Abstract
Abnormal cell proliferation and invasion of vascular smooth muscle cells are among the primary causes of cardiovascular disease. Studies have shown that microRNA(miR)-342-5p participates in the development of cardiovascular diseases. The current study aimed to explore the role of miR-342-5p in the proliferation and differentiation of mouse aortic vascular smooth muscle (MOVAS) cells. MOVAS cells were transfected with miR-342-5p mimics, miR-342-5p inhibitor or their respective negative controls, and co-transfected with small interfering (si)RNA targeting phosphatidylinositol 3-kinase regulatory subunit α (PIK3R1) and miR-342-5p inhibitor. The cell proliferation of MOVAS cells was detected using the Cell Counting Kit-8, while cell migration and cell invasion were investigated using a wound healing and Transwell assays, respectively. Target genes for miR-342-5p were confirmed using reverse transcription-quantitative PCR (RT-qPCR) and dual luciferase reporter assay. The relative mRNA and protein expression levels of miR-342-5p were measured using RT-qPCR and western blot analysis. MOVAS cells were treated with a PI3K inhibitor (LY294002) to explore the role of miR-342-5p on the Akt pathway. The results revealed that miR-342-5p mimics promoted cell viability, migration and invasion, and increased the expression of vimentin and phosphorylated-Akt but reduced a-smooth muscle actin (α-SMA) and PIK3R1 expression. However, miR-342-5p inhibitor produced the opposite effects. PIK3R1 was the target gene for miR-342-5p and the effect of siPIK3R1 on MOVAS cells was similar to that of miR-342-5p mimics, while siPIK3R1 partially reversed the effect of miR-342-5p inhibitor on MOVAS cells. The Akt signaling pathway was activated by miR-342-5p mimics or siPIK3R1. Moreover, miR-342-5p mimics partially activated the Akt signaling pathway inhibited by LY294002. MiR-342-5p could promote the proliferation and differentiation of MOVAS and phenotypic transformation. The mechanism behind these processes may be associated with the activation of the Akt signaling pathway induced by PIK3R1 inhibition.
Keywords: miR-342-5p, vascular smooth muscle cells, phenotypic conversion, Akt signaling pathway
Introduction
Cardiovascular disease, cancer, diabetes and other chronic non-communicable diseases are the most frequent causes of death worldwide (1). Cardiovascular diseases such as stroke, ischemic heart disease, atherosclerosis and congestive heart failure are the major causes of death in China, with the number of cardiovascular disease related deaths expected to rise by 39 million between 2016 and 2030(2). Vascular smooth muscle cells (VSMCs) are important components of blood vessels. Atherosclerosis, hypertension and restenosis are associated with increased VSMC proliferation, invasion and migration (3,4). VSMCs maintain an organized, differentiated and contractile phenotype under normal physiological conditions, and can be converted into a proliferative, migratory and synthetic phenotype under various stimuli, such as mechanical injury, activation of growth factors (such as transforming growth factor-β and platelet-derived growth factor), ligand-receptor signaling and increased hemodynamics, eventually leading to an increase in proliferation, migration and secretory activities of VSMCs (5).
MicroRNAs (miRNAs/miRs) are non-coding RNAs that degrade mRNA or impede its translation to regulate the levels of target genes (6,7). miRNAs play an important role in the occurrence and development of cardiovascular diseases, such as miR-665 which suppresses VSMC proliferation, invasion and migration through targeting FGF9 and MEF2D (8), and it is hypothesized that miRNAs could be used as novel targets in the treatment of cardiovascular diseases (9,10). Studies have confirmed that miRNAs, such as miR-541 promote VSMC proliferation and miR-146b-5p promote VSMC proliferation and migration (11,12), while miR-124 and miR-503 inhibit the proliferation of VSMCs (13,14). A previous study demonstrated that macrophage-derived miR-342-5p contributed to atherosclerosis by inhibiting Akt1 expression (15). Another study revealed that the levels of miR-342-5p in patients with coronary heart disease were increased (16). Furthermore, Yan et al (17) revealed that miR-342-5p promoted endothelial cell migration and reduced angiogenesis by targeting endoglin. Although the proliferation and differentiation of VSMCs play a key role in the progression of cardiovascular disease (18), the role of miR-342-5p in VSMCs remains to be elucidated. Therefore, the effects and mechanism of action of miR-342-5p on VSMC proliferation and differentiation were explored in the current study.
Materials and methods
Cell culture
Mouse aortic vascular smooth muscle (MOVAS) cells (American Type Culture Collection; 2x105 cells/well) were cultured in 24-well plate with DMEM (Sigma-Aldrich; Merck KGaA) containing 10% FBS (Sigma-Aldrich; Merck KGaA), 100 U/m penicillin (Invitrogen; Thermo Fisher Scientific, Inc.) and 100 U/m streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C in a humidified incubator with 5% CO2.
Cell transfection
Cells were divided into specific groups as follows for transfection: i) Control group (cells with no transfected material); mock group [50 nM miR-342-5p mimics negative control (NC); GeneCopoeia, Inc.]; mimics group (50 nM miR-342-5p mimics; GeneCopoeia, Inc.); NC group (50 nM miR-342-5p inhibitor NC; GeneCopoeia, Inc.) and inhibitor group (50 nM miR-342-5p inhibitor; GeneCopoeia, Inc.); ii) control + phosphatidylinositol 3-kinase regulatory subunit α (PIK3R1)-3'-untranslated region (UTR) group (50 ng/µl PIK3R1-3'-UTR expression plasmid), miR-342-5p + PIK3R1-3'-UTR group (50 ng/µl PIK3R1-3'-UTR expression plasmid and miR-342-5p mimics) and miR-342-5p + PIK3R1-3'-UTR mutant (mut) group (miR-342-5p mimics and 50 ng/µl mut PIK3R1-3'-UTR); iii) control group (cells with no transfected material), small interfering (si)NC group (siPIK3R1 NC; Sigma-Aldrich; Merck KGaA) and siPIK3R1 group (siPIK3R1; Sigma-Aldrich; Merck KGaA); iv) NC group (miR-342-5p inhibitor NC), inhibitor group (miR-342-5p inhibitor), NC + siPIK3R1 group (miR-342-5p inhibitor NC and 50 nM siPIK3R1) and inhibitor + siPIK3R1 group (miR-342-5p inhibitor and 50 nM siPIK3R1); and v) mock group (miR-342-5p mimics NC), mimics group (miR-342-5p mimics), mock + LY294002 (Cayman Chemical Company) group (transfected with miR-342-5p mimics NC and treated with 10 µmol/l LY294002) and mimics + LY294002 group (transfected with miR-342-5p mimics and treated with 10 µmol/l LY294002).
Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) was used to transfect the aforementioned groups into the cells and incubated for 24 h at 37˚C in a humidified incubator with 5% CO2. Sequences for miRNA and siRNA used were as follows: miR-342-5p mimics, 5'-AGGGGUGCUAUCUGUGAUUGAG-3'; miR-342-5p mimics NC, 5'-UUUGUACUACACAAAAGUACUG-3'; miR-342-5p inhibitor NC, 5'-CAGUACUUUUGUGUAGUACAA-3'; siPIK3R1, 5'-GCAGAGGCACTCCTGATATATGATTT-3'; and siNC, 5'-AATTCACTCCAAGTCTCTTCC-3'.
Cell proliferation assay
Cell viability was measured using a Cell Counting Kit (CCK)-8 cell proliferation assay kit (Nanjing Jiancheng Bioengineering Institute), according to the manufacturer's protocol. Following transfection and culturing of cells (3x103 cells/well) for 0, 24, 48 and 72 h, CCK-8 solution was added into each well and cells were incubated in the dark at 37˚C in a humidified incubator with 5% CO2 for 2 h. A microplate reader (Bio-Rad Laboratories, Inc.) was used to determine the optical density of each well at a wavelength of 450 nm.
