Abstract
ETS variant 1 (ETV1) is an oncogenic transcription factor. However, its role in colorectal cancer has remained understudied. The present study demonstrated that ETV1 downregulation led to reduced HCT116 colorectal cancer cell growth and clonogenic activity. Furthermore, the ETV1 mRNA levels were enhanced in colorectal tumors and were associated with disease severity. In addition, ETV1 directly bound to Jumonji C domain-containing (JMJD) 1A, a histone demethylase known to promote colon cancer. ETV1 and JMJD1A, but not a catalytically inactive mutant thereof, cooperated in inducing the matrix metalloproteinase (MMP)1 gene promoter that was similar to the cooperation between ETV1 and another histone demethylase, JMJD2A. RNA-sequencing revealed multiple potential ETV1 target genes in HCT116 cells, including the FOXQ1 and TBX6 transcription factor genes. Moreover, JMJD1A co-regulated FOXQ1 and other ETV1 target genes, but not TBX6, whereas JMJD2A downregulation had no impact on FOXQ1 as well as TBX6 transcription. Accordingly, the FOXQ1 gene promoter was stimulated by ETV1 and JMJD1A in a cooperative manner, and both ETV1 and JMJD1A bound to the FOXQ1 promoter. Notably, the overexpression of FOXQ1 partially reversed the growth inhibitory effects of ETV1 ablation on HCT116 cells, whereas TBX6 impaired HCT116 cell growth and may thereby dampen the oncogenic activity of ETV1. The latter also revealed for the first time, to the best of our knowledge, a potential tumor suppressive function of TBX6. Taken together, the present study uncovered a ETV1/JMJD1A-FOXQ1 axis that may drive colorectal tumorigenesis.
Keywords: colorectal cancer, ETS protein, FOXQ1, gene transcription, Jumonji C domain-containing protein
Introduction
ETS variant 1 (ETV1; previously also known as ER81) is a transcription factor endowed with an ETS domain that binds to DNA sequences with a GGA(A/T) core (1-3). Nucleotides flanking this core or neighboring DNA-binding sites for interaction partners determine which ETS protein can bind to a specific gene regulatory element (4,5). However, ETV1 has two close homologs, ETV4 and ETV5, and they can bind potentially to the same sites throughout the genome. However, tissue-specific expression and possibly different interactomes endow each of these three transcription factors with a specific role during development (6). Indeed, compared to ETV4 and ETV5 ablation, ETV1 knockout leads to different phenotypic alterations in mice, such as deficient connection between muscle sensory and spinal motor neurons, abnormal numbers of spindles in limb muscles, the lack of Pacinian corpuscle limb mechanoreceptors, cardiac conduction defects, and the abnormal development of the ventricular conduction system (7-10). Furthermore, the activity of ETV1 is heavily regulated by post-translational modification, which includes phosphorylation, acetylation and ubiquitylation (11-18); however, the mechanisms thorough which these post-translational modifications affect the developmental functions of ETV1 remain elusive.
ETV1 has been implicated in tumorigenesis, most prominently in prostate cancer (6). Chromosomal translocations occur in ~5-10% of all human prostate tumors that lead to the overexpression of ETV1 (19). Furthermore, ETV1 overexpression is associated with an increased Gleason score and disease recurrence, suggesting that ETV1 promotes the development of aggressive prostate cancer in particular (20-22). Transgenic mice that prostate-specifically overexpress ETV1 develop prostatic intraepithelial neoplasia and the simultaneous homozygous loss of the tumor suppressor, Pten, leads to the development of invasive adenocarcinomas (21-23). Likewise, ETV1 transgenic mice develop prostate carcinoma when Jumonji C domain-containing (JMJD)2A is concurrently over-expressed in a heterozygous Pten knockout background (24). JMJD2A is a member of the JMJD protein family that encompasses numerous, diverse histone demethylases (25-27). The JMJD2A enzyme particularly demethylates tri-methylated lysine 9 and 36 on histone H3, and can thereby induce changes in gene transcription (28-30). It also binds directly to ETV1 and serves the function of an ETV1 coactivator (24,31).
Notably, JMJD2A is robustly expressed in several established colorectal cancer cell lines (32). In addition, the downregulation of JMJD2A in HCT116, DLD-1 and HT-29 human colorectal cancer cells has been shown to reduce their proliferative potential and increase apoptosis (33), while a small molecule inhibitor of JMJD2A catalytic activity has been shown to suppress the growth of HCT116 colorectal cancer cells (34). These data suggest that JMJD2A promotes colorectal cancer growth. Furthermore, ETV1 is expressed in human colorectal cancer cell lines and tumors (35,36). Hence, the present study examined the possible association between ETV1 and JMJD2A in colon cancer. Another histone demethylase, JMJD1A (37,38), was also included in the present study, since this enzyme is reportedly expressed in HCT116 cells and is overexpressed in human colorectal tumors (39-42).
Materials and methods
Cell culture and transfection
HCT116 cells (CCL-247; American Type Culture Collection), 293T cells (CRL-3216; American Type Culture Collection) and LNCaP cells (CRL-1740; American Type Culture Collection) were cultured at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (43). The calcium phosphate coprecipitation method was employed to transfect 293T cells cultured in poly-L-lysine coated 6-cm plates (44), while transfection of HCT116 cells grown in 12-wells was accomplished with the help of 2 µg polyethylenimine (45).
Retrovirus
In order to downregulate a gene, respective shRNA was cloned into the pSIREN-RetroQ retroviral vector (Clontech Laboratories, Inc.). The sequences targeted by ETV1 shRNAs were as previously published (46), while JMJD1A shRNA#1 targeted the sequence 5′-GCAGGUGUCAAUAGUGAUA-3′ and shRNA#2 the sequence 5′-GUAGACCUAGUUAAUUGUA-3′. The sequence of the control shRNA was 5′-CAACAAGAU GAAGAGCACCAA-3′, which has at least 5 mismatches to known human genes. The production of retrovirus in 293T cells has been previously described (47). HCT116 cells were then infected two or three times, temporally spaced by 12 h, with the indicated retrovirus, grown for 24 h in fresh media and then selected with 1 µg/ml puromycin for 2-3 days. Changes in protein expression were determined by western blot analysis (48); for this, cells were lysed in Laemmli sample buffer, resulting protein extracts run on SDS polyacrylamide gels and proteins transferred to PVDF membranes. For overexpression experiments, HA-tagged FOXQ1 and TBX6 were cloned into the pQCXIH retroviral vector (Clontech). HCT116 cells were infected twice within 12 h, grown for 24 h with fresh media, selected with 200 µg/ml hygromycin B for 3 days and then expanded under continuous hygromycin B selection. To detect expression of proteins in retro-virally transduced cells, rabbit polyclonal antibodies directed against ETV1 [#959; as previously described (46); 1:1,000 dilution], actin (A2066; Sigma-Aldrich; Merck KGaA; 1:5,000 dilution) or JMJD1A (NB100-77282; Novus Biologicals; 1:1,500 dilution) and goat polyclonal antibodies directed against FOXQ1 (NB100-1283; Novus Biologicals; 1:1,000 dilution) or TBX6 (AF4744; R&D Systems; 1:1,000 dilution) were used. Secondary goat anti-rabbit (170-6515; Biorad) or mouse anti-goat (sc-2768; Santa Cruz Biotechnology, Inc.) antibodies coupled to horse-radish peroxidase were used at a 1:3,000 dilution. All antibody incubations were performed in TRIS-buffered saline/0.05% Tween-20 with 4% non-fat dry milk. Detection was done with enhanced chemiluminescence using an ECL kit (GERPN2106; Sigma-Aldrich; Merck KGaA).
Cell growth and clonogenic assays
A total of 2,000 (for Fig. 1) or 2,400 (for Fig. 9) cells were seeded in 96-wells (49) and growth was determined using the PrestoBlue Cell Viability kit (A13262, Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's recommendations. For clonogenic assays, 3,000 (for Fig. 1) or 2,400 (for Fig. 9) cells were spread into 6-wells (50) and the formation of colonies was assayed after 8 days by staining with 0.4% crystal violet (C0775; Sigma-Aldrich; Merck KGaA) in 10% methanol/10% glacial acetic acid for 10 min at room temperature.
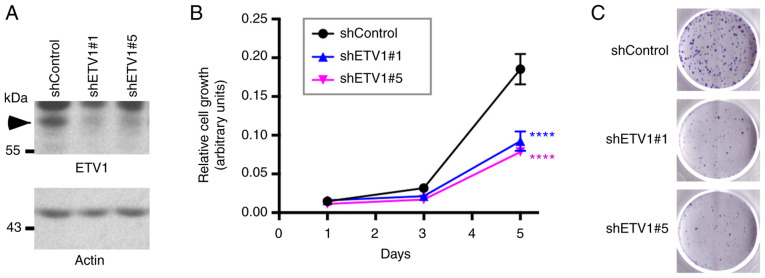
Figure 1.
Effect of ETV1 on HCT116 colorectal cells. (A) Downregulation of ETV1 with two different shRNAs in HCT116 cells was evaluated by western blot analysis. Arrowhead points to ETV1. (B) Analysis of HCT116 cell growth. Statistical significance was assessed with two-way ANOVA with post hoc Dunnett's multiple comparisons test; n=3; ****P<0.0001 vs. control. (C) Clonogenic assay. Representative images from 3 independent experiments.
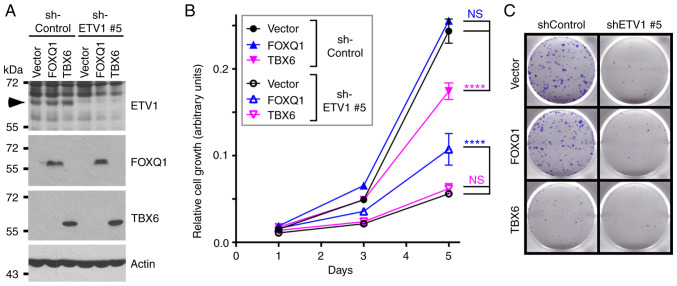
Figure 9.
Rescue of growth defects. HCT116 cells were stably transduced with pQCXIH vector control or retrovirus expressing HA-tagged FOXQ1 or TBX6. Following selection with hygromycin B, cells were additionally infected with retrovirus either expressing control shRNA or ETV1 #5 shRNA. (A) Western blots. Arrowhead indicates ETV1. (B) Growth assays. Two-way ANOVA with post hoc Tukey's multiple comparisons test (n=3). NS, not significant; ****P<0.0001. (C) Representative images for clonogenic assays (n=3).
Analysis of mRNA expression levels in public databases
All respective bioinformatics analyses were performed using the Oncomine tool (51). Data for mRNA expression levels were downloaded through the website www.oncomine.org and imported into GraphPad Prism 6 for Mac OS X for analysis.
