Summary
Background
Intratumoural heterogeneity has been previously shown to be related to clonal evolution and genetic instability and associated with tumour progression. Phenotypically, it is reflected in the diversity of appearance and morphology within cell populations. Computer-extracted features relating to tumour cellular diversity on routine tissue images might correlate with outcome. This study investigated the prognostic ability of computer-extracted features of tumour cellular diversity (CellDiv) from haematoxylin and eosin (H&E)-stained histology images of non-small cell lung carcinomas (NSCLCs).
Methods
In this multicentre, retrospective study, we included 1057 patients with early-stage NSCLC with corresponding diagnostic histology slides and overall survival information from four different centres. CellDiv features quantifying local cellular morphological diversity from H&E-stained histology images were extracted from the tumour epithelium region. A Cox proportional hazards model based on CellDiv was used to construct risk scores for lung adenocarcinoma (LUAD; 270 patients) and lung squamous cell carcinoma (LUSC; 216 patients) separately using data from two of the cohorts, and was validated in the two remaining independent cohorts (comprising 236 patients with LUAD and 335 patients with LUSC). We used multivariable Cox regression analysis to examine the predictive ability of CellDiv features for 5-year overall survival, controlling for the effects of clinical and pathological parameters. We did a gene set enrichment and Gene Ontology analysis on 405 patients to identify associations with differentially expressed biological pathways implicated in lung cancer pathogenesis.
Findings
For prognosis of patients with early-stage LUSC, the CellDiv LUSC model included 11 discriminative CellDiv features, whereas for patients with early-stage LUAD, the model included 23 features. In the independent validation cohorts, patients predicted to be at a higher risk by the univariable CellDiv model had significantly worse 5-year overall survival (hazard ratio 1·48 [95% CI 1·06–2·08]; p=0·022 for The Cancer Genome Atlas [TCGA] LUSC group, 2·24 [1·04–4·80]; p=0·039 for the University of Bern LUSC group, and 1·62 [1·15–2·30]; p=0·0058 for the TCGA LUAD group). The identified CellDiv features were also found to be strongly associated with apoptotic signalling and cell differentiation pathways.
Interpretation
CellDiv features were strongly prognostic of 5-year overall survival in patients with early-stage NSCLC and also associated with apoptotic signalling and cell differentiation pathways. The CellDiv-based risk stratification model could potentially help to determine which patients with early-stage NSCLC might receive added benefit from adjuvant therapy.
Funding
National Institue of Health and US Department of Defense.
Introduction
Tumour cellular heterogeneity has been shown to be a hallmark of all cancers, with a diverse group of cell populations including cancer cells, immune cells, mesenchymal cells, and the like making up a heterogeneous solid tumour.1-3 Several studies have shown that tumour progression and carcinogenesis are related to clonal evolution and genetic instability, with highly aggressive tumours being far more heterogeneous than less aggressive variants. The presence of genetic sub-clonal populations in cancers, termed intratumoural heterogeneity, has been shown to be an independent prognostic factor of outcome in several different cancer types such as breast cancer3 and head and neck cancer,4 with high intratumoural heterogeneity having markedly worse patient survival and implicated in drug resistance.5
Nuclear morphological differences or nuclear pleomorphism have traditionally been known to be pathognomonic of cancer and a marker for tumour differentiation.6 This genetic intratumoural heterogeneity is reflected in the morphological makeup of the tissue, with more rapidly growing or aggressive cancers showing a greater cellular heterogeneity among cancer cells compared with a relatively indolent tumour.7 Tumours with higher intratumoural heterogeneity and higher nuclear diversity have been shown to have a poorer prognosis than cancers with low intratumoural heterogeneity and less nuclear diversity. Thus, quantifying this sub-visual morphological diversity in tissues would be a good surrogate for genetic intratumoural heterogeneity.
In early-stage (stage I and II) non-small cell lung carcinomas (NSCLCs), surgical resection is the treatment of choice, but almost 40–55% of these tumours recur after surgery.8 Several genomic-based prognostic biomarkers of outcome exist in early-stage NSCLC, but these are typically developed from a single biopsy and thus might not comprehensively account for genetic and morphological intratumoural heterogeneity present in NSCLC. Features relating to tumour cellular diversity and local morphological heterogeneity within tissue slides might be driven by the genomic and epigenetic alterations and could potentially provide a tissue non-destructive way of predicting disease outcome.
Here, we present a new histogenomic approach—local cellular morphological diversity (referred to as CellDiv)—to interrogate the diversity of nuclear morphology in the epithelium region, and employ it in conjunction with a Cox proportional hazards model to predict overall survival in early-stage NSCLC. A deep-learning neural network model based on U-net9 was first employed to segment the epithelium region and nuclei in the image for downstream calculation of the nuclear morphological features. Because early-stage NSCLC can be broadly differentiated into the two major groups—lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD)—with varying driver mutations and epigenetic pathways,10 we independently analysed the two subpopulations, keeping in mind the intrinsic differences between them. Our histogenomic analysis involved investigating the associations of these computerised CellDiv features with biological pathways implicated in carcinogenesis as well as studying the underexpression and overexpression of biological pathways associated with the CellDiv-derived prognostic risk groups.
Methods
Study design
The experimental design of this study has five key steps: data acquisition, local cellular diversity computation, calculation of the cellular diversity-based risk score, survival analysis, and histogenomic analysis (appendix 1 pp 9–10). Digitised tissue micro-arrays (TMAs) and whole slide images (WSIs) were obtained from four independent cohorts, which were divided into two training cohorts, and two independent validation cohorts. The nuclei identified in the haematoxylin and eosin (H&E)-stained images were segmented by an automatic method and a local nuclear graph (LNG) was constructed based on nuclear proximity. CellDiv features were then extracted from each LNG and used to calculate the cellular diversity-based risk score. We used the Least Absolute Shrinkage and Selection Operator (LASSO) method to discover the top features for constructing risk score, for LUAD and LUSC specifically, using a Cox proportional hazard model on the training cohorts. After locking down the Cox model, a risk score was generated for each patient in the two independent validation cohorts and survival analysis was done to evaluate the pre-trained Cox model. We compared the CellDiv model with existing models based on clinical variables in terms of precision-recall area under the receiver operating characteristic curve (AUC). Finally, we did histogenomic analysis to explore the association of morphological tumour cellular diversity with biological pathways.
Datasets
Formalin-fixed paraffin-embedded H&E-stained WSIs and TMAs collated from four independent and well characterised NSCLC cohorts were included in this study, representing 2213 patients. We required routine H&E-stained diagnostic images from patients with overall stage I and II cancer, for whom overall survival information was available (appendix 1 p 8). We excluded patients with locally advanced and metastatic (stage III and IV) tumours (n=1155), TMA spots that were not usable due to a lack of sufficient tissue for analysis (n=62), and slides with artifacts such as tissue folding and bubbles (n=37). Of the 1057 patients retained for this study, 506 had early-stage LUAD and 551 had early-stage LUSC (appendix 1 p 8).
The four cohorts are represented by D1 (n=395), D2(n=91), D3 (n=473), and D4 (n=98). D1 comprised TMA samples from the Cleveland Clinic, resected between 2004 and 2014, with a mean follow-up of 53· 8 months (SD 9·6). D2 comprised TMA samples from Yale Medical School, resected between 1988 and 2003, with a mean follow-up of 41·7 months (11 ·5). D3 comprised diagnostic WSIs from The Cancer Genome Atlas (TCGA). D4 comprised TMA samples from the University of Bern,11 resected between 2000 and 2013, with a mean follow-up of 29·1 months (SD 1838). All cohorts featured patients with LUAD and LUSC, except for the University of Bern cohort, which featured LUSC only. Scanning details of the different cohorts are included in appendix 1 (p 5). Clinicopathological and outcome information for patients in D1, D2, and D4 was obtained from Insitutional Review Board-approved retrospective chart review from the respective institutions. The corresponding information for patients in D3 was obtained from the TCGA. Cohorts D1 and D2 were used for feature discovery and model training, whereas D3 and D4 were used for independently validating the trained model.
