Abstract
The exact mechanisms underlying hypertrophic scarring is yet to be fully understood. However, excessive collagen deposition by fibroblasts has been demonstrated to result in hypertrophic scar formation, and collagen synthesis in dermal fibroblasts is regulated by the transforming growth factor-β1/Smad signaling pathway. In view of this, a Smad-binding decoy was designed and its effects on hypertrophic scar-derived human skin fibroblasts was evaluated. The results of the present study revealed that the Smad decoy attenuates the total amount of collagen, collagen I and Smad2/3 expression in scar fibroblasts. Data from RNA sequencing indicated that the Smad decoy induced more than 4-fold change in 178 genes, primarily associated with to the extracellular matrix, compared with the untreated control. In addition, results from quantitative real-time polymerase chain reaction further confirmed that the Smad decoy significantly attenuated the expression of extracellular matrix-related genes, including COL1A1, COL1A2 and COL3A1. Furthermore, the Smad decoy reduced transforming growth factor-β1-induced collagen deposition in scar fibroblasts. Data generated from the present study provide evidence supporting the use of the Smad decoy as a potential hypertrophic scar treatment.
Keywords: hypertrophic scar, collagen, Smad, transforming growth factor-β1
Introduction
Hypertrophic scarring (HS), caused by various cutaneous injuries, including surgery, insect bites, vaccination and folliculitis, is a highly prevalent clinical condition (1). For example, HS has been reported to occur following 70% of burns (2). Patients with HS suffer from pain, itching and loss of joint mobility (3). Although the exact mechanisms underpinning HS formation are not yet fully understood, the excessive collagen deposition at the end of wound healing has been widely demonstrated to result in HS formation (4). At the end of normal cutaneous wound healing, dermal fibroblasts stop generating more extracellular matrix (ECM; mainly collagen) and the excessive ECM is degraded by collagenases; failure of this process results in overabundance of ECM and triggers the formation of HS (4). Overexpression of ECM is therefore a hallmark of hypertrophic scar fibroblasts. Therefore, blockage of the collagen production from dermal fibroblasts at an appropriate time point in the healing process may reduce the formation of HS.
Transforming growth factor-β1 (TGF-β1) has been demonstrated to serve essential roles in HS formation as TGF-β1 stimulates the proliferation and collagen deposition of dermal fibroblasts (5). Evidence indicates that the expression of TGF-β1 is higher in HS tissues compared with normal skin tissues (6). The Smad protein family serves important roles in regulating various signaling pathways. Smad2, 3 and 4 are particularly crucial in the TGF-β1/Smad signaling pathway (7), although there are also Smad-independent signaling cascade (8). Binding of TGF-β1 and its receptor (TβR) activates the phosphorylation of Smad2/3 (9). Phosphorylated Smad 2/3 further binds to Smad4 and this complex eventually translocates into nucleus to initiate the transcription of collagens (10). It has been reported that the expression of TβR and Smad3 are increased in HS tissues, indicating the roles of Smad in HS formation (11). In addition, suppression of the TGF-β1/Smad signaling pathway has been demonstrated to attenuate the formation of HS (12).
As human Smad 3 and 4 have been demonstrated to specifically bind an 8 bp palindromic sequence to activate transcription (13), we hypothesized that a double-stranded DNA decoy that binded Smad 3 and 4 would decrease the expression of Smad pathway-associated genes, including collagen (9), by sequestering Smad, while not interfering with the non-Smad signaling pathways of the TGF-β1 pathway. In the present study, a Smad decoy was generated and its effects on collagen deposition, collagen I and Smad2/3 protein expression, and the gene expression of hypertrophic scar-derived human skin fibroblasts (HSF) were evaluated. In addition, effects of the Smad decoy on the collagen production and gene expression of TGF-β1-stimulated HSF were also examined.
Materials and methods
Cell culture
The HSFs (primary cells derived from 3 patients: 106, 107 and 108) were purchased from Cell Research Corporation (Singapore), with ethical approval obtained from the A STAR Institute of Medical Biology (IRB: B-16-135E). Primary human keratinocytes (Kc) were collected from the Asian Skin Bank, Institute of Medical Biology, A STAR, Singapore following ethical approval (IRB: B-16-135E). HSF was cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc.) containing 4.5 g/l D-Glucose and 110 mg/l sodium pyruvate, supplemented with 10% foetal calf serum (FCS; Thermo Fisher Scientific, Inc.) and 1% v/v penicillin/streptomycin solution (Thermo Fisher Scientific, Inc.) at 37°C in an incubator with 5% CO2. Kc was cultured in Green's medium containing 10% FCS, according to a previously described protocol (14). The cell culture medium was changed every 2–3 days.
Preparation of the reagents
The Smad decoy was a double-stranded DNA with the following sequence: GGAGTATGTCTAGACTGACAATGTAC. The underlined palindromic sequence is the cognate recognition sequence for Smad3 and 4. The Smad decoy was annealed by adding 100 µM of the sequence and its reverse complement in water, prior to heating to 95°C and gradually cooling at 0.5°C per second to 25°C. The negative control sequence has the same nucleotide balance, but a scrambled sequence: GATGAAGTTCGAATCTGACATAGTAC. A cell-penetrating peptide (PepC) was synthesized by Pepscan as the delivery vehicle for the Smad decoy based on previous studies with modification (15,16). TGF-β1 was purchased from Merck KGaA and used at 5 ng/ml for the stimulation of HSF.
Collagen production
Effects of the Smad decoy on HSF (with or without TGF-β1 stimulation) collagen production were assayed using Sirius red staining (Sigma-Aldrich; Merck KGaA), as previously described (17). In brief, HSFs (4×104) were seeded onto a 24-well culture plate for 24 h. Smad decoy (to reach a final concentration in 500 µl medium of 30, 100 and 300 nM) was mixed with PepC at a mass ratio of 1:1 and then applied to the cell cultures. Following exposure to the Smad decoy for 48 and 72 h, Sirius red was added to each well and the plates were incubated at 37°C for a further 90 min. Following drying overnight, the staining was dissolved in 0.1 M sodium hydroxide (NaOH; Sigma-Aldrich; Merck KGaA) and the absorbance was measured at 540 nm using a SpectraMax M5 Multi-Mode microplate reader (Molecular Devices, LLC).
Western blotting
Expression of collagen I and Smad 2/3 in HSF following Smad decoy treatment was detected using western blotting using secondary antibodies with fluorescence. HSFs were treated with 100 nM Smad decoy for 24 h and the whole cell lysate was collected in RIPA buffer (Merck KGaA), containing protease inhibitor cocktail (PIC; Sigma-Aldrich), 10 mM sodium fluoride (NaF; Sigma-Aldrich) and 2 mM sodium vanadate (Na3VO4; Sigma-Aldrich). The protein concentrations were quantified using Bradford protein assay (Bio-Rad Laboratories, Inc.). Equal amounts of protein (10 µg) from each cell lysate were prepared and separated using NuPage 4–12% gradient Bis-tris protein gel (Thermo Fisher Scientific, Inc.), prior to being transferred onto nitrocellulose membranes (Bio-Rad Laboratories, Inc.). The membranes were incubated with primary antibodies at 4°C overnight in Odyssey blocking buffer (LI-COR Biosciences). Primary antibodies included rabbit anti-collagen I (cat. no. 21286; Abcam; dilution 1:1,000), rabbit anti-Smad 2/3 (cat. no. 8685; Cell Signaling Technology, Inc.; dilution 1:1,000) and mouse anti-GAPDH (cat. no. G8795; Sigma-Aldrich; Merck KgaA; dilution 1:10,000). Secondary antibodies used were anti-rabbit Alexa Fluor 680 (cat. no. A-21076; Invitrogen; Thermo Fisher Scientific, Inc.; dilution 1:10,000), anti-mouse Alexa Fluor 790 (cat. no. A11371; Invitrogen; Thermo Fisher Scientific, Inc.; dilution 1:10,000) against anti-Smad2/3 and anti-collagen, and anti-GAPDH respectively. Images were captured and analysed using the Odyssey Infrared Imaging system and Image Studio Version 5.2 software (LI-COR Biosciences).