Wound healing assay
Cell migration was determined using a wound healing assay. Cells were plated at a density of 4x105 cells/well in a six-well plate. A pipette tip was used to create a straight wound in each well. Cells were washed twice with PBS (Gibco; Thermo Fisher Scientific, Inc) to remove floating cells and the remaining cells were cultured in DMEM at 37˚C in a humidified incubator with 5% CO2 for 0 and 24 h. Subsequently, cell migration was observed under an inverted phase contrast microscope (magnification, x200; Olympus Corporation). The distance of cell migration was measured by Image ProPlus v6.0 analysis software (Media Cybernetics, Inc.). The relative migration distance = (0 h migration distance -24 h migration distance)/0 h migration distance.
Transwell assay
Transwell assay was performed to detect cell invasion. Transwell chambers (8-µm pores; Corning, Inc.) were placed in a 24-well plate. Cells (1x105) were re-suspended in serum-free medium and plated into the upper chamber which pre-coated with Matrigel® (BD Biosciences). DMEM containing 10% FBS was added into the lower chamber and cells were cultured at 37˚C in a humidified incubator with 5% CO2 for 24 h. The Transwell chambers were moved to another 24-well plate, washed with PBS three times and then fixed in 4% methanol solution for 30 min at room temperature and stained with 0.1% crystal violet for 25 min at room temperature. Subsequently, the transwell chambers were washed with PBS twice and non-migrating cells in the upper chamber were removed using a cotton swab. The chambers were then placed under an inverted fluorescence microscope (Olympus Corporation) to observe cells and capture images (magnification, x200).
Target gene prediction for miR-342-5p
The target gene online prediction databases TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org/) and miRBase (http://www.mirbase.org/) were used to predict potential target genes and binding sites of miR-342-5p. Potential target genes of miR-342-5p were further confirmed using dual-luciferase reporter assays.
Dual luciferase reporter assay
The sequences of PIK3R1-3'-UTR was amplified from cDNA derived from total RNA and PIK3R1-3'-UTR-mut was constructed using a Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.) according to the manufacturer's protocol. The two target gene fragments were cloned into pmirGLO vectors (Promega Corporation) and transfection was performed using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc), as aforementioned. Cells were harvested 24 h after the transfection, and firefly and Renilla luciferase activity was measured using a Dual-Luciferase® Reporter Assay System (Promega Corporation). Firefly luciferase activity was normalized to that of Renilla luciferase activity.
RT-qPCR
Total RNA was isolated from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Briefly, the cells were mixed with Trizol reagent and subsequently transferred to an Eppendrof™ tube and incubated on the bench at room temperature. Then 200 µl chloroform was added, and mixed with the cell/Trizol mixture and placed on ice for 5 min. The tubes were centrifuged at 12,000 x g at 4˚C for 15 min, following which the upper aqueous phase was transferred to a clean EppendorfTM tube and 500 µl isopropanol was added, and the tubes were incubated on ice for 10 min. Pre-cooled 75% ethanol was subsequently added and the tubes were centrifuged for a second time at 12,000 x g at 4˚C for 10 min. The supernatant was discarded and the RNA pellet was air dried for 3 min, following which pre-cooled 20 µl DEPC was added. A spectrophotometer (ND-1000; NanoDrop Technologies; Thermo Fisher Scientific, Inc.) was used to determine RNA purity and concentration at 260/280 nm. Total RNA (2 µg) was reverse transcribed into cDNA for mRNA detection using a PrimeScript RT master mix kit (Takara Biotechnology Co., Ltd.) at 37˚C for 60 min, then 85̊C for 5 min and the samples were stored at 4˚C until further experimentation. qPCR was performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) on the ABI7500 Real-Time PCR System (Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95˚C for 5 sec; 40 cycles of 95˚C for 10 sec, 60˚C for 30 sec; and a final extension step at 72˚C for 10 sec, with storage of the samples at 4˚C. The relative expression of genes was calculated using the 2-ΔΔCq method (19). U6, GAPDH and β-actin (GeneCopoeia) served as internal controls. The following primer pairs were used for the qPCR: miR-342-5p forward, 5'-CGGAGGGGTGCTATCTGTGA-3' and reverse, 5'-AGTCGGCAATTGCACTGGAT-3'; U6 forward, 5'-CTCAGAGCGTGGTTCTCCGTCAC-3' and reverse, 5'-TATAAATCTTTACCCTGTTGGCAGT-3'; GAPDH forward, 5'-AGGTCGGTGTGAACGGATTTG-3' and reverse, 5'-TGTAGACCATGTAGTTGAGGTCA-3'; α-smooth muscle actin (α-SMA) forward, 5'-CATCCGTAAAGACCTCTATGCCAAC-3' and 5'-ATGGAGCCACCGATCCACAA-3' and reverse, 5'-TCGGATACTTCAGCGTCAGGA-3'; vimentin forward, 5'-GAGAACTTTGCCGTTGAAGC-3' and reverse, 5'-GCTTCCTGTAGGTGGCAATC-3'; apelin (APLN) forward, 5'-TGCTCTGGCTCTCCTTGACT-3' and reverse, 5'-ATGGGTCCCTTATGGGAGAG-3'; zinc finger and BTB domain-containing protein 39 (ZBTB39) forward, 5'-CGTGTTAACTAGGTCCCCTTTG-3' and reverse, 5'-GTTCCTTTAATCCAGAAGGGCT-3'; PIK3R1 forward, 5'-AGCATTGGGACCTCACATTACACA-3' and reverse, 5'-ACTGGAAACACAGTCCATGCACATA-3'; myocyte-specific enhancer factor 2D (MEF2D) forward, 5'-CGCGAATTCACCATGGGGAGGAAAAAGATT-3' and reverse, 5'-TGGCTCGAGTCACTTTAATGTCCAGGT-3'; and neurogenic locus notch homolog protein 2 (NOTCH2) forward, 5'-CCCCTTGCCCTCTATGTACCA-3' and reverse, 5'-GGTAGGTGGGAAAGCCACACT-3'; β-actin forward, 5'-GCTGCGTGTGGCCCCTGAG-3' and reverse, 5'-ACGCAGGATGGCATGAGGGA-3'.
Western blot analysis
Cell lysates were extracted using RIPA buffer (Beyotime Institute of Biotechnology) and the protein concentration was determined with a Pierce bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.). 10% SDS-PAGE was used to separate the proteins (30 µg) according to the molecular weight of protein. Samples were transferred to PVDF membranes (Bio-Rad Laboratories, Inc.), which were subsequently blocked with 5% skimmed milk for 1 h. PVDF membranes were incubated with the following primary antibodies: Anti-GAPDH (1:1,000; cat. no. ab8245); anti-Akt (1:1,000; cat. no. ab8805); anti-α-SMA (1:200; cat. no. ab5694); anti-vimentin (1:1,000, cat. no. ab92547); anti-phosphorylated (p)-Akt (1:500; cat. no. ab38449); and anti-PIK3R1 (1:1,000; cat. no. ab86714; all Abcam) overnight at 4˚C. Following primary incubation, membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG) (H+L) or goat anti-mouse IgG (H+L) secondary antibodies (1:1,000; cat. nos. A0208 and A0216; Beyotime Institute of Biotechnology) for 1 h at room temperature. Protein bands were visualized using the BeyoECL Plus kit (Beyotime Institute of Biotechnology) and quantified using ImageJ (version 5.0; National Institutes of Health). The expression levels of GAPDH served as an internal control.