Co-immunoprecipitation and GST pulldown experiments
Human 293T cells were transfected with the following amounts of plasmids: 3 µg Flag-tagged JMJD expression vector or empty vector pEV3S (52), 1 µg 6Myc-tagged ETV1 expression vector or empty vector pCS3+-6Myc (kindly provided by Professor Tony Hunter, Salk Institute), and 5 µg pBluescript KS+ (Stratagene; Agilent Technologies, Inc.). At 2 days following transfection, cells were lysed in 650 µl of 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.5% Igepal CA-630, 50 mM NaF, 0.1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 2 µg/ml aprotinin, 10 µg/ml leupeptin, 1 µg/ml pepstatin A, 0.1 mM dithiothreitol and immunoprecipitations were performed with 1 µg of anti-Flag M2 (F1804; Sigma-Aldrich; Merck KGaA) mouse monoclonal antibody (53); the nature of the beads used, incubation times and washing procedures, including centrifugation steps, for the immunoprecipitations were as previously described (54). Following western blot analysis, as described above, with mouse monoclonal anti-Myc 9E10 antibody (M4439; Sigma-Aldrich; Merck KGaA; 1:3,000 dilution) and secondary goat anti-mouse antibodies coupled to horseradish peroxidase (170-6516, Bio-Rad Laboratories, Inc.; 1:3,000 dilution), the blots were developed utilizing chemiluminescence (55). For endogenous coimmunoprecipitation, ETV1 was immunoprecipitated with 0.5 µg of rabbit poly-clonal antibody H-70 (sc-28681; Santa Cruz Biotechnology, Inc.) and JMJD1A was detected with rabbit polyclonal anti-body NB100-77282 (Novus Biologicals; 1:1,500 dilution). Glutathione S-transferase (GST) pulldown experiments were performed essentially as described (56) utilizing as a binding and washing buffer phosphate-buffered saline supplemented with 0.05% Tween-20, 1 mM dithiothreitol, 0.2 mM phenyl-methylsulfonyl fluoride, 2 µg/ml aprotinin, 10 µg/ml leupeptin and 1 µg/ml pepstatin A. The GST-ETV1 fusion protein or the GST moiety was expressed in Escherichia coli and puri-fied with the help of glutathione agarose, while Flag- and His-tagged JMJD1A was produced in baculovirus-infected Sf9 cells and purified with the help of Ni2+-NTA agarose (24).
Luciferase assays
The following amounts of plasmids were used to transfect the HCT116 cells: 0.25 µg matrix metallo-proteinase (MMP)1(-525/+15) luciferase reporter plasmid (12), 0.75 µg pBluescript KS+ (Stratagene; Agilent Technologies, Inc.), 20 ng ETV1 expression vector or empty vector pEV3S (52), and 50 ng Flag-tagged JMJD expression vector or empty vector pEV3S (52). For experiments comparing pGL2-Basic to the FOXQ1(-2000/+124) luciferase reporter, 100 ng pEV3S or ETV1 expression vector, as well as 75 ng pEV3S or JMJD1A expression vector were employed. Cells were washed once with phosphate-buffered saline 8 h after transfection and lysed 40 h later as previously described (57). Subsequently, light emission following the addition of D-luciferin was measured using a luminometer (Berthold Lumat LB9507), as previously described (58). Please note that, similar to recent publications (59-62), no internal control plasmid was used to normalize for transfection efficiency, since the authors have observed that such internal control plasmids are often themselves affected upon transcription factor overexpression, furthering the notion that the use of internal control plasmids can lead to artefacts (63-66).
RNA analyses
RNA was isolated from the HCT116 cells using TRIzol reagent (67) and reverse transcribed into cDNA using the GoScript Reverse Transcription kit (A5000; Promega Corporation) utilizing random p(dN)6 primers according to the manufacturer's recommendations. The resultant cDNA was then amplified by polymerase chain reaction, as previously described (68). The temperature program was as follows: 97°C for 1 min; 9 cycles of 97°C for 25 sec, 65°C (−1°C per cycle) for 25 sec, 72°C for 25 sec; 16-29 cycles of 97°C for 25 sec, 56°C for 25 sec, 72°C for 25 sec; 72°C for 4 min followed by cooling down to 4°C (69). The PCR products were visualized on ethidium bromide-stained agarose gels (70). Alternatively, qPCR was performed with the AccuPower 2X Greenstar qPCR kit (Bioneer K-6251) with initial denaturation at 95°C for 6 min followed by 32 cycles of 95°C for 15 sec, 56°C for 15 sec and 72°C for 35 sec. Relative quantification was performed with the ΔΔCq method (71). The primer sequences are listed in Table SI.
RNA-sequencing was performed in the Targeted DNA Methylation and Mitochondrial Heteroplasmy Core of the Nathan Shock Center of Excellence in the Biology of Aging in Oklahoma City. Briefly, Illumina Truseq Stranded HT library generation was performed according to the manufacturer's instructions. For strand-specificity, the incorporation of dUTP instead of dTTP in the second strand cDNA synthesis did not allow amplification past this dUTP with the polymerase. Following cDNA synthesis, each product underwent end repair process, the addition of a single A-base and ligation of adpaters. The cDNA products were further purified and enriched using PCR to make the final library for sequencing. Library sizing was performed by TapeStation (Agilent Technologies, Inc.) and the RNA-seq libraries were sequenced using an Illumina NextSeq (2×75 bp) instrument. Following sequencing, reads were trimmed and aligned against the GRCh38 build of the human genome, and differential expression statistics and correlation analyses were performed with the help of the Strand NGS software package (Avadis). Normalization was performed with the DESeq algorithm and a z-test was used to determine differential expression. P-values were adjusted according to the Benjamini and Hochberg method and deemed significant for a value <0.05 (72). No biological/technical replicates were used. Ingenuity Pathway Analysis (Qiagen GmbH) was employed to identify pathways affected upon ETV1 downregulation. Sequencing data were deposited in Gene Expression Omnibus (GEO) under accession number GSE158294.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays were performed on formaldehyde-treated HCT116 cells essentially as previously described (73). ETV1 and JMJD1A were precipitated with the rabbit polyclonal antibodies, ab81086 (Abcam) and NB100-77282 (Novus Biologicals), respectively, while control rabbit IgG was purchased from Santa Cruz Biotechnology, Inc. (sc-2027). Approximately 2 µg of the antibodies were employed for the immunoprecipitations that were performed at 4°C over-night (74). Genomic DNA was amplified in two steps with nested primers (75). The first PCR encompassed the following temperature steps: 97°C for 1 min; 9 cycles of 97°C for 20 sec, 65°C (-1°C per cycle) for 20 sec, 72°C for 40 sec; 20 cycles of 97°C for 20 sec, 56°C for 20 sec, 72°C for 40 sec; 72°C for 4 min followed by cooling down to 4°C. The second PCR was performed in the same manner except that 31 instead of 20 cycles were used. The primer sequences are presented in Table SII.
Statistical analyses
Means with standard deviations are presented in all applicable figures. Statistical tests that were used are indicated in the figure legends and included an unpaired, two-tailed Student's t-test, as well as one-way or two-way ANOVA with post hoc tests (Dunnett's, Sidak's or Tukey's) for multiple comparisons. All calculations were performed with GraphPad Prism 6 for Mac OS X. A P-value <0.05 was considered to reflect a statistical significance difference.
Results
Effect of ETV1 on HCT116 cells
Previously, it was reported that ETV1 downregulation reduced the viability of HT-29 colorectal cancer cells (76). The present study wished to assess whether ETV1 is also a promoter of HCT116 colorectal cancer cell growth; this particular cell line was selected for use in the present study, as ETV1, JMJD1A and JMJD2A were all robustly expressed in HCT116 cells (see below). Indeed, the downregulation of ETV1 with two different shRNAs significantly decreased the growth of HCT116 cells (Fig. 1A and B). In addition, the cell clonogenic activity was markedly reduced in the presence of ETV1 shRNA (Fig. 1C). Hence, similar to JMJD2A (33) or JMJD1A (39,41,42), ETV1 is a promoter of HCT116 colorectal cancer cell growth; this may therefore serve as a model system to study the potential cooperation of ETV1 with JMJD histone demethylases.
ETV1 and JMJD2A mRNA levels in colorectal cancer
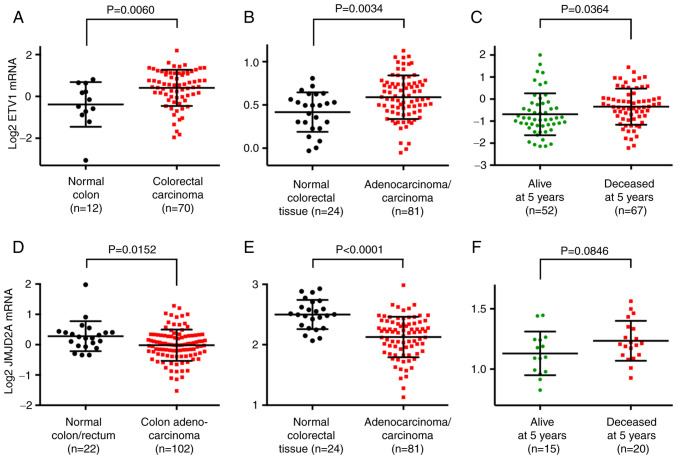
In order to substantiate that ETV1 is overexpressed in colorectal tumors, bioinformatics analyses were performed through the Oncomine website (www.oncomine.org). Utilizing published microarray data (77,78), it was found that ETV1 mRNA expression was significantly upregulated in colorectal tumors compared with normal tissue (Fig. 2A and B). Moreover, a high ETV1 expression was significantly associated with a reduced survival at 5 years [Fig. 2C; data from a previous study (79)], 3 years and 1 year (Fig. S1A) following diagnosis and was also positively associated with the stage of the disease (Fig. S1B), the latter being consistent with the reported presence of ETV1 in lymph node metastases (80). These data strongly support the notion that ETV1 promotes colorectal tumor formation and may be a predictor of an unfavorable outcome.
Figure 2.
Expression of ETV1 and JMJD2A mRNA in colorectal cancer. (A) Upregulation of ETV1 mRNA in colorectal tumors. Data were from the study by Hong et al (77). (B) Analogous in microarray data from the study published by Skrzypczak et al (78). (C) Higher ETV1 expression was associated with a reduced 5-year survival according to the microarray data from the study by Smith et al (79). (D) JMJD2A mRNA levels were downregulated in colorectal carcinomas. Data were from the Cancer Genome Atlas (81). (E) Analogous in microarray data published in the study by Skrzypczak et al (78). (F) Decreased survival (5 years) was associated with increased JMJD2A mRNA levels. Microarray data retrieved from the study by Smith et al (79). Statistical significance was assessed with unpaired, two-tailed Student's t test. JMJD, Jumonji C domain-containing protein.