This study conforms to Health Insurance Portability and Accountability Act guidelines and was approved by the Institutional Review Board at University Hospitals Cleveland Medical Center (number 02–13–42C). Informed consent requirement was waived as the study used archival tissue. Usage of the University of Bern cohort was approved by the local Ethics Commission (KEK 200/14), which waived the requirement for written informed consent.
Automatic characterisation of cellular diversity
A U-net-based convolutional neural network model9 was employed to detect the epithelial region from the digitised H&E-stained images, and then to detect and segment the nuclei. Once nuclei were detected and segmented, LNGs were constructed on the basis of the proximity of the individual nuclei (appendix 1 p 11). The intuition behind using LNGs was to capture the dissimilarity of proximally situated nuclei. The process of construction of an LNG involves first representing the centroids of the individual nuclei as nodes of a graph. Using the approach described by Foulkes12 and Corredor and colleagues,13 each node is then connected to the other nodes according to the Euclidean distance, a weighting function that favours the connectivity between proximal nodes. After this process, multiple disconnected subgraphs or clusters of nuclei are generated (appendix 1 p 2).
A set of 11 nuclear morphologic features, quantifying nuclear shape and appearance, were first extracted from the H&E-stained images to quantify the nuclear shape and appearance based on the presegmented nuclei (appendix 1 pp 20–22). Each individual nuclear feature was discretised into five levels. We explored the discretisation criterion ω for values ranging from 3 to 7. Setting ω=3 will lead to a small co-occurrence matrix of size 3 × 3, which limits the spectrum of the diversity that can be captured, whereas setting ω=7 will lead to a very sparse co-occurrence matrix. Thus, empirically, we identified ω=5 to be the ideal level for discretising the nuclear diversity features. To explore the local tumour cellular diversity in terms of different shape and texture attributes, corresponding co-occurrence matrices based on the 11 extracted nuclear features were constructed (appendix 1 pp 2–4). 13 high-order statistical features (eg, entropy, energy)14 were then extracted from each of the 11 co-occurrence matrices. Thus, each of the M different LNGs in each WSI represented by Gu, where u belongs to the set {1, 2, …., M}, is uniquely represented by a total of 11 different 13-dimensional feature vectors Hk=[hk,1,…,hk,13], where k ranges from one to 11. The final CellDiv signature (715-dimensional vector) for each single WSI is formed by the first-order statistics (mean, SD, kurtosis, skewness, and range) aggregated across all Gu.
Cox proportional hazard model
A Cox proportional hazard model, henceforth referred to as the Cox model, was trained using the top CellDiv features identified from D1 and D2 to generate continuous risk scores for all patients. We chose this model because it considers the time-to-event duration as well as censoring information to construct the model. The top discriminant CellDiv features were identified using LASSO with the Cox model as the cost function. The LASSO model was fitted under a ten-fold cross-validation scheme. The risk score for each patient was then calculated as the linear combination of the weights, β, of the top CellDiv features and associated values. The median value Topt of all the risk scores in training cohorts D1 and D2 was locked down as the optimal threshold for separating patients by risk level, with any value higher than the median categorised as high risk and median or lower categorised as low risk. We constructed the image models specifically for early-stage LUAD and LUSC and evaluated them separately.
The performance of the locked-down Cox model was evaluated in a blinded fashion on the independent validation test sets D3 and D4. The locked-down Cox model generated a risk score for each patient in the validation test set. The optimal threshold Topt learnt from the training cohorts was then applied to these risk scores to separate the patients into low risk and high risk.
Survival analysis
We chose overall survival as our endpoint because it is considered the gold standard in outcome for clinical trials and studies. We focused on 5-year overall survival because studies have shown that in early-stage NSCLC, 5-year and 10-year overall survival were equivalent.15 Overall survival was defined as the time interval between the date of diagnosis and the date of death. Patients who were still alive at the last reported date were labelled as censored.
We used Kaplan-Meier survival analysis to examine the difference in overall survival between patients categorised as high risk or low risk by the model, and the difference of overall survival in each group was assessed by the log-rank test. Univariable Cox regression analysis was calculated to examine the prognostic ability of CellDiv features and other clinical and pathological parameters including age (>65 years vs ≤65 years), sex (male vs female), race (white vs other), smoking status (ever smoker vs never smoker), overall stage (II vs I), T stage (T2/T2a/T2b/T3 vs T1/T1a/T1b), and N stage (N1 vs N0). Multivariable Cox regression analysis was calculated to examine the predictive ability of CellDiv risk group when controlling for the effects of clinical and pathological parameters including age, smoking status, overall stage, T stage, and N stage. Mantel-Haenszel hazard ratios were calculated in univariable and multivariable analysis. p values were two sided and p<0·05 was considered to be statistically significant.
Histogenomics analysis
We first used CellDiv features to construct a machine learning classifier for KRAS mutational status, using data from the 236 patients with LUAD with data on KRAS mutational status (appendix 1 pp 4–5). We evaluated the association of CellDiv-identified prognostic risk groups and differentially expressed pathways, to help to elucidate the relationship between the histological image phenotype and the corresponding genotype. For the TCGA LUAD and LUSC cohorts, normalised mRNA expression data were available for 405 patients (195 with LUAD and 210 with LUSC), obtained from the Genomic Data Commons portal. These transcriptomic data (IlluminaHiSeq), which consisted of 20 531 annotated genes, were used to investigate the underlying biological pathways of the risk scores derived from the pathological image analysis. First, all the normalised genes were recorded based on their association with the CellDiv risk group, with patients categorised as high risk or low risk. Based on an assumption that gene expression values are not normally distributed,16 genes that differentially express across patients in the two risk categories were selected using the Wilcoxon rank sum test, using a statistically significant threshold of 0·5. The Benjamini and Hochberg method was used to adjust p values and control for the false discovery rate in multiple testing.17 The most differentially expressed genes, which were significantly associated with the risk score, were then used in Gene Ontology analysis to identify distinct Gene Ontology-based biological processes.18 Gene Ontology provides structured, controlled vocabularies and classifications that cover several domains of molecular and cellular biology. Gene Ontology analysis highlights the most over-represented genes and finds the systematic linkages between those genes and biological processes.
The next step in the histogenomic analysis involved selecting a set of pathways that were representative of biological processes and doing single-sample gene set enrichment analysis (ssGSEA). ssGSEA, an extension of GSEA, is a computational method that determines whether a predefined set of genes shows significant, concordant differences between two biological states (eg, phenotypes),19 and calculates an enrichment score for every patient in the cohort. Each ssGSEA enrichment score represents the degree to which the genes in a particular gene set are coordinately upregulated or downregulated within a sample. The predefined sets of genes for the Gene Ontology-based biological processes were acquired from the Molecular Signatures Database. In our case, ssGSEA was used to find pathway associations with tumour cellular diversity-defined phenotypes individually for LUSC and LUAD. This helps to overcome limitations of single-gene analysis which often misses important biological pathways that tend to affect a set of genes acting together, rather than a single gene-based analysis.19 Significant differentially expressing pathways with respect to CellDiv features that contributed to the risk score were then selected using the Wilcoxon rank sum test.
Role of the funding source
The funders of the study played no role in study design, data collection, data analysis, data interpretation, or writing of the report. All authors had full access to all the data in the study and the corresponding author had final responsibility for the decision to submit for publication.
Results
Clinical and pathological data for all four cohorts are summarised in table 1. Patients were primarily white men in their mid-60s, and about 80% of patients were current or former smokers.