Statistical analysis
Data are presented as the mean ± SD. Data were analyzed using SPSS v21.0 software (IBM Corp.). Any significant difference among different groups was analyzed by one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference. All experiments were performed in triplicate.
Results
miR-342-5p mimics and inhibitors was successfully expressed in MOVAS cells
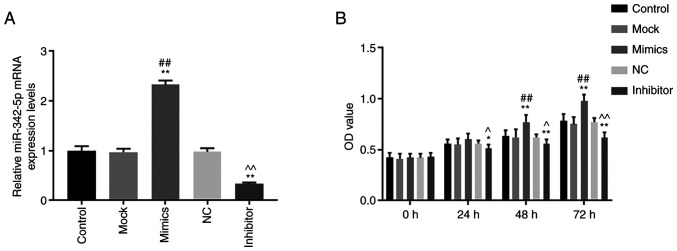
The transfection efficiency of miR-342-5p was determined using RT-qPCR. The levels of miR-342-5p mRNA were significantly increased in the mimics group compared with that in the control and mock groups (Fig. 1A), while mRNA levels of miR-342-5p were significantly decreased in the inhibitor group compared with that in the control and NC groups (Fig. 1A). The data showed that MOVAS cells with high and low expression levels of miR-342-5p were successfully constructed.
Figure 1.
Transfection efficiency of miR-342-5p in MOVAS cells and its effect on cell proliferation. (A) MOVAS cells were transfected with control, mock, mimics, NC and inhibitor and the mRNA expression levels of miR-342-5p were determined using reverse transcription-quantitative PCR. (B) Cell proliferation was determined using a Cell Counting Kit-8 assay. *P<0.05 and **P<0.01 vs. control; ##P<0.01 vs. mock; ^P<0.05 and ^^P<0.01 vs. NC. MOVAS, mouse aortic vascular smooth muscle; miR, microRNA; mock, miR-342-5p mimic negative control; NC, miR-342-5p inhibitor negative control.
miR-342-5p promotes cell viability, migration and invasion
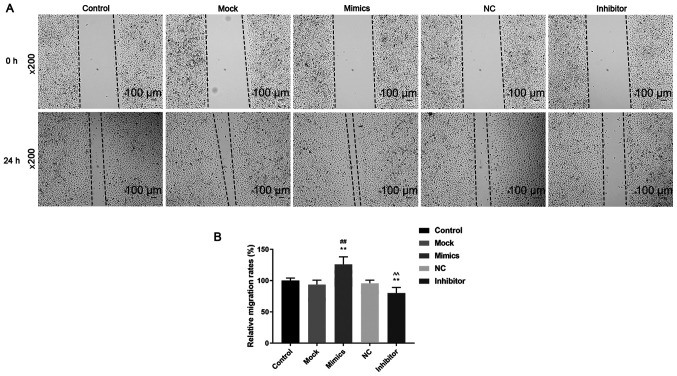
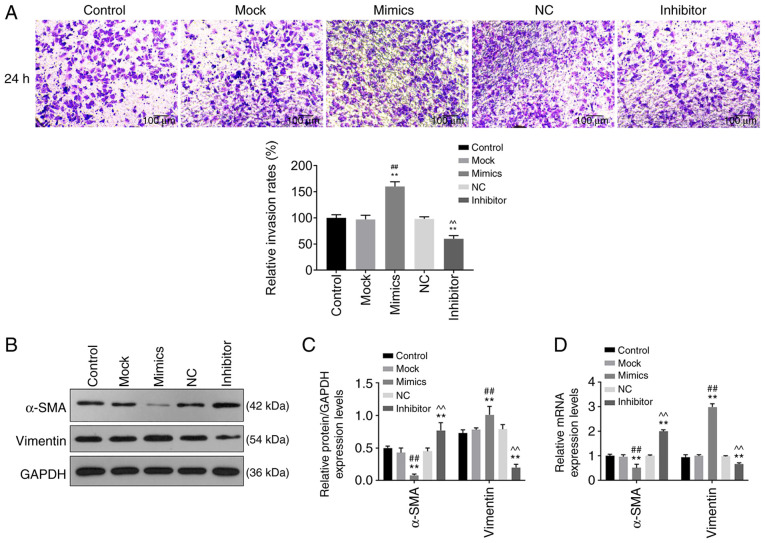
The cell viability, migration and invasion of cells were determined using CCK-8, wound healing and Transwell assays, respectively. Cell viability was significantly higher in the mimics group compared with that in the control and mock groups 48 and 72 h after transfection (P<0.05; Fig. 1B), while cell viability in the inhibitor group was significantly lower compared with control and NC groups at 24, 48 and 72 h after transfection (P<0.05; Fig. 1B). Cell migration (Fig. 2A and B) and invasion (Fig. 3A) was significantly increased in the mimics group (P<0.05) compared with that in the control and mock groups, whereas they were significantly decreased in the inhibitor group (P<0.05) compared with that in the control and NC groups. These results suggest that miR-342-5p increased cell viability and promoted cell migration and invasion in MOVAS cells.
Figure 2.
Role of miR-342-5p in cell migration. Cell migration was investigated in MOVAS cells that were transfected with control, mock, mimics, NC and inhibitor using a wound healing assay. (A) Cell migration images of the 5 experimental groups were obtained using an inverted phase contrast microscope at x200 magnification. (B) Relative migration rate was analyzed statistically. **P<0.01 vs. control; ##P<0.01 vs. mock; ^^P<0.01 vs. NC. miR, microRNA; mock, miR-342-5p mimic negative control; NC, miR-342-5p inhibitor negative control; MOVAS, mouse aortic vascular smooth muscle.
Figure 3.
Role of miR-342-5p in cell invasion and its effects on relative mRNA expression of vimentin and α-SMA. MOVAS cells were transfected with control, mock, mimics, NC and inhibitor, following which (A) cell invasion, and (B and C) protein and (D) mRNA expression levels of target genes were determined using Transwell assay, western blot analysis and reverse transcription-quantitative PCR, respectively. (A) Representative images of cell invasion in each experimental group and quantitative analysis. Magnification, x200. α-SMA and vimentin protein expression levels were determined using (B) western blotting and (C) quantified and analyzed statistically. (D) The mRNA levels of α-SMA and vimentin. **P<0.01 vs. control; ##P<0.01 vs. mock; ^^P<0.01 vs. NC. miR-342-5p, microRNA-342-5p; α-SMA, α-smooth muscle actin; mock, miR-342-5p mimic negative control; NC, miR-342-5p inhibitor negative control; MOVAS, mouse aortic vascular smooth muscle.
Protein and mRNA levels of a-SMA and vimentin are regulated by miR-342-5p
The relative protein and mRNA levels of α-SMA and vimentin were determined using western blot analysis and RT-qPCR, respectively. Protein and mRNA expression levels of α-SMA in the mimics group were downregulated compared with that in the control and mock groups, while protein and mRNA expression levels of vimentin were upregulated compared with that in the control and mock groups (Fig. 3B-D). While the protein and mRNA levels of α-SMA in the inhibitor group were upregulated compared with that in the control and NC groups, and the protein and mRNA levels of vimentin were downregulated compared with that in the control and NC groups (Fig. 3B-D). These data revealed that miR-342-5p mimics downregulated α-SMA expression and upregulated vimentin expression, and the effects were reversed by the miR-342-5p inhibitor.
