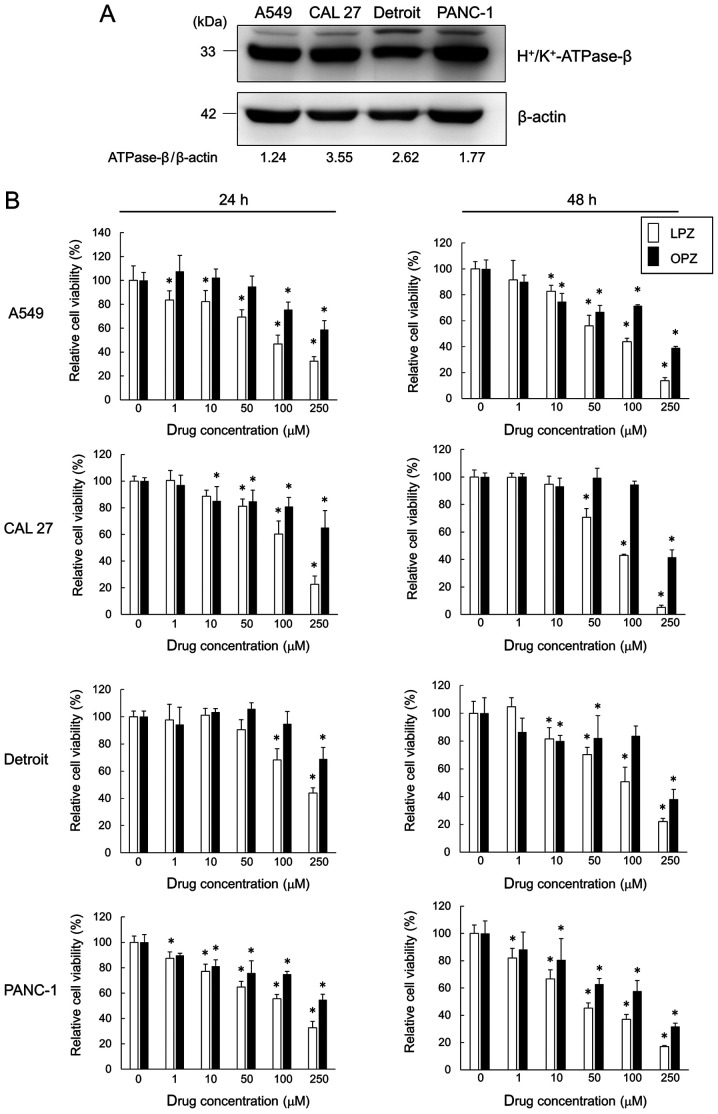
Western blot results from all the tested cell lines revealed the presence of H+/K+-ATPase-β subunit protein, a PPI target (Fig. 1A). Cells were treated with varying concentrations (0, 1, 10, 50, 100 and 250 µM) of LPZ and OPZ for 24 and 48 h, and the cell viability was assessed. LPZ and OPZ inhibited the growth of all cell lines in both time- and dose-dependent manners (Fig. 1B), with a greater growth inhibition observed with LPZ-treated cells. It has been suggested that PPI-treatment increases the pH of the tumor microenvironment (3,4). However, no pH differences between LPZ- or OPZ-treated and untreated culture media were observed after monitoring for over 72 h (data not shown). In addition, changing the culture media pH from 6.0 to 8.0 did not affect the PPI cytotoxicity profile of tumor cell lines (data not shown). Hence, subsequent experiments with LPZ were conducted with cells cultured in pH 7.3 media.
Figure 1.
PPIs inhibit the growth of H+/K+-ATPase-β-expressing cancer cell lines. (A) Immunoblots revealing H+/K+-ATPase-β expression. Cancer cell line proteins were separated on an 11.25% SDS-PAGE gel and probed with anti-H+/K+-ATPase-β Ab. (B) A549, CAL 27, Detroit, and PANC-1 cells were incubated with LPZ or OPZ at various concentrations (0, 1, 10, 50, 100, 250 µM) for 24 and 48 h. Cell viability was assessed using a CellTiter Blue viability assay, as described in the Materials and methods section, and the viability of cells (control) without drug treatment was set as 100%. (n=5; mean ± SD). *P<0.05 vs. the control. PPIs, proton pump inhibitors; LPZ, lansoprazole; OPZ, omeprazole.