Sequencing
HSFs were seeded onto a 12-well plate at 1×105 cells per well. Following a 24-h incubation, the cells were left untreated or transfected with 100 nM of the Smad decoy mixed with PepC (Pepscan). Following a 72-h transfection, the cells were harvested and the total RNA was isolated using DirectZol kit (Zymo Research Corp.). The barcoded mRNA sequencing library was subsequently prepared using TruSeq RNA Library Prep v2 (Illumina, Inc.), according to the manufacturer's protocol. The resultant libraries were pooled and sequenced on a HiSeq 2000 sequencer (Illumina, Inc.). Following removal of the adaptor sequences using Prinseq, the sequences were mapped to the human transcriptome (hg19 genome using TOPHAT (18) and the aligned BAM file was used to read counts for each gene using HTSeq (19). Subsequent analysis was performed using normalised counts and standard deviations derived from the 3-fold-changes of individual genes between the untreated and treated samples. Gene ontology analysis was performed by analysing the 178 most differentially regulated genes using the DAVID analysis algorithm v6.8 hosted on the website (https://david.ncifcrf.gov) using the GOTERM_CC_DIRECT (cellular component) annotation (20).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
HSFs or Kc were plated onto a 24-well plate at 5×104 cells per well. Following a 24-h incubation, the cells were left untreated, treated with the scrambled Smad decoy oligonucleotide or the Smad decoy mixed with PepC. A total of 48 h after transfection, RNA from the cells were harvested using DirectZol kit (Zymo Research Corp.) according to the manufacturer's protocols, and reverse transcribed using MMLV reverse transcriptase (Promega Corporation) using random nanomers (Integrated DNA Technologies, Inc.), according to the manufacturer's protocol. The amplicons were then assayed by qPCR using Maxima SyBr Green Mastermix (Thermo Fisher Scientific, Inc.) using the primers listed in Table I with the following protocol: 95°C for 5 min, followed by 50 cycles of 95°C for 10 sec, 58°C for 8 sec and 60°C for 30 sec. The samples were quantified using the StepOnePlus software (Thermo Fisher Scientific, Inc.) using the relative standard curve method with automatic Ct calling. The relative expression of each gene was quantified against a relative standard curve made up of serial dilution of a few of the samples pooled together and then normalised to GAPDH expression by dividing the relative expression of a query gene by GAPDH expression relative expression. The ratios were then normalised to 1 for the untreated control compared between sample groups (21).
Table I.
Primers used in reverse transcription-quantitative polymerase chain reaction.
| Gene | Forward and reverse primers |
|---|---|
| COL1A1 | F: 5′-TCTGCGACAACGGCAAGGTG-3′ |
| R: 5′-GACGCCGGTGGTTTCTTGGT-3′ | |
| COL1A2 | F: 5′-ACACGTCTGGCTAGGAGAAAC-3′ |
| R: 5′-GGCATAGTTGGCCAGCAGGC-3′ | |
| COL3A1 | F: 5′-ACCAGGAGAGAAGGGATCGC-3′ |
| R: 5′-TTCCCCTAGGACCTGGCATG-3′ | |
| TAGLN | F: 5′-CCGTGGAGATCCCAACTGG-3′ |
| R: 5′-CCATCTGAAGGCCAATGACAT-3′ | |
| SPARC | F: 5′-ACATCGCCCTGGATGAGTGG-3′ |
| R: 5′-CGGTACTGTGGAAGGAGTGG-3′ | |
| MMP1 | F: 5′-GCCGACAGAGATGAAGTCCG-3′ |
| R: 5′-CTTGGGGTATCCGTGTAGCAC-3′ | |
| MMP3 | F: 5′-CTCCAACCGTGAGGAAAATCG-3′ |
| R: 5′-TGGGAAAGCCTGGCTCCATG-3′ | |
| SERPINB2 | F: 5′-TTGCCGATGTGTCCACTGGC-3′ |
| R: 5′-GTCTTTGCTGGTCCACTTGTTG-3′ | |
| MMP2 | F: 5′-GATGCCGCCTTTAACTGGAGC-3′ |
| R: 5′-TCCAGGCATCTGCGATGAGC-3′ | |
| GAPDH | F: 5′-AAGGTGAAGGTCGGAGTCAA-3′ |
| R: 5′-GAAGATGGTGATGGGATTTC-3′ | |
| TIMP1 | F: 5′-TCAACCAGACCACCTTATACC-3′ |
| R: 5′-GTAGACGAACCGGATGTCAGC-3′ | |
| FN14 | F: 5′-CTGGACAAGTGCATGGACTGC-3′ |
| R: 5′-CCAAGGATGGGCCAAAGCAG-3′ |
F, forward; R, reverse.
Statistical analysis
All experiments were performed 3 times using cells from 3 different patients. The data are expressed as the percentage of the control group. One-way analysis of variance, followed by Tukey's post hoc test, were used to analyse the statistical difference. P<0.05 was considered to indicate a statistically significant difference.
Results
The Smad decoy inhibits the total amount of collagen produced in HSF
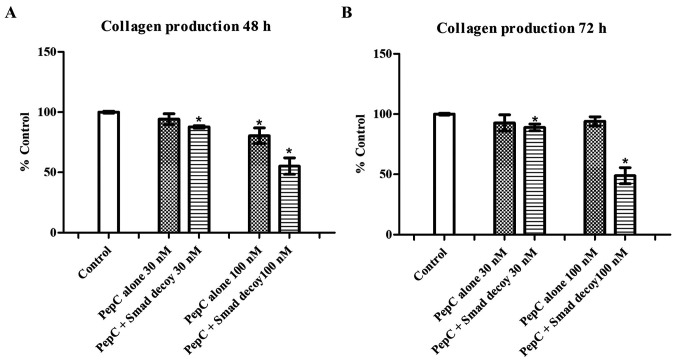
To evaluate the effects of the Smad decoy on the total amount of collagen produced in HSF, 3 different concentrations of the Smad decoy mixed with PepC, a novel peptide transfection reagent, were applied to HSF. The same concentrations of PepC alone were used as the vehicle control. As demonstrated in Fig. 1, PepC alone at 30 nM showed no effects on HSF collagen production at 48 and 72 h. However, the Smad decoy mixed with PepC at 30 nM significantly decreased HSF collagen production by 12.2±1.0 and 11.0±2.8% at 48 and 72 h, respectively, compared with the control (P<0.05). PepC alone at 100 nM decreased HSF collagen production by 19.5±6.5% at 48 h compared with the control (P<0.05); however, no inhibitory effects of PepC on HSF collagen production was detected at 72 h. The Smad decoy mixed with PepC at 100 nM significantly attenuated HSF collagen production by 44.8±6.8 and 51.0±6.6% at 48 and 72 h, respectively, compared with the control (P<0.05). The data suggested that the Smad decoy inhibits HSF collagen production in a dose-dependent manner. In addition, the Smad decoy mixed with PepC had no effects on cell viability at 72 h (Fig. S1), suggesting that the downregulation of collagen induced by the Smad decoy is not simply due to the cytotoxicity.