PIK3R1 is a target gene of miR-342-5p
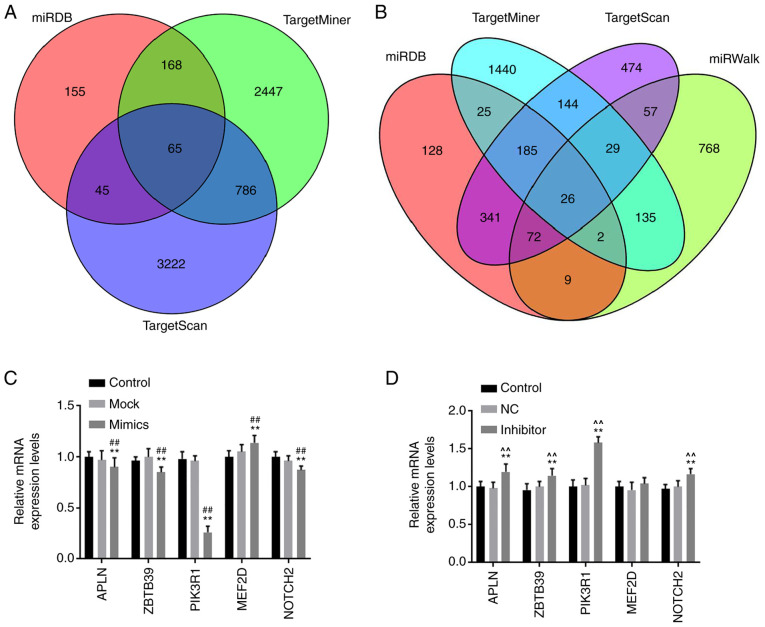
Online databases was applied to predict the target gene of miR-342-5p (Fig. 4A-B) and it has been reported that the APLN, ZBTB39, PIK3R1, MEF2D and NOTCH2 genes may be associated with cardiovascular disease (8,20-23), therefore infer that APLN, ZBTB39, PIK3R1, MEF2D and NOTCH2 may were potential target genes of miR-342-5p. The role of miR-342-5p in the aforementioned target genes was examined using RT-qPCR. The mRNA levels of APLN, ZBTB39, PIK3R1 and NOTCH2 in the mimics group were significantly lower compared with that in the control and mock groups, while MEF2D levels were significantly higher (P<0.05; Fig. 4C), notably PIK3R1 was the most significantly associated (P<0.01). The mRNA levels of APLN, ZBTB39, PIK3R1 and NOTCH2 were significantly upregulated in the inhibitor group compared with that in the control and NC groups (Fig. 4D), and the upregulation in expression was more significant for PIK3R1 (P<0.01). Based on the significant results from the mRNA expression levels, to further determine whether PIK3R1 was a target of miR-342-5p, target gene online prediction databases were used to predict the binding sites of PIK3R1 and miR-342-5p (Fig. 5A). The binding sites were further verified using a dual luciferase reporter assay. The results revealed that compared with the control + PIK3R1-3'-UTR and miR-342-5p + PIK3R1-3'-UTR mut groups, the relative luciferase activity in the miR-342-5p + PIK3R1-3'-UTR group was significantly decreased (P<0.05; Fig. 5B).
Figure 4.
miR-342-5p target genes were predicted using target gene prediction websites. (A) Venn diagrams of miR-342-5p target genes, the target genes were predicted using miRDB, TargetMiner and TargetScan. (B) Venn diagrams of miR-342-5p target genes, the target genes were predicted using miRDB, TargetMiner, TargetScan and miRWalk. MOVAS cells were transfected with (C) control, mock or miR-342-5p mimic and (D) control, NC or miR-342-5p inhibitor and the relative mRNA expression levels of APLN, ZBTB39, PIK3R1, MEF2D and NOTCH2 were determined using reverse transcription-quantitative PCR. **P<0.01 vs. control; ##P<0.01 vs. mock; ^^P<0.01 vs. NC. miR, microRNA; mock, miR-342-5p mimic negative control; NC, miR-342-5p inhibitor negative control; APLN, apelin; ZBTB39, zinc finger and BTB domain-containing protein 39; PIK3R1, phosphatidylinositol 3-kinase regulatory subunit α; MEF2D, myocyte-specific enhancer factor 2D; NOTCH2, neurogenic locus notch homolog protein 2.
Figure 5.
Binding capacity between miR-342-5p and PIK3R1-3'-UTR detected using a dual-luciferase assay. (A) Binding sites between miR-342-5p and PIK3R1-3'-UTR. (B) Detection of fluorescence activity using a dual luciferase assay. **P<0.01 vs. control + PIK3R1-3'-UTR; ##P<0.01 vs. miR-342-5p + PIK3R1-3'-UTR mut. miR, microRNA; PIK3R1, phosphatidylinositol 3-kinase regulatory subunit α; UTR, untranslated region; mut, mutant.
siPIK3R1 results in lower expression of PIK3R1 in MOVAS cells
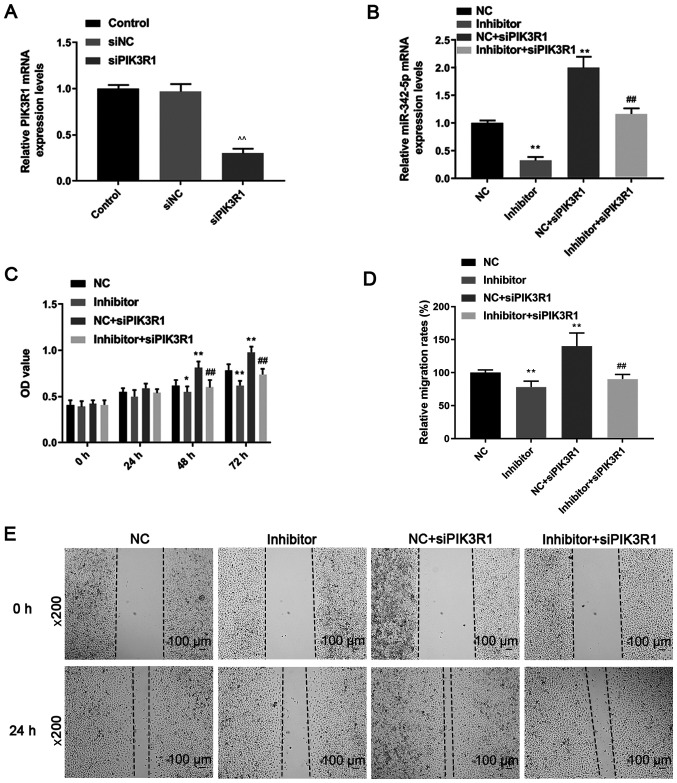
RT-qPCR was performed to determine the mRNA expression levels of PIK3R1 following transfection with siPIK3R1. PIK3R1 mRNA expression was downregulated in the siPIK3R1 group compared with that in the control and siNC groups (P<0.05; Fig. 6A). The results indicated that MOVAS cells with low PIK3R1 expression were successfully constructed.
Figure 6.
Role of siPIK3R1, miR-342-5p inhibitor and the combination of miR-342-5p inhibitor with siPIK3R1 on cell proliferation and migration. (A) Transfection efficiency of siPIK3R1 was determined using RT-qPCR. (B) The mRNA expression levels of miR-342-5p were detected using RT-qPCR following transfection. (C) A Cell Counting Kit-8 assay was used to determine cell proliferation. (D) Quantification of cell migration rates in 5 experimental groups. (E) Representative cell migration in 5 experimental groups detected using a wound healing assay. Magnification, x200. ^^P<0.01 vs. siNC; *P<0.05, **P<0.01 vs. NC; ##P<0.01 vs. NC + siPIK3R1. miR, microRNA; siPIK3R1, si, small interfering; PIK3R1, phosphatidylinositol 3-kinase regulatory subunit α; NC, miR-342-5p inhibitor negative control; RT-qPCR, reverse transcription-quantitative PCR.
siPIK3R1 reverses the effects of miR-342-5p inhibitor on cell proliferation, migration and invasion
Cells were co-transfected with miR-342-5p inhibitor and siPIK3R1 and the RT-qPCR results (Fig. 6B) revealed that the miR-342-5p mRNA expression levels in the inhibitor group were downregulated compared with that in the NC group, while miR-342-5p levels were upregulated in NC + siPIK3R1 compared with that in the NC group. The mRNA expression levels of miR-342-5p in the inhibitor + siPIK3R1 group was significantly lower compared with that in the NC + siPIK3R1 group (P<0.05; Fig. 6B).
