Abstract.
The incidence and spread of dengue virus (DENV) have increased rapidly in recent decades. Dengue is underreported in Africa, but recent outbreaks and seroprevalence data suggest that DENV is widespread there. A lack of ongoing surveillance limits knowledge about its spatial reach and hinders disease control planning. We sought to add data on dengue distribution in Kenya through diagnostic testing of serum specimens from persons with an acute febrile illness (AFI) attending an outpatient clinic in rural western Kenya (Asembo) during rainy seasons. Patients with symptoms not likely to be misclassified as dengue (e.g., diarrhea and anemia), those with a positive diagnostic laboratory results which explained their febrile illness, or those with serum collected more than 5 days after fever onset were excluded. However, febrile patients with a positive malaria smear were included in the study. We used reverse transcription polymerase chain reaction (RT-PCR) to test for DENV and IgM anti-DENV to test for recent infection. Of the 615 serum specimens available for testing, none were dengue positive by either RT-PCR or IgM anti-DENV testing. Dengue did not appear to be a cause of febrile illness in this area of western Kenya, although our relatively small sample size may not have identified DENV infections occurring at low incidence. A more widespread AFI surveillance system that includes dengue diagnostic testing by RT-PCR and antibody-based methods is required to more definitively gauge the size and geographic distribution of DENV infection in western Kenya.
INTRODUCTION
Over the past several decades, the incidence of dengue virus (DENV) infections has increased rapidly to an estimated 390 million each year.1 Originally thought to occur mainly in Southeast Asia and South America, DENV infection is now present in at least 128 countries, including those in Africa and the eastern Mediterranean, placing more than half the world’s population at risk of acquiring DENV infection.2
Dengue is caused by any of four types of a single-stranded RNA virus, which are primarily transmitted by the mosquitoes Aedes aegypti and Aedes albopictus. Dengue virus infections are usually self-limited and asymptomatic (56–80% according to recent estimates),1,3 but can cause symptoms ranging from mild illness (headaches and low fever) to high fevers, myalgia, and encephalitis. Recovery from DENV infection confers long-lasting immunity against the infecting DENV type.4 However, subsequent infection with a different DENV type places a person at increased risk of severe dengue, which is associated with plasma leakage, hemorrhagic symptoms, and death.5–7 There is currently no pharmacologic treatment for dengue, but skilled management of a patient with severe dengue can reduce mortality from as high as 20% to < 1%.8 Vaccine development is complicated by the need to confer protection against all four DENV types simultaneously so as to avoid increasing the risk of severe dengue, and vaccines are only now being tested in clinical trials.9–11 Vector control efforts have shown potential, but there are few high-quality studies and only very weak evidence that they reduce dengue incidence.12 Although dengue is a major cause of acute febrile illness (AFI) worldwide, healthcare workers in settings with no history of DENV circulation are unlikely to consider dengue as a diagnosis, which may hinder appropriate care of febrile patients. Given the spread of DENV, determining whether the virus is present in regions with high incidence of AFI may lead to improved treatment outcomes.
Dengue is likely underreported in Africa because of limited availability of diagnostics and the routine presumption that AFI is due to malaria.13,14 However, recent outbreaks in Angola, Kenya, Gabon, and Somalia, combined with serological studies that indicate ongoing endemic transmission, suggest that dengue is widespread on the continent.15–18 Specifically in Kenya, outbreaks have been reported in the inland northeast (Mandera County) during 2011 and eastern coastal regions (Mombasa county) in 2013.3,19,20 Serological studies have shown a wide range in the prevalence of DENV or general flavivirus infections throughout Kenya, from 1.1% to 26.2%.21,22 Entomological data and models support these findings by demonstrating the presence of Ae. aegypti in these settings.23 As dengue incidence has increased in Africa over the past decade, malaria incidence has been decreasing, and widespread deployment of the Haemophilus influenzae type b and pneumococcal vaccines promises to reduce the burden of fever caused by pneumonia.24,25 Healthcare workers and planners must adapt to the changing distribution of AFI etiologies. However, a lack of systematic, ongoing surveillance limits knowledge about the spatial reach of dengue and hinders disease control planning. We sought to estimate the extent to which DENV infection contributes to AFI by testing for the virus among febrile patients in a western Kenya outpatient setting.