Table 1:
Summary of clinical and pathological data by cohort
| Entire cohort | D1: Cleveland Clinic | D2: Yale Medical School | D3: TCGA |
D4: University of Bern (LUSC only; n=98) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| LUSC (n=551) |
LUAD (n=506) |
LUSC (n=152) |
LUAD (n=243) |
LUSC (n=64) |
LUAD (n=27) |
LUSC (n=237) |
LUAD (n=236) |
||
| Age, years | 66·8 (9·4) | 65·8 (10·4) | 66·3 (9·9) | 67·5 (9·9) | 64·7 (8·7) | 63·6 (11·6) | 67·0 (9·5) | 66·3 (9·7) | 69·2 (7·9) |
| Sex | |||||||||
| Male | 360 (65%) | 238 (47%) | 82 (54%) | 128 (53%) | 54 (84%) | 21 (78%) | 146 (62%) | 89 (38%) | 78 (80%) |
| Female | 191 (35%) | 268 (53%) | 70 (46%) | 115 (47%) | 10 (16%) | 6 (22%) | 91 (38%) | 147 (62%) | 20 (20%) |
| Race | |||||||||
| White | 492 (89%) | 429 (85%) | 133 (88%) | 213 (88%) | 59 (92%) | 23 (85%) | 202 (85%) | 193 (82%) | 98 (100%) |
| Other | 47 (9%) | 64 (13%) | 18 (12%) | 30 (12%) | 5 (8%) | 4 (15%) | 24 (10%) | 30 (13%) | 0 |
| Smoking status | |||||||||
| Ever | 436 (79%) | 389 (77%) | 139 (91%) | 186 (77%) | NA | NA | 211 (89%) | 203 (86%) | 86 (88%) |
| Never | 24 (4%) | 66 (13%) | 1 (1%) | 36 (15%) | NA | NA | 23 (10%) | 30 (13%) | 0 |
| T stage | |||||||||
| T1/T1a/T1b | 181 (33%) | 213 (42%) | 64 (42%) | 126 (52%) | 8 (13%) | 3 (11%) | 86 (36%) | 84 (36%) | 23 (23%) |
| T2/T2a/T2b | 309 (56%) | 242 (48%) | 68 (45%) | 96 (40%) | 31 (48%) | 12 (44%) | 135 (57%) | 134 (57%) | 75 (77%) |
| T3 | 41 (7%) | 43 (8%) | 20 (13%) | 21 (9%) | 5 (8%) | 4 (15%) | 16 (7%) | 18 (8%) | 0 |
| N stage | |||||||||
| N0 | 414 (75%) | 391 (77%) | 127 (84%) | 205 (84%) | 32 (50%) | 11 (41%) | 180 (76%) | 175 (74%) | 75 (77%) |
| N1 | 87 (16%) | 103 (20%) | 25 (16%) | 38 (16%) | 12 (19%) | 8 (30%) | 27 (11%) | 57 (24%) | 23 (23%) |
| Overall stage | |||||||||
| I/IA/IB | 309 (56%) | 278 (55%) | 75 (49%) | 108 (44%) | 39 (61%) | 15 (56%) | 146 (62%) | 155 (66%) | 49 (50%) |
| II/IIA/IIB | 177 (32%) | 107 (21%) | 15 (10%) | 14 (6%) | 22 (34%) | 12 (44%) | 91 (38%) | 81 (34%) | 49 (50%) |
Data are n (%) or mean (SD). NA=not applicable. LUAD=lung adenocarcinoma. LUSC=lung squamous cell carcinoma. TCGA=The Cancer Genome Atlas.
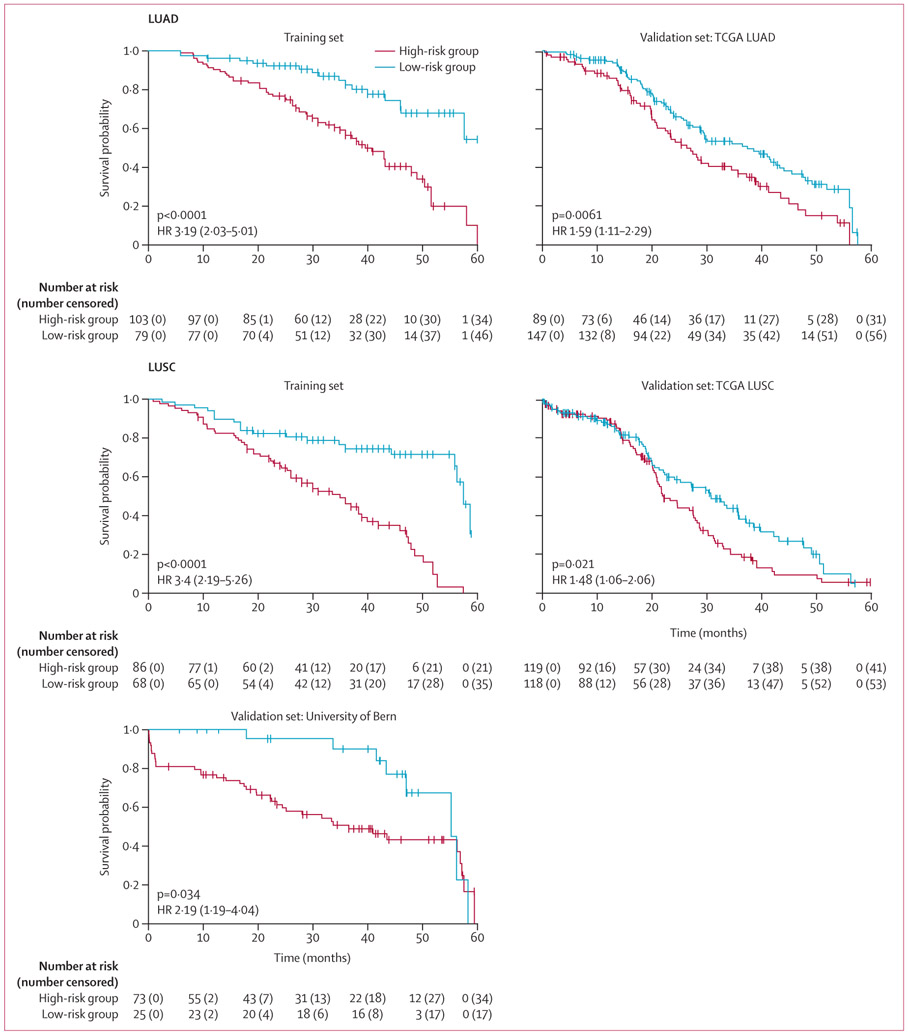
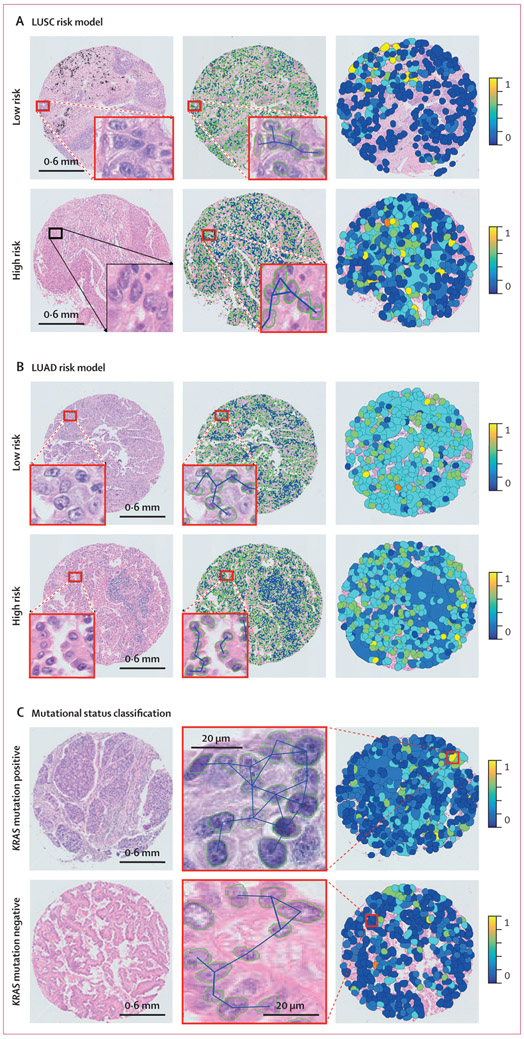
For early-stage LUSC prognostication, the CellDiv LUSC model included the 11 most discriminative CellDiv features, which were related to the nuclear shape (ie, major axis length of nuclei) and the nuclear intensity; the full list of feature names and their associated weight is presented in appendix 1 (p 23). In the univariable analysis of LUSC in D3, the CellDiv model was prognostic of 5-year overall survival, whereas none of the included clinical and pathological factors were significant (table 2). CellDiv LUSC was prognostic of overall survival in D4 as well, along with sex (table 2). In multivariable analysis, while controlling for clinicopathological factors, CellDiv was independently prognostic of overall survival in both validation test sets (table 2). This was supported by the Kaplan-Meier analysis (figure 1). When considering two representative cases of patients with LUSC who were identified as high risk and low risk by CellDiv with feature maps overlaid, the model determined that the low-risk tissue image had more local cell clusters (represented by the coloured patches) than the high-risk tissue image, with relatively lower CellDiv (figure 2).