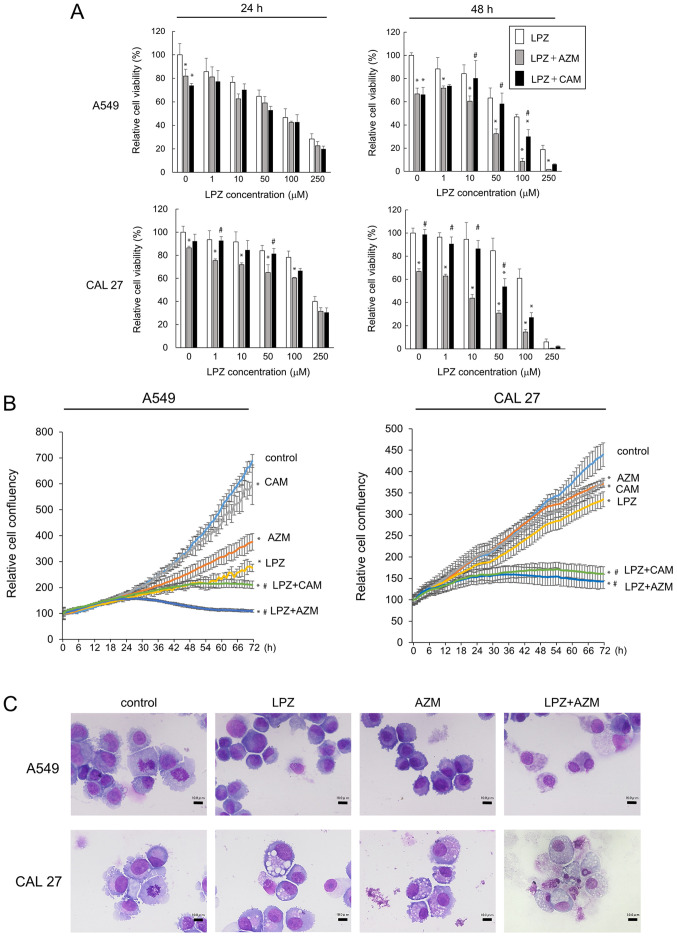
Next, we examined the drug combination treatment effects by adding either LPZ + CAM, LPZ + AZM, or LPZ alone. Combination treatment potentiated the cytotoxicity effects as compared to that by LPZ alone (Fig. 2A). The increased cytotoxicity after 48-h incubation with LPZ + AZM or LPZ + CAM was observed in CAL 27 and A549 cells, and this effect was confirmed by cell confluency assays with the IncuCyte ZOOM (Fig. 2B). In addition, the enhancing effect of AZM was greater than that of CAM (Fig. 2A and B). We examined the morphology of May-Grünwald-Giemsa-stained A549 and CAL 27 cells after LPZ and AZM treatments to evaluate the cytotoxic vs. cytostatic effect. A549 and CAL 27 cells did not exhibit the typical apoptotic morphological features of cells, such as condensed chromatin, nuclear fragments, and apoptotic bodies; however, they exhibited cytoplasmic and nuclear swelling with reduced plasma membrane integrity (Fig. 2C), thus indicating a non-apoptotic mode of cell death. Cell cycle analysis after treatment with LPZ and AZM revealed no cytostatic effects (data not shown).
Figure 2.
Growth inhibition of cancer cell lines after LPZ combination treatment with CAM or AZM. (A) A549 and CAL 27 cells were treated with LPZ (0-250 µM) with or without AZM (50 µM) or CAM (50 µM). Cell growth was assessed by a CellTiter Blue assay. The viability of cells (control) without drug treatment was set as 100%. (n=5; mean ± SD). *P<0.05 vs. the control; #P<0.05 vs. LPZ+AZM. (B) The IncuCyte cell imaging system tracked cell confluency in five fields of view in each culture plate as a measure of cell growth over 72 h following the addition of LPZ (100 µM), CAM (50 µM), AZM (50 µM), or LPZ + CAM/AZM combinations. *P<0.05 vs. the control at 72-h exposure; #P<0.05 vs. each drug alone. (C) May-Grünwald-Giemsa staining of A549 and CAL 27 cytospins after treatment with LPZ (100 µM), AZM (50 µM), or LPZ + AZM for 48 h. Scale bar, 10 µm. LPZ, lansoprazole; CAM, clarithromycin; AZM, azithromycin.
Combination treatment with LPZ and AZM causes accumulation of autolysosomes containing undigested cytoplasmic debris
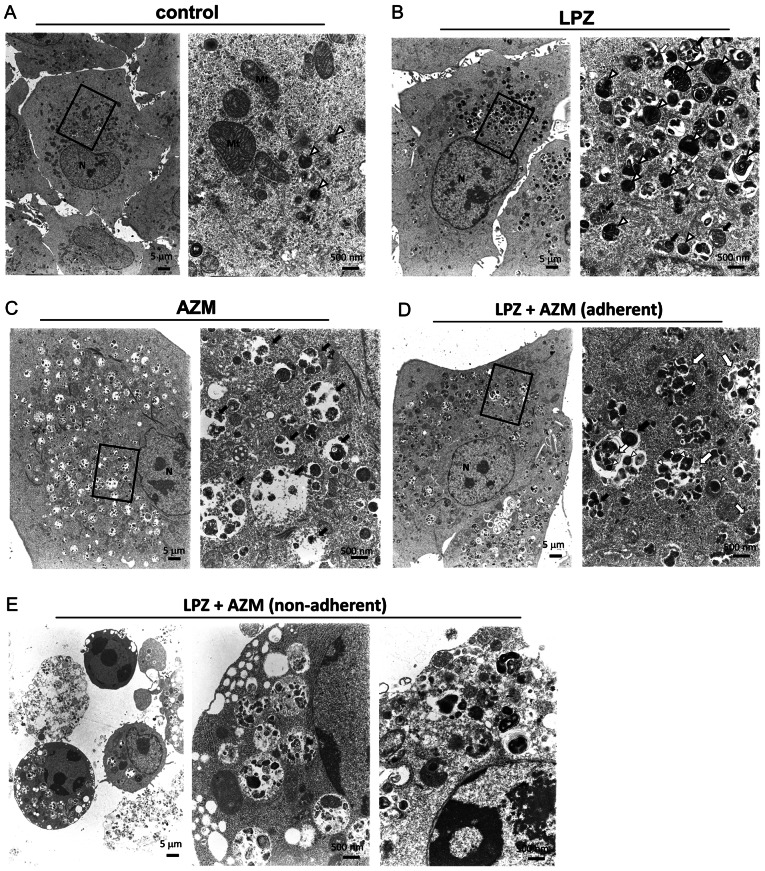
The cell-death phenotype after LPZ and AZM treatment was verified by conducting TEM of A549 cells incubated with either AZM (50 µM), LPZ (100 µM), or both for 48 h. AZM-treated cells exhibited a significantly increased number of swollen autolysosomes in the cytoplasm (Fig. 3A and C). Notably, in LPZ-treated cells, the lysosomes and autolysosomes were localized to the perinuclear regions, whereas a majority of the swollen autolysosomes were distributed evenly in the cytoplasm of AZM-treated cells (Fig. 3B and C). In addition, the autolysosomes in LPZ-treated cells were smaller than those in AZM-treated cells. The combination treatment with LPZ + AZM caused an increased number of swollen autolysosomes containing undigested cellular components, including autophagosomes/lysosomes, and were distributed in the cytoplasm of adherent cells (Fig. 3D). Non-adherent A549 cells, mostly dead cells that lost their ability to adhere, contained the largest autolysosomes containing undigested materials (Fig. 3E), and some cells also had condensed chromatin. However, the dying cells were swollen and had enlarged organelles and reduced plasma membrane integrity. These observations are consistent with the necrotic cell death processes along with increased autolysosome numbers, although no morphological changes associated with apoptosis, such as fragmented nuclei and apoptotic bodies, were observed (26).