MATERIALS AND METHODS
Study setting.
This research was conducted within the population-based disease surveillance (PBIDS) program implemented by the Kenya Medical Research Institute (KEMRI) with support from the CDC-Kenya. Population-based disease surveillance is located in Asembo, a rural site in Siaya County in western Kenya near Lake Victoria, which has been operating since late 2005. The area has holoendemic malaria transmission and a high prevalence of HIV (17% in adults aged ≥ 18 years). Asembo stands at an altitude of 1,100 m above sea level and experiences seasonal rainy periods from March through May and in October and December.26–28
The approximately 26,000 participants in PBIDS live within 5 km of Lwak Mission Hospital (LMH) and are eligible for free treatment for acute illness at the clinic.27 Healthcare workers use protocols to take a history and perform a physical examination, with results recorded on standardized forms. In addition to the standardized syndromic diagnoses, healthcare workers also record clinical diagnoses based on their assessment. Syndromes under surveillance include acute respiratory disease, fever, diarrhea, and jaundice. All clinic patients with a presenting temperature of ≥ 38.0°C receive a malaria blood smear, and a subset undergo a series of other diagnostic tests depending on other presenting symptoms, including blood culture, urine antigen testing for pneumococcus, naso- and oropharyngeal swabs, and stool cultures. Sample collection and testing methods have been previously described.29
All participants provide written informed consent to participate. Parents or guardians provided informed written consent on behalf of minors. The protocol and consent forms were reviewed and approved by the ethical review boards of the KEMRI, #1899, and the Institutional Review Board of the U.S. CDC, #4566.
Serum sample selection.
To increase the likelihood of detecting acute DENV infections among febrile patients in Asembo, we applied the following inclusion criteria: patients of all ages with fever (≥ 38.0°C) at the time of visit, a clinic visit during or shortly after a rainy season during a time of dengue outbreaks in other parts of Kenya (September–December 2011 and March–July 2013), and a serum sample collected at the time of visit. For syndromic surveillance, serum samples were collected from the first five patients aged 5 years or older and the first five patients aged younger than 5 years who visited LMH with fever each day. Patients with fever and severe acute respiratory illness also had a serum specimen collected regardless of when in the day they visited. Of the sera collected from febrile patients, we included specimens from patients with a positive malaria smear, a standardized surveillance diagnosis of conditions with symptoms that often overlap with dengue symptoms (measles, oral candidiasis, otitis media, rash, rubella, tonsillitis, upper respiratory tract infection, and viral syndrome), or a surveillance diagnosis of a condition that is not likely to be the cause of fever (anemia, conjunctivitis, diarrhea, intestinal worms, scabies, and wheezing). We excluded sera from patients with a syndromic diagnosis of a condition for which symptoms do not largely overlap with dengue symptoms (amebiasis, burn, dysentery, mumps, pneumonia, pulmonary tuberculosis, sexually transmitted infections, and varicella), a clinical diagnosis for conditions not likely to be conflated with dengue (namely, two diagnoses of rotavirus), a positive result from laboratory diagnosis of other samples (bacteremia, urine antigen test for pneumococcus, or naso- and oropharyngeal swabs for influenza), and patients with serum that was collected more than 5 days after date of fever onset (the typical viremic period for dengue).
Dengue virus testing.