Figure 1.
Effects of the Smad decoy on collagen production in HSF. HSF was treated with different concentrations of PepC with or without the Smad decoy for (A) 48 h and (B) 72 h. Collagen production was measured using Sirius red staining. Data are expressed as the average percentage of the control and are pooled from 3 replicate experiments. Error bars indicate the mean ± standard error of the mean (n=3). *P<0.05 vs. control. Statistical analysis was performed using one-way analysis of variance and Tukey's post hoc test. PepC, cell-penetrating peptide.
The Smad decoy attenuates the expression of collagen I and Smad 2/3 in HSF
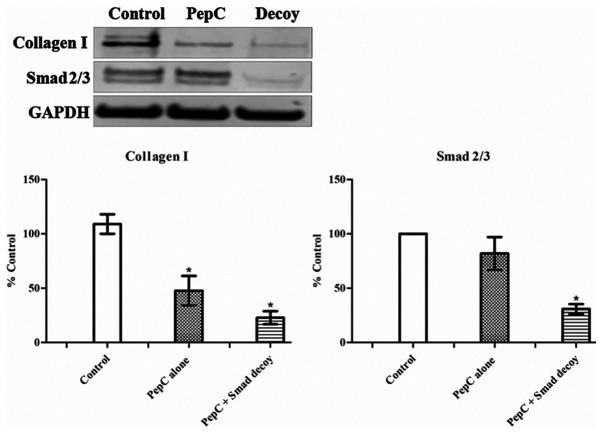
To further investigate the effects of the Smad decoy on the TGF-β1/Smad signaling pathway, expression of collagen I and Smad2/3 in HSF following treatment with the Smad decoy was analysed using western blotting (Fig. 2). PepC alone and Smad decoy mixed with PepC at 100 nM attenuated the expression of collagen I in HSF. Although the western blotting indicated that the Smad decoy has a greater inhibitory effect on collagen I expression compared with PepC alone, the difference between these two groups was not statistically significant. PepC alone showed no effects on the expression of Smad 2/3 in HSF; however, the Smad decoy significantly downregulated the expression of Smad2/3 in HSF compared with the control. These results suggested that the Smad decoy is capable of regulating Smad2/3 expression, thereby resulting in a decrease in collagen I.
Figure 2.
Effects of the Smad decoy on the expression of Collagen I and Smad2/3 in HSF. The expression of proteins was detected using the LI-COR Odyssey Fc imaging system. GAPDH was used as a loading control. Representative images of western blots are present. Quantitative analysis was performed using ImageJ software. Protein bands were first normalised to GAPDH and then converted to a percentage of the untreated control. Data were pooled from the 3 replicate experiments. Error bars indicate standard error of the mean (n=3). *P<0.05 vs. control. Statistical analysis was performed using one-way analysis of variance and Tukey's post hoc test. HSF, human skin fibroblasts; PepC, cell-penetrating peptide.
The Smad decoy induces an alteration of gene expression in HSF and human primary keratinocytes (Kc)
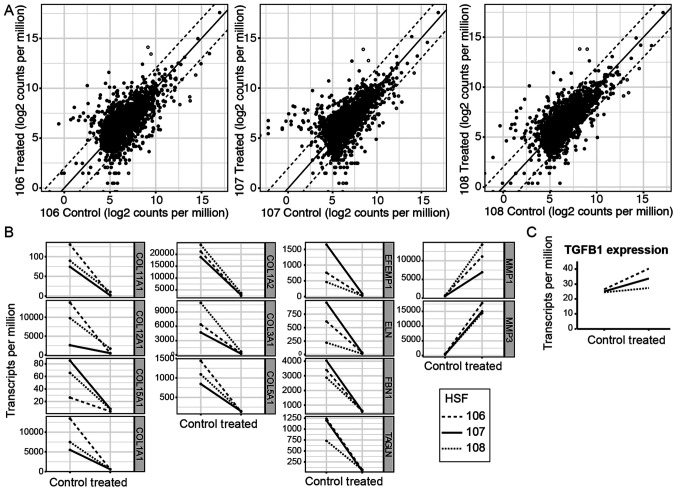
To ascertain the genes affected, HSFs were treated with the Smad decoy, put through RNA sequencing and compared individually with untreated matched controls. The top 5,000 expressed genes were then plotted against each other on a logarithmic plot. The majority of the genes fell within a 4-fold difference in expression between untreated and treated samples, with 178 genes exceeding this (Fig. 3A and Table II). The 178 genes were analyzed with cell component gene ontology analysis using DAVID (20) to determine which cellular component the most differentially expressed genes functioned in Table III. The list suggests that the Smad decoy primarily affects ECM-associated functions and the present study aimed to highlight the ECM expression changes with selected upregulated and downregulated genes (Fig. 3B) corresponding to the hollow (upregulated) and grey (downregulated) dots in Fig. 3A. Notably, the Smad decoy does not appear to affect the expression of the TGFB1 gene, which codes for TGF-β1 (Fig. 3C). Therefore, it is unlikely that the changes triggered by the Smad decoy are directly attributable to unintended perturbation of TGF-β1 levels rather than direct perturbation of the Smad signaling pathway. The effect on the ECM is similar to the results of a previous study, which used a microarray of dermal fibroblasts stimulated with TGF-β1 (22). In particular, COL1A2, COL3A1 and FBN1 were downregulated by inhibition of Smad, corresponding to upregulation when TGF-β1 was added. However, several genes upregulated by increase in TGF-β1 levels, including TIMP1, TIMP3 and COL6A1, were notably not downregulated by inhibition by the Smad decoy. Furthermore, while Verrecchia et al (2001) noted upregulation of matrix metalloproteinase-1 (MMP1) and MMP3 by TGF-β1, the present study reported marked increases in MMP1 and MMP3 expression by Smad inhibition (22). The divergence in the results suggests that, in the present study, it was possible to isolate the Smad pathway from the other TGF-β1 signaling pathways using the Smad decoy.
Figure 3.
Sequencing analysis of the Smad decoy-induced gene changes in HSF. The 3 different HSF lines were transfected with 100 nM Smad decoy and harvested 72 h after transfection for RNA-seq. (A) Log2 scatter plot of 5,000 of the top expressed genes in control and treated matched HSF lines with dotted lines indicating a 4-fold difference in relative expression. (B) Selected genes highlighted in hollow dots (upregulated) and grey dots (downregulated) in (A) shown to be differentially expressed in control and treated across the different HSF lines. (C) TGF-β1 expression is similar across control and treated HSFs and is unlikely to cause expression changes in the Smad pathway. HSF, human skin fibroblasts; TGF, transforming growth factor.
Table II.