Figure 3.
Transmission electron microscopy of A549 cells treated with LPZ or AZM or both. A549 cells were cultured for 48 h in complete culture medium containing (A) 0.1% DMSO as a control, treated with (B) 50 µM AZM, or with (C) 100 µM LPZ for 48 h. (D) Adherent A549 cells after 48 h-treatment with AZM (50 µM) and LPZ (100 µM). (E) Non-adherent A549 cells after 48 h-treatment with the LPZ + AZM combination. Scale bar represents the magnification. The right panels reveal enlarged images of the section indicated by the square box in the left panels. N, nucleus; Mt, mitochondria; open (white) arrowhead, lysosome; closed (black) arrow, autolysosome; open (white) arrow, autophagosome. LPZ, lansoprazole; AZM, azithromycin; DMSO, dimethyl sulfoxide.
Combination treatment with LPZ and AZM exhibits atypical cell death phenotypes
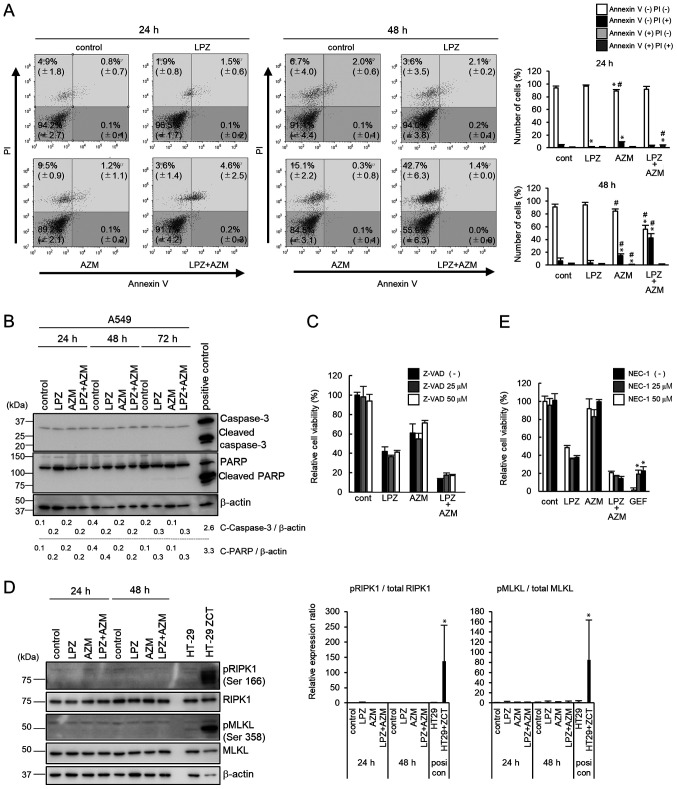
To confirm that cells treated with the combination of LPZ and AZM undergo non-apoptotic cell death, flow cytometric analysis of PI and Annexin V-stained cells was performed. As revealed in Fig. 4A, after LPZ + AZM treatment, there were more PI+/Annexin V− cells, as compared to PI−/Annexin V+ cells, thus indicating cells did not undergo early apoptosis (26). Immunoblots of protein expression did not detect caspase-3 or PARP cleavage (Fig. 4B). Furthermore, treatment with Z-VAD-fmk, a pan-caspase inhibitor, did not rescue cells from LPZ + AZM treatment-induced cytotoxicity (Fig. 4C). Collectively, these results suggest that LPZ + AZM treatment induced a non-apoptotic form of cell death. Hence, it was investigated whether LPZ + AZM treatment induces necroptosis instead. However, following LPZ + AZM treatment, phosphorylation of RIPK1 and MLKL was undetectable (Fig. 4D). Additionally, cell death could not be prevented by co-culturing cells with the RIPK1 inhibitor NEC-1, although NEC-1 exhibited the significant cancellation of GEF-induced cell death under amino-acid-depleted culture conditions, which was used as a positive control for RIPK1-dependent cell death induction (22) (Fig. 4E). Thus, the involvement of necroptosis in LPZ + AZM-induced cell death can also be excluded.
Figure 4.