Table 2:
Univariable and multivariable analysis for 5-year overall survival on the validation test sets D3 and D4
|
D3: TCGA (LUSC) |
D3: TCGA (LUAD) |
D4: University of Bern (LUSC) |
||||
|---|---|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | HR (95% CI) | p value | |
| Univariable Cox model analysis for overall survival | ||||||
| Age: >65 years vs ≤65 years | 1·13 (0·81–1·57) | 0·488 | 1·00 (0·72–1·41) | 0·982 | 1·71 (0·84–3·50) | 0·139 |
| Sex: male vs female | 0·81 (0·58–1·13) | 0·219 | 1·30 (0·92–1·86) | 0·142 | 3·62 (1·30–10·12) | 0·014 |
| Race: white vs other | 1·60 (0·93–2·74) | 0·087 | 0·81 (0·53–1·24) | 0·326 | NA | NA |
| Smoking status: ever vs never | 1·29 (0·79–2·10) | 0·303 | 1·36 (0·79–2·34) | 0·266 | NA | NA |
| T stage: T2/T2a/T2b/T3 vs T1/T1a/T1b | 1·24 (0·88–1·74) | 0·213 | 1·22 (0·86–1·72)) | 0·273 | 1·26 (0·60–2·61) | 0·542 |
| N stage: N1 vs N0 | 1·35 (0·90–2·01) | 0·146 | 1·92 (1·18–3·13) | 0·0083 | 1·19 (0·60–2·35) | 0·614 |
| Overall stage: II vs I | 1·22 (0·86–1·73) | 0·259 | 1·25 (0·85–1·84) | 0·257 | 1·05 (0·59–1·85) | 0·867 |
| Image model: high risk vs low risk | 1·48 (1·06–2·08) | 0·022 | 1·62 (1·15–2·30) | 0·006 | 2·24 (1·04–4·80) | 0·039 |
| Multivariable Cox model analysis controlling for clinical and pathological variables | ||||||
| Age: >65 years vs ≤65 years | 1·14 (0·81–1·61) | 0·451 | 1·12 (0·78–1·60) | 0·540 | 1·80 (0·84–3·85) | 0·131 |
| Smoking status: ever vs never | 1·36 (0·83–2·23) | 0·221 | 1·14 (0·64–2·01) | 0·661 | NA | NA |
| Overall stage: II vs I | 1·13 (0·66–1·94) | 0·651 | 1·86 (1·04–3·32) | 0·037 | 1·13 (0·66–1·94) | 0·708 |
| T stage: T2/T2a/T2b/T3 vs T1/T1a/T1b | 1·26 (0·85–1·87) | 0·244 | 1·25 (0·85–1·85) | 0·263 | 1·39 (0·60–3·19) | 0·438 |
| N stage: N1 vs N0 | 1·36 (0·77–2·41) | 0·292 | 3·11 (1·55–6·23) | 0·0012 | 1·17 (0·52–2·65) | 0–709 |
| Image model: high risk vs low risk | 1·52 (1·08–2·13) | 0·016 | 1·55 (1·09–2·22) | 0·015 | 2·34 (1·07–5·14) | 0·034 |
Mantel-Haenszel HRs are provided. TCGA=The Cancer Genome Atlas. LUSC=lung squamous cell carcinoma. LUAD=lung adenocarcinoma. HR=hazard ratio. NA=not applicable.
Figure 1: Kaplan-Meier 5-year overall survival according to risk category.
HR=hazard ratio. LUAD=lung adenocarcinoma. LUSC=lung squamous cell carcinoma. NA=not applicable. TCGA=The Cancer Genome Atlas.
Figure 2: Cellular diversity feature maps in LUSC risk model (A), LUAD risk model (B), and mutational status classification (C).
(A) Representative cases of LUSC and CellDiv feature map illustration. (B) Representative cases of LUAD and CellDiv feature map illustration. In (A) and (B), the first column shows haematoxylin and eosin-stained images with low-risk and high-risk patients as identified by the CellDiv model. The segmented nuclei contour and connecting edges are shown in the second column. The third column shows CellDiv features that capture the CellDiv in terms of nuclear shape (ie, area in panel A and eccentricity in panel B). Each colour patch represents individual LNGs in the image, where the blue and yellow colours represent the low and high normalised feature values. (C) Representative cases of KRAS mutation positive versus KRAS mutation negative, and the corresponding CellDiv feature map. LNG=local nuclear graph. LUAD=lung adenocarcinoma. LUSC=lung squamous cell carcinoma.
For early-stage LUAD prognostication, the CellDiv-LUAD model included the 23 most discriminative CellDiv features, which were related to the nuclear shape (eg, the solidity of nuclei) and the nuclear intensity; the full list of feature names and their associated weight is presented in appendix 1 (p 24). In the univariable analysis of LUAD in D3, the CellDiv model was prognostic of 5-year overall survival, while N stage was also significant (table 2). In multivariable analysis, CellDiv was independently prognostic of overall survival, along with overall stage and N stage (table 2). This was supported by the Kaplan-Meier analysis (figure 1). When considering two representative cases with local nuclear shape diversity feature maps overlaid, the high-risk example had a higher expression of the CellDiv feature relating to nuclear shape than the low-risk example (figure 2). In precision-recall AUC analysis, the CellDiv model outperformed existing clinical variable-based models for LUSC and LUAD (appendix 1 p 26).
We obtained a mean AUC of 0·63 in classification of KRAS status (60 KRAS mutation positive vs 176 KRAS mutation negative) using top six discriminative CellDiv features under five-fold cross-validation over 100 iterations (appendix 1 pp 4–5).