Since ETV1 and JMJD2A synergize in prostate cancer (24), the present study also examined the mRNA levels of the JMJD2A histone demethylase. Surprisingly, it was discovered that the JMJD2A mRNA levels were downregulated in colorectal cancer [Fig. 2D and E; data from previous studies (78,81)]. Notably, high JMJD2A mRNA levels were associated with a reduced survival (Figs. 2F and S1C) and were associated with an increased disease stage (Fig. S1D). These data suggest that JMJD2A is not required for the initiation of disease, but rather for the progression to the more aggressive and lethal end stages of colorectal cancer.
Physical interaction between ETV1 and JMJD1A
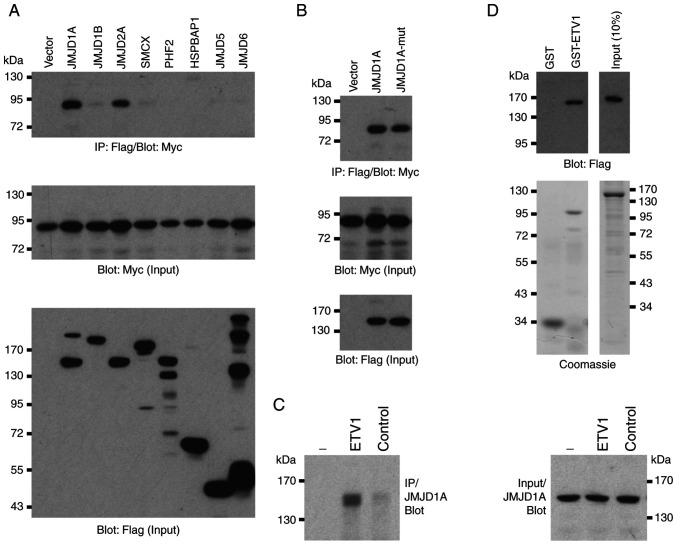
It has been previously reported that JMJD1A is overexpressed in colorectal tumors (39,40). Since it has been found that ETV1 is upregulated in colorectal cancer and as ETV1 has been shown to physically bind to JMJD2A (24), the present study wished to determine whether JMJD1A may also form complexes with ETV1. Hence, the present study investigated the potential interaction between these two proteins in co-immunoprecipitation assays. Indeed, overexpressed ETV1 robustly co-immunoprecipitated with JMJD1A to a similar degree as with JMJD2A (Fig. 3A), but not with several other JMJD proteins (JMJD1B, SMCX, PHF2, HSPBAP1, JMJD5 and JMJD6). This complex formation of JMJD1A with ETV1 was independent of catalytic activity, as the H1120A/D1122G catalytic mutant of JMJD1A was indistinguishable from the wild-type in its ability to co-immunoprecipitate with ETV1 (Fig. 3B). Furthermore, endogenous JMJD1A co-immunopre-cipitated with endogenous ETV1 in HCT116 cells (Fig. 3C). In addition, recombinant JMJD1A purified from baculovirus bound to bacterially expressed, purified GST-ETV1, but not to GST (Fig. 3D). Taken together, these data demonstrate that ETV1 and JMJD1A can directly bind to each other in vitro and in vivo.
Figure 3.
Interaction of JMJD1A with ETV1. (A) Flag-tagged JMJD proteins were co-expressed with Myc-tagged ETV1. Shown are immunoprecipitations (IP) with anti-Flag antibody followed by western blot analysis with anti-Myc antibody (top panel), as well as input levels of ETV1 (middle panel) and JMJD proteins (bottom panel). (B) Comparable degree of co-immunoprecipitation of ETV1 with wild-type JMJD1A and its H1120A/D1122G catalytic mutant (mut). (C) Endogenous coimmunoprecipitation in HCT116 cells. No, anti-ETV1 or a control antibody was utilized for immunoprecipitation. Right panel shows equal input levels of JMJD1A. (D) Interaction of purified proteins was tested by GST pull-down assays (top panel). Purity of recombinant proteins (GST, GST-ETV1 and Flag-His-JMJD1A) was assessed by Coomassie staining (bottom panel). JMJD, Jumonji C domain-containing protein.
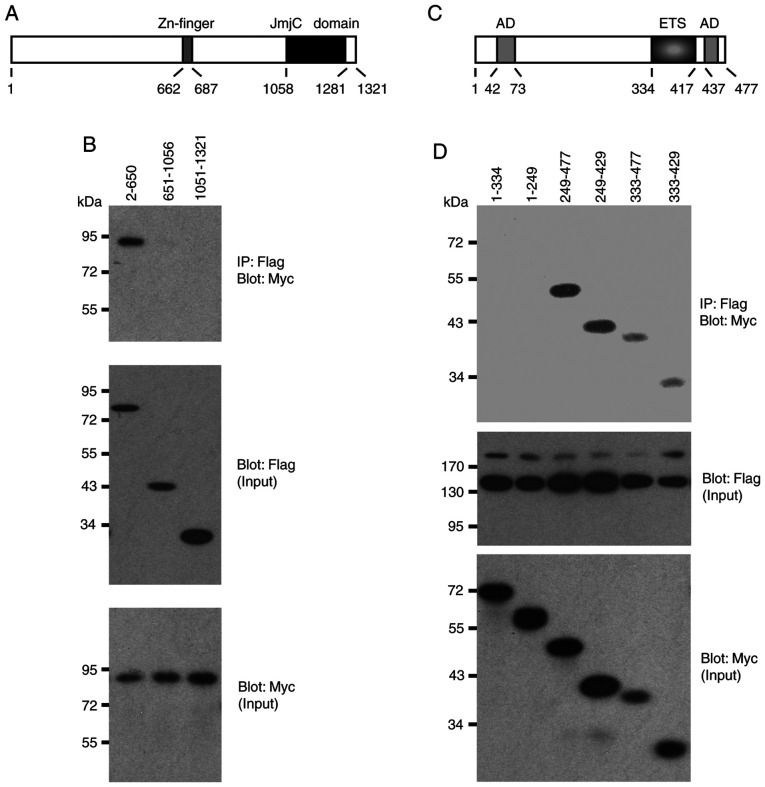
Domain mapping experiments
Two structural domains were identified in JMJD1A, a central Zn-finger and the C-terminal Jumonji C domain that is its catalytic center (Fig. 4A). The present study then examined whether any of these domains are involved in the interaction with ETV1 by utilizing truncation mutants of JMJD1A. ETV1 co-immunoprecipitated with the N-terminal 650 JMJD1A amino acids, whereas the 651-1056 and 1051-1321 truncations, which encompass the Zn-finger or Jumonji C domain, did not form complexes with ETV1 (Fig. 4B). Likewise, the Jumonji C domain of JMJD2A was reportedly not required for binding to ETV1 (24), which may be the reason why ETV1 did not promiscuously interact with all of the Jumonji C domain-containing proteins tested in Fig. 3A.
Figure 4.
Domain mapping experiments. (A) Scheme of the human JMJD1A protein. JmjC, Jumonji C domain that is the catalytic center. (B) Indicated Flag-tagged JMJD1A amino acids were co-expressed with 6Myc-tagged ETV1 and coimmunoprecipitation experiments performed. Top panel shows the immunoprecipitates, while the bottom two panels show input levels of Flag- and Myc-tagged proteins. (C) Scheme of human ETV1. AD, acidic domain that is part of an N-terminal or C-terminal transactivation domain. ETS, DNA-binding domain. (D) Similar as in panel (B) co-immunoprecipitation experiments with indicated 6Myc-tagged ETV1 amino acids and Flag-JMJD1A. JMJD, Jumonji C domain-containing protein.
Conversely, the present study assessed which domains in ETV1 mediate its interaction with JMJD1A. Neither amino acids 1-334 nor 1-249, which encompass a strong N-terminal transactivation domain of ETV1 (3), co-immunoprecipitated with JMJD1A (Fig. 4C and D). In contrast, all other truncations tested formed complexes with JMJD1A. The smallest fragment that still interacted with JMJD1A encompassed ETV1 amino acids 333-429, suggesting that the ETS DNA-binding domain of ETV1 mediates the interaction with JMJD1A. Similarly, the ETS domain was reportedly crucial for binding of ETV1 to JMJD2A (24), suggesting that the DNA-binding ETS domain of ETV1 is responsible for recruitment of either JMJD1A or JMJD2A. This also suggests that the highly homologous ETV4 and ETV5 proteins are likely to bind to JMJD1A and JMJD2A through their ETS domains.
Cooperation between ETV1 and JMJD histone demethylases
To assess whether ETV1 and JMJD1A (or JMJD2A) function-ally cooperate in gene transcription in colon cancer cells, the MMP1 gene promoter that is a bona fide target of ETV1 (12) was examined. When an MMP1 luciferase reporter gene was transfected into HCT116 cells, expectedly, a significant stimulation of luciferase activity was observed upon ETV1 co-transfection (Fig. 5). Notably, JMJD1A, but not JMJD2A co-transfection alone also significantly increased MMP1 luciferase activity; this different behavior may be attributable to the fact that endogenous levels of ETV1 were only sufficient in the case of JMJD1A to detect transactivation. It should be noted that JMJD1A and JMJD2A were over-expressed at comparable levels (Fig. S2A). When JMJD1A or JMJD2A was co-expressed with ETV1, a cooperative transcriptional activation was observed (Fig. 5). As previously reported in prostate cells (24), the JMJD2A-H188A catalytic mutant did not cooperate with ETV1, and neither did the JMJD1A-H1120A/D1122G catalytic mutant (Fig. 5), indicating that the catalytic activity of JMJD1A is required for the coactivation of ETV1. Similarly, JMJD1A was capable of cooperating with ETV1 in LNCaP prostate cancer cells, although in this case, JMJD2A was much more potent (Fig. S2B). Taken together, these data reveal the novel finding (to the best of our knowledge) that enzymatically active JMJD1A can serve as a co-activator of ETV1.
Figure 5.