Genes differentially expressed in treated and untreated HSFs.
| Treated/untreated, log2 fold-change | Treated/untreated, log2 fold-change | Treated/untreated, log2 fold-change | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | 106 | 107 | 108 | Gene | 106 | 107 | 108 | Gene | 106 | 107 | 108 | |
| RSAD2 | 7.52 | 6.49 | 5.73 | LIF | 3.75 | 4.01 | 0.24 | AKR1C1 | 3.44 | 2.03 | 2.74 | |
| IL24 | 3.71 | 6.91 | 7.14 | IFI27 | 2.19 | 4.14 | 2.98 | DDX60 | 2.7 | 3.16 | 2.52 | |
| SERPINB2 | 6.67 | 6.1 | 6.03 | RND3 | 2.57 | 3.71 | 3.41 | MX2 | 2.75 | 3.31 | 1.89 | |
| PTGS2 | 5.1 | 6.81 | 5.68 | OAS2 | 2.55 | 4.12 | 2.44 | IFITM1 | 2.69 | 3.08 | 2.43 | |
| TFPI2 | 5.92 | 5.83 | 6.22 | IL11 | 3.7 | 3.27 | 2.54 | MT2A | 2.99 | 2.37 | 2.71 | |
| THBD | 5.46 | 6.41 | 5.07 | HSPA1B | 3.07 | 2.45 | 3.79 | STC1 | 2.38 | 2.39 | 3.16 | |
| OASL | 6.44 | 5.2 | 3.91 | DUSP10 | 4.23 | 1.72 | 2.39 | PID1 | 2.94 | 2.4 | 2.63 | |
| ESM1 | 5.72 | 5.77 | 4.74 | INHBA | 3.89 | 2.65 | 2.54 | SAMD9 | 3.3 | 2 | 2.37 | |
| MMP10 | 4.81 | 5.92 | 5.31 | SAT1 | 3.16 | 3.5 | 2.73 | SQSTM1 | 2.82 | 2.35 | 2.58 | |
| MMP3 | 4.97 | 5.42 | 5.62 | HMGA2 | 3.19 | 3.53 | 2.63 | ANGPTL4 | 2.22 | 2.81 | 2.69 | |
| BMP2 | 5.46 | 5.79 | 4.57 | FAM167A | 2.58 | 3.3 | 3.28 | PITPNC1 | 3.35 | 2.26 | 1.59 | |
| MX1 | 3.15 | 6.72 | 3.07 | TNFAIP3 | 2.57 | 3.61 | 2.61 | SLC5A3 | 2.49 | 1.49 | 3.25 | |
| MMP12 | 5.08 | 5.52 | 4.44 | ID1 | 3.76 | 2.41 | 2.33 | DDX58 | 2.92 | 2.6 | 2.09 | |
| IL1B | 5.65 | 5.2 | 3.5 | TBX3 | 2.7 | 3.51 | 2.14 | HSPA1A | 2.6 | 1.39 | 3.13 | |
| CMPK2 | 5.13 | 5.28 | 3.87 | IFI44 | 3.22 | 2.96 | 2.36 | IL6 | 1.48 | 3.21 | 2.3 | |
| CXCL8 | 3.86 | 5.52 | 4.58 | SPRY2 | 2.93 | 2.42 | 3.21 | DUSP5 | 2.16 | 2.77 | 2.43 | |
| ITGA2 | 5.29 | 4.38 | 4.27 | AKR1C1 | 3.44 | 2.03 | 2.74 | CXCL1 | 1.99 | 3.12 | 1.95 | |
| CXCL5 | 4.36 | 5.33 | 2.57 | DDX60 | 2.7 | 3.16 | 2.52 | IER3 | 2.46 | 2.59 | 2.19 | |
| OAS1 | 3.74 | 5.33 | 3.49 | MX2 | 2.75 | 3.31 | 1.89 | TNFAIP6 | 1.84 | 2.93 | 2.25 | |
| IFIT1 | 3.91 | 5.32 | 3.24 | IFITM1 | 2.69 | 3.08 | 2.43 | GNG11 | 2.58 | 1.89 | 2.46 | |
| IFIT2 | 5.03 | 4.47 | 3.21 | MT2A | 2.99 | 2.37 | 2.71 | HMOX1 | 3.14 | 1.71 | 1.61 | |
| IFIT3 | 4.59 | 4.84 | 3.5 | STC1 | 2.38 | 2.39 | 3.16 | JARID2 | 2.35 | 2.41 | 2.24 | |
| CXCL3 | 3.78 | 5.28 | 3.26 | PID1 | 2.94 | 2.4 | 2.63 | NPC1 | 2.13 | 1.9 | 2.7 | |
| MMP1 | 3.95 | 3.71 | 4.82 | SAMD9 | 3.3 | 2 | 2.37 | ABL2 | 1.55 | 2.77 | 2.26 | |
| PHLDA1 | 4.03 | 4.35 | 4.06 | SQSTM1 | 2.82 | 2.35 | 2.58 | FOSL1 | 2.28 | 2.1 | 2.4 | |
| IFI44L | 3.13 | 5.19 | 2.64 | ANGPTL4 | 2.22 | 2.81 | 2.69 | ITPRIP | 2.43 | 2.52 | 1.67 | |
| IFI6 | 1.83 | 5.3 | 2.44 | PITPNC1 | 3.35 | 2.26 | 1.59 | FGF5 | 0.93 | 2.58 | 2.55 | |
| DUSP6 | 3.95 | 4.28 | 3.24 | SLC5A3 | 2.49 | 1.49 | 3.25 | NT5E | 2.16 | 2.41 | 1.94 | |
| HERC6 | 2.93 | 4.7 | 3.41 | DDX58 | 2.92 | 2.6 | 2.09 | KLHL21 | 2.39 | 1.94 | 2.15 | |
| PLIN2 | 3.17 | 3.93 | 4.11 | HSPA1A | 2.6 | 1.39 | 3.13 | PTGS1 | 2.41 | 1.82 | 2.21 | |
| NAMPT | 3.99 | 3.55 | 3.76 | IL6 | 1.48 | 3.21 | 2.3 | PARP12 | 2.16 | 2.24 | 2.06 | |
| IFIH1 | 3.44 | 4.43 | 2.86 | DUSP5 | 2.16 | 2.77 | 2.43 | DCBLD2 | 2.11 | 2.52 | 1.65 | |
| TMEM158 | 3.71 | 4.37 | 2.56 | CXCL1 | 1.99 | 3.12 | 1.95 | PLSCR1 | 2.05 | 2.28 | 1.96 | |
| CDCP1 | 3.01 | 4.24 | 3.43 | IER3 | 2.46 | 2.59 | 2.19 | PRDM1 | 2.14 | 2.42 | 1.47 | |