As part of our histogenomics analysis, we did an empirical analysis of the 20 531 annotated genes across the D3 LUSC and LUAD cohorts, which resulted in 299 and 207 differentially expressing genes (DEGs), respectively, between CellDiv-defined low-risk and high-risk groups based on 5-year overall survival (the full list of DEGs is presented in appendix 2). Our Gene Ontology analysis using these DEGs identified 23 significant biological pathways for LUSC and 15 for LUAD (a complete list of pathways is presented in appendix 2). These significant pathways were chosen on the basis of their biological significance in regulating tumour cellular diversity and carcinogenesis. In LUSC and LUAD, these pathways were broadly concerned with cell signalling, adhesion, division, localisation, apoptosis, and replication. Specifically, in LUAD, dendritic cell cytokine production, mast cell proliferation, regulation of apoptosis, pathways leading to DNA replication, and nucleus development were overexpressed in high-risk patients with higher cellular diversity. In LUSC, pathways of apoptotic signalling by p53, regulation of protein imports into the nucleus, cell adhesion and negative regulation of cellular differentiation, and cell signalling were differentially expressed between the CellDiv risk groups. The fold enrichment changes and strength of association between the CellDiv risk groups and significant biological processes in LUAD and LUSC are shown in appendix 2.
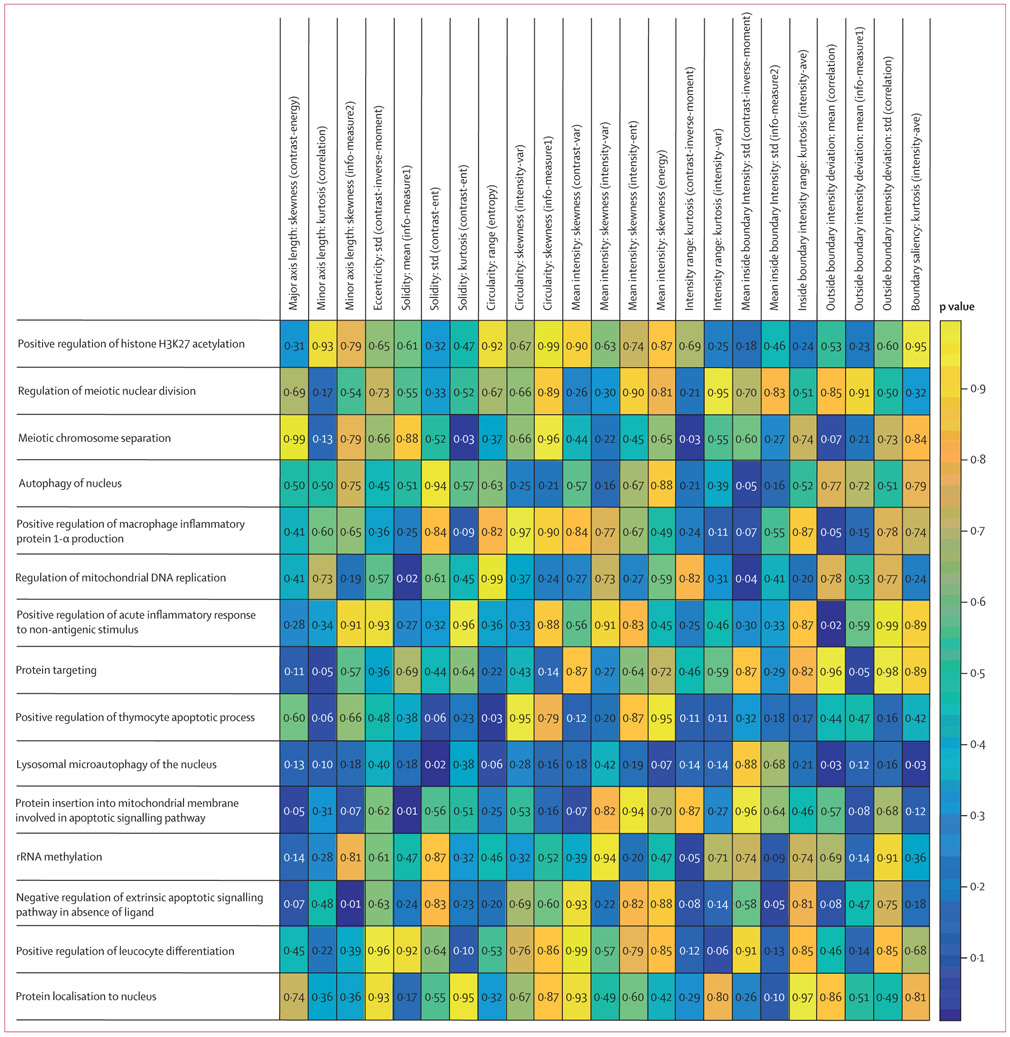
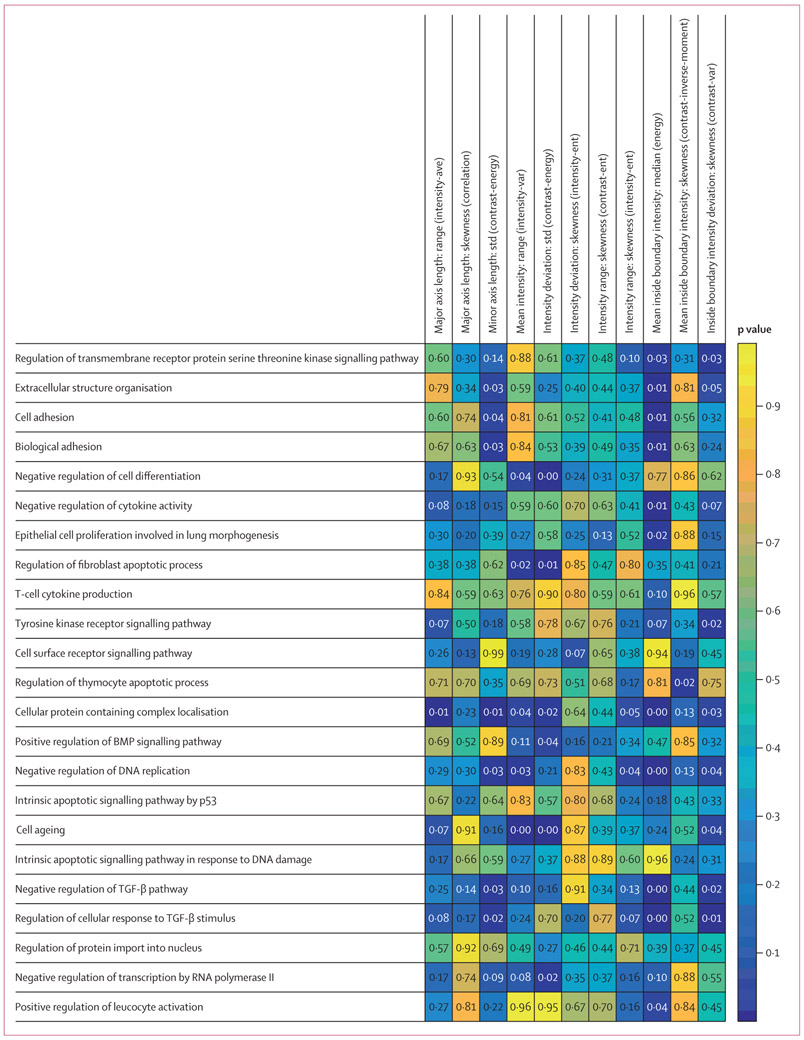
For a comprehensive histogenomics analysis, we evaluated the molecular underpinning of the prognostic CellDiv features by studying the corresponding association with ssGSEA. Gene set annotations for the 15 and 23 biological processes that were found significant in Gene Ontology analysis were used to calculate ssGSEA scores for each of the 23 most discriminative tumour cellular diversity features for LUAD and the 11 features for LUSC, respectively. In LUAD, CellDiv features were strongly associated with gene sets corresponding to apoptotic signalling, DNA replication, acute inflammatory response, and chromosome separation in meiosis pathways (figure 3). Meanwhile, in LUSC, CellDiv features were strongly correlated with pathways related to adhesion, cytokine activity, cell differentiation, leucocyte activation, and apoptotic signalling, among others (figure 4). A complete list of these differentially expressing genes can be found in appendix 2. In the case of LUSC, the local CellDiv features in terms of nuclear intensity (eg, mean intensity and mean inside boundary intensity: median [energy], which measure the nuclear intensity diversity in a local region) were strongly associated with cell ageing, adhesion, localisation, replication, apoptosis, and cytokine production. CellDiv features related to nuclear shape (eg, length of minor axis) were similarly strongly associated with pathways regulating cellular differentiation, cell signalling including bone morphogenetic protein signalling, and extracellular organisation. Similarly, in the case of LUAD, the local CellDiv in terms of shape (solidity and circularity) were strongly associated with pathways controlling histone acetylation, nuclear division, apoptosis, cellular differentiation, and nuclear autophagy among others. CellDiv features related to nuclear intensity meanwhile was found to be strongly correlated with apoptosis, nuclear autophagy, inflammatory response, nuclear division, and protein targeting.