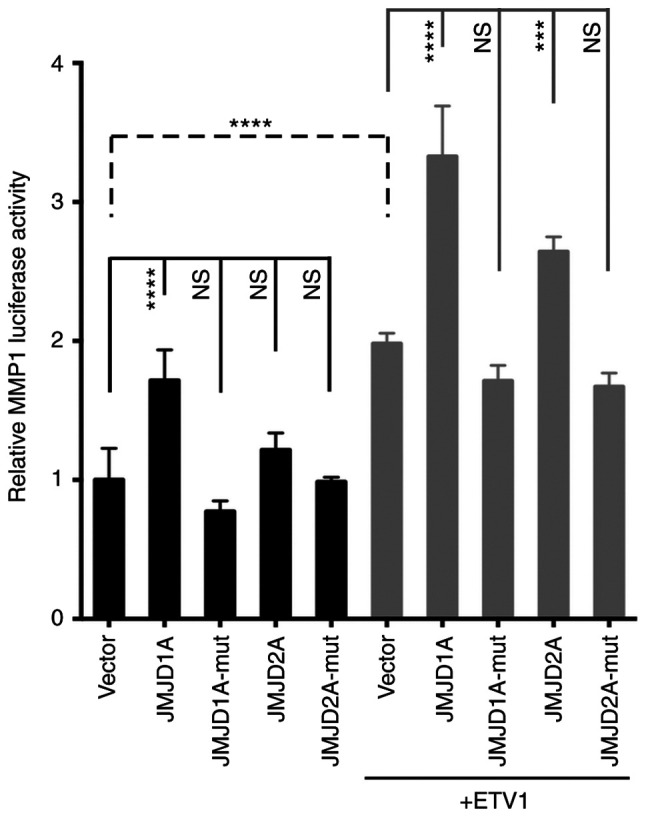
Activation of the MMP1 gene promoter by JMJD1A. Indicated expression vectors were co-transfected with an MMP1 luciferase reporter gene into HCT116 cells. Catalytically inactive JMJD proteins are indicated by the suffix 'mut'. ANOVA (Tukey's multiple comparisons test; n=4). NS, not significant; ***P<0.001; ****P<0.0001. JMJD, Jumonji C domain-containing protein; MMP1, matrix metalloproteinase 1.
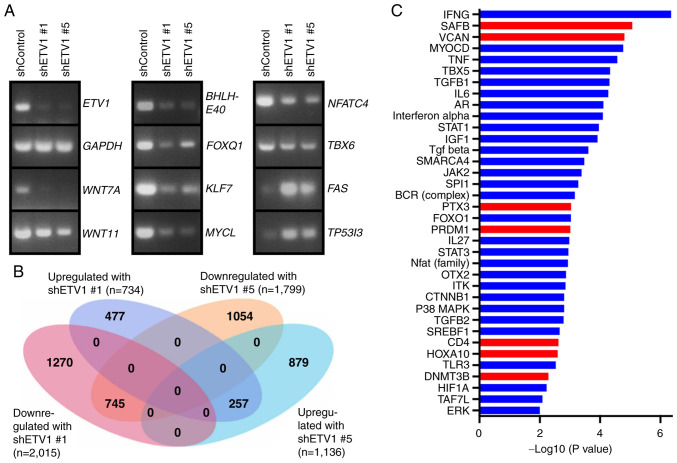
Regulation of the transcriptome by ETV1
To gain deeper insight into the mechanisms through which ETV1 may affect colon cancer cells, ETV1 expression was downregulated with shRNA in HCT116 cells and RNA-sequencing was performed (Fig. 6A and B). A total of 745 genes were downregulated upon the expression of either of the two ETV1 shRNAs used, and 257 genes were upregulated, indicating that ETV1 regulates approximately a thousand genes in HCT116 cells. Amongst the downregulated genes, RT-qPCR confirmed the reduced expression of signaling molecules (WNT7A and WNT11) and transcription factors (BHLHE40, FOXQ1, KLF7, MYCL, NFATC4 and TBX6), whereas two examples of validated upregulated genes were FAS and TP53I3 (Fig. 6A). WNT7A is a secreted morphogen that is overexpressed in colorectal cancer and may activate cancer-associated fibroblasts, which enhances tumor aggressiveness (82,83). Likewise, WNT11 is overexpressed in colon cancer and may promote colorectal cancer cell growth and invasion (84,85). Hence, the upregulation of WNT7A and WNT11 may be involved in the mechanisms through which ETV1 overexpression contributes to colon tumorigenesis.
Figure 6.
RNA-sequencing analyses. (A) RT-qPCR analyses showing mRNA levels of indicated genes in HCT116 cells expressing control shRNA, shRNA ETV1 #1 or shRNA ETV1 #5. (B) Venn diagram depicting genes that were >2-fold up- or downregulated in HCT116 cells upon expression of the two different ETV1 shRNAs compared to control shRNA. (C) Ingenuity Pathway Analysis for upstream regulator pathways. Blue color indicates inhibition and red color activation in the presence of ETV1 shRNAs. Inclusion criteria were the following: i) Absolute value of the activation z-score was larger than 1.5, and (ii) P-value of overlap <0.01.
This may also pertain to the upregulation of FOXQ1, as the encoded transcription factor is overexpressed in colorectal cancer and promotes tumor initiation and metastasis (86-89). By contrast, the authors were unable to find substantial evidence in the literature that BHLHE40, KLF7, MYCL, NFATC4 or TBX6 promote colon cancer. However, the downregulation of the FAS cell surface death receptor has been reported in colon tumors and seems to be associated with a reduced survival, likely due to the fact that FAS downregulation facilitates escape from immune surveillance and thus, also resistance to immune checkpoint inhibitors (90,91). Accordingly, the suppression of FAS expression by ETV1 could contribute to colon cancer aggressiveness. By contrast, the suppression of the expression of the oxidoreduc-tase TP53I3 by ETV1 cannot account for the pro-growth role of ETV1 in HCT116 cells, since TP53I3 is also a pro-growth factor (92). This indicates that results from RNA-sequencing, although they can provide some insight into how ETV1 facilitates its oncogenic effects, should be treated with caution and identifying seminal downstream effectors of ETV1 requires experimental validation (see below).
The present study also applied Ingenuity Pathway Analysis to better comprehend ETV1 function. This revealed multiple upstream regulatory pathways that were affected by ETV1 downregulation (Fig. 6C). Amongst the downregulated path-ways, there were interleukin (IL)-6, signal transducer and activator of transcription (STAT)3 and β-catenin (CTNNB1). Given that IL6/STAT3 as well as β-catenin are potent oncogenic agents in colorectal cancer (93,94), this outlines how ETV1 overexpression may facilitate tumorigenesis by stimulating the IL6/STAT3 and β-catenin pathways.
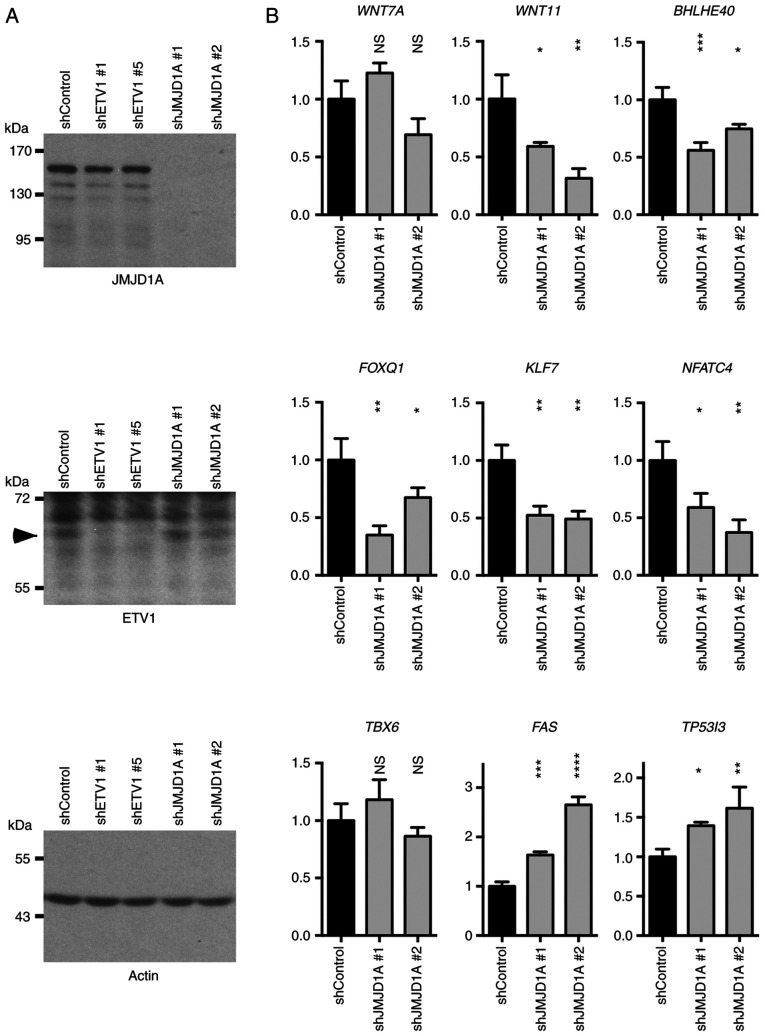
Subsequently, the authors wished to determine whether JMJD1A may also regulate the newly identified ETV1 target genes. To this end, two different JMJD1A shRNAs were transfected into HCT116 cells. This robustly ablated JMJD1A expression, but did not alter the ETV1 levels, and neither did ETV1 downregulation affect JMJD1A levels (Fig. 7A). The mRNA levels of WNT7A, WNT11, BHLHE40, FOXQ1, KLF7, NFATC4, TBX6, FAS and TP53I3 were then measured (Fig. 7B). As observed with ETV1 downregulation, JMJD1A shRNA led to a decrease in the WNT11, BHLHE40, FOXQ1, KLF7 and NFATC4 mRNA levels, while the FAS and TP53I3 levels were increased. However, in contrast to ETV1, WNT7A and TBX6 mRNA levels were not significantly affected by JMJD1A shRNAs. Collectively, these results suggest that JMJD1A cooperates with ETV1 in the regulation of several, but not all of ETV1 target genes.
Figure 7.
Co-regulation of ETV1 target genes by JMJD1A in HCT116 cells. (A) Downregulation of JMJD1A or ETV1 with shRNA; arrowhead indicates ETV1. Shown are indicated western blots. (B) Gene expression upon JMJD1A downregulation was determined by RT-qPCR. All mRNA levels were normalized to those of GAPDH. One-way ANOVA with post hoc Dunnett's multiple comparisons test; n=3; NS, not significant; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001. JMJD, Jumonji C domain-containing protein.