Mechanism of cell death after treatment with LPZ or AZM or both in A549 cells. (A) Flow cytometry of Annexin V/PI double-stained A549 cells 24 and 48 h after treatment with 0.1% DMSO (control), LPZ (100 µM), AZM (50 µM), or LPZ + AZM combination. The vertical axis indicates the log fluorescence intensity of PI, and the horizontal axis indicates the log fluorescence intensity of Annexin V. The numbers indicate the percentage of cells in each area. The percentage of cells in each area were summarized and presented in the right panel. (n=3; mean ± SD). *P<0.05 vs. the control, #P<0.05 vs. LPZ. (B) Immunoblots of caspase-3 and PARP expression in A549 cells cultured in control medium, LPZ (100 µM), AZM (50 µM), or LPZ + AZM for 24, 48, and 72 h. CAL 27 cells cultured in complete culture medium with 1 µM staurosporine for 4 h were used as the positive control. Band intensities were standardized by β-actin. (C) The effect of Z-VAD-fmk (25 and 50 µM) on the viability of A549 cells treated with or without LPZ (100 µM) or AZM (50 µM) or both for 48 h, as measured by a CellTiter Blue viability assay. The viability of cells (control) without drug treatment was set as 100%. (n=3, mean ± SD). (D) Immunoblots of phospho-RIPK1, RIPK1, phospho-MLKL, and MLKL expression in A549 cells cultured in control medium, LPZ (100 µM), AZM (50 µM), or LPZ + AZM for 24 and 48 h. HT-29 cells treated with Z-VAD-fmk, cycloheximide, and TNF-α indicated as ZCT were used as a positive necroptosis control as previously described (22). Relative band intensities of pRIPK1/RIPK1 and pMLKL/MLKL were summarized in the right panel. (n=3; mean ± SD). *P<0.05 vs. the control. (E) The effect of NEC-1 (25 and 50 µM) on the viability of A549 cells treated with or without LPZ (100 µM) or AZM (50 µM) or both for 48 h. GEF-treated A549 cells in amino acid-depleted culture conditions were used as positive controls for RIPK1-dependent cell death as previously described (22). The viability of cells (control) without rug treatment was set as 100%. (n=3, mean ± SD). *P<0.05 vs. without NEC-1. LPZ, lansoprazole; AZM, azithromycin; DMSO, dimethyl sulfoxide; PI, propidium iodide; RIPK1, receptor-interacting serine/threonine-protein kinase 1; MLKL, mixed lineage kinase domain-like protein; NEC-1, necrostatin-1; GEF, gefitinib,
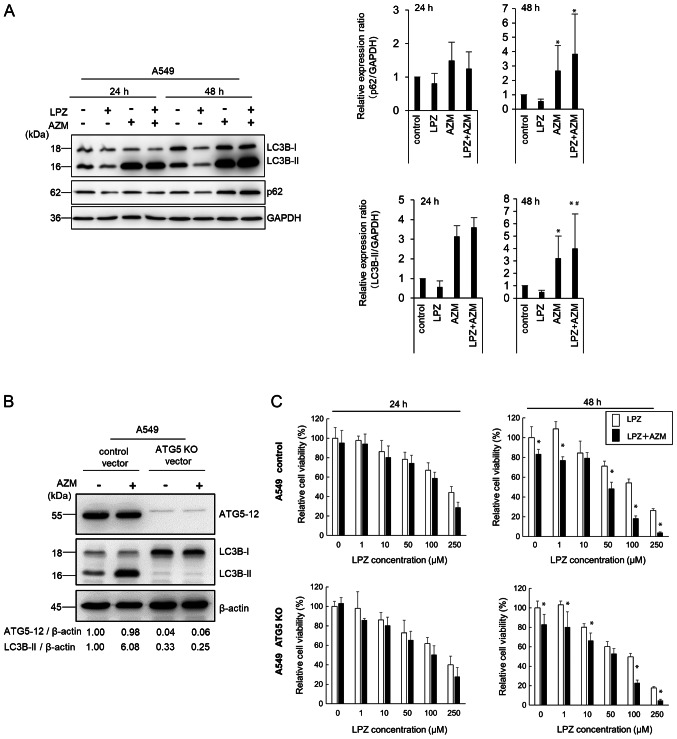
We previously reported that macrolide antibiotics, such as AZM and CAM, suppress autophagic flux (15,17), whereas EPZ was revealed to induce autophagy in melanoma cells as a survival response to oxidative stress (14). However, recent studies revealed that PPIs suppress autophagy in various cancer types (12,13). As revealed in Fig. 5A, AZM induced the autophagic substrate p62 accumulation and increased the autophagosome marker LC3B-II expression in A549 cells (27). These changes indicated that AZM treatment blocked the autophagic flux, leading to autophagosome/autolysosome accumulation, as previously demonstrated (15,17), and is consistent with the TEM observations revealing accumulation of swollen autolysosomes (Fig. 3C). After LPZ treatment, the expression levels of p62 and LC3B-II were almost equivalent to those in control cells. Co-administration of AZM and LPZ for 48 h further increased LC3B-II expression compared with AZM alone. Thus, the present results indicated that LPZ exhibited a minimal effect on autophagic induction.
Figure 5.
Comparison of LPZ or AZM cytotoxic effects on A549 cells with or without autophagosome forming ability. (A) Immunoblots revealing the protein expression of LC3B, p62, and GAPDH in A549 cells treated with or without LPZ (100 µM) or AZM (50 µM) for 24 and 48 h. Band intensities were measured and summarized in the right panel. (n=3; mean ± SD). *P<0.05 vs. the control; #P<0.05 vs. AZM. (B) Immunoblots revealing the protein expression of ATG5, LC3B, and β-actin in A549-ATG5 KO and control cell lines treated with LPZ (100 µM) or AZM (50 µM) for 24 and 48 h. Band intensities were standardized by β-actin. (C) Treatment-associated growth inhibition of A549-ATG5 KO and control cell lines was assessed by a CellTiter Blue assay. The viability of cells (control) without drug treatment was set as 100%. (n=3; mean ± SD). *P<0.05 LPZ alone vs. LPZ+AZM. LPZ, lansoprazole; AZM, azithromycin; KO, knockout.
Since the number of autolysosomes increased in A549 cells after treatment with LPZ + AZM (Fig. 3D and E), the ATG5-knockout A549 cell line was next established to exclude autophagy involvement (Fig. 5B). The cytotoxic response to LPZ + AZM combination treatment did not differ between ATG5-knockout and the parental cell lines (Fig. 5C), indicating a lack of involvement of autophagic cell death in drug cytotoxicity. Collectively, LPZ and AZM combination treatment induced a potent antitumor cytotoxic response independent of apoptosis, necroptosis, or autophagy-dependent cell death.