Figure 3: Association between biological processes and the CellDiv features used to construct the prognostic models for LUAD.
The strength of association of biological processes, shown in rows, with the CellDiv features, shown in columns, by ssGSEA analysis. Wilcoxon rank sum test p values are shown, where p<0·05 shows an association between histomorphometric features used in the CellDiv models and certain pathways (while p<0·05). LUAD=lung adenocarcinoma. ssGSEA=single-sample gene set enrichment analysis.
Figure 4: Association between biological processes and the CellDiv features used to construct the prognostic models for LUSC.
The strength of association of biological processes, shown in rows, with the CellDiv features, shown in columns, by ssGSEA analysis. Wilcoxon rank sum test p values are shown, where p<0·05 shows an association between histomorphometric features used in the CellDiv models and certain pathways (while p<0·05). LUSC=lung squamous cell carcinoma. BMP=bone morphogenetic protein. TGF=transforming growth factor. ssGSEA=single-sample gene set enrichment analysis.
Discussion
Definitive resection in early-stage NSCLC is potentially curative and the standard of care, yet almost half of these patients experience recurrence following surgery. While adjuvant chemotherapy is routinely used in patients with stage II NSCLC, it is currently not recommended in patients with stage IA disease and there is controversy regarding its use in stage IB NSCLC due to contradictory results from prospective clinical trials.20 There is thus a need to develop a prognostic biomarker that can identify which patients with stage I NSCLC have more aggressive disease and can derive potential benefit from additional therapy following resection. Subsequently, a prognostic biomarker would also work to eliminate unnecessary chemotherapy for patients with low-risk stage II disease who would do well with surgery alone.
Existing prognostic biomarkers in NSCLC mostly rely on molecular or multigene-based assays.21 These tend to be expensive, time consuming, and tissue destructive while also not accounting for the inherent intratumoural heterogeneity present in tissues. For instance, Sandoval and colleagues22 presented a prognostic five-gene DNA methylation signature analysing 450 000 CpG sites from the tumoural DNA for stage I NSCLC. On an independent test cohort of 143 patients with stage I disease, the signature had a hazard ratio of 3·24 (95% CI 1·61–6·54; p<0·001) in prognosticating recurrence-free survival. Chen and colleagues23 presented a five-gene signature panel using RT-PCR that was prognostic of recurrence-free survival and overall survival on an independent test cohort of 42 patients with early-stage NSCLC, with a hazard ratio of 3·36 (1·35–8·35; p=0·009). Several studies have also shown the usefulness of single gene-based biomarkers including p53,24 ERBB2,25 RRM1,26 and BRCA for prognosticating survival in early-stage NSCLC.21
In this work, a risk score leveraging quantitative pathomorphometric features related to nuclear and morphologic diversity (CellDiv) was used to prognosticate overall survival in early-stage (stage I and II) NSCLC. Accounting for the well explored differences both morphologically and in the genetic makeup between LUSC and LUAD, independent CellDiv models were developed for each histological subtype to maximise model performance and to showcase the different biological underpinning behind the CellDiv features depending on tumour subtype. The developed CellDiv models were independently validated on a large multi-institutional cohort from the TCGA as well as an independent and blinded test cohort from the University of Bern.
Previous work in the area of computational pathology-based prognostic predictors for early-stage NSCLC includes works by Corredor and colleagues,13 Wang and colleagues,27 Saltz and colleagues,28 and Yu and colleagues.29 While not explicitly capturing cellular diversity, these approaches involved characterising the spatial arrangement and appearance of tumour-infiltrating lymphocytes and nuclei and relating these measurements with the likelihood of disease recurrence and progression. Andor and colleagues6 showed that diversity in nuclear intensity and shape were correlated with intratumoural heterogeneity in four different cancer types (LUAD, head and neck squamous cell carcinoma, and bladder and renal cell carcinomas, in 382 patients). Coudray and colleagues30 showed that a deep learning model is able to classify NSCLC into LUAD, LUSC, and normal with AUC of 0·97. In addition, the trained deep learning model can predict the ten most commonly mutated genes in LUAD, with AUCs from 0·73 to 0·86. Unlike the approach presented in this work, which relied on computationally derived intuitive features representing local cellular diversity, Coudray and colleagues30 used so-called black-box deep learning features with little explainability. Additionally, their work considered associations with single-gene driver mutations, whereas we explicitly looked at genome-level representations. Similarly, the work of Kather and colleagues31 showed an association between deep learning representations from WSI for gastrointestinal cancer and microsatellite instability. Note that while we did employ deep learning, it was solely used for tissue partitioning and nuclear segmentation, the pre-processing steps for the subsequent feature extraction.
Our work differed from these studies by developing and validating CellDiv features that represent morphological intratumoural heterogeneity and are representative of gene expression, and are also prognostic of survival in early-stage NSCLC. The CellDiv features employed a mathematical and computational model to capture local morphological heterogeneity. Given that there are both morphological and biological differences between LUAD and LUSC, independent dedicated prognostic models for LUAD and LUSC were separately constructed. The present work also encompasses histogenomics analysis by investigating the molecular and biological pathways that might drive these histomorphometric prognostic features by Gene Ontology and ssGSEA analysis. Additionally, we believe this is the first work to show that computer-extracted histomorphometric features were not only strongly prognostic of overall survival but associated with underlying morphological and biological pathway correlations.
In this work, we also explored the molecular underpinning of the CellDiv-defined prognostic risk groups on the TCGA dataset with available mRNA sequencing data. We showed the associations between specific CellDiv features and the significant biological pathways determined by ssGSEA. In adenocarcinomas, for instance, the selected CellDiv features in terms of nuclear solidity and mean inside boundary showed higher expression of genes related to the biological pathway of DNA replication32 and nucleus development.33 With the CellDiv features essentially capturing the degree of heterogeneity and diversity in shape, size, and texture of cancer nuclei, this seems to suggest that higher expression of those developmental pathways leads to more disordered or chaotic nuclei. Meanwhile, in LUSC, the family of bone morphogenetic protein and transforming growth factor β receptors, which have been already shown to be implicated in lung cancer carcinogenesis,34 were found to be associated with CellDiv features that measuring nuclear shape and intensity, clearly suggesting that the diversity features are being driven by the cellular differentiating and adhesion pathways.35 This was possibly reflective of the increased differentiation present in high-risk tumours as analysed by the CellDiv risk groups. Additionally, CellDiv features were also found to be associated with KRAS mutational status in adenocarcinomas, with a classification AUC of 0·63. Unlike deep learning-based methods presented by Coudray30 and Kather,31 CellDiv features explicitly capture morphologic heterogeneity in terms of cellular diversity, as opposed to more opaque representations that are not as intuitive or explainable.5
In LUAD, the apoptotic signalling pathway was found to be significantly associated with the degree of the solidity of the nuclei, possibly suggesting that the degree of nuclear diversity is dependent on the targeted cellular destruction of cancer nuclei leading to more disordered cellular structure in more aggressive cancers.36 Nuclear autophagy, which could be another potential reason for the disordered and more heterogeneous cellular structure,37 was also correlated with a CellDiv feature relating to the nuclear boundary intensity, reflecting textural heterogeneity in nuclei. For LUSC, biological pathways connected with the regulation of DNA replication and cell differentiation38 were strongly associated with the CellDiv feature analysing nuclear textural heterogeneity. This seems to suggest that more aggressive cancers are represented by a more heterogeneous cellular and nuclear organisation and architecture.