Total RNA was extracted from 100 µL of each serum sample using the MagNA Pure 96 System with the DNA and Viral Nucleic Acid Small Volume Kit according to the manufacturer’s instructions (Roche, Mannheim, Germany) and stored at −80°C for later processing. Dengue viral RNA was quantitated by real-time RT-PCR using reagents from the AgPath-ID One-Step RT-PCR Kit (Life Technologies, Carlsbad, CA) and primers from the CDC DENV-1–4 real-time RT-PCR assay.30 Real-time RT-PCR reaction mixtures were formulated according to the manufacturer’s instructions with 400 nM each of the forward and reverse primers and 120 nM of the TaqMan probe. Samples were run on an Applied Biosystems 7500 Fast Real-Time PCR System (Life Technologies) using the following reaction conditions: 45°C for 10 minutes, 95°C for 10 minutes, then 45 cycles of alternating between 95°C for 15 seconds, and 55°C for 1 minute, with TaqMan probe fluorescence measuring each cycle during the 55°C step. Data were analyzed using the Applied Biosystems 7500 SDS Software (Life Technologies). RNA quality was validated using a human RNase P control, which ruled out inhibition of the RT-PCR reaction. Purified DENV genomic RNA was used as a positive control.
IgM anti-DENV testing.
Sera from the study subjects were tested using an IgM capture assay that is specific for antibody to DENV. This was performed according to the manufacturer’s instructions (DENV Detect™ IgM Capture ELISA kit; InBios International, Inc., Seattle, WA).31 The serum samples were incubated in microtiter plates pre-coated by the manufacturer with antihuman IgM antibodies. After subsequent incubation with serum specimens and washing, the wells were treated with a specific conjugate and thereafter with substrate. A stop solution was then added, and the absorbance measurements were read using the ELx800 absorbance microplate reader (BioTek, Winooski, VT) to determine the optical density.
Definition of dengue.
We defined DENV infection as either a positive RT-PCR result or a positive IgM anti-DENV result.
RESULTS
During the study period (September–December 2011 and March–July 2013), there were 11,773 unique visit records. The median patient age among all visits was 11.4 years, and 59.0% were female (Table 1). Almost 60% of patients reported recent fever, and 16.9% were febrile at the time of visit. The most common clinical diagnoses among all patients were malaria (40.3%) and upper respiratory tract infection (40.1%). Of those tested, 41.7% had a positive malaria smear result.
Table 1.
Demographics of clinic visits to Lwak Mission Hospital, Asembo, Kenya
| Demographic | All clinic visits (N = 11,773) | Included patients (N = 615) |
|---|---|---|
| Age (years), median (interquartile range) | 11.4 (22.0) | 4.8 (6.6) |
| Female, n (%) | 6,942 (59.0)* | 327 (53.2) |
| Febrile, n (%) | ||
| Reported | 6,975 (59.5) | 613 (99.7) |
| At the time of visit | 1,984 (16.9) | 615 (100) |
| Common diagnoses (> 5% visits), n (%) | ||
| Diarrhea | 596 (5.1) | 29 (4.7) |
| Malaria | 4,476 (40.3) | 475 (77.2) |
| Pharyngitis | 714 (6.1) | 19 (3.1) |
| Pneumonia | 947 (8.0) | 0 (0.0) |
| Upper respiratory tract infection | 4,726 (40.1) | 4 (0.7) |
| Malaria smear positive | 4,013 (41.7)† | 442 (77.1)‡ |
n = 11,771.
n = 9,630.
n = 573.