Combination treatment with LPZ and AZM induces lysosomal membrane permeabilization in A549 cells
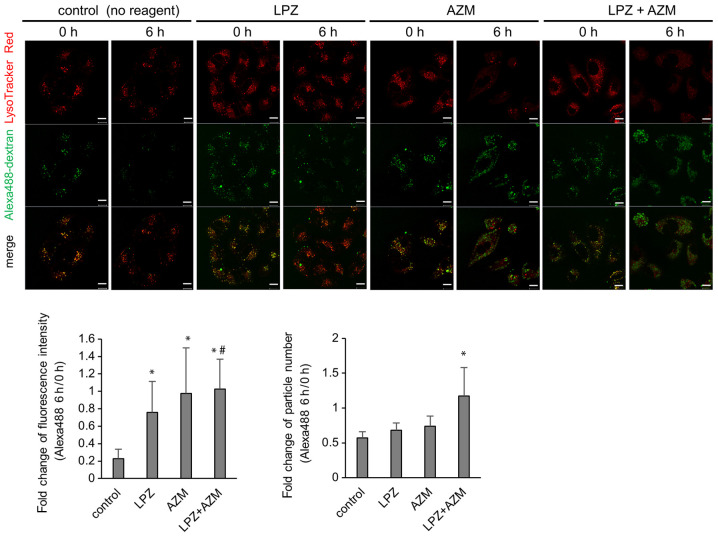
To identify the molecular mechanism responsible for combination treatment-induced cytotoxicity, TEM images were obtained revealing the necrosis-like phenotype with increased autolysosomes containing undigested debris (Fig. 3E). Alexa 488-labeled dextran was used to monitor the lysosomal degradation processes. In control cells, numerous Alexa 488-dextran particles co-localized with LysoTracker Red, and the Alexa 488 signal diminished after 6 h (Fig. 6). This represents the process by which Alexa 488-dextran is endocytosed, fused with lysosomes, and subsequently digested by acidic hydrolases as part of the lysosomal content. The cytoplasmic LysoTracker intensity was reduced in the presence of AZM, and Alexa 488-dextran particles were visible 6 h after AZM addition. The intensity and/or the number of undigested Alexa 488-dextran particles was increased when cells were treated with a combination of AZM + LPZ. These results are consistent with the TEM images that revealed that the number of autolysosomes increased in A549 cells after treatment with LPZ + AZM and strongly suggest the impairment of lysosomal function along with increased lysosomal pH (Fig. 3D).
Figure 6.
LysoTracker Red- and Alexa 488-dextran-stained A549 cells treated with LPZ or AZM. Alexa Fluor 488-dextran was added at 50 µg/ml to the culture medium. Eight hours later, the fluorescent dextran-containing medium was replaced with fresh medium containing 50 nM of LysoTracker Red and cultured subsequently with or without either LPZ (100 µM) or AZM (50 µM) for up to 6 h. LysoTracker Red represents the acidic state of late endosome/lysosome, where Alexa Flour 488-dextran is degraded by lysosomal hydrolases. Scale bar, 10 µm. Fold change of Alexa488 fluorescence intensity or number of particles in each cell was calculated and summarized in the lower panel. A total of 29-49 cells from 6 fields in each condition were used for the analysis. (mean ± SD). *P<0.05 vs. the control; #P<0.05 vs. LPZ. LPZ, lansoprazole; AZM, azithromycin.
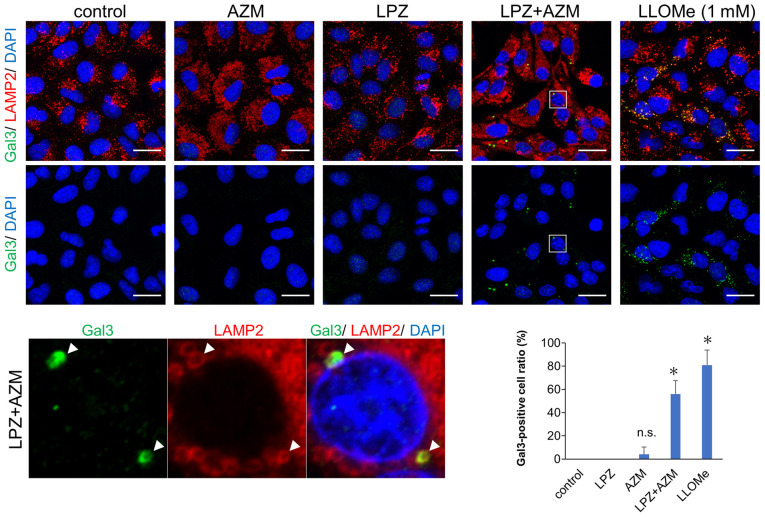
Recent studies have revealed leakage of intra-lysosomal hydrolases such as cathepsins due to several types of stress-induced lysosomal membrane permeabilization (LMP), resulting in lysosome-dependent cell death (LDCD) (28,29). Thus, it is likely that accumulation of autolysosomes with undigested contents induced LMP-associated cell death. A galectin puncta assay involving double-immunostaining with anti-galactin-3 and anti-LAMP-2 Abs revealed that LMP, which is shown as colocalized puncta of galectin-3 and LAMP-2, was induced by LPZ + AZM combination treatment (Fig. 7) (25). Notably, treatment with either LPZ or AZM alone did not affect lysosomal galectin-3 puncta expression. Therefore, the induction of non-apoptotic cell death by LPZ + AZM drug combination appears to be mediated by LMP-associated necrosis.
Figure 7.