FOXQ1 as a downstream effector of ETV1 and JMJD1A
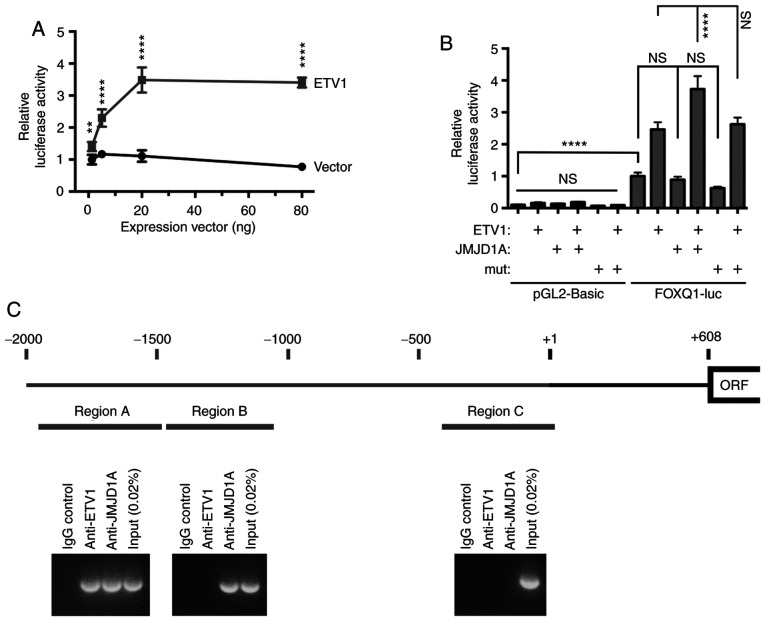
Out of the validated ETV1-regulated genes, FOXQ1 was selected for further analysis. The reasons were that the pro-oncogenic association of FOXQ1 with colorectal cancer has been established (87,89) and it could be corroborated that FOXQ1 was upregulated in colorectal tumors (Fig. S3A-C). In addition, FOXQ1 mRNA levels were regulated by JMJD1A (Fig. 7B), but not by JMJD2A (Fig. S4), suggesting that ETV1 and JMJD1A cooperate in the transcriptional control of FOXQ1.
Thus, the human FOXQ1 promoter from -2,000 to +124 was cloned and it was demonstrated that it was dose-dependently inducible by ETV1 (Fig. 8A). Accordingly, multiple potential ETV1 binding sites were identified within these 2.1 kbp of genomic DNA (Fig. S5). Subsequently, JMJD1A was expressed and it was observed that JMJD1A, but not the H1120A/D1122G catalytic mutant, was capable of inducing the FOXQ1 gene promoter only in the presence of ectopic ETV1 (Fig. 8B); as a control, the parental luciferase construct pGL2-Basic was not affected by ETV1 or JMJD1A. In addition, as determined by chromatin immunoprecipitation experiments, both ETV1 and JMJD1A bound to the FOXQ1 promoter within region A (Fig. 8C), but not within region C. Of note, JMJD1A, but not ETV1, also bound to region B (Fig. 8C), suggesting that a transcription factor(s) other than ETV1 likewise recruits JMJD1A to the FOXQ1 gene promoter. Taken together, these data strongly support the notion that FOXQ1 is a valid target of the ETV1/JMJD1A complex.
Figure 8.
Analysis of the FOXQ1 gene promoter. (A) ETV1-mediated activation of a luciferase gene controlled by the FOXQ1 gene promoter. Indicated amounts of empty vector pEV3S or ETV1 expression vector were used for transient transfection of HCT116 cells. Two-way ANOVA with post hoc Sidak's multiple comparisons test; n=6; **P<0.01; ****P<0.0001. (B) Comparison of control pGL2-Basic to the FOXQ1 luciferase reporter; 'mut' signifies the JMJD1A-H1120A/D1122G catalytic mutant. One-way ANOVA with post hoc Tukey's multiple comparisons test; n=4; ****P<0.0001; NS, not significant. (C) Chromatin immunoprecipitation experiments assessing binding of ETV1 and JMJD1A to three different upstream regions of the human FOXQ1 promoter. Shown are agarose gels with amplified promoter regions. The transcription start site is marked by +1, and the open reading frame (ORF) begins at +608.
Finally, the present study wished to determine whether FOXQ1 is seminal for ETV1 function in HCT116 cells. To this end, ETV1 was downregulated and FOXQ1 was overexpressed simultaneously. As also shown above (Fig. 1), ETV1 downregulation suppressed HCT116 cell growth and clonogenic activity (Fig. 9). The overexpression of FOXQ1 had no effect in the presence of control shRNA, suggesting that endogenous levels of FOXQ1 were already sufficient for a maximal growth-stimulatory effect of this transcription factor in the presence of normal ETV1 levels. However, FOXQ1 overexpression partially reversed the growth-suppressive effect of ETV1 shRNA on HCT116 cells, although not the clonogenic activity. These data demonstrate that FOXQ1 is indeed one of the crucial target genes of ETV1.
The present study also attempted to rescue ETV1 ablation by TBX6 overexpression, since our interest was piqued by the fact that we did not find any literature convincingly linking the TBX6 transcription factor to cancer at all. This was despite conflicting bioinformatics results demonstrating either the up- or downregulation of TBX6 mRNA in colorectal cancer (Fig. S3D-F) and JMJD1A as well as JMJD2A not regulating TBX6 gene transcription (Figs. 7B and S4). Notably, TBX6 overexpression significantly impaired HCT116 cell growth and clonogenic activity in the presence of control shRNA, but had no effect in the presence of ETV1 shRNA (Fig. 9). This indicates that TBX6 transcriptional upregulation does not contribute to the pro-growth activity of ETV1, but may instead suppress it. In addition, to the best of our knowledge, these data suggest for the first time that TBX6 may perform a tumor-suppressive activity in colorectal cancer cells.
Discussion
The present study provided evidence that the ETS transcription factor, ETV1, may contribute to the development of colorectal cancer. This is primarily based on ETV1 being overexpressed in respective tumors, its expression levels being associated with the aggressiveness of the disease, and being required for the efficient growth and clonogenic activity of HCT116 colorectal cancer cells. Moreover, it was discovered that ETV1 binds directly to the JMJD1A histone demethylase that appears to coregulate many ETV1 target genes. Given that JMJD1A itself is overexpressed in colorectal cancer and has been implicated as a respective tumor promoter (39-42,95), this interaction may have lethal consequences. Furthermore, the relevance of the ETV1-JMJD1A complex may extend to other malignancies, including prostate cancer, where ETV1 is known to be a driver of tumorigenesis (21-23,96) and some in vitro evidence points to a similar role for JMJD1A (97-100). Consistently, JMJD1A is upregulated in prostate tumors and even more so at metastatic sites (Fig. S6).
The depletion of ETV1 in HCT116 cells resulted in the inhibition of multiple regulatory pathways (Fig. 6C), including IL6/STAT3 and β-catenin signaling that facilitate colon cancer (93,94). Accordingly, the stimulation of these pathways upon ETV1 overexpression could contribute to tumorigenesis. Seemingly inconsistent with the oncogenic role of ETV1 in colon cancer, interferon-γ (IFNG) signaling (Fig. 6C), whose tumor suppressive role has long been recognized (101), was inhibited upon ETV1 downregulation. However, IFNG can also contribute to the progression of cancer, in particular by promoting the immunoevasion of tumor cells, which may occur through the upregulation of e.g., PD-L1 or the increased secretion of immunosuppressive molecules that is associated with a reduced survival (102). Another pathway diminished by ETV1 downregulation was TGF-β signaling (Fig. 6C). TGF-β normally suppresses the growth of epithelial cells, but it can also promote epithe-lial-to-mesenchymal transition and thereby metastasis (103). Hence, it is plausible that the stimulation of TGF-β signaling by ETV1 may particularly be relevant during the progression from localized to disseminated tumors. Conversely, from the RNA-sequencing data in the present study, it was predicted that ETV1 causes the inhibition of scaffold attachment factor B (SAFB) signaling (Fig. 6C). SAFB is downregulated in colorectal tumors, being associated with a reduced survival, and its overexpression leads to the inhibition of metastasis (104), outlining another mechanism through which ETV1 could potentially drive colon cancer. However, whether the aforementioned pathways are indeed crucial for the role of ETV1 in colorectal cancer needs to be validated in the future.
FOXQ1 is overexpressed in colorectal tumors (86-88,105) and this transcription factor can promote in vitro migration, invasion, angiogenesis and proliferation, as well as the xenograft tumor growth of colorectal cancer cells (87-89). The present study discovered FOXQ1 was a target gene of ETV1 and JMJD1A. Notably, FOXQ1 overexpression partially reversed the growth suppressive effect of ETV1 shRNA on HCT116 cells, strongly arguing that FOXQ1 is a seminal downstream effector of ETV1 in colorectal and possible other cancers.
However, albeit FOXQ1 is in general considered as an oncoprotein, a recent study demonstrated that FOXQ1 can also act as a tumor suppressor in melanomas (106). It is possible that the same holds true for prostate cancer, as it was found that FOXQ1 was downregulated in prostate tumors, and this down-regulation was even accentuated upon metastasis (Fig. S7). Accordingly, FOXQ1 may not always be a crucial downstream effector of ETV1, and it remains to be determined whether and why ETV1 would potentially be ineffectual to induce FOXQ1 transcription in prostate tumors.
FOXQ1 overexpression only partially reversed ETV1 ablation, indicating that other crucial ETV1 downstream effectors await discovery. For instance, one of these could be the transcription factor, MYCL, as MYCL transcription was reduced upon ETV1 shRNA expression. The MYCL gene has been found amplified and/or overexpressed in small cell lung cancer, ovarian carcinomas and prostate cancer (107-109). In mice in which both the TP53 and RB1 tumor suppressor genes were deleted, the overexpression of MYCL promoted, while the knockout of MYCL suppressed small cell lung cancer formation, indicating an oncogenic activity for MYCL (110,111). This was further substantiated by the fact that transgenic mice over-expressing MYCL in T cells developed lymphoid tumors (112). Consistent with being another downstream effector of ETV1, MYCL was found overexpressed in both colon and prostate cancer (Fig. S8).
It was also unraveled that TBX6 transcription was stimulated by ETV1, but not by JMJD1A. The T-box transcription factor TBX6 is essential for embryonic development (113), yet its role in cancer has, to our knowledge, not yet been investigated. TBX6 overexpression reduced HCT116 cell growth and clonogenicity, suggesting that TBX6 exerts a tumor suppressive function, an important finding in its own right. Accordingly, TBX6 upregulation may dampen the oncogenic activity of ETV1. Since it is conceivable that TBX6 gene activity is modulated by transcription factors other than ETV1, TBX6 downregulation may occur in a number of instances in colorectal cancer and promote tumorigenesis. Similar to numerous cancer-critical transcription factors, TBX6 has been shown to be associated with congenital developmental diseases, including Müllerian aplasia, scoliosis, anomalies of the kidney and urinary tract, and cervical vertebral malformations (114-117). One possible mechanism through which TBX6 could affect development is through TBX6-mediated repression of the stem cell factor SOX2 (118,119). The same mechanism may apply for TBX6-mediated tumor suppression; consistently, the elevated expression of SOX2 is associated with increased metastasis, recurrence and lethality in colorectal cancer patients (120-122).