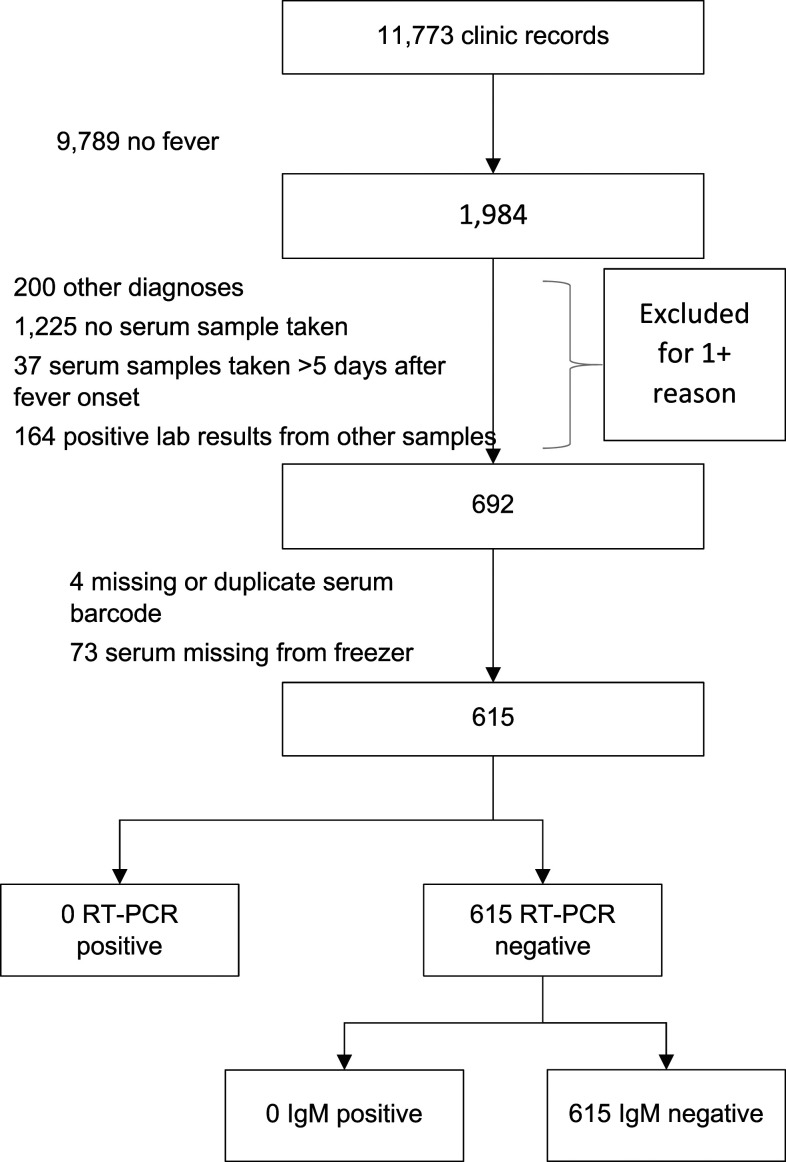
During the sampling period, there were 1,984 visits to the clinic where the patient currently had a current fever. A total of 693 febrile patients met the inclusion criteria, of whom 615 (88.7%) had samples available for testing (Figure 1). A majority (465, 75.6%) of the samples came from 2013, and most (86.0%) were collected within 2 days of fever onset. Patients included in the analysis were younger (median 4.8 years), and a smaller proportion were female (53.2%) than patients at all clinic visits (Table 1). The most common clinical diagnosis was malaria (77.2%), and 442 (77.1%) patients had a positive malaria smear result (Table 1).
Figure 1.
Selection of febrile patients for dengue diagnostic testing.
There were no positive results for dengue using either RT-PCR or IgM anti-DENV testing.
DISCUSSION
In this study, no DENV infections were detected using molecular diagnostic testing, despite samples being collected during acute febrile episodes when patients would be expected to be viremic. In addition, there was no evidence of current or recent DENV infections when using IgM antibody-based testing. Dengue does not appear to be a cause of AFI among this rural population in western Kenya during the time period sampled.
The question of the actual etiology of the patients’ fevers remains unanswered. It is possible that the malaria smear results and clinical diagnoses represent the true cause of disease, although relying on these methods alone is likely to result in significant misdiagnosis because of the presence of asymptomatic parasitemia in older children and adults.32,33 The nonspecific nature of AFI and the variety of etiologies found in similar settings highlight the need for more advanced molecular diagnostics available at the point of care that can direct clinicians toward an etiologic agent and appropriate treatment.34
There are several possible explanations for DENV infections not being detected in this study of AFI patients, the most straightforward being a true absence of DENV infection in this area of western Kenya. This contrasts with previous studies from adjacent or nearby districts that documented low levels of DENV seroprevalence and seroincidence.35–38 However, these studies relied on IgG DENV antibody ELISA tests, which are known to have high cross-reactivity with infections by other flaviviruses and possibly antibodies to yellow fever vaccine, likely overestimating the presence of prior DENV infections.39 Direct comparison between these studies and our findings is therefore not possible.