Immunofluorescence images of Gal3 and LAMP-2 expression in AZM and LPZ-treated cells for assessing lysosomal membrane permeability. Fluorescence images of galectin-3 puncta and LAMP-2 expression and distribution in A549 cells treated with AZM (50 µM) or LPZ (100 µM) for 24 h. A549 cells treated with 1 mM LLOMe for 3 h were used as LMP-positive control as previously described (29). Scale bar, 10 µm. *P<0.05 vs. untreated A549 cells. The summarized Gal3-positive cell ratio in 6 fields is presented in the bottom right panel. (n=6; mean ± SD). *P<0.05 vs. the control. Gal3, galectin-3; LAMP-2, lysosome-associated membrane protein 2; AZM, azithromycin; LPZ, lansoprazole; LLOMe, L-Leucyl-L-Leucine methyl ester (hydrochloride); LMP, lysosomal membrane permeabilization.
Discussion
In the present study, it is reported that LPZ + AZM combination treatment induced a potent cytotoxic effect in several cancer cell lines. Since AZM alone was not cytotoxic, AZM enhanced LPZ-induced cell death acting as an agonist. However, the cell death induced by this drug combination was not due to apoptosis, necroptosis, or autophagy-dependent cell death and appears to be unique. Necrosis along with a substantial number of autolysosomes and lysosomes containing undigested materials were identified. The impaired lysosomal function was also supported by the long-term retention of Alexa 488-dextran in the endosomes. Additionally, LMP was also induced.
Lysosomes recycle cellular components and contain over 50 different acid hydrolases. These acid hydrolases are active at relatively low pH (approximately 4-5) and can degrade most cellular macromolecules (30). Cytosolic galectins act as sensors for lysosomal damage by binding to lysosomal β-galactosidases and are localized to the luminal side of the lysosomal membrane and become accessible to galectins during LMP (25,28,29). LMP and the consequent cytosolic release of lysosomal acid hydrolases result in an uncontrolled breakdown of cell components, which leads to cell death by necrosis and LDCD (28). Thus, the cell death phenotype observed in the present study could be categorized as 'LDCD', LDCD can be invoked when the cell death execution is dependent on cathepsin activity. Hence, cell death induction should be suppressed by pharmacologic or genetic blockade of cathepsin activities (28,29). However, no difference in LPZ + AZM-mediated cytotoxicity was observed when CA-074, a cathepsin B inhibitor, was added to the cultures (data not shown). Since lysosomes contain numerous different acid hydrolases other than cathepsin B, their role in the cell death processes by inhibiting other cathepsins require further evaluation. Hence, the involvement of necrosis along with LMP in the treatment-associated cytotoxicity was surmised.
Next, it was determined whether LPZ + AZM combination treatment enhances LMP-associated necrosis. As demonstrated in the results, a galectin-3 puncta assay revealed detectable LMP after concurrent treatment with AZM and LPZ, but not with AZM or LPZ alone. In addition, significant lysosomal dysfunction was observed after treatment with both drugs. AZM blocks the later stage of autophagic flux and leads to the accumulation of cytoplasmic autolysosomes (17,18). However, ATG5 knockout in the A549 cell line resulted in complete inhibition of autophagosome formation and did not affect LPZ + AZM treatment-induced cytotoxicity. In addition, unlike previous studies (12-14), we could not detect any autophagic changes in LPZ-treated cells, regardless of whether it promotes or suppresses autophagy. Therefore, the present results suggest that autophagy did not play a significant role in the LPZ+AZM treatment cytotoxicity.
Notably, AZM treatment increased the number of LAMP-2-positive vesicles, which suggests that lysosomal biogenesis was upregulated. AZM impaired lysosomal function and increased lysosomal pH. PPIs, including LPZ, have been reported to inhibit lysosomal enzyme activities, including acid phosphatase and β-N-acetylglucosaminidase, both in vitro and in vivo (31,32). The combination of enzyme activity inhibition with lysosomal alkalization leads to lysosomal impairment (33). Using the LysoTracker Red reagent, decreased lysosomal acidification in cells after AZM or LPZ + AZM treatments, were detected but not after treatment with LPZ alone. Thus, the LPZ + AZM combination induced significant accumulation of damaged lysosomes, leading to LMP. Additionally, the cytosolic lysosomal membrane surface acts as a signaling platform for the interaction of the mammalian target of rapamycin complex 1 (mTORC1) with its cofactors in response to stress and other cellular factors (34,35). Impaired lysosomal accumulation signals mTORC1 release from the lysosomal membranes. Subsequently, dephosphorylation of the master regulator of lysosomal biogenesis transcription factor EB (TFEB) occurs, causing its translocation to the nucleus, transcriptional activation, and de novo lysosomal biogenesis (30). Concurrently, the damaged lysosomes are removed by lysophagy (36). Thus, the present results indicated that after AZM blocked the autophagic flux, the feedback loop described below may explain the resulting increase in impaired lysosome numbers, leading to LMP and necrosis: LPZ and AZM induced lysosomal damage → lysosomal biogenesis by TFEB → lysosomal accumulation due to AZM blocking lysophagy → a considerable number of lysosomes with LMP → pronounced LMP-mediated necrosis induction. Although further studies are required to elucidate the underlying molecular mechanisms, this hypothesis adequately explains our TEM findings.
In the present study, it was reported that AZM potently enhanced the antitumor effects of LPZ in various cancer cell lines via necrosis induction and LMP. The present results indicate the potential of AZM and LPZ for use in cancer therapeutics by the induction of LMP-mediated tumor cell death. Although these drugs are in clinical use, caution is advised to minimize adverse events such as necrosis-induced inflammatory response. Further studies on cancer cell specificity and to reduce the likelihood of the non-specific targeting of normal cells are warranted.
Acknowledgments
We thank Ms Yumiko Yamada, Ms Ayako Hirota, and Ms Hiromi Kazama (Department of Biochemistry, Tokyo Medical University, Tokyo, Japan) for their technical assistance, helpful advice, and fruitful discussions.