In conclusion, the data of the present study revealed plausible mechanisms through which an ETV1-JMJD1A protein complex could facilitate tumorigenesis in the colon, including by upregulating the FOXQ1 gene. This knowledge suggests that interfering with ETV1, JMJD1A and/or FOXQ1 function may be a valid strategy with which to combat metastatic colorectal cancer.
Supplementary Data
Acknowledgments
Not applicable.
Funding
The present study was in part funded by grants R01 CA154745, R03 CA223615 and R01 CA233613 from the National Cancer Institute of the USA (to RJ) and a team science seed grant from the Stephenson Cancer Center (to RJ and WMF). In addition, RJ was supported in part by the Oklahoma Tobacco Settlement Endowment Trust through an award made to the University of Oklahoma/Stephenson Cancer Center. Additional assistance was provided by the Stephenson Cancer Center Molecular Biology Core that was supported by NIH grants P30 CA225520 and P20 GM103639. The funding sources had no involvement in the study design, data collection, analysis, interpretation and the writing of this manuscript. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the granting agencies.
Availability of data and materials
Original data and material will be made available upon reasonable request. RNA-sequencing data have been deposited in Gene Expression Omnibus (GSE158294).
Authors' contributions
SO, HS and RJ designed and performed the experiments. SO, HS, WMF, SS and RJ analyzed and interpreted the data. RJ supervised the study and wrote the manuscript with input from all other authors. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Brown TA, McKnight SL. Specificities of protein-protein and protein-DNA interaction of GABP alpha and two newly defined ets-related proteins. Genes Dev. 1992;6:2502–2512. doi: 10.1101/gad.6.12b.2502. [DOI] [PubMed] [Google Scholar]
- 2.Monte D, Coutte L, Baert JL, Angeli I, Stehelin D, de Launoit Y. Molecular characterization of the ets-related human transcription factor ER81. Oncogene. 1995;11:771–779. [PubMed] [Google Scholar]
- 3.Janknecht R. Analysis of the ERK-stimulated ETS transcription factor ER81. Mol Cell Biol. 1996;16:1550–1556. doi: 10.1128/MCB.16.4.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei GH, Badis G, Berger MF, Kivioja T, Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29:2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh S, Shin S, Janknecht R. ETV1, 4 and 5: An oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta. 2012;1826:1–12. doi: 10.1016/j.bbcan.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arber S, Ladle DR, Lin JH, Frank E, Jessell TM. ETS gene Er81 controls the formation of functional connections between group Ia sensory afferents and motor neurons. Cell. 2000;101:485–498. doi: 10.1016/S0092-8674(00)80859-4. [DOI] [PubMed] [Google Scholar]
- 8.Kucera J, Cooney W, Que A, Szeder V, Stancz-Szeder H, Walro J. Formation of supernumerary muscle spindles at the expense of Golgi tendon organs in ER81-deficient mice. Dev Dyn. 2002;223:389–401. doi: 10.1002/dvdy.10066. [DOI] [PubMed] [Google Scholar]
- 9.Sedy J, Tseng S, Walro JM, Grim M, Kucera J. ETS transcription factor ER81 is required for the pacinian corpuscle development. Dev Dyn. 2006;235:1081–1089. doi: 10.1002/dvdy.20710. [DOI] [PubMed] [Google Scholar]
- 10.Shekhar A, Lin X, Liu FY, Zhang J, Mo H, Bastarache L, Denny JC, Cox NJ, Delmar M, Roden DM, et al. Transcription factor ETV1 is essential for rapid conduction in the heart. J Clin Invest. 2016;126:4444–4459. doi: 10.1172/JCI87968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papoutsopoulou S, Janknecht R. Phosphorylation of ETS transcription factor ER81 in a complex with its coactivators CREB-binding protein and p300. Mol Cell Biol. 2000;20:7300–7310. doi: 10.1128/mcb.20.19.7300-7310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosc DG, Goueli BS, Janknecht R. HER2/Neu-mediated activation of the ETS transcription factor ER81 and its target gene MMP-1. Oncogene. 2001;20:6215–6224. doi: 10.1038/sj.onc.1204820. [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Janknecht R. Regulation of the ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1 and protein kinase A. J Biol Chem. 2002;277:42669–42679. doi: 10.1074/jbc.M205501200. [DOI] [PubMed] [Google Scholar]
- 14.Xie L, Gazin C, Park SM, Zhu LJ, Debily MA, Kittler EL, Zapp ML, Lapointe D, Gobeil S, Virbasius CM, Green MR. A synthetic interaction screen identifies factors selectively required for proliferation and TERT transcription in p53-deficient human cancer cells. PLoS Genet. 2012;8:e1003151. doi: 10.1371/journal.pgen.1003151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janknecht R. Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1) Oncogene. 2003;22:746–755. doi: 10.1038/sj.onc.1206185. [DOI] [PubMed] [Google Scholar]
- 16.Goel A, Janknecht R. Acetylation-mediated transcriptional activation of the ETS protein ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 2003;23:6243–6254. doi: 10.1128/MCB.23.17.6243-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baert JL, Monte D, Verreman K, Degerny C, Coutte L, de Launoit Y. The E3 ubiquitin ligase complex component COP1 regulates PEA3 group member stability and transcriptional activity. Oncogene. 2010;29:1810–1820. doi: 10.1038/onc.2009.471. [DOI] [PubMed] [Google Scholar]
- 18.Vitari AC, Leong KG, Newton K, Yee C, O'Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 19.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 20.Attard G, Clark J, Ambroisine L, Mills IG, Fisher G, Flohr P, Reid A, Edwards S, Kovacs G, Berney D, et al. Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer. 2008;99:314–320. doi: 10.1038/sj.bjc.6604472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin S, Kim TD, Jin F, van Deursen JM, Dehm SM, Tindall DJ, Grande JP, Munz JM, Vasmatzis G, Janknecht R. Induction of prostatic intraepithelial neoplasia and modulation of androgen receptor by ETS variant 1/ETS-related protein 81. Cancer Res. 2009;69:8102–8110. doi: 10.1158/0008-5472.CAN-09-0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baena E, Shao Z, Linn DE, Glass K, Hamblen MJ, Fujiwara Y, Kim J, Nguyen M, Zhang X, Godinho FJ, et al. ETV1 directs androgen metabolism and confers aggressive prostate cancer in targeted mice and patients. Genes Dev. 2013;27:683–698. doi: 10.1101/gad.211011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomlins SA, Laxman B, Dhanasekaran SM, Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–599. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 24.Kim TD, Jin F, Shin S, Oh S, Lightfoot SA, Grande JP, Johnson AJ, van Deursen JM, Wren JD, Janknecht R. Histone demethylase JMJD2A drives prostate tumorigenesis through transcription factor ETV1. J Clin Invest. 2016;126:706–720. doi: 10.1172/JCI78132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 26.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: Epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh S, Shin S, Janknecht R. The small members of the JMJD protein family: Enzymatic jewels or jinxes? Biochim Biophys Acta Rev Cancer. 2019;1871:406–418. doi: 10.1016/j.bbcan.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 29.Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 30.Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun. 2007;353:973–977. doi: 10.1016/j.bbrc.2006.12.147. [DOI] [PubMed] [Google Scholar]
- 31.Kim TD, Oh S, Lightfoot SA, Shin S, Wren JD, Janknecht R. Upregulation of PSMD10 caused by the JMJD2A histone demethylase. Int J Clin Exp Med. 2016;9:10123–10134. [PMC free article] [PubMed] [Google Scholar]
- 32.Kim TD, Fuchs JR, Schwartz E, Abdelhamid D, Etter J, Berry WL, Li C, Ihnat MA, Li PK, Janknecht R. Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon cancer cells and identification of curcuminoids as JMJD2 inhibitors. Am J Transl Res. 2014;6:236–247. [PMC free article] [PubMed] [Google Scholar]
- 33.Kim TD, Shin S, Berry WL, Oh S, Janknecht R. The JMJD2A demethylase regulates apoptosis and proliferation in colon cancer cells. J Cell Biochem. 2012;113:1368–1376. doi: 10.1002/jcb.24009. [DOI] [PubMed] [Google Scholar]
- 34.Lee HJ, Kim BK, Yoon KB, Kim YC, Han SY. Novel inhibitors of lysine (K)-specific demethylase 4A with anticancer activity. Invest New Drugs. 2017;35:733–741. doi: 10.1007/s10637-017-0496-2. [DOI] [PubMed] [Google Scholar]
- 35.Crawford HC, Fingleton B, Gustavson MD, Kurpios N, Wagenaar RA, Hassell JA, Matrisian LM. The PEA3 subfamily of Ets transcription factors synergizes with beta-catenin-LEF-1 to activate matrilysin transcription in intestinal tumors. Mol Cell Biol. 2001;21:1370–1383. doi: 10.1128/MCB.21.4.1370-1383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horiuchi S, Yamamoto H, Min Y, Adachi Y, Itoh F, Imai K. Association of ets-related transcriptional factor E1AF expression with tumour progression and overexpression of MMP-1 and matrilysin in human colorectal cancer. J Pathol. 2003;200:568–576. doi: 10.1002/path.1387. [DOI] [PubMed] [Google Scholar]
- 37.Yoo J, Jeon YH, Cho HY, Lee SW, Kim GW, Lee DH, Kwon SH. Advances in histone demethylase KDM3A as a cancer therapeutic target. Cancers (Basel) 2020;12:1098. doi: 10.3390/cancers12051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sui Y, Gu R, Janknecht R. Crucial functions of the JMJD1/KDM3 epigenetic regulators in cancer. Mol Cancer Res. 2020 Jun 30; doi: 10.1158/1541-7786.MCR-20-0404. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uemura M, Yamamoto H, Takemasa I, Mimori K, Hemmi H, Mizushima T, Ikeda M, Sekimoto M, Matsuura N, Doki Y, Mori M. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: In vivo identification from hypoxic tumor cells. Clin Cancer Res. 2010;16:4636–4646. doi: 10.1158/1078-0432.CCR-10-0407. [DOI] [PubMed] [Google Scholar]