Our study had some limitations. First, the CellDiv prognostic model was developed and validated using retrospective data for prognosticating patient outcome, but was not validated for predicting the added benefit of adjuvant therapy. Future work will entail validation of the CellDiv model with access to the appropriate early-stage NSCLC clinical trial datasets (eg, one arm with surgery alone, the other arm with surgery plus adjuvant therapy). Similarly, another future direction to explore would be evaluating the ability of the CellDiv features to predict response to therapies such as checkpoint inhibitors. In addition, the correlative analysis between morphology and gene expression was done only on TCGA patients. Another limitation of the study was the evaluation of the CellDiv feature solely on TMA spot images, but not the WSIs. In the precision-recall AUC analysis, a marked improvement was seen in the TCGA LUSC cohort with CellDiv features only compared with the clinical variable-based model (0·85 vs 0·84; appendix 1 p 26). However, despite the limited tissue area, the CellDiv features were still able to prognosticate patient outcomes.
To summarise, we presented a histogenomics approach that attempts to capture CellDiv in the tissue. The CellDiv feature-based classifier was evaluated in H&E-stained image cohorts. The CellDiv features showed a strong correlation with overall survival in early-stage NSCLC, were associated with biological pathways of cellular differentiation, apoptosis, and signalling, and could distinguish KRAS status (in LUAD). CellDiv needs to be clinically validated, first on archived clinical samples from completed clinical trials in early-stage NSCLC (eg, the International Adjuvant Lung Cancer Trial and SWOG Cancer Research Network’s JBR10 trial) for generating level 1 evidence before it can be deployed clinically. Our goal following validation is to provide oncologists with a risk score to guide their treatment decision making. Those patients with early-stage lung cancer but identified by the CellDiv feature classifier as high risk might be good candidates for adjuvant chemotherapy, whereas those identified as being at low risk are likely to do well with surgery alone. Additionally, following validation on archived clinical trials, we will deploy CellDiv as a biomarker to guide therapy in a prospective clinical trial setting, where CellDiv scores will be used to randomly assign patients to either adjuvant chemotherapy or surgery alone for early-stage NSCLC.
Supplementary Material
Research in context.
Evidence before this study
We searched the PubMed database for research articles published between June 25, 2009, and Sept 25, 2019, containing the words “lung cancer” and “pathology”, and one of the phrases “machine learning”, “artificial intelligence”, or “deep learning”. We reviewed the titles and abstracts of the 496 results.
Many studies have explored machine learning-based approaches for lung cancer risk assessment. Findings from many of these studies suggest that computerised descriptors of tumour morphology are prognostic, but many studies rely on abstract black-box features that do not clearly map to and hence are disassociated from tumour morphology. Additionally, these studies do not appear to appreciate or account for the distinctive morphology associated with adenocarcinomas versus squamous cell carcinomas. Also, many of these studies lack large-scale independent validation of their approaches across multiple different sites.
Added value of this study
In this study, we show that computer-derived morphological features reflecting the diversity in cellular features from haematoxylin and eosin-stained non-small cell lung carcinomas (NSCLCs) are associated with disease outcome, and can estimate risk of 5-year overall survival in early-stage cancers.
Procedures to make the model robust to sample preparation variation yielded a model that works across a large study population prepared and digitised across many different institutions. The image-based risk model, based on local tumour cellular diversity features, showed added value over clinical risk factors in clinically low-risk patients (ie, stage I and II) and was prognostic independent of cancer grade and smoking status. Additionally, the hand-crafted local tumour cellular diversity features provide an intuitive way to associate phenotype with the underlying tumour genotype.
Our study differs from other approaches in its focus on prognosis of early-stage NSCLC, as well as the size of the validation set and the plurality of sites from which the validation set is drawn for independent testing of the histomorphometric image signature. We also attempt to explain the relationship between the prognostic histological image phenotype and the corresponding genotype, which has not been done in most related works.
Implications of all the available evidence
The prognostic ability of computer-extracted features of tumour cellular diversity derived from images warrants further study into its potential to supplement or replace molecular tests, to identify which patients with early-stage NSCLC stand to receive added benefit from adjuvant therapy.
Acknowledgments
Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health (NIH) under award numbers 1U24CA199374–01, R01CA202752–01A1, R01CA208236–01A1, R01 CA216579–01A1, R01 CA220581–01A1, and 1U01 CA239055–01; National Center for Research Resources under award number 1-C06-RR12463–01VA; Merit Review Award IBX004121A from the US Department of Veterans Affairs Biomedical Laboratory Research and Development Service; the Department of Defense (DOD) Breast Cancer Research Program Breakthrough Level 1 Award (W81XWH-19–1-0668); the DOD Prostate Cancer Idea Development Award (W81XWH-15–1-0558); the DOD Lung Cancer Investigator-Initiated Translational Research Award (W81XWH-18–1-0440); the DOD Peer Reviewed Cancer Research Program (W81XWH-16–1-0329); The Ohio Third Frontier Technology Validation Fund; The Wallace H Coulter Foundation Program in the Department of Biomedical Engineering; and The Clinical and Translational Science Award Program at Case Western Reserve University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH, the US Department of Veterans Affairs, the DOD, or the US Government. Tissues of the University of Bern cohort were provided by the Tissue Bank Bern. CL is partially supported by the the DoD Breast Cancer Research Program Breakthrough Level 1 Award (W81XWH-19–1-0668). JX is supported by the National Natural Science Foundation of China (grant number 61771249).
Footnotes
Declaration of interests
CL, KB, and AM have a pending patent (Predicting Cancer Recurrence Using Local Co-occurrence of Cell Morphology). AM is an equity holder in Elucid Bioimaging and in Inspirata, to whom his technology has been licensed. He is currently a scientific advisory board member at Aiforia. He is also involved in a National Institutes of Health U24 grant with PathCore and is involved in four grants with Inspirata. His work has received sponsored research funding from Bristol Myers Squibb, AstraZeneca, and Philips, outside of the submitted work. AJ has patents 10528848 and 9111179 issued, and patents 20190266726, 20190251687, 20180129911, and 20160307305 pending (US patents, registered in Case Western Reserve University). VV reports grants and personal fees from Merck, Bristol Myers Squibb, Genentech, AstraZeneca, Celgene, Novartis, Amgen, Fulgent Genetics, Reddy Labs, Alkermes, Nektar Therapeutics, Novocure, and Foundation Medicine, outside of the submitted work; and advisory or consulting fees from Genentech, Merck, Bristol Myers Squibb, AstraZeneca, Foundation Medicine, Nektar Therapeutics, Alkermes, Reddy Labs, and Millennium Pharma, outside of the submitted work. All other authors declare no competing interests.
Data sharing
Access to datasets from the Yale Medical School, Cleveland Clinic Foundation, and University of Bern (used with permission for this study) should be requested directly from these institutions via their data access request forms. Subject to the institutional review boards’ ethical approval, deidentified data can be made available as a test subset. All relevant data used for the validation cohorts during the current study are available through the Genomic Data Commons portal. These datasets were generated by TCGA Research Network and have been made publicly available. The source code for tumour cellular diversity feature extracted can be accessed online.
Contributor Information
Cheng Lu, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA.