Abbreviations
- PPIs
proton pump inhibitors
- LPZ
lansoprazole
- EPZ
esomeprazole
- PPZ
pantoprazole
- OPZ
omeprazole
- VCR
vincristine
- AZM
azithromycin
- CAM
clarithromycin
- NSCLC
non-small cell lung cancer
- DMSO
dimethyl sulfoxide
- RIPK1
receptor-interacting serine/threonine-protein kinase 1
- PARP
poly(ADP-ribose) polymerase
- LMP
lysosomal membrane permeabilization
- LDCD
lysosome-dependent cell death
- mTORC1
mammalian target of rapamycin complex 1
- TFEB
transcription factor EB
Funding
The present study was supported by funds provided by the Strategic Research Foundation at Private Universities (grant no. S1411011, 2014-2018) from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan awarded to KM, and by JSPS KAKENHI grants (no. 17K08771) to KM, (no. 17K15031) to NT, and (no. 18K15031) to HH.
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
NT and KM designed the experiments. AT performed most of the experiments and analyzed the data. HK performed trans-mission electron microscopy. NT, HH, SM, and AA assisted AT for acquisition and analysis of data. NT, MH, and KT were involved in the conception and mentoring, as well as in critical interpretation and evaluation of the data. AT, NT, MH, and KM were involved in writing, reviewing, and editing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient's consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Kato Y, Ozawa S, Miyamoto C, Maehata Y, Suzuki A, Maeda T, Baba Y. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013;13:89. doi: 10.1186/1475-2867-13-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luciani F, Spada M, De Milito A, Molinari A, Rivoltini L, Montinaro A, Marra M, Lugini L, Logozzi M, Lozupone F, et al. Effect of proton pump inhibitor pretreatment on resistance of solid tumors to cytotoxic drugs. J Natl Cancer Inst. 2004;96:1702–1713. doi: 10.1093/jnci/djh305. [DOI] [PubMed] [Google Scholar]
- 3.Lu ZN, Tian B, Guo X. Repositioning of proton pump inhibitors in cancer therapy. Cancer Chemother Pharmacol. 2017;80:925–937. doi: 10.1007/s00280-017-3426-2. [DOI] [PubMed] [Google Scholar]
- 4.Spugnini EP, Fais S. Drug repurposing for anticancer therapies. A lesson from proton pump inhibitors. Expert Opin Ther Pat. 2020;30:15–25. doi: 10.1080/13543776.2020.1704733. [DOI] [PubMed] [Google Scholar]
- 5.Ihraiz WG, Ahram M, Bardaweel SK. Proton pump inhibitors enhance chemosensitivity, promote apoptosis, and suppress migration of breast cancer cells. Acta Pharm. 2020;70:179–190. doi: 10.2478/acph-2020-0020. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Ling T, Zhaxi P, Cao Y, Qian L, Zhao D, Kang W, Zhang W, Wang L, Xu G, Zou X. Proton pump inhibitor pantoprazole inhibits gastric cancer metastasis via suppression of telomerase reverse transcriptase gene expression. Cancer Lett. 2019;452:23–30. doi: 10.1016/j.canlet.2019.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Spugnini EP, Citro G, Fais S. Proton pump inhibitors as Anti-vacuolar-ATPases drugs: A novel anticancer strategy. J Exp Clin Cancer Res. 2010;29:44. doi: 10.1186/1756-9966-29-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geeviman K, Babu D, Prakash Babu P. Pantoprazole induces mitochondrial apoptosis and attenuates NF-κB signaling in glioma cells. Cell Mol Neurobiol. 2018;38:1491–1504. doi: 10.1007/s10571-018-0623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He J, Shi XY, Li ZM, Pan XH, Li ZL, Chen Y, Yan SJ, Xiao L. Proton pump inhibitors can reverse the YAP mediated paclitaxel resistance in epithelial ovarian cancer. BMC Mol Cell Biol. 2019;20:49. doi: 10.1186/s12860-019-0227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu ZN, Shi ZY, Dang YF, Cheng YN, Guan YH, Hao ZJ, Tian B, He HW, Guo XL. Pantoprazole pretreatment elevates sensitivity to vincristine in drug-resistant oral epidermoid carcinoma in vitro and in vivo. Biomed Pharmacother. 2019;120:109478. doi: 10.1016/j.biopha.2019.109478. [DOI] [PubMed] [Google Scholar]
- 11.Tan Q, Wang M, Yu M, Zhang J, Bristow RG, Hill RP, Tannock IF. Role of autophagy as a survival mechanism for hypoxic cells in tumors. Neoplasia. 2016;18:347–355. doi: 10.1016/j.neo.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Tan Q, Joshua AM, Wang M, Bristow RG, Wouters BG, Allen CJ, Tannock IF. Up-regulation of autophagy is a mechanism of resistance to chemotherapy and can be inhibited by pantoprazole to increase drug sensitivity. Cancer Chemother Pharmacol. 2017;79:959–969. doi: 10.1007/s00280-017-3298-5. [DOI] [PubMed] [Google Scholar]