| OAS3 | 2.61 | 4.51 | 2.82 | TNFAIP6 | 1.84 | 2.93 | 2.25 | PDGFD | −5.58 | −6.17 | −4.33 | |
| DUSP4 | 3.32 | 4.19 | 2.85 | ID1 | 3.76 | 2.41 | 2.33 | SPARC | −3.26 | −3.33 | −2.39 | |
| ISG15 | 3.68 | 4.09 | 2.26 | TBX3 | 2.7 | 3.51 | 2.14 | FIBIN | −2.09 | −2.95 | −2.21 | |
| PTGES | 4.34 | 2.78 | 2.79 | IFI44 | 3.22 | 2.96 | 2.36 | RCAN2 | −4.34 | −4.49 | −4.9 | |
| PODXL | 4.05 | 3.29 | 2.94 | SPRY2 | 2.93 | 2.42 | 3.21 | COL5A1 | −3.43 | −2.54 | −2.89 | |
| ADAMTSL1 | −1.86 | −1.55 | −3.81 | SYNPO2 | −5.52 | −4.89 | −2.84 | WISP1 | −2.33 | −3.34 | −2.97 | |
| GAS6 | −2.37 | −2.65 | −2.01 | COL11A1 | −5.12 | −4.97 | −2.99 | NEDD9 | −5.79 | −1.32 | −1.52 | |
| ADAM33 | −2.47 | −2.15 | −2.36 | ELN | −4.05 | −4.99 | −3.98 | COL15A1 | −2.4 | −3.2 | −2.93 | |
| LRIG3 | −3.76 | −2.29 | −0.88 | DKK2 | −4.05 | −4.56 | −4.03 | DAPK1 | −2.16 | −3.89 | −2.43 | |
| INHBB | −3.54 | −1.68 | −1.71 | EFEMP1 | −4.3 | −4.14 | −4.1 | KIF26B | −2.49 | −3.92 | −2.04 | |
| ANGPT1 | −2.7 | −2.4 | −1.81 | COL3A1 | −4.27 | −4.22 | −3.97 | COL1A2 | −3.04 | −2.7 | −2.69 | |
| LOXL4 | −2.14 | −1.96 | −2.79 | HMCN1 | −4.05 | −3.51 | −4.76 | SORT1 | −4.06 | −2.28 | −2.05 | |
| ITIH5 | −3.02 | −2.21 | −1.63 | POSTN | −3.67 | −5 | −3.49 | SGCD | −2.76 | −3.29 | −2.29 | |
| SERAC1 | −2.73 | −2.27 | −1.83 | CNN1 | −3.36 | −3.38 | −5.12 | CARMN | −3.43 | −3.1 | −1.78 | |
| FBLN2 | −2.25 | −2.5 | −2.05 | WNT2 | −3.72 | −5.31 | −2.52 | CTGF | −4.64 | −2.39 | −1.27 | |
| PDE1C | −2.71 | −2.18 | −1.88 | COL1A1 | −4.57 | −3.22 | −3.74 | VCAN | −2.9 | −2.84 | −2.52 | |
| TENM2 | −3.97 | −1.08 | −1.71 | TAGLN | −4.05 | −4 | −3.37 | ACTA2 | −3.92 | −2.51 | −1.67 | |
| GADD45B | −3.18 | −2.08 | −1.5 | LMOD1 | −3.6 | −4.93 | −2.61 | STARD4-AS1 | −2.8 | −2.59 | −2.65 | |
| SLIT3 | −2.63 | −1.31 | −2.81 | OXTR | −4.21 | −4.5 | −2.1 | PRUNE2 | −2.69 | −3.13 | −2.2 | |
| COMP | −2.49 | −1.56 | −2.64 | ITGA11 | −4.43 | −3.34 | −2.95 | PODN | −1.91 | −3.61 | −2.44 | |
| TNFRSF11B | −4 | −1.93 | −0.69 | CHN1 | −3.26 | −4.56 | −2.81 | NTN4 | −4.04 | −3.24 | −0.68 | |
| FBLN1 | −2.08 | −1.96 | −2.44 | ITGBL1 | −3 | −4.29 | −3.21 | VSIR | −2.1 | −2.78 | −2.94 | |
| PYCR1 | −2.45 | −1.77 | −2.25 | MFAP4 | −2.51 | −5.12 | −2.84 | ALDH1B1 | −3.35 | −2.48 | −1.95 | |
| RHOBTB1 | −2.51 | −1.94 | −2.01 | DPT | −2.46 | −4.36 | −3.57 | TNFRSF19 | −2.31 | −3 | −2.4 | |
| CCL2 | −2.26 | −0.9 | −3.22 | MXRA5 | −3.98 | −2.99 | −2.97 | FBN1 | −2.64 | −2.77 | −2.2 | |
| MYL9 | −1.47 | −2.96 | −1.92 | COL12A1 | −5.02 | −2.38 | −2.52 | NTN1 | −1.35 | −3.06 | −3.15 | |
| CCDC80 | −2.01 | −2.46 | −1.86 | FBN2 | −3.85 | −3.31 | −2.49 | RDH10 | −4.59 | −1.8 | −1.17 | |
| DHCR24 | −2.04 | −2.88 | −1.35 | LMCD1 | −4.14 | −3.72 | −1.75 | RNF150 | −2.41 | −2.45 | −2.63 | |
| COL8A1 | −2.68 | −1.21 | −2.35 | IGFBP5 | −4.33 | −4.2 | −1.07 | THY1 | −1.92 | −2.78 | −2.79 | |
| GLT8D2 | −1.28 | −3.02 | −1.92 | NREP | −3.12 | −4.11 | −2.17 | CNN2 | −2.56 | −2.6 | −2.31 | |
| SERPINH1 | −1.48 | −2.62 | −2.07 | SVEP1 | −3.84 | −2.86 | −2.64 | APCDD1 | −1.96 | −2.29 | −3.16 | |
| RN7SL5P | −0.18 | −0.89 | −5.07 | GFRA1 | −3.56 | −3.2 | −2.57 | DIO2 | −0.43 | −4.8 | −2.11 | |
| COL5A2 | −2.1 | −2.23 | −1.78 | MFAP5 | −2.4 | −4.27 | −2.34 | ALPK2 | −3.12 | −3.02 | −1.13 | |
| TP53I11 | −1.51 | −2.11 | −2.47 | |||||||||
Underlined genes are highlighted in Fig. 3B.
Table III.
Gene classification of differentially expressed genes.