- 40.Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork K, Ramadoss S, Ding X, Li X, Wang CY. KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/β-catenin signalling. Nat Commun. 2017;8:15146. doi: 10.1038/ncomms15146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng K, Su G, Ji J, Yang X, Miao M, Mo P, Li M, Xu J, Li W, Yu C. Histone demethylase JMJD1A promotes colorectal cancer growth and metastasis by enhancing Wnt/β-catenin signaling. J Biol Chem. 2018;293:10606–10619. doi: 10.1074/jbc.RA118.001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li X, Oh S, Song H, Shin S, Zhang B, Freeman WM, Janknecht R. A potential common role of the Jumonji C domain-containing 1A histone demethylase and chromatin remodeler ATRX in promoting colon cancer. Oncol Lett. 2018;16:6652–6662. doi: 10.3892/ol.2018.9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dowdy SC, Mariani A, Janknecht R. HER2/Neu- and TAK1-mediated up-regulation of the transforming growth factor beta inhibitor Smad7 via the ETS protein ER81. J Biol Chem. 2003;278:44377–44384. doi: 10.1074/jbc.M307202200. [DOI] [PubMed] [Google Scholar]
- 44.Mooney SM, Goel A, D'Assoro AB, Salisbury JL, Janknecht R. Pleiotropic effects of p300-mediated acetylation on p68 and p72 RNA helicase. J Biol Chem. 2010;285:30443–30452. doi: 10.1074/jbc.M110.143792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim TD, Shin S, Janknecht R. ETS transcription factor ERG cooperates with histone demethylase KDM4A. Oncol Rep. 2016;35:3679–3688. doi: 10.3892/or.2016.4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh S, Shin S, Lightfoot SA, Janknecht R. 14-3-3 proteins modulate the ETS transcription factor ETV1 in prostate cancer. Cancer Res. 2013;73:5110–5119. doi: 10.1158/0008-5472.CAN-13-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berry WL, Shin S, Lightfoot SA, Janknecht R. Oncogenic features of the JMJD2A histone demethylase in breast cancer. Int J Oncol. 2012;41:1701–1706. doi: 10.3892/ijo.2012.1618. [DOI] [PubMed] [Google Scholar]
- 48.Kim J, Shin S, Subramaniam M, Bruinsma E, Kim TD, Hawse JR, Spelsberg TC, Janknecht R. Histone demethylase JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem Biophys Res Commun. 2010;401:412–416. doi: 10.1016/j.bbrc.2010.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim TD, Oh S, Shin S, Janknecht R. Regulation of tumor suppressor p53 and HCT116 cell physiology by histone demethylase JMJD2D/KDM4D. PLoS One. 2012;7:e34618. doi: 10.1371/journal.pone.0034618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berry WL, Kim TD, Janknecht R. Stimulation of β-catenin and colon cancer cell growth by the KDM4B histone demethylase. Int J Oncol. 2014;44:1341–1348. doi: 10.3892/ijo.2014.2279. [DOI] [PubMed] [Google Scholar]
- 51.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matthias P, Muller MM, Schreiber E, Rusconi S, Schaffner W. Eukaryotic expression vectors for the analysis of mutant proteins. Nucleic Acids Res. 1989;17:6418. doi: 10.1093/nar/17.15.6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X, Moon G, Shin S, Zhang B, Janknecht R. Cooperation between ETS variant 2 and Jumonji domain-containing 2 histone demethylases. Mol Med Rep. 2018;17:5518–5527. doi: 10.3892/mmr.2018.8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rossow KL, Janknecht R. The Ewing's sarcoma gene product functions as a transcriptional activator. Cancer Res. 2001;61:2690–2695. [PubMed] [Google Scholar]
- 55.Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- 56.Knebel J, De Haro L, Janknecht R. Repression of transcription by TSGA/Jmjd1a, a novel interaction partner of the ETS protein ER71. J Cell Biochem. 2006;99:319–329. doi: 10.1002/jcb.20945. [DOI] [PubMed] [Google Scholar]
- 57.Goel A, Janknecht R. Concerted activation of ETS protein ER81 by p160 coactivators, the acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J Biol Chem. 2004;279:14909–14916. doi: 10.1074/jbc.M400036200. [DOI] [PubMed] [Google Scholar]
- 58.Janknecht R, Hunter T. Activation of the Sap-1a transcription factor by the c-Jun N-terminal kinase (JNK) mitogen-activated protein kinase. J Biol Chem. 1997;272:4219–4224. doi: 10.1074/jbc.272.7.4219. [DOI] [PubMed] [Google Scholar]
- 59.Yang YL, Wang PW, Wang FS, Lin HY, Huang YH. miR-29a modulates GSK3β/SIRT1-linked mitochondrial proteostatic stress to ameliorate mouse non-alcoholic steatohepatitis. Int J Mol Sci. 2020;21:E6884. doi: 10.3390/ijms21186884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin X, Feng D, Li P, Lv Y. LncRNA LINC00857 regulates the progression and glycolysis in ovarian cancer by modulating the Hippo signaling pathway. Cancer Med. 2020 Sep 12; doi: 10.1002/cam4.3322. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen X, Liu X, Gao Y, Lin J, Liu X, Pang X, Lin J, Chen L. Application of firefly luciferase (Luc) as a reporter gene for the chemoautotrophic and acidophilic Acidithiobacillus spp. Curr Microbiol. 2020;77:3724–3730. doi: 10.1007/s00284-020-02195-w. [DOI] [PubMed] [Google Scholar]
- 62.Ou Y, Liao C, Li H, Yu G. LncRNA SOX2OT/Smad3 feed-back loop promotes myocardial fibrosis in heart failure. IUBMB Life. 2020 Sep 21; doi: 10.1002/iub.2375. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 63.Sims RJ, III, Liss AS, Gottlieb PD. Normalization of luciferase reporter assays under conditions that alter internal controls. Biotechniques. 2003;34:938–940. doi: 10.2144/03345bm07. [DOI] [PubMed] [Google Scholar]
- 64.Shifera AS, Hardin JA. Factors modulating expression of Renilla luciferase from control plasmids used in luciferase reporter gene assays. Anal Biochem. 2010;396:167–172. doi: 10.1016/j.ab.2009.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu GQ, Wang X, Zhou HY, Chai KQ, Xue Q, Zheng AH, Zhu XM, Xiao JY, Ying XH, Wang FW, et al. Evidence for transcriptional interference in a dual-luciferase reporter system. Sci Rep. 2015;5:17675. doi: 10.1038/srep17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Repele A, Manu Robust normalization of luciferase reporter data. Methods Protoc. 2019;2:62. doi: 10.3390/mps2030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shin S, Oh S, An S, Janknecht R. ETS variant 1 regulates matrix metalloproteinase-7 transcription in LNCaP prostate cancer cells. Oncol Rep. 2013;29:306–314. doi: 10.3892/or.2012.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shin S, Bosc DG, Ingle JN, Spelsberg TC, Janknecht R. Rcl is a novel ETV1/ER81 target gene upregulated in breast tumors. J Cell Biochem. 2008;105:866–874. doi: 10.1002/jcb.21884. [DOI] [PubMed] [Google Scholar]
- 69.Oh S, Shin S, Song H, Grande JP, Janknecht R. Relationship between ETS transcription factor ETV1 and TGF-β-regulated SMAD proteins in prostate cancer. Sci Rep. 2019;9:8186. doi: 10.1038/s41598-019-44685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oh S, Janknecht R. Histone demethylase JMJD5 is essential for embryonic development. Biochem Biophys Res Commun. 2012;420:61–65. doi: 10.1016/j.bbrc.2012.02.115. [DOI] [PubMed] [Google Scholar]
- 71.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 72.Sui Y, Li X, Oh S, Zhang B, Freeman WM, Shin S, Janknecht R. Opposite roles of the JMJD1A interaction partners MDFI and MDFIC in colorectal cancer. Sci Rep. 2020;10:8710. doi: 10.1038/s41598-020-65536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goueli BS, Janknecht R. Upregulation of the catalytic telomerase subunit by the transcription factor ER81 and oncogenic HER2/Neu, Ras, or Raf. Mol Cell Biol. 2004;24:25–35. doi: 10.1128/MCB.24.1.25-35.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goueli BS, Janknecht R. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene. 2003;22:8042–8047. doi: 10.1038/sj.onc.1206847. [DOI] [PubMed] [Google Scholar]
- 75.DiTacchio L, Bowles J, Shin S, Lim DS, Koopman P, Janknecht R. Transcription factors ER71/ETV2 and SOX9 participate in a positive feedback loop in fetal and adult mouse testis. J Biol Chem. 2012;287:23657–23666. doi: 10.1074/jbc.M111.320101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orlando G, Law PJ, Cornish AJ, Dobbins SE, Chubb D, Broderick P, Litchfield K, Hariri F, Pastinen T, Osborne CS, et al. Promoter capture Hi-C-based identification of recurrent noncoding mutations in colorectal cancer. Nat Genet. 2018;50:1375–1380. doi: 10.1038/s41588-018-0211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A 'metastasis-prone' signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27:83–90. doi: 10.1007/s10585-010-9305-4. [DOI] [PubMed] [Google Scholar]
- 78.Skrzypczak M, Goryca K, Rubel T, Paziewska A, Mikula M, Jarosz D, Pachlewski J, Oledzki J, Ostrowski J. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5:e13091. doi: 10.1371/journal.pone.0013091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith JJ, Deane NG, Wu F, Merchant NB, Zhang B, Jiang A, Lu P, Johnson JC, Schmidt C, Bailey CE, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138:958–968. doi: 10.1053/j.gastro.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xie N, Yao Y, Wan L, Zhu T, Liu L, Yuan J. Next-generation sequencing reveals lymph node metastasis associated genetic markers in colorectal cancer. Exp Ther Med. 2017;14:338–343. doi: 10.3892/etm.2017.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Y, Wei J, Zhang S, Li G, Zhang T, Yu X, Chen H, Liu M. Overexpression of Wnt7α protein predicts poor survival in patients with colorectal carcinoma. Tumour Biol. 2015;36:8781–8787. doi: 10.1007/s13277-015-3633-6. [DOI] [PubMed] [Google Scholar]
- 83.Avgustinova A, Iravani M, Robertson D, Fearns A, Gao Q, Klingbeil P, Hanby AM, Speirs V, Sahai E, Calvo F, Isacke CM. Tumour cell-derived Wnt7a recruits and activates fibroblasts to promote tumour aggressiveness. Nat Commun. 2016;7:10305. doi: 10.1038/ncomms10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nishioka M, Ueno K, Hazama S, Okada T, Sakai K, Suehiro Y, Okayama N, Hirata H, Oka M, Imai K, et al. Possible involvement of Wnt11 in colorectal cancer progression. Mol Carcinog. 2013;52:207–217. doi: 10.1002/mc.21845. [DOI] [PubMed] [Google Scholar]