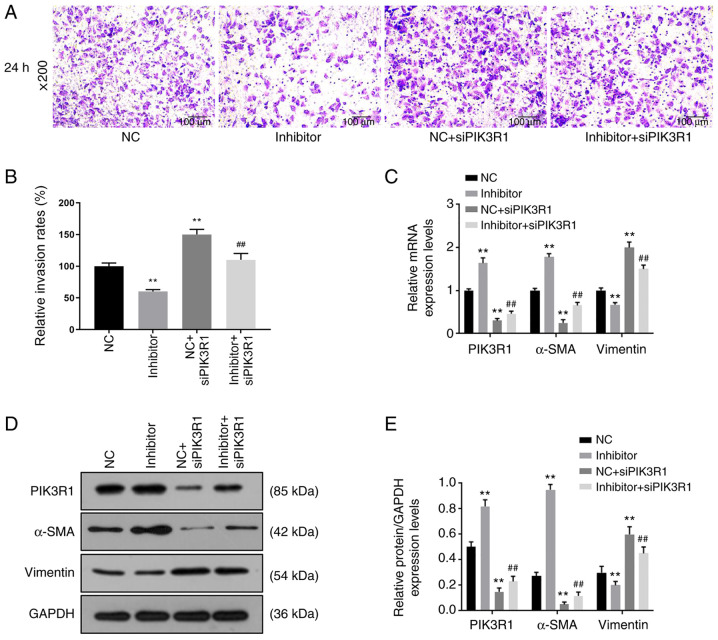
Cell proliferation, migration and invasion were determined by CCK-8, wound healing and Transwell assays, respectively. Following transfection for 48 and 72 h, cell proliferation was higher in the NC + siPIK3R1 group compared with that in the NC group, while it was significantly lower in the inhibitor group compared with that in the NC group (P<0.05; Fig. 6C). The cell proliferation in the inhibitor + siPIK3R1 group was significantly lower compared with that in the NC + siPIK3R1 group (P<0.05; Fig. 6C). Compared with that in the NC group, cell migration was decreased in the inhibitor group, but increased in the NC + siPIK3R1 group (P<0.05; Fig. 6D and E). Cell migration in the inhibitor + siPIK3R1 group was significantly lower compared with that in the NC + siPIK3R1 group, but was slightly higher compared with that in the inhibitor group (P<0.05; Fig. 6D and E). Cell invasion decreased in the inhibitor group but increased in the NC + siPIK3R1 group compared with that in the NC group (P<0.05; Fig. 7A and B). In addition, cell invasion was lower in the inhibitor + siPIK3R1 group compared with that in the NC + siPIK3R1 group (P<0.05; Fig. 7A and B). The results of the current study showed that PIK3R1 silencing increased miR-342-5p expression. siPIK3R1 antagonized the effects of miR-342-5p inhibitor on cells, increasing cell viability and promoting cell migration and invasion.
Figure 7.
Roles of siPIK3R1, miR-342-5p inhibitor and the combination of miR-342-5p inhibitor with siPIK3R1 in cell invasion and the relative expression of target genes. MOVAS cells were transfected with NC, miR-342-5p inhibitor, NC + siPIK3R1 or miR-342-5p inhibitor + siPIK3R1, following which (A and B) cell invasion, and (C) mRNA and (D and E) protein expression levels of target genes were determined using Transwell assay, reverse transcription-quantitative PCR and western blot analysis, respectively. (A) Representative cell invasion images of the 5 experimental groups using an inverted fluorescence microscope at x200 magnification. (B) Quantitative analysis of the rate of cell invasion. The (C) mRNA and (D) protein expression levels of α-SMA, PIK3R1 and vimentin. (E) The quantification of the protein levels in 4 experimental groups. **P<0.01 vs. NC; ##P<0.01 vs. NC + siPIK3R1. miR-342-5p, microRNA-342-5p; α-SMA, α-smooth muscle actin; PIK3R1, phosphatidylinositol 3-kinase regulatory subunit α; siPIK3R1, small interfering RNA targeting PI3K1; siNC, siPIK3R1 negative control; NC, miR-342-5p inhibitor negative control.
siPIK3R1 reverses the effects of miR-342-5p inhibitor on the expression of a-SMA and vimentin
The mRNA and protein expression levels of PIK3R1, α-SMA and vimentin were analyzed using RT-qPCR and western blot analysis, respectively, following transfection with miR-342-5p inhibitor and siPIK3R1. Compared with that in the NC group, the mRNA and protein expression levels of PIK3R1 were upregulated in the inhibitor group, but downregulated in the NC + siPIK3R1 and inhibitor + siPIK3R1 groups (P<0.05; Fig. 7C-E). However, compared with that in the NC + siPIK3R1 group, the mRNA and protein expression levels of PIK3R1 were higher in the inhibitor + siPIK3R1 group, but lower compared with that in the inhibitor group. The mRNA and protein expression levels of α-SMA in the NC + siPIK3R1 and the inhibitor + siPIK3R1 groups were downregulated compared with the NC group, but were upregulated in the inhibitor group compared with that in the NC group (P<0.05; Fig. 7C-E). By contrast, the mRNA and protein expression levels of α-SMA in the inhibitor + siPIK3R1 group were higher compared with that in the NC + siPIK3R1 group (P<0.05; Fig. 7C-E). The mRNA and protein expression levels of vimentin were upregulated in NC + siPIK3R1 and inhibitor + siPIK3R1 groups, and downregulated in the inhibitor group, when compared with that in the NC group, while the mRNA and protein expression levels of vimentin in the inhibitor + siPIK3R1 group were lower compared with that in the NC + siPIK3R1 group (P<0.05; Fig. 7C-E). These results showed that PIK3R1 silencing caused decreased PIK3R1 and α-SMA expression, and increased vimentin expression. siPIK3R1 partially reversed the effects of miR-342-5p inhibitor on the expressions of α-SMA, PIK3R1 and vimentin.
siPIK3R1 reverses the effects of miR-342-5p inhibitor on p-Akt protein levels
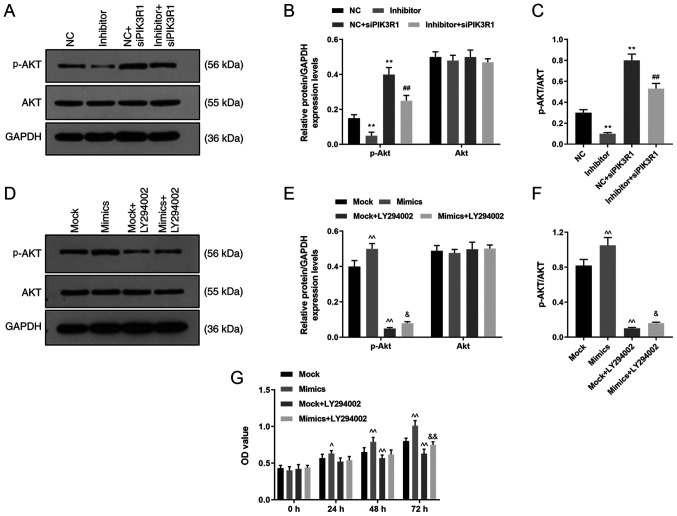
The protein expression levels of p-Akt and Akt were detected using western blot analysis following transfection with miR-342-5p inhibitor and siPIK3R1. p-Akt protein levels were lower in the inhibitor group compared with that in the NC group (P<0.05; Fig. 8A and B), but was upregulated in the NC + siPIK3R1 and inhibitor + siPIK3R1 groups compared with that in the NC group (P<0.01; Fig. 8A and B). The p-Akt protein levels in inhibitor + siPIK3R1 group was lower compared with that in the NC + siPIK3R1 group. These results revealed that p-Akt protein levels were downregulated after inhibiting miR-342-5p expression, and that p-Akt expression was upregulated and the Akt signaling pathway was activated following PIK3R1 silencing. In addition, siPIK3R1 partially reversed the inhibitory effects of miR-342-5p inhibitor on p-Akt protein expression levels. The ratio of p-Akt/Akt was reduced in inhibitor group, while increased in NC + siPIK3R1 group compared with NC group; and the ratio of p-Akt/Akt was lower in inhibitor + siPIK3R1 group than that in NC + siPIK3R1 group (P<0.01; Fig. 8C).