In addition, even if the incidence of DENV infection was truly low rather than absent, we likely had an insufficient sample size to detect them. Assuming that the care-seeking rate for AFI caused by dengue was the same as reported for other AFIs in this region (13.5%),40 we would have needed an apparent dengue incidence rate of 226/100,000 person-years to have detected one case of symptomatic DENV infection. This incidence rate is similar to regions with high dengue incidence, such as Singapore and Brazil,41,42 and much higher than would be expected in the study setting. Using data from the 2013 outbreak in coastal Kenya, the most relevant context from which to draw on, we calculated that if a similar outbreak had been occurring during the sampling period, only 135 infected individuals would have presented at the Asembo clinic (assuming 13% infected and 44% symptomatic).3 Given that there did not appear to be an outbreak, the chances of detecting isolated cases in the population was very low, despite designing the inclusion criteria to enhance the probability of selecting patients with dengue. In this study, the sample size was constrained by available resources. Future studies should be resourced appropriately to maximize the chance of accurately estimating DENV levels in a population.
Another possible explanation for the results is the rural nature of the study site. The population density in Asembo is relatively low (325 persons/km2), and the primary economic activity is agriculture.27 The main DENV vectors, Ae. aegypti and Ae. albopictus, are predominantly urban and peri-urban in nature, and therefore may be limited to nearby towns and cities. Although we were unable to identify any published entomologic data from the study region, Ae. aegypti has been found in nearby Kisumu.43 Future studies should focus on febrile patients in these more urban settings.
This study was one of the first in Africa to use RT-PCR to test for DENV infection rather than antibody-based methods alone. Of two previous instances where a combined testing regimen was used, one found moderate evidence of circulating DENV in Abidjan, Côte d’Ivoire, and the other took place during an outbreak on the Kenyan coast.3,44 Combining the two detection approaches allows for greater certainty that DENV is the etiologic agent of fever.45–48
Besides what was probably too small a sample size to make a robust estimate of dengue incidence in this population, there were other limitations to this study. Our selection criteria may have systematically eliminated subjects with DENV infections. The most likely reason was restricting samples to those collected during or shortly after the rainy season. Although dengue incidence tends to increase with increasing rainfall, the effect typically lags by 1–2 months, or even longer in some models.41,49,50 Although we included cases from the month following the traditional rainy periods, the lag between rainfall and dengue incidence has not been established in western Kenya, and a 1-month lag period may have been too short.
A further challenge to finding incident DENV is that most infections are asymptomatic, although the proportion varies because of several factors, including incidence in recent years.51 Infected patients without fever are unlikely to have presented to clinic and therefore will have remained undiagnosed. The proportion of DENV infections that are clinically silent is unclear in an area of emerging infections but may be higher than that in endemic areas, where secondary infections are more common.51 The best estimate of the ratio of asymptomatic to symptomatic DENV infections in Kenya comes from a seroincidence survey conducted in Mombasa during the 2013 outbreak. Investigators there found that 44% of people with current or recent DENV infections reported having had recent fever.3 Although some of the reported fevers in that study would likely have been caused by malaria,52 these data suggest a substantial proportion of DENV infections in Asembo would present at the clinic with fever if DENV was circulating in the study area.
Although dengue is increasingly being recognized as a problem in sub-Saharan Africa, our finding of no dengue in rural western Kenya suggests that DENV may not be well established in this area. Combined use of RT-PCR and antibody detection methods can provide a more accurate picture of disease incidence than serology alone. This approach, combined with greater use of rapid diagnostic tests at the point of care, will be needed as part of a widespread surveillance system to accurately gauge the size and geographic spread of dengue and DENV infections.