Kaustav Bera, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA.
Xiangxue Wang, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA.
Prateek Prasanna, Department of Biomedical Informatics, Stony Brook University, New York.
Jun Xu, Jiangsu Key Laboratory of Big Data Analysis Technique, Nanjing University of Information Science and Technology, Nanjing, China.
Andrew Janowczyk, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA; Precision Oncology Center, Lausanne University Hospital, Lausanne, Switzerland.
Niha Beig, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA.
Michael Yang, University of Colorado Anschutz Medical Campus, Aurora, CO, USA.
Pingfu Fu, Department of Population and Quantitative Health Sciences, Case Western Reserve University, Cleveland, OH, USA.
James Lewis, Department of Pathology, Microbiology, and Immunology, Vanderbilt University Medical Center, Nashville, TN, USA.
Humberto Choi, Department of Pulmonary Medicine, Cleveland Clinic, Cleveland, OH, USA.
Ralph A Schmid, Division of General Thoracic Surgery, Inselspital University Hospital Bern, Bern, Switzerland.
Sabina Berezowska, The Institute of Pathology, University of Bern, Bern, Switzerland.
Kurt Schalper, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
David Rimm, Department of Pathology, Yale University School of Medicine, New Haven, CT, USA.
Vamsidhar Velcheti, Perlmutter Cancer Center, New York University, NY, USA.
Anant Madabhushi, Department of Biomedical Engineering, Case Western Reserve University, Cleveland, OH, USA; Louis Stokes Cleveland Veterans Administration Medical Center, Cleveland, OH, USA.
References
- 1.Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol Mech Dis 2013; 8: 277–302. [DOI] [PubMed] [Google Scholar]
- 2.Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus 2014; 37: E11. [DOI] [PubMed] [Google Scholar]
- 3.Yuan Y, Failmezger H, Rueda OM, et al. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med 2012; 4: 157ra143. [DOI] [PubMed] [Google Scholar]
- 4.Mroz EA, Rocco JW. Intra-tumor heterogeneity in head and neck cancer and its clinical implications. World J Otorhinolaryngol-Head Neck Surg 2016; 2: 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bera K, Schalper KA, Rimm DL, Velcheti V, Madabhushi A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019; 16: 703–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andor N, Graham TA, Jansen M, et al. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2016; 22: 105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes MJ, Sood A, Sevinsky C, Pris AD, Zavodszky MI, Ginty F. Emerging understanding of multiscale tumor heterogeneity. Front Oncol 2014; 4: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oppedijk V, van der Gaast A, van Lanschot JJB, et al. Patterns of recurrence after surgery alone versus preoperative chemoradiotherapy and surgery in the CROSS trials. J Clin Oncol Off J Am Soc Clin Oncol 2014; 32: 385–91. [DOI] [PubMed] [Google Scholar]
- 9.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. ArXiv 2015; published online May 18 http://arxiv.org/abs/1505.04597 (preprint).
- 10.Yang Y, Wang M, Liu B. Exploring and comparing of the gene expression and methylation differences between lung adenocarcinoma and squamous cell carcinoma: J Cell Physiol 2019; 234: 4454–59. [DOI] [PubMed] [Google Scholar]
- 11.Keller MD, Neppl C, Irmak Y, et al. Adverse prognostic value of PD-L1 expression in primary resected pulmonary squamous cell carcinomas and paired mediastinal lymph node metastases. Mod Pathol Off J U S Can Acad Pathol Inc 2018; 31: 101–10. [DOI] [PubMed] [Google Scholar]
- 12.Foulkes WD. Inherited susceptibility to common cancers. N Engl J Med 2008; 359: 2143–53. [DOI] [PubMed] [Google Scholar]
- 13.Corredor G, Wang X, Zhou Y, et al. Spatial architecture and arrangement of tumor-infiltrating lymphocytes for predicting likelihood of recurrence in early-stage non-small cell lung cancer. Clin Cancer Res Off J Am Assoc Cancer Res 2019; 25: 1526–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haralick RM, Shanmugam K, Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern 1973; 3: 610–21. [Google Scholar]
- 15.Martini N, Rusch VW, Bains MS, et al. Factors influencing ten-year survival in resected stages I to IIIA non-small cell lung cancer. J Thorac Cardiovasc Surg 1999; 117: 32–38. [DOI] [PubMed] [Google Scholar]
- 16.Marko NF, Weil RJ. Non-Gaussian distributions affect identification of expression patterns, functional annotation, and prospective classification in human cancer genomes. PLoS One 2012; 7: e46935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995; 57: 289–300. [Google Scholar]
- 18.The Gene Ontology Consortium. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 2017; 45: D331–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 2005; 102: 15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Artal Cortés Á, Calera Urquizu L, Hernando Cubero J. Adjuvant chemotherapy in non-small cell lung cancer: state-of-the-art. Transl Lung Cancer Res 2015; 4: 191–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burotto M, Thomas A, Subramaniam D, Giaccone G, Rajan A. Biomarkers in early-stage non-small cell lung cancer: current concepts and future directions. J Thorac Oncol Off Publ Int Assoc Study Lung Cancer 2014; 9: 1609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandoval J, Mendez-Gonzalez J, Nadal E, et al. A prognostic DNA methylation signature for stage I non–small-cell lung cancer. J Clin Oncol 2013; 31: 4140–47. [DOI] [PubMed] [Google Scholar]
- 23.Chen H-Y, Yu S-L, Chen C-H, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med 2007; 356: 11–20. [DOI] [PubMed] [Google Scholar]
- 24.Barletta JA, Yeap BY, Chirieac LR. Prognostic significance of grading in lung adenocarcinoma. Cancer 2010; 116: 659–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takenaka M, Hanagiri T, Shinohara S, et al. The prognostic significance of HER2 overexpression in non-small cell lung cancer. Anticancer Res 2011; 31: 4631–36. [PubMed] [Google Scholar]
- 26.Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G. DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med 2007; 356: 800–08. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Janowczyk A, Zhou Y, et al. Prediction of recurrence in early stage non-small cell lung cancer using computer extracted nuclear features from digital H&E images. Sci Rep 2017; 7: 13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saltz J, Gupta R, Hou L, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep 2018; 23: 181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu K-H, Zhang C, Berry GJ, et al. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun 2016; 7: 12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non–small cell lung cancer histopathology images using deep learning. Nat Med 2018; 24: 1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019; 25: 1054–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Q, Zhang H, Kong M, Mao X, Cao X. Hub genes and key pathways of non-small lung cancer identified using bioinformatics. Oncol Lett 2018; 16: 2344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai B, Ren L, Han X, Liu D. Bioinformatics analysis reveals 6 key biomarkers associated with non-small-cell lung cancer. J Int Med Res 2019; published online June 20 10.1177/2F0300060519887637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langenfeld EM, Calvano SE, Abou-Nukta F, Lowry SF, Amenta P, Langenfeld J. The mature bone morphogenetic protein-2 is aberrantly expressed in non-small cell lung carcinomas and stimulates tumor growth of A549 cells. Carcinogenesis 2003; 24: 1445–54. [DOI] [PubMed] [Google Scholar]
- 35.Chen M, Liu X, Du J, Wang X-J, Xia L. Differentiated regulation of immune-response related genes between LUAD and LUSC subtypes of lung cancers. Oncotarget 2017; 8: 133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao J, Qiu X, Xi G, et al. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in non-small cell lung cancer. Oncol Rep 2018; 40: 1971–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Wu L, Ao H, et al. Prognostic implications of autophagy-associated gene signatures in non-small cell lung cancer. Aging 2019; 11: 11440–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, Wang Z, Niu X, et al. Identification of seven-gene signature for prediction of lung squamous cell carcinoma. OncoTargets Ther 2019; 12: 5979–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.