Figure 8.
Role of siPIK3R1 and LY294002 in the Akt signaling pathway. MOVAS cells were transfected with NC, miR-342-5p inhibitor, NC + siPIK3R1 or miR-342-5p inhibitor + siPIK3R1 and the p-Akt and Akt protein expression levels were determined using (A) western blot analysis and (B) quantified, and analyzed statistically. (C) The ratio of p-AKT/AKT was quantified in each experiment group. MOVAS cells were transfected with mock, mimics, mock + LY294002 or mimics + LY294002 and the p-Akt and Akt protein levels were analyzed using (D) western blot analysis and (E) quantified, and analyzed statistically. (F) The ratio of p-AKT/AKT was quantified in each experiment group. (G) Cell Counting Kit-8 was used to determine cell proliferation following transfection. **P<0.01 vs. NC; ##P<0.01 vs. NC + siPIK3R1; ^P<0.05, ^^P<0.01 vs. mock; and &P<0.05, &&P<0.01 vs. mock + LY294002. p, phosphorylated; NC, miR-342-5p inhibitor negative control; si, small interfering; PIK3R1, phosphatidylinositol 3-kinase regulatory subunit α; mock, miR-342-5p mimic negative control; OD, optical density; NC, miR-342-5p inhibitor negative control.
miR-342-5p activates the Akt signaling pathway and reverses the effect of LY294002
The cells were transfected with miR-342-5p mimics or NC, and treated with PI3K inhibitor (LY294002) to confirm the role of miR-342-5p in the Akt signaling pathway and the p-Akt and Akt protein expression levels were subsequently determined using western blot analysis. The data revealed that p-Akt protein levels were higher in the mimics group compared with that in the mock group (P<0.05; Fig. 8D and E), but were downregulated in mock + LY294002 and mimics + LY294002 groups compared with that in the mock group (P<0.05; Fig. 8D and E). Moreover, the p-Akt protein level was upregulated in the mimics + LY294002 group compared with that in the mock + LY294002 group (P<0.05; Fig. 8D and E). However, there were no significant differences in the total Akt protein expression levels among the four groups (P<0.05; Fig. 8D and E). The ratio of p-Akt/Akt was increased in the mimic group, while reduced in the mock + LY294002 group compared with the mock group; and the ratio of p-Akt/Akt was higher in the mimic + LY294002 group than that in the mock + LY294002 group (P<0.05; Fig. 8F). Thus, miR-342-5p upregulated p-Akt levels and activated the Akt pathway and miR-342-5p could activate the AKT pathway, which was inhibited by LY294002.
To investigate the role of the Akt pathway in cells, cell proliferation was examined using a CCK-8 assay. Following 24 and 48 h of transfection, cell proliferation increased in the miR-342-5p mimics group, but decreased in the mock + LY294002 group compared with that in the mock group after 48 h of transfection (P<0.05; Fig. 8E). In addition, cell proliferation was also increased in the mimics + LY294002 group after 72 h compared with that in the mock + LY294002 group (P<0.05; Fig. 8E). Inhibition of the Akt signaling pathway decreased cell proliferation, and this decrease could be reversed by miR-342-5p transfection.
Discussion
The proliferation and migration of VSMCs can trigger vascular lesions, leading to abnormal functioning of the heart or cause diseases such as hypertension and diabetes (24). A recent study suggested that some miRNAs (such as miR-665 and miR-143/-145) may play important roles in the development of cardiovascular diseases (8,25). In the present study, miR-342-5p was shown to improve cell migration and invasion, promote cell proliferation and the phenotypic transformation of VSMCs (as shown by the altered α-SMA and vimentin expression profiles). PIK3R1 was found to be a target gene for miR-342-5p, and miR-342-5p could regulate the proliferation and differentiation of VSMCs via activation of the Akt signaling pathway through inhibition of PIK3R1.
In previous studies, Yan et al (17) found that miR-342-5p was involved in mediating angiogenesis in human umbilical cord tissue samples; Ahmadi et al (16) found that the mRNA expression level of miR-342-5p in peripheral blood mononuclear cells in patients with coronary heart disease was increased and Ge et al (26) concluded that serum miR-342-5p levels might be a novel biomarker for pertussis. To further investigate the role of miR-342-5p in MOVAS cells, in vitro transfection was performed to increase or suppress the expression of miR-342-5p in the cells for subsequent experiments. miR-342-5p mimics significantly upregulated miR-342-5p levels in MOVAS cells compared with that in the control group, whereas miR-342-5p inhibitor downregulated miR-342-5p levels. Abnormally increased proliferation of VSMCs plays a key role in the in the development of atherosclerosis and in restenosis in some diseases (27-29), and miR-92 regulates vascular smooth muscle cell function by targeting KLF4 during vascular restenosis and injury (29). In VSMC injury, VSMCs undergo phenotype switching by transforming from a differentiated and contractile phenotype to a proliferative, migratory and synthetic state. (30) This can lead to increased proliferation, migration and secretory abilities and contributes to the development of cardiovascular diseases (31). In the present study, high expression of miR-342-5p in MOVAS cells increased cell viability, migration and invasion, which, could be reversed by inhibiting the expression of miR-342-5p. These results indicated that miR-342-5p might promote the proliferation and phenotypic transformation of VSMCs.
Increases in VSMCs proliferation causes cell migration, expression of chemokines and regulation of extracellular substrates (32,33). VSMCs are considered to have two phenotypes: Contractile (differentiated) phenotype and synthetic (undifferentiated or dedifferentiated) phenotype (34). Contractile VSMCs belongs to the mature type, which has poor proliferation and migration ability, and elevated protein expression levels of contractile markers, such as α-SMA, calponin H1 and smooth muscle protein 22-α (35). However, the expression levels of these contractile markers were reversed in the synthetic VSMC phenotype, which also present with a simultaneous increase in the protein expression of synthetic phenotype markers, such as vimentin and osteopontin (36,37). Thus, phenotypic transformation of VSMCs is characterized by expressional changes of these markers. Yan et al (17) demonstrated that overexpression of miR-342-5p upregulated mesenchymal phenotype markers, such as atlastin 1, vimentin, Twist-related protein 1, β-catenin and α-SMA. The results from the present study showed that overexpression of miR-342-5p increased vimentin and reduced α-SMA mRNA and protein expression levels, and regulate VSMC phenotypic conversion; however, low expression levels of miR-342-5p decreased vimentin and increased α-SMA expression. This indicates that overexpression of miR-342-5p inhibited the expression of contractile markers but promoted the expression of synthetic phenotype markers, thus possibly inducing the phenotypic transformation of VSMCs from a contractile phenotype to a synthetic phenotype. Conversely, low expression of miR-342-5p could inhibit phenotypic transformation of VSMCs.