| Gene Ontology analysis for differentially expressed genes | Count | % of genes out of 178 | P-value |
|---|---|---|---|
| Proteinaceous extracellular matrix | 30 | 16.9 | 1.7×10−22 |
| Extracellular region | 61 | 34.3 | 1.7×10−21 |
| Extracellular space | 52 | 29.2 | 2.3×10−18 |
| Extracellular matrix | 25 | 14.0 | 8.4×10−16 |
| Endoplasmic reticulum lumen | 14 | 7.9 | 3.8×10−8 |
| Collagen trimer | 10 | 5.6 | 2.2×10−7 |
| Basement membrane | 8 | 4.5 | 1.0×10−5 |
| Microfibril | 4 | 2.2 | 9.6×10−5 |
| Elastic fiber | 3 | 1.7 | 5.3×10−4 |
| Perinuclear region of cytoplasm | 14 | 7.9 | 6.4×10−3 |
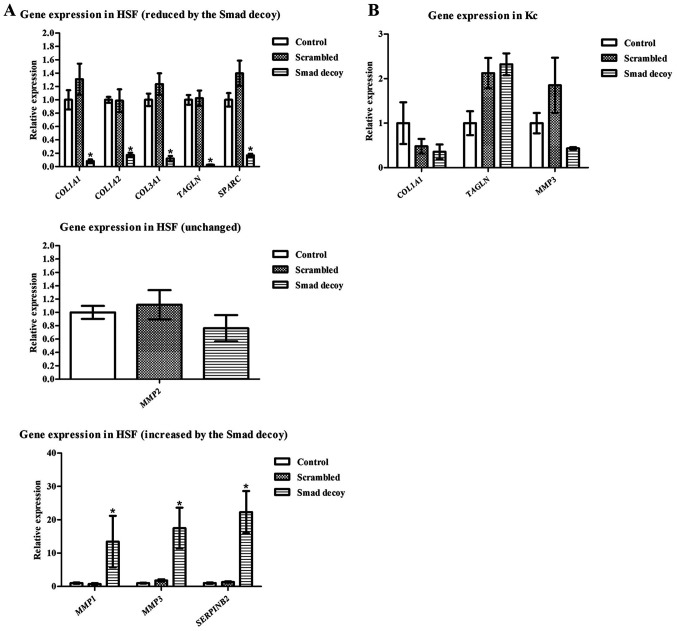
To validate the sequencing results, RT-qPCR analysis was performed of a number of genes, which are highly associated with HS formation, on HSF treated for 3 days with the Smad decoy, using a scrambled decoy as a negative control. Corresponding to the sequencing data, COL1A1, COL1A2, COL3A1, TAGLN and SPARC were significantly downregulated following exposure to the Smad decoy, but not with the scrambled decoy (Fig. 4A). Similarly, MMP1, MMP3 and SERPINB2 were significantly upregulated in the Smad decoy treated group compared with the scrambled and control groups. To ascertain that genes unchanged in the sequencing data were also unchanged in the independent experiment, MMP2, another matrix metalloprotease was also assayed using RT-qPCR. Data from the sequencing assay suggested that MMP2 is not affected by the Smad decoy (treated/untreated in sequencing=0.93±0.03); consistently, no significant changes in MMP2 expression were observed in RT-qPCR. This suggests that the collagen-inhibiting ability of the Smad decoy relies on its regulation of the expression of various genes, including COL1A1, COL3A1 and MMPs.
Figure 4.
RT-qPCR analysis of the Smad decoy-induced gene changes in HSF and Kc. (A) gene expression in HSF. (B) gene expression in Kc. Cells were treated with the Smad decoy or scrambled oligonucleotide at 100 nM for 48 h and then the total RNA was collected. Following RNA extraction, first-strand cDNA was synthesized. The cDNA sample was amplified by RT-qPCR. The expression of the target genes was first normalized to GAPDH and then further converted to the percentage of the control. Error bars indicate the mean ± standard error of the mean (n=3). *P<0.05 vs. the control. Statistical analysis was performed using one-way analysis of variance and Tukey's post hoc test. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HSF, human skin fibroblasts; Kc, keratinocytes.
Additionally, effects of the Smad decoy on the gene expression of Kc were evaluated as Kc also serves essential roles in HS formation (23). Application of the Smad decoy did not appear to affect COL1A1 nor MMP3 significantly, but it increased TAGLN expression slightly (Fig. 4B). However, the effect appears to be associated with PepC-based delivery as the scrambled oligonucleotide also resulted in increased TAGLN expression.
Effects of the Smad decoy on TGF-β1-stimulated HSF
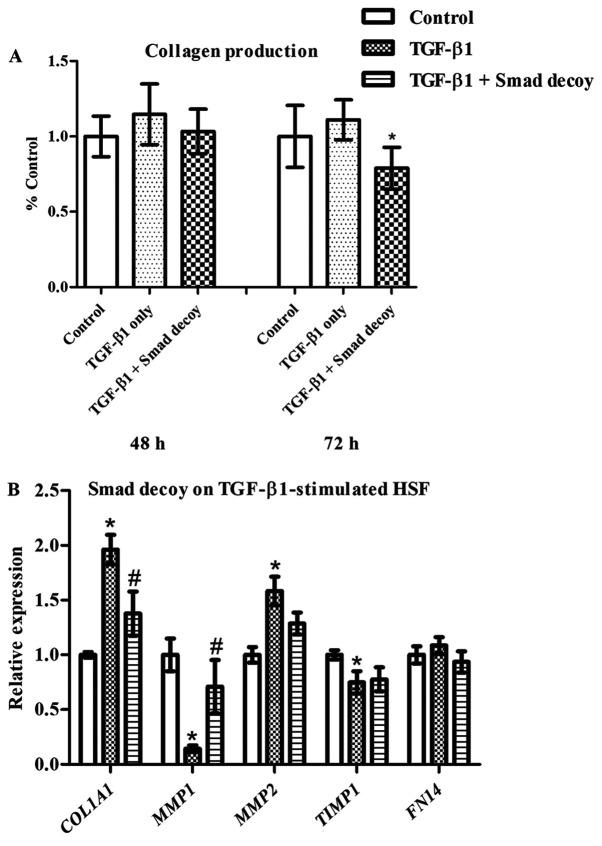
As TGF-β1 has been demonstrated to serve essential roles in HS formation and overabundant TGF-β1 expression is frequently observed in HS tissues (6), the present study investigated the effects of the Smad decoy on TGF-β1-stimulated HSF. The Smad decoy was initially delivered with PepC into the cells followed by the addition of TGF-β1 3 h later. Although collagen production was modestly increased by TGF-β1, Smad decoy inhibition of collagen was markedly less (Fig. 5A). TGF-β1 has been revealed to significantly increase the expression of COL1A1 and MMP2, and reduce the expression of MMP1 and TIMP1 in HSF, as demonstrated in Fig. 5B. The Smad decoy significantly inhibited TGF-β1-induced upregulation of COL1A1 and downregulation of MMP1 in HSF. Notably, FN14, reported to be upregulated by TGF-β1 in dermal fibroblasts (24), was not affected by the addition of TGF-β1 in the present study. This demonstrates that the Smad decoy alters TGF-β1-induced ECM expression in HSF.
Figure 5.
Effects of the Smad decoy on TGF-β1-stimulated HSF. (A) Total amount of collagen deposition. (B) Expression of genes. HSF was treated with the Smad decoy and then TGF-β1 (5 ng/ml) was added to the cell culture 3 h later. The total amount of collagen was measured using Sirius red staining and the gene expression was detected using reverse transcription-quantitative polymerase chain reaction. The expression of the target genes was first normalized to GAPDH and then further converted to the percentage of the control. Error bars indicate the mean ± standard error of the mean (n=3). *P<0.05 vs. the control; #P<0.05 vs. TGF-β1 group. Statistical analysis was performed using one-way analysis of variance and Tukey's post hoc test. TGF, transforming growth factor; HSF, human skin fibroblasts.
Discussion
HS remains a challenging problem for both patients and clinicians. The underlying mechanisms for HS formation remain elusive. However, excessive collagen deposition by fibroblasts has been widely demonstrated to result in HS formation (4). Numerous therapies are available in clinic, but no single therapy can guarantee the improvement of HS. For example, silicone dressings have been used to treat HS since 1983 (25), but require frequent changes and cause skin rashes (26). Laser therapy is becoming more popular in the clinic for managing HS, but high recurrence is observed in patients receiving laser therapy (27). Therefore, novel therapies with more accurate pathological target are of particular relevance and may present patients with more options. Targeted therapy is becoming increasingly important in healthcare as many traditional therapies lack selectivity and specificity (28). One of the major advantages of small molecule drugs to treat HS is that they are typically cell permeable, thereby blocking the activities of targeted proteins and regulating the downstream signaling pathways (29). For example, a TGF-β1 inhibitor has been reported to prevent HS formation when it is applied during wound healing (30), as increased TGF-β1 results in the formation of HS (6). The mechanism underpinning TGF-β1-induced HS formation is that TGF-β1 activates the phosphorylation of Smad 2/3, which then initiates collagen expression (9,10). The aim of the present study was to investigate if a Smad decoy that sequesters the Smad protein may inhibit collagen production and deposition.