REFERENCES
- 1.Bhatt S, et al. 2013. The global distribution and burden of dengue. Nature 496: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI, 2012. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis 6: e1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis EM, et al. 2015. A household serosurvey to estimate the magnitude of a dengue outbreak in Mombasa, Kenya, 2013. PLoS Negl Trop Dis 9: e0003733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmons CP, Farrar JJ, Nguyen VV, Wills B, 2012. Dengue. N Engl J Med 366: 1423–1432. [DOI] [PubMed] [Google Scholar]
- 5.Gubler DJ, 2004. The changing epidemiology of yellow fever and dengue, 1900 to 2003: full circle? Comp Immunol Microbiol Infect Dis 27: 319–330. [DOI] [PubMed] [Google Scholar]
- 6.Huy NT, Van Giang T, Thuy DH, Kikuchi M, Hien TT, Zamora J, Hirayama K, 2013. Factors associated with dengue shock syndrome: a systematic review and meta-analysis. PLoS Negl Trop Dis 7: e2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizumoto K, Ejima K, Yamamoto T, Nishiura H, 2014. On the risk of severe dengue during secondary infection: a systematic review coupled with mathematical modeling. J Vector Borne Dis 51: 153–164. [PubMed] [Google Scholar]
- 8.World Health Organization , 2012. Dengue and Severe Dengue. Fact Sheet. Geneva, Switzerland: WHO; Available at: http://www.who.int/mediacentre/factsheets/fs117/en/index.html. Accessed April 26, 2017. [Google Scholar]
- 9.Capeding MR, et al. 2014. Clinical efficacy and safety of a novel tetravalent dengue vaccine in healthy children in Asia: a phase 3, randomised, observer-masked, placebo-controlled trial. Lancet 384: 1358–1365. [DOI] [PubMed] [Google Scholar]
- 10.Hadinegoro SR, et al. 2015. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med 373: 1195–1206. [DOI] [PubMed] [Google Scholar]
- 11.Villar L, et al. 2015. Efficacy of a tetravalent dengue vaccine in children in Latin America. N Engl J Med 372: 113–123. [DOI] [PubMed] [Google Scholar]
- 12.Ballenger-Browning KK, Elder JP, 2009. Multi-modal Aedes aegypti mosquito reduction interventions and dengue fever prevention. Trop Med Int Heal 14: 1542–1551. [DOI] [PubMed] [Google Scholar]
- 13.Amarasinghe A, Kuritsk JN, Letson GW, Margolis HS, 2011. Dengue virus infection in Africa. Emerg Infect Dis 17: 1349–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Were F, 2012. The dengue situation in Africa. Paediatr Int Child Heal 32 (Suppl 1): 18–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DENGUE/DHF UPDATE (33): ASIA, AFRICA, PACIFIC , 2013. ProMed Mail. Available at: http://www.promedmail.org/direct.php?id=20130428.1676860. Accessed April 26, 2017. [Google Scholar]
- 16.Caron M, Grard G, Paupy C, Mombo IM, Bikie Bi Nso B, Kassa Kassa FR, Nkoghe D, Leroy EM, 2013. First evidence of simultaneous circulation of three different dengue virus serotypes in Africa. PLoS One 8: e78030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention , 2013. Ongoing dengue epidemic–Angola, June 2013. MMWR Morb Mortal Wkly Rep 62: 504–507. [PMC free article] [PubMed] [Google Scholar]
- 18.Kyobe Bosa H, Montgomery JM, Kimuli I, Lutwana JJ, 2014. Dengue fever outbreak in Mogadishu, Somalia 2011: co-circulation of three dengue virus serotypes. Int J Infect Dis 21: 3. [Google Scholar]