- 13.Tan Q, Joshua AM, Saggar JK, Yu M, Wang M, Kanga N, Zhang JY, Chen X, Wouters BG, Tannock IF. Effect of pantoprazole to enhance activity of docetaxel against human tumour xenografts by inhibiting autophagy. Br J Cancer. 2015;112:832–840. doi: 10.1038/bjc.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Marino ML, Fais S, Djavaheri-Mergny M, Villa A, Meschini S, Lozupone F, Venturi G, Della Mina P, Pattingre S, Rivoltini L, et al. Proton pump inhibition induces autophagy as a survival mechanism following oxidative stress in human melanoma cells. Cell Death Dis. 2010;1:e87. doi: 10.1038/cddis.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moriya S, Che XF, Komatsu S, Abe A, Kawaguchi T, Gotoh A, Inazu M, Tomoda A, Miyazawa K. Macrolide antibiotics block autophagy flux and sensitize to bortezomib via endoplasmic reticulum stress-mediated CHOP induction in myeloma cells. Int J Oncol. 2013;42:1541–1550. doi: 10.3892/ijo.2013.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Renna M, Schaffner C, Brown K, Shang S, Tamayo MH, Hegyi K, Grimsey NJ, Cusens D, Coulter S, Cooper J, et al. Azithromycin blocks autophagy and may predispose cystic fibrosis patients to mycobacterial infection. J Clin Invest. 2011;121:3554–3563. doi: 10.1172/JCI46095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirasawa K, Moriya S, Miyahara K, Kazama H, Hirota A, Takemura J, Abe A, Inazu M, Hiramoto M, Tsukahara K, Miyazawa K. Macrolide antibiotics exhibit cytotoxic effect under amino Acid-depleted culture condition by blocking autophagy flux in head and neck squamous cell carcinoma cell lines. PLoS One. 2016;11:e0164529. doi: 10.1371/journal.pone.0164529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukai S, Moriya S, Hiramoto M, Kazama H, Kokuba H, Che XF, Yokoyama T, Sakamoto S, Sugawara A, Sunazuka T, et al. Macrolides sensitize EGFR-TKI-induced non-apoptotic cell death via blocking autophagy flux in pancreatic cancer cell lines. Int J Oncol. 2016;48:45–54. doi: 10.3892/ijo.2015.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugita S, Ito K, Yamashiro Y, Moriya S, Che XF, Yokoyama T, Hiramoto M, Miyazawa K. EGFR-independent autophagy induction with gefitinib and enhancement of its cytotoxic effect by targeting autophagy with clarithromycin in non-small cell lung cancer cells. Biochem Biophys Res Commun. 2015;461:28–34. doi: 10.1016/j.bbrc.2015.03.162. [DOI] [PubMed] [Google Scholar]
- 20.Komatsu S, Miyazawa K, Moriya S, Takase A, Naito M, Inazu M, Kohno N, Itoh M, Tomoda A. Clarithromycin enhances Bortezomib-induced cytotoxicity via endoplasmic reticulum Stress-mediated CHOP (GADD153) induction and autophagy in breast cancer cells. Int J Oncol. 2012;40:1029–1039. doi: 10.3892/ijo.2011.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang JC, Lu CW, Lin CJ. Treatment of Helicobacter pylori infection: Current status and future concepts. World J Gastroenterol. 2014;20:5283–5293. doi: 10.3748/wjg.v20.i18.5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito Y, Moriya S, Kazama H, Hirasawa K, Miyahara K, Kokuba H, Hino H, Kikuchi H, Takano N, Hiramoto M, et al. Amino acid starvation culture condition sensitizes EGFR-expressing cancer cell lines to gefitinib-mediated cytotoxicity by inducing atypical necroptosis. Int J Oncol. 2018;52:1165–1177. doi: 10.3892/ijo.2018.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Prey J, Sakamaki J, Baudot AD, New M, Van Acker T, Tooze SA, Long JS, Ryan KM. Application of CRISPR/Cas9 to autophagy research. Methods Enzymol. 2017;588:79–108. doi: 10.1016/bs.mie.2016.09.076. [DOI] [PubMed] [Google Scholar]
- 24.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;11:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aits S, Kricker J, Liu B, Ellegaard AM, Hämälistö S, Tvingsholm S, Corcelle-Termeau E, Høgh S, Farkas T, Holm Jonassen A, et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy. 2015;11:1408–1424. doi: 10.1080/15548627.2015.1063871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. Molecular mechanisms of cell death: Recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 2018;25:486–541. doi: 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 28.Wang F, Gómez-Sintes R, Boya P. Lysosomal membrane permeabilization and cell death. Traffic. 2018;19:918–931. doi: 10.1111/tra.12613. [DOI] [PubMed] [Google Scholar]
- 29.Aits S, Jäättelä M. Lysosomal cell death at a glance. J Cell Sci. 2013;126:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- 30.Andrea B, Juan SB. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Wang Y, Li SJ. Lansoprazole induces apoptosis of breast cancer cells through inhibition of intracellular proton extrusion. Biochem Biophys Res Commun. 2014;448:424–429. doi: 10.1016/j.bbrc.2014.04.127. [DOI] [PubMed] [Google Scholar]
- 32.Liu W, Baker SS, Trinidad J, Burlingame AL, Baker RD, Forte JG, Virtuoso LP, Egilmez NK, Zhu L. Inhibition of lysosomal enzyme activities by proton pump inhibitors. J Gastroenterol. 2013;48:1343–1352. doi: 10.1007/s00535-013-0774-5. [DOI] [PubMed] [Google Scholar]
- 33.Česen MH, Pegan K, Spes A, Turk B. Lysosomal pathways to cell death and their therapeutic applications. Exp Cell Res. 2012;318:1245–1251. doi: 10.1016/j.yexcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 34.Zhitomirsky B, Yunaev A, Kreiserman R, Kaplan A, Stark M, Assaraf YG. Lysosomotropic drugs activate TFEB via lysosomal membrane fluidization and consequent inhibition of mTORC1 activity. Cell Death Dis. 2018;9:1191. doi: 10.1038/s41419-018-1227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ballabio A, Bonifacino JS. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 36.Papadopoulos C, Kravic B, Meyer H. Repair or lysophagy: Dealing with damaged lysosomes. J Mol Biol. 2020;432:231–239. doi: 10.1016/j.jmb.2019.08.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.