In the current study, it was found that PIK3R1 was a target gene for miR-342-5p, and silencing PIK3R1 could reverse the effect of miR-342-5p inhibitors on the VSMCs. Akt is a threonine protein kinase and its signaling pathway is involved in the regulation of a variety of biological processes, including proliferation, growth, metabolism, angiogenesis and metastasis (38,39). Several studies have confirmed that the PI3K/Akt signaling pathway plays an important role in the regulation of VSMC phenotypic transformation (40-42). The results from the present study revealed that the Akt pathway was activated by silencing PIK3R1. However, inhibition of miR-342-5p mRNA expression could suppress the Akt pathway, which was partially reversed by siPIK3R1. The results revealed that reduction of PIK3R1 could activate the Akt signaling pathway. The cell proliferation of VSMCs and protein expression levels of p-Akt were significantly inhibited following treatment with LY294002, indicating that the Akt pathway was inhibited by LY294002. Under the effects of high miR-342-5p expression, cell proliferation increased and the Akt pathway was partially activated.
The present study demonstrated that miR-342-5p could enhance cell proliferation, and promote cell migration, invasion and phenotypic transformation of MOVAS cells, which may be associated with the activation of the Akt pathway induced by PIK3R1 inhibition. Other studies have also confirmed the role of miR-342-5p in atherosclerosis or angiogenesis using animal studies, in vitro functional experiments and in human tissue samples (15-17). However, whether miR-342-5p can be used as a marker in vascular diseases remains to be further elucidated.
In conclusion, miR-342-5p could improve cell proliferation, promote cell migration and invasion, increase vimentin and decrease α-SMA expression. PIK3R1 was a target gene of miR-342-5p, and decreased PIK3R1 expression could improve cell proliferation, promote cell migration and invasion, and activate the Akt signaling pathway. Thus, miR-342-5p may promote the proliferation and differentiation of VSMCs via regulating the Akt signaling pathway through targeting PIK3R1. The results from the present study may provide a novel insight into the treatment of cardiovascular diseases.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SB made substantial contributions to the conception and design of the study, and drafted and revised the manuscript for important intellectual content. QP, WL, CZ and ZL acquired the data, and performed analysis and interpretation. All authors have read and approved the final manuscript and agree to be accountable for the accuracy and integrity of the study.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lisy K, Campbell JM, Tufanaru C, Moola S, Lockwood C. The prevalence of disability among people with cancer, cardiovascular disease, chronic respiratory disease and/or diabetes: A systematic review. Int J Evid Based Healthc. 2018;16:154–166. doi: 10.1097/XEB.0000000000000138. [DOI] [PubMed] [Google Scholar]
- 2.Stevens W, Peneva D, Li JZ, Liu LZ, Liu G, Gao R, Lakdawalla DN. Estimating the future burden of cardiovascular disease and the value of lipid and blood pressure control therapies in China. BMC Health Serv Res. 2016;16(175) doi: 10.1186/s12913-016-1420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang CH, Ciou JS, Chen ST, Kok VC, Chung Y, Tsai JJ, Kurubanjerdjit N, Huang CF, Ng KL. Identify potential drugs for cardiovascular diseases caused by stress-induced genes in vascular smooth muscle cells. PeerJ. 2016;4(e2478) doi: 10.7717/peerj.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tykocki NR, Boerman EM, Jackson WF. Smooth muscle ion channels and regulation of vascular tone in resistance arteries and arterioles. Compr Physiol. 2017;7:485–581. doi: 10.1002/cphy.c160011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang W, Chen S, Zhang Z, Wang C, Liu C. FAM3B mediates high glucose-induced vascular smooth muscle cell proliferation and migration via inhibition of miR-322-5p. Sci Rep. 2017;7(2298) doi: 10.1038/s41598-017-02683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nazari-Jahantigh M, Egea V, Schober A, Weber C. MicroRNA-specific regulatory mechanisms in atherosclerosis. J Mol Cell Cardiol. 2015;89:35–41. doi: 10.1016/j.yjmcc.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 7.Andreou I, Sun X, Stone PH, Edelman ER, Feinberg MW. miRNAs in atherosclerotic plaque initiation, progression, and rupture. Trends Mol Med. 2015;21:307–318. doi: 10.1016/j.molmed.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li K, Pan J, Wang J, Liu F, Wang L. MiR-665 regulates VSMCs proliferation via targeting FGF9 and MEF2D and modulating activities of Wnt/β-catenin signaling. Am J Transl Res. 2017;9:4402–4414. [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen MA, Karunakaran D, Rayner KJ. Unlocking the door to new therapies in cardiovascular disease: MicroRNAs hold the key. Curr Cardiol Rep. 2014;16(539) doi: 10.1007/s11886-014-0539-7. [DOI] [PubMed] [Google Scholar]
- 10.Zhu R, Liu X, Zhu Y, He Z. MiRNAs: Potential diagnostic and therapeutic targets for cerebral ischaemia. Neurol Res. 2016;38:86–92. doi: 10.1080/01616412.2015.1136103. [DOI] [PubMed] [Google Scholar]
- 11.Jia YY, Zhao JY, Li BL, Gao K, Song Y, Liu MY, Yang XJ, Xue Y, Wen AD, Shi L. miR-592/WSB1/HIF-1α axis inhibits glycolytic metabolism to decrease hepatocellular carcinoma growth. Oncotarget. 2016;7:35257–35269. doi: 10.18632/oncotarget.9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Jiang M, Xu Z, Huang H, Gong P, Zhu H, Ruan C. miR-146b-5p promotes VSMC proliferation and migration. Int J Clin Exp Pathol. 2015;8:12901–12907. [PMC free article] [PubMed] [Google Scholar]
- 13.Choe N, Kwon DH, Shin S, Kim YS, Kim YK, Kim J, Ahn Y, Eom GH, Kook H. The microRNA miR-124 inhibits vascular smooth muscle cell proliferation by targeting S100 calcium-binding protein A4 (S100A4) FEBS Lett. 2017;591:1041–1052. doi: 10.1002/1873-3468.12606. [DOI] [PubMed] [Google Scholar]
- 14.Bi R, Ding F, He Y, Jiang L, Jiang Z, Mei J, Liu H. miR-503 inhibits platelet-derived growth factor-induced human aortic vascular smooth muscle cell proliferation and migration through targeting the insulin receptor. Biomed Pharmacother. 2016;84:1711–1716. doi: 10.1016/j.biopha.2016.10.081. [DOI] [PubMed] [Google Scholar]
- 15.Wei Y, Nazari-Jahantigh M, Chan L, Zhu M, Heyll K, Corbalán-Campos J, Hartmann P, Thiemann A, Weber C, Schober A. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127:1609–1619. doi: 10.1161/CIRCULATIONAHA.112.000736. [DOI] [PubMed] [Google Scholar]
- 16.Ahmadi R, Heidarian E, Fadaei R, Moradi N, Malek M, Fallah S. miR-342-5p expression levels in coronary artery disease patients and its association with inflammatory cytokines. Clin Lab. 2018;64:603–609. doi: 10.7754/Clin.Lab.2017.171208. [DOI] [PubMed] [Google Scholar]