- 19.Obonyo M, Fidhow A, Ofula V, 2018. Investigation of laboratory confirmed dengue outbreak in north-eastern Kenya, 2011. PLoS One 13: e0198556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutomiah J, et al. 2016. Dengue outbreak in Mombasa city, Kenya, 2013–2014: entomologic investigations. PLoS Negl Trop Dis 10: e0004981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simo FBN, Bigna JJ, Kenmoe S, Ndangang MS, Temfack E, Moundipa PF, Demanou M, 2019. Dengue virus infection in people residing in Africa: a systematic review and meta-analysis of prevalence studies. Sci Rep 9: 13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hortion J, Mutuku FM, Eyherabide AL, Vu DM, Boothroyd DB, Grossi-Soyster EN, King CH, Ndenga BA, LaBeaud AD, 2019. Acute flavivirus and alphavirus infections among children in two different areas of Kenya, 2015. Am J Trop Med Hyg 100: 170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Attaway DF, Jacobsen KH, Falconer A, Manca G, Rosenshein Bennett L, Waters NM, 2014. Mosquito habitat and dengue risk potential in Kenya: alternative methods to traditional risk mapping techniques. Geospat Heal 9: 119–130. [DOI] [PubMed] [Google Scholar]
- 24.O’Meara WP, Mangeni JN, Steketee R, Greenwood B, 2010. Changes in the burden of malaria in sub-Saharan Africa. Lancet Infect Dis 10: 545–555. [DOI] [PubMed] [Google Scholar]
- 25.Madhi SA, Levine OS, Hajjeh R, Mansoor OD, Cherian T, 2008. Vaccines to prevent pneumonia and improve child survival. Bull World Heal Organ 86: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalal W, et al. 2013. Home-based HIV testing and counseling in rural and urban Kenyan communities. J Acquir Immune Defic Syndr 62: e47–e54. [DOI] [PubMed] [Google Scholar]
- 27.Feikin DR, Olack B, Bigogo GM, Audi A, Cosmas L, Aura B, Burke H, Njenga MK, Williamson J, Breiman RF, 2011. The burden of common infectious disease syndromes at the clinic and household level from population-based surveillance in rural and urban Kenya. PLoS One 6: e16085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odhiambo FO, et al. 2012. Profile: the KEMRI/CDC health and demographic surveillance system--western Kenya. Int J Epidemiol 41: 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feikin DR, et al. 2012. Etiology and incidence of viral and bacterial acute respiratory illness among older children and adults in rural western Kenya, 2007–2010. PLoS One 7: e43656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, Medina F, Colon C, Margolis H, Munoz-Jordan JL, 2013. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis 7: e2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, Sengvilaipaseuth O, Chanthongthip A, Phonemixay O, Vongsouvath M, Phouminh P, Blacksell SD, Newton PN, Dubot-Pérès A, 2019. Comparison of two commercial ELISA kits for the detection of anti-dengue IgM for routine dengue diagnosis in Laos. Trop Med Infect Dis 4: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nkengasong JN, Nsubuga P, Nwanyanwu O, Gershy-Damet GM, Roscigno G, Bulterys M, Schoub B, DeCock KM, Birx D, 2010. Laboratory systems and services are critical in global health: time to end the neglect? Am J Clin Pathol 134: 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gwer S, Newton CR, Berkley JA, 2007. Over-diagnosis and co-morbidity of severe malaria in African children: a guide for clinicians. Am J Trop Med Hyg 77 (Suppl 6): 6–13. [PMC free article] [PubMed] [Google Scholar]
- 34.D’Acremont V, Kilowoko M, Kyungu E, Philipina S, Sangu W, Kahama-Maro J, Lengeler C, Cherpillod P, Kaiser L, Genton B, 2014. Beyond malaria--causes of fever in outpatient Tanzanian children. N Engl J Med 370: 809–817. [DOI] [PubMed] [Google Scholar]
- 35.Blaylock JM, Maranich A, Bauer K, Nyakoe N, Waitumbi J, Martinez LJ, Lynch J, 2011. The seroprevalence and seroincidence of dengue virus infection in western Kenya. Travel Med Infect Dis 9: 246–248. [DOI] [PubMed] [Google Scholar]