As human Smad 3 and 4 have been identified to specifically bind a 8 bp palindromic sequence to activate transcription (13), a double-stranded DNA decoy that contains this sequence was designed. To evaluate the effects of this Smad decoy on HSF, the present study aimed to make use of a non-toxic reagent for transfection. Transfection efficiency is limited by the biological barriers of the cells, including cellular uptake, intracellular trafficking, metabolic degradation and nuclear entry (31). Lipofectamine is the most frequently used transfection reagent due to its high transfection efficiency of DNA and RNA on various cells, including those considered to be difficult-to-transfect cells (31). However, Lipofectamine is cytotoxic (16). A novel CPP-based peptide (PepC), similar to the PF14 peptide that was recently reported to be more effective in transfecting splice-correcting oligonucleotides into HeLa pLuc705 cells with significantly lower cytotoxicity than Lipofectamine (16), was used for delivery of the Smad decoy. PepC had no effects on the collagen production in HSF at 72 h; by contrast, the Smad decoy delivered by PepC significantly inhibited HSF collagen production at 72 h. These results not only demonstrated that PepC is a suitable transfection reagent for HSF, but also suggested that the Smad decoy may potentially be used as a novel anti-scarring therapy. As collagen type I is the most abundant collagen type in humans (32) and is mediated by the TGF-β1/Smad signaling pathway (33), the present study also investigated the effects of the Smad decoy on the expression of collagen I and Smad2/3 in HSF. The results suggested that the Smad decoy markedly attenuates the expression of collagen I and Smad2/3, suggesting its roles in regulating the TGF-β1/Smad signaling pathway. Notably, PepC was revealed to decrease collagen I expression in HSF based on western blot analysis, albeit to a lesser extent than the Smad decoy (Fig. 2). This appears to contradict the results in Fig. 1 in which PepC does not decrease total collagen. This difference may be partially explained by the fact that only type I collagen was measured by western blotting and this was performed 24 h after treatment; while Sirius red staining in Fig. 1 quantified the total amount of collagen production and was performed at 72 h post-treatment. The data suggested that PepC may have short-term inhibitory effects on HSF collagen I deposition, but the effect size is much less than the combined effect of Smad and PepC together, particularly after 72 h. Limiting the western blots to only collagen I and Smad 2/3 provides only a small snapshot of the full picture. Insight into how the Smad decoy affects the expression of different types of collagen found in the extracellular matrix and the precise effect of these changes on hypertrophic scaring would aid in clarifying the mechanisms of how the Smad decoy may ameliorate hypertrophic scaring. This, in turn, could aid in refining a therapeutic plan to improve on the results reported in the present study.
At the cellular level, the collagen-reducing ability of the Smad decoy makes it a potential novel HS treatment as excessive collagen deposition is the hallmark of HS formation. However, the exact mechanisms of HS formation remain elusive and numerous other factors contribute toward the formation of HS. For example, Kc reform a functional epidermis (re-epithelialization) to protect exposed dermal tissue at the end of wound healing (34) and delayed re-epithelialization has been revealed to induce HS formation (35). In addition, MMPs are well known for their roles in regulating tissue remodeling (36). Decreased expression of MMP-1 is associated with HS formation (37). Therefore, the present study investigated the effects of the Smad decoy on HSF and Kc at the transcriptional level using sequencing and RT-qPCR. A total of 178 genes were reported to exceed a 4-fold difference in HSF treated with the Smad decoy compared with the untreated control. Certain representative genes were further analyzed (Fig. 3B) and it was revealed that the expression of these genes has similar trend in cells from 3 separate patient cell lines. Notably, the majority of those genes are associated with the extracellular matrix and are highly relevant to HS formation. For example, COL1A1, COL1A2, COL3A1, COL5A1, COL11A1, COL12A1 and COL15A1, genes encoding various collagen types, have been found to be downregulated by the Smad decoy, indicating the mechanism of the Smad decoy-induced collagen reduction in HSF. In particular, COL1A1, COL1A2 and COL3A1 are key genes encoding collagen I and collagen III in HS formation (38). In addition, EGF-containing fibulin-like extracellular matrix protein 1, encoded by EFEMP1, has been found to be highly expressed in HS and keloids (39,40). The results of the present study suggested that the Smad decoy attenuates the expression of EFEMP1 in HSF from all 3 individual patients. A previous study also suggested that elastin (encoded by ELN) and fibrillin-1 (encoded by FBN1) are differentially expressed in HS compared with normal tissues, suggesting their roles in HS formation (41). The results of the present study showed that the Smad decoy significantly decreases the expression of ELN and FBN1 in HSF, compared with the control. Transgelin, encoded by TAGLN, is an actin-binding protein in smooth muscle and fibroblasts (42). Although the function of transgelin remains largely unknown, one study reported that transgelin may be involved in the calcium-independent smooth muscle contraction (43). The increased fibroblast contraction has been reported to be associated with the over-abundant expression of collagen (17), a downregulated TAGLN expression may therefore improve the reduction in HS. The MMPs are widely known for their roles in tissue remodeling by degrading collagen and other ECM during wound healing (44). In particular, MMP-1 is decreased in HS tissue in vivo and HSF in vitro, compared with normal skin tissue or dermal fibroblasts, suggesting that the decreased expression of MMP-1 contributes toward the formation of HS (37). Similarly to MMP-1, another important condition for HS formation is the lack of MMP-3 (45), a metalloprotease responsible for the degradation of collagen. The results of the present study showed that the Smad decoy significantly enhances the expression of MMP1 and MMP3 in HSF, offering further evidence supporting its use as an anti-HS reagent. It may be worthwhile to investigate the effects of the Smad decoy on other Smads in future studies as other Smads also participate in the regulation the TGF/collagen signalling pathway, although Smads have different sequence specificity (46). For example, Smad1/5 are anti-fibrotic proteins, which antagonize Smad2/3, and Smad 7 has been demonstrated to inhibit the collagen synthesis (47).
TGF-β1 is a cytokine known to be involved in cell proliferation, differentiation and apoptosis on top of its effects in mediating ECM expression in dermal fibroblasts, as described previously (5). Although part of its activity is mediated through Smad3/4, there are numerous Smad-independent pathways involved. We hypothesized that the Smad decoy may facilitate the isolation of the TGF-β1-dependent ECM modulation pathway for wound healing, which was supported by the Gene Ontology assignment of the differentially expressed genes (Table III). Therefore, while TGF-β1 has been proposed to be useful for wound healing applications, a Smad decoy delivered locally to the HS site may be a better option in chronic wound applications.