- 17.Yan XC, Cao J, Liang L, Wang L, Gao F, Yang ZY, Duan JL, Chang TF, Deng SM, Liu Y, et al. miR-342-5p is a notch downstream molecule and regulates multiple angiogenic pathways including notch, vascular endothelial growth factor and transforming growth factor β signaling. J Am Heart Assoc. 2016;5(e003042) doi: 10.1161/JAHA.115.003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivard A, Andres V. Vascular smooth muscle cell proliferation in the pathogenesis of atherosclerotic cardiovascular diseases. Histol Histopathol. 2000;15:557–571. doi: 10.14670/HH-15.557. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Zhong JC, Zhang ZZ, Wang W, McKinnie SMK, Vederas JC, Oudit GY. Targeting the apelin pathway as a novel therapeutic approach for cardiovascular diseases. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1942–1950. doi: 10.1016/j.bbadis.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Yu G, Luo Z, Zhou Y, Zhang L, Wu Y, Ding L, Shi Y. Uncovering the pharmacological mechanism of Carthamus tinctorius L. On cardiovascular disease by a systems pharmacology approach. Biomed Pharmacother. 2019;117(109094) doi: 10.1016/j.biopha.2019.109094. [DOI] [PubMed] [Google Scholar]
- 22.Cai W, Zhang J, Yang J, Fan Z, Liu X, Gao W, Zeng P, Xiong M, Ma C, Yang J. doi: 10.1016/j.numecd.2019.03.002. MicroRNA-24 attenuates vascular remodeling in diabetic rats through PI3K/Akt signaling pathway. Nutr Metab Cardiovasc Dis 621-632, 2019. [DOI] [PubMed] [Google Scholar]
- 23.Meester JAN, Verstraeten A, Alaerts M, Schepers D, Van Laer L, Loeys BL. Overlapping but distinct roles for NOTCH receptors in human cardiovascular disease. Clin Genet. 2019;95:85–94. doi: 10.1111/cge.13382. [DOI] [PubMed] [Google Scholar]
- 24.Reddy MA, Das S, Zhuo C, Jin W, Wang M, Lanting L, Natarajan R. Regulation of vascular smooth muscle cell dysfunction under diabetic conditions by miR-504. Arterioscler Thromb Vasc Biol. 2016;36:864–873. doi: 10.1161/ATVBAHA.115.306770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao W, Zhao SP, Zhao YH. MicroRNA-143/-145 in cardiovascular diseases. Biomed Res Int. 2015;2015(531740) doi: 10.1155/2015/531740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ge Y, Zhao K, Qi Y, Min X, Shi Z, Qi X, Shan Y, Cui L, Zhou M, Wang Y, et al. Serum microRNA expression profile as a biomarker for the diagnosis of pertussis. Mol Biol Rep. 2013;40:1325–1332. doi: 10.1007/s11033-012-2176-9. [DOI] [PubMed] [Google Scholar]
- 27.Chistiakov DA, Orekhov AN, Bobryshev YV. Vascular smooth muscle cell in atherosclerosis. Acta Physiol (Oxf) 2015;214:33–50. doi: 10.1111/apha.12466. [DOI] [PubMed] [Google Scholar]
- 28.Psaltis PJ, Simari RD. Vascular wall progenitor cells in health and disease. Circ Res. 2015;116:1392–1412. doi: 10.1161/CIRCRESAHA.116.305368. [DOI] [PubMed] [Google Scholar]
- 29.Deng S, Zhang Y, Wang Y, Lu X, Jiang Q. MicroRNA-92 regulates vascular smooth muscle cell function by targeting KLF4 during vascular restenosis and injury. Int J Clin Exp Pathol. 2019;12:4253–4262. [PMC free article] [PubMed] [Google Scholar]
- 30.Kingsley K, Huff JL, Rust WL, Carroll K, Martinez AM, Fitchmun M, Plopper GE. ERK1/2 mediates PDGF-BB stimulated vascular smooth muscle cell proliferation and migration on laminin-5. Biochem Biophys Res Commun. 2002;293:1000–1006. doi: 10.1016/S0006-291X(02)00331-5. [DOI] [PubMed] [Google Scholar]
- 31.Chiong M, Cartes-Saavedra B, Norambuena-Soto I, Mondaca-Ruff D, Morales PE, García-Miguel M, Mellado R. Mitochondrial metabolism and the control of vascular smooth muscle cell proliferation. Front Cell Dev Biol. 2014;2(72) doi: 10.3389/fcell.2014.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park ES, Kang SI, Yoo KD, Lee MY, Yoo HS, Hong JT, Shin HS, Kim B, Yun YP. Camptothecin inhibits platelet-derived growth factor-BB-induced proliferation of rat aortic vascular smooth muscle cells through inhibition of PI3K/Akt signaling pathway. Exp Cell Res. 2013;319:982–991. doi: 10.1016/j.yexcr.2012.12.024. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Zhan Y, Izumi Y, Yasumoto H, Yano M, Iwao H. In vivo activation of rat aortic platelet-derived growth factor and epidermal growth factor receptors by angiotensin II and hypertension. Arterioscler Thromb Vasc Biol. 2000;20:2539–2545. doi: 10.1161/01.atv.20.12.2539. [DOI] [PubMed] [Google Scholar]
- 34.Pei C, Qin S, Wang M, Zhang S. Regulatory mechanism of human vascular smooth muscle cell phenotypic transformation induced by NELIN. Mol Med Rep. 2015;12:7310–7316. doi: 10.3892/mmr.2015.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Jacquet L, Karamariti E, Xu Q. Origin and differentiation of vascular smooth muscle cells. J Physiol. 2015;593:3013–3030. doi: 10.1113/JP270033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salabei JK, Cummins TD, Singh M, Jones SP, Bhatnagar A, Hill BG. PDGF-mediated autophagy regulates vascular smooth muscle cell phenotype and resistance to oxidative stress. Biochem J. 2013;451:375–388. doi: 10.1042/BJ20121344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong W, Li B, Yang P, Chen R, Wang C, Wang Z, Shao C, Yuan W, Yan J. CD137-CD137L interaction modulates neointima formation and the phenotype transformation of vascular smooth muscle cells via NFATc1 signaling. Mol Cell Biochem. 2018;439:65–74. doi: 10.1007/s11010-017-3136-4. [DOI] [PubMed] [Google Scholar]
- 38.Ersahin T, Tuncbag N, Cetin-Atalay R. The PI3K/AKT/mTOR interactive pathway. Mol Biosyst. 2015;11:1946–1954. doi: 10.1039/c5mb00101c. [DOI] [PubMed] [Google Scholar]
- 39.Xu T, Zhu H, Li D, Lang Y, Cao L, Liu Y, Wu W, Chen D. Luteolin inhibits angiotensin II-stimulated VSMC proliferation and migration through downregulation of Akt phosphorylation. Evid Based Complement Alternat Med. 2015;2015(931782) doi: 10.1155/2015/931782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown DJ, Rzucidlo EM, Merenick BL, Wagner RJ, Martin KA, Powell RJ. Endothelial cell activation of the smooth muscle cell phosphoinositide 3-kinase/Akt pathway promotes differentiation. J Vasc Surg. 2005;41:509–516. doi: 10.1016/j.jvs.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 41.Ponnusamy A, Sinha S, Hyde GD, Borland SJ, Taylor RF, Pond E, Eyre HJ, Inkson CA, Gilmore A, Ashton N, et al. FTI-277 inhibits smooth muscle cell calcification by up-regulating PI3K/Akt signaling and inhibiting apoptosis. PLoS One. 2018;13(e0196232) doi: 10.1371/journal.pone.0196232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fang H, Yang S, Luo Y, Zhang C, Rao Y, Liu R, Feng Y, Yu J. Notoginsenoside R1 inhibits vascular smooth muscle cell proliferation, migration and neointimal hyperplasia through PI3K/Akt signaling. Sci Rep. 2018;8(7595) doi: 10.1038/s41598-018-25874-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.