- 36.Mease LE, Coldren RL, Musila LA, Prosser T, Ogolla F, Ofula VO, Schoepp RJ, Rossi CA, Adungo N, 2011. Seroprevalence and distribution of arboviral infections among rural Kenyan adults: a cross-sectional study. Virol J 8: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sutherland LJ, Cash AA, Huang YJ, Sang RC, Malhotra I, Moormann AM, King CL, Weaver SC, King CH, LaBeaud AD, 2011. Serologic evidence of arboviral infections among humans in Kenya. Am J Trop Med Hyg 85: 158–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vu DM, Mutai N, Heath CJ, Vulule JM, Mutuku FM, Ndenga BA, LaBeaud AD, 2017. Unrecognized dengue virus infections in children, western Kenya, 2014–2015. Emerg Infect Dis 23: 1915–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peeling RW, et al. 2010. Evaluation of diagnostic tests: dengue. Nat Rev Microbiol 8 (Suppl 12): S30–S38. [DOI] [PubMed] [Google Scholar]
- 40.Bigogo G, Audi A, Aura B, Aol G, Breiman RF, Feikin DR, 2010. Health-seeking patterns among participants of population-based morbidity surveillance in rural western Kenya: implications for calculating disease rates. Int J Infect Dis 14: e967–e973. [DOI] [PubMed] [Google Scholar]
- 41.Hii YL, Zhu H, Ng N, Ng LC, Rocklov J, 2012. Forecast of dengue incidence using temperature and rainfall. PLoS Negl Trop Dis 6: e1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teixeira MG, Siqueira JB, Jr., Ferreira GL, Bricks L, Joint G, 2013. Epidemiological trends of dengue disease in Brazil (2000–2010): a systematic literature search and analysis. PLoS Negl Trop Dis 7: e2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mireji PO, Keating J, Hassanali A, Mbogo CM, Nyambaka H, Kahindi S, Beier JC, 2008. Heavy metals in mosquito larval habitats in urban Kisumu and Malindi, Kenya, and their impact. Ecotoxicol Environ Saf 70: 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.L’Azou M, Succo T, Kamagate M, Ouattara A, Gilbernair E, Adjogoua E, Luxemburger C, 2015. Dengue: etiology of acute febrile illness in Abidjan, Cote d’Ivoire, in 2011–2012. Trans R Soc Trop Med Hyg 109: 717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teoh B-T, Sam S-S, Tan K-K, Johari J, Abd-Jamil J, Hooi P-S, AbuBakar S, 2016. The use of NS1 rapid diagnostic test and qRT-PCR to complement IgM ELISA for improved dengue diagnosis from single specimen. Sci Rep 6: 27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunsperger EA, et al. 2016. Performance of dengue diagnostic tests in a single-specimen diagnostic algorithm. J Infect Dis 214: 836–844. [DOI] [PubMed] [Google Scholar]
- 47.Huang CH, et al. 2013. Laboratory diagnostics of dengue fever: an emphasis on the role of commercial dengue virus nonstructural protein 1 antigen rapid test. J Microbiol Immunol Infect 46: 358–365. [DOI] [PubMed] [Google Scholar]
- 48.Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV, 1992. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 30: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomes AF, Nobre AA, Cruz OG, 2012. Temporal analysis of the relationship between dengue and meteorological variables in the city of Rio de Janeiro, Brazil, 2001–2009. Cad Saude Publica 28: 2189–2197. [DOI] [PubMed] [Google Scholar]
- 50.Karim MN, Munshi SU, Anwar N, Alam MS, 2012. Climatic factors influencing dengue cases in Dhaka city: a model for dengue prediction. Indian J Med Res 136: 32–39. [PMC free article] [PubMed] [Google Scholar]
- 51.Grange L, Simon-Loriere E, Sakuntabhai A, Gresh L, Paul R, Harris E, 2014. Epidemiological risk factors associated with high global frequency of inapparent dengue virus infections. Front Immunol 5: 280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Snow RW, Kibuchi E, Karuri SW, Sang G, Gitonga CW, Mwandawiro C, Bejon P, Noor AM, 2015. Changing malaria prevalence on the Kenyan coast since 1974: climate, drugs and vector control. PLoS One 10: e0128792. [DOI] [PMC free article] [PubMed] [Google Scholar]