- 85.He D, Yue Z, Liu L, Fang X, Chen L, Han H. Long noncoding RNA ABHD11-AS1 promote cells proliferation and invasion of colorectal cancer via regulating the miR-1254-WNT11 pathway. J Cell Physiol. 2019;234:12070–12079. doi: 10.1002/jcp.27877. [DOI] [PubMed] [Google Scholar]
- 86.Kaneda H, Arao T, Tanaka K, Tamura D, Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y, et al. FOXQ1 is overexpressed in colorectal cancer and enhances tumorigenicity and tumor growth. Cancer Res. 2010;70:2053–2063. doi: 10.1158/0008-5472.CAN-09-2161. [DOI] [PubMed] [Google Scholar]
- 87.Abba M, Patil N, Rasheed K, Nelson LD, Mudduluru G, Leupold JH, Allgayer H. Unraveling the role of FOXQ1 in colorectal cancer metastasis. Mol Cancer Res. 2013;11:1017–1028. doi: 10.1158/1541-7786.MCR-13-0024. [DOI] [PubMed] [Google Scholar]
- 88.Peng X, Luo Z, Kang Q, Deng D, Wang Q, Peng H, Wang S, Wei Z. FOXQ1 mediates the crosstalk between TGF-β and Wnt signaling pathways in the progression of colorectal cancer. Cancer Biol Ther. 2015;16:1099–1109. doi: 10.1080/15384047.2015.1047568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liu JY, Wu XY, Wu GN, Liu FK, Yao XQ. FOXQ1 promotes cancer metastasis by PI3K/AKT signaling regulation in colorectal carcinoma. Am J Transl Res. 2017;9:2207–2218. [PMC free article] [PubMed] [Google Scholar]
- 90.Möller P, Koretz K, Leithäuser F, Brüderlein S, Henne C, Quentmeier A, Krammer PH. Expression of APO-1 (CD95), a member of the NGF/TNF receptor superfamily, in normal and neoplastic colon epithelium. Int J Cancer. 1994;57:371–377. doi: 10.1002/ijc.2910570314. [DOI] [PubMed] [Google Scholar]
- 91.Xiao W, Ibrahim ML, Redd PS, Klement JD, Lu C, Yang D, Savage NM, Liu K. Loss of Fas expression and function Is coupled with colon cancer resistance to immune checkpoint inhibitor immunotherapy. Mol Cancer Res. 2019;17:420–430. doi: 10.1158/1541-7786.MCR-18-0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park SJ, Kim HB, Kim J, Park S, Kim SW, Lee JH. The oncogenic effects of p53-inducible gene 3 (PIG3) in colon cancer cells. Korean J Physiol Pharmacol. 2017;21:267–273. doi: 10.4196/kjpp.2017.21.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: Potential therapeutic strategies in treating colorectal cancer. Int J Oncol. 2014;44:1032–1040. doi: 10.3892/ijo.2014.2259. [DOI] [PubMed] [Google Scholar]
- 94.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 95.Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–353. doi: 10.1128/MCB.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zong Y, Xin L, Goldstein AS, Lawson DA, Teitell MA, Witte ON. ETS family transcription factors collaborate with alternative signaling pathways to induce carcinoma from adult murine prostate cells. Proc Natl Acad Sci USA. 2009;106:12465–12470. doi: 10.1073/pnas.0905931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan L, Peng G, Sahgal N, Fazli L, Gleave M, Zhang Y, Hussain A, Qi J. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene. 2016;35:2441–2452. doi: 10.1038/onc.2015.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson S, Fan L, Sahgal N, Qi J, Filipp FV. The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells. Oncotarget. 2017;8:30328–30343. doi: 10.18632/oncotarget.15681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fan L, Zhang F, Xu S, Cui X, Hussain A, Fazli L, Gleave M, Dong X, Qi J. Histone demethylase JMJD1A promotes alter-native splicing of AR variant 7 (AR-V7) in prostate cancer cells. Proc Natl Acad Sci USA. 2018;115:E4584–E4593. doi: 10.1073/pnas.1802415115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang DE, Dai Y, Fan LL, Geng XY, Fu DX, Jiang HW, Xu SH. Histone demethylase JMJD1A promotes tumor progression via activating Snail in prostate cancer. Mol Cancer Res. 2020;18:698–708. doi: 10.1158/1541-7786.MCR-19-0889. [DOI] [PubMed] [Google Scholar]
- 101.Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13:95–109. doi: 10.1016/S1359-6101(01)00038-7. [DOI] [PubMed] [Google Scholar]
- 102.Mojic M, Takeda K, Hayakawa Y. The dark side of IFN-ү: Its role in promoting cancer immunoevasion. Int J Mol Sci. 2017;19:89. doi: 10.3390/ijms19010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jung B, Staudacher JJ, Beauchamp D. Transforming growth factor β superfamily signaling in development of colorectal cancer. Gastroenterology. 2017;152:36–52. doi: 10.1053/j.gastro.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiao HL, Ye YP, Yang RW, Sun HY, Wang SY, Wang YX, Xiao ZY, He LQ, Cai JJ, Wei WT, et al. Downregulation of SAFB sustains the NF-κB pathway by targeting TAK1 during the progression of colorectal cancer. Clin Cancer Res. 2017;23:7108–7118. doi: 10.1158/1078-0432.CCR-17-0747. [DOI] [PubMed] [Google Scholar]
- 105.Weng W, Okugawa Y, Toden S, Toiyama Y, Kusunoki M, Goel A. FOXM1 and FOXQ1 are promising prognostic biomarkers and novel targets of tumor-suppressive miR-342 in human colorectal cancer. Clin Cancer Res. 2016;22:4947–4957. doi: 10.1158/1078-0432.CCR-16-0360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bagati A, Bianchi-Smiraglia A, Moparthy S, Kolesnikova K, Fink EE, Lipchick BC, Kolesnikova M, Jowdy P, Polechetti A, Mahpour A, et al. Melanoma suppressor functions of the carcinoma oncogene FOXQ1. Cell Rep. 2017;20:2820–2832. doi: 10.1016/j.celrep.2017.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nau MM, Brooks BJ, Battey J, Sausville E, Gazdar AF, Kirsch IR, McBride OW, Bertness V, Hollis GF, Minna JD. L-myc, a new myc-related gene amplified and expressed in human small cell lung cancer. Nature. 1985;318:69–73. doi: 10.1038/318069a0. [DOI] [PubMed] [Google Scholar]
- 108.Wu R, Lin L, Beer DG, Ellenson LH, Lamb BJ, Rouillard JM, Kuick R, Hanash S, Schwartz DR, Fearon ER, Cho KR. Amplification and overexpression of the L-MYC proto-onco-gene in ovarian carcinomas. Am J Pathol. 2003;162:1603–1610. doi: 10.1016/S0002-9440(10)64294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Boutros PC, Fraser M, Harding NJ, de Borja R, Trudel D, Lalonde E, Meng A, Hennings-Yeomans PH, McPherson A, Sabelnykova VY, et al. Spatial genomic heterogeneity within localized, multifocal prostate cancer. Nat Genet. 2015;47:736–745. doi: 10.1038/ng.3315. [DOI] [PubMed] [Google Scholar]
- 110.Huijbers IJ, Bin Ali R, Pritchard C, Cozijnsen M, Kwon MC, Proost N, Song JY, de Vries H, Badhai J, Sutherland K, et al. Rapid target gene validation in complex cancer mouse models using re-derived embryonic stem cells. EMBO Mol Med. 2014;6:212–225. doi: 10.1002/emmm.201303297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim DW, Wu N, Kim YC, Cheng PF, Basom R, Kim D, Dunn CT, Lee AY, Kim K, Lee CS, et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev. 2016;30:1289–1299. doi: 10.1101/gad.279307.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Möröy T, Fisher P, Guidos C, Ma A, Zimmerman K, Tesfaye A, DePinho R, Weissman I, Alt FW. IgH enhancer deregulated expression of L-myc: Abnormal T lymphocyte development and T cell lymphomagenesis. EMBO J. 1990;9:3659–3666. doi: 10.1002/j.1460-2075.1990.tb07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chapman DL, Papaioannou VE. Three neural tubes in mouse embryos with mutations in the T-box gene Tbx6. Nature. 1998;391:695–697. doi: 10.1038/35624. [DOI] [PubMed] [Google Scholar]
- 114.Sandbacka M, Laivuori H, Freitas E, Halttunen M, Jokimaa V, Morin-Papunen L, Rosenberg C, Aittomaki K. TBX6, LHX1 and copy number variations in the complex genetics of Mullerian aplasia. Orphanet J Rare Dis. 2013;8:125. doi: 10.1186/1750-1172-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, Shen Y, Yu G, Liu J, Xie H, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Verbitsky M, Westland R, Perez A, Kiryluk K, Liu Q, Krithivasan P, Mitrotti A, Fasel DA, Batourina E, Sampson MG, et al. The copy number variation landscape of congenital anomalies of the kidney and urinary tract. Nat Genet. 2019;51:117–127. doi: 10.1038/s41588-018-0281-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ren X, Yang N, Wu N, Xu X, Chen W, Zhang L, Li Y, Du RQ, Dong S, Zhao S, et al. Increased TBX6 gene dosages induce congenital cervical vertebral malformations in humans and mice. J Med Genet. 2020;57:371–379. doi: 10.1136/jmedgenet-2019-106333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell-Badge R, Papaioannou VE, Kondoh H. Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature. 2011;470:394–398. doi: 10.1038/nature09729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sadahiro T, Isomi M, Muraoka N, Kojima H, Haginiwa S, Kurotsu S, Tamura F, Tani H, Tohyama S, Fujita J, et al. Tbx6 induces nascent mesoderm from pluripotent stem cells and temporally controls cardiac versus somite lineage diversification. Cell Stem Cell. 2018;23:382–395.e5. doi: 10.1016/j.stem.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Saigusa S, Tanaka K, Toiyama Y, Yokoe T, Okugawa Y, Ioue Y, Miki C, Kusunoki M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann Surg Oncol. 2009;16:3488–3498. doi: 10.1245/s10434-009-0617-z. [DOI] [PubMed] [Google Scholar]
- 121.Ong CW, Kim LG, Kong HH, Low LY, Iacopetta B, Soong R, Salto-Tellez M. CD133 expression predicts for non-response to chemotherapy in colorectal cancer. Mod Pathol. 2010;23:450–457. doi: 10.1038/modpathol.2009.181. [DOI] [PubMed] [Google Scholar]
- 122.Neumann J, Bahr F, Horst D, Kriegl L, Engel J, Luque RM, Gerhard M, Kirchner T, Jung A. SOX2 expression correlates with lymph-node metastases and distant spread in right-sided colon cancer. BMC Cancer. 2011;11:518. doi: 10.1186/1471-2407-11-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data and material will be made available upon reasonable request. RNA-sequencing data have been deposited in Gene Expression Omnibus (GSE158294).