High throughput RNA sequencing revealed the potential underlying mechanism of the Smad decoy-induced collagen reduction in HSF. To further investigate the findings, cells were treated with the Smad decoy or the scrambled oligonucleotide and the expression of selected genes was measured using RT-qPCR. Consistent with the sequencing data, the Smad decoy downregulated the expression of COL1A1, COL1A2, COL3A1 and TAGLN and upregulated the expression of MMP1 and MMP3 in HSF. The scrambled oligonucleotide had no effect on the expression of those genes. In addition, the secreted protein acidic and rich in cysteine (SPARC) encoded by SPARC is a well-known protein which serves essential roles in various biological activities, including wound healing and angiogenesis (48). In particular, increased SPARC has been reported to upregulate the expression of collagen type I in fibroblasts (49). The results of the present study suggested that the expression of SPARC was significantly attenuated in HSF following the Smad decoy treatment. Furthermore, although the roles of plasminogen activator inhibitor-2 (SERPINB2) in wound healing and HS formation are unknown, there is evidence to suggest that the expression of SERPINB2 is detected in macrophages and Kc, and it may participate in regulating the inflammation of wound healing (50). The present study was the first to demonstrate that SERPINB2 may contribute toward the formation of HS and that the Smad decoy significantly attenuates the expression of SERPINB2 in HSF. MMP-2 is a key regulator for cell migration and re-epithelialization during wound healing, and increased MMP-2 has been reported to induce HS formation (51). Notably, the results of the present study demonstrated that the Smad decoy has no effects on the expression of MMP2. In addition to the gene expression in HSF, effects of the Smad decoy on Kc gene expression were also investigated as Kc also serves important roles in HS formation (35). These results indicated that the expression of COL1A1, TAGLN and MMP3 in Kc were not affected following exposure to the Smad decoy, indicating that the Smad decoy has minor effects on Kc compared with those on HSF. These results further support the use of the Smad decoy for HS treatment, as delayed re-epithelialization causes HS formation (35). An ideal HS therapy should be able to decrease the excessive collagen deposition of fibroblasts without affecting the function of Kc.
After investigating the effects of the Smad decoy on HSF and Kc, the present study investigated the effects of the Smad decoy on TGF-β1-stimulated HSF as overabundant TGF-β1 expression has been demonstrated to induce HS formation (6). Previous studies have reported that additional TGF-β1 alters the expression of COL1A1 (17), MMP2 (52), TIMP1 (53) and FN14 (24) in HSF. Although the total amount of collagen was not increased in HSF following TGF-β1 stimulation, the Smad decoy was reported to significantly attenuate the collagen production compared with TGF-β1-stimulated HSF. In addition, increased COL1A1 and decreased MMP1 were detected in HSF stimulated with TGF-β1, which is consistent with previous studies (17). The Smad decoy, however, significantly decreased TGF-β1-induced upregulation of COL1A1 and increased TGF-β1-induced downregulation of MMP1 in HSF, suggesting its ability to inhibit the effects of TGF-β1 on HSF. Notably, although TGF-β1 has been reported to significantly increase MMP2 and decrease TIMP1 expression in HSF, no effects of the Smad decoy on the expression of MMP2 and TIMP1 were detected compared with the TGF-β1-treated group. Fibroblast growth factor-inducible molecule 14, encoded by FN14, has been reported to be a downstream target of the TGF-β signaling pathway and serves essential roles in HS formation (24). Notably, neither TGF-β1 nor the Smad decoy was revealed to affect FN14 expression in HSF.
In addition to decreasing collagen production, when the transcriptomic changes induced by the Smad decoy were investigated, results from sequencing and RT-qPCR suggested that the primary effect of the Smad decoy was to decrease ECM and membrane components and increase their degradation through the upregulation of MMP1 and MMP3. This limits the side effects that may be associated TGF-β inhibition beyond the ECM components. Notably, MMP-2, which had been reported to be regulated through p38 MAPK signaling rather than Smad (54), is thought to be important in collagen remodeling, while MMP-1 activity is more biased towards a reduction of collagen I (55). Therefore, the ability to maintain TGF-β-induced upregulation of MMP-2, while decreasing collagen I and increasing MMP-1 through Smad inhibition may be beneficial to the wound-healing process. Therefore, the application of Smad inhibitors may be more beneficial than TGF-β inhibition therapies, including with anti-TGF-β antibodies (56). An interesting phenomenon identified in the present study was that the Smad decoy has effects independent of exogenous addition of TGF-β1 (Fig. 5). This is probably due to the fact that TGFB1 mRNA is produced in all HSFs from 3 patients and was detected in the top 5,000 genes expressed in these cells, which suggests that TGF-β1 is autoregulated in these fibroblasts. In fact, TGF-β1 has been previously demonstrated to be produced in HS fibroblasts (57); therefore, it makes sense that the Smad decoys would work despite the lack of exogenously added TGF-β1.
In conclusion, a novel Smad decoy was designed, which inhibits the total amount of collagen production, including collagen type I in HSF, by altering the expression of various genes. In particular, the Smad decoy has been reported to change the expression of COL1A1, COL1A2, COL3A1, TAGLN, SPARC, MMP1 and MMP3 in HSF, which is associated with collagen deposition and HS formation. In addition, the Smad decoy is able to attenuate TGF-β1-induced upregulation of COL1A1 and increase TGF-β1-induced downregulation of MMP1 in HSF. The results of the present study support the use of this Smad decoy as a potential novel HS therapy. However, this in vitro study is limited in its scope and further studies are required to establish the safety of PepC and the decoy in animal models prior to its clinical application. The efficacy of spray-on local delivery for a nucleic acid Smad decoy drug is also unknown as no previous studies are known to have utilized a simple nozzle to deliver complexed nucleic acid drugs, although nucleic acid drugs have been nebulized. It is also not known if Smad-specific inhibition of the ECM proteins will be sufficient for the inhibition of HS formation, particularly since little is known about HS formation. In conclusion, the Smad decoy may offer patients another option to combat HS.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This work was funded by core funding for the Molecular Engineering Laboratory and the Molecular Therapeutics Programme grant (grant no. IAF-PP/H17/01/a0/012) and Wound Care Innovation for the Tropics [grant no. IAF-PP/2017 (HBMS) H17/01/a0/009].
Availability of data and materials
The datasets used and/or analysed during the present study are available from the corresponding author upon reasonable request.
Author contributions
YS designed the Smad decoy. CF and YS designed the experimental protocol. SEA and TL provided PepC and engaged in discussions on the optimal use of PepC. CF, YS and KWK performed the experiments. CF and YS analysed the data. CF and YS wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The HSF (primary cells derived from 3 patients: 106, 107 and 108) was purchased from Cell Research Corporation (Singapore), with ethical approval obtained from A STAR Institute of Medical Biology (IRB: B-16-135E). Primary human keratinocytes (Kc) were collected from Asian Skin Bank, Institute of Medical Biology, A STAR, with ethical approval (IRB: B-16-135E).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ogawa R. Keloid and hypertrophic scars are the result of chronic inflammation in the reticular dermis. Int J Mol Sci. 2017;18:606. doi: 10.3390/ijms18030606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bombaro KM, Engrav LH, Carrougher GJ, Wiechman SA, Faucher L, Costa BA, Heimbach DM, Rivara FP, Honari S. What is the prevalence of hypertrophic scarring following burns? Burns. 2003;29:299–302. doi: 10.1016/S0305-4179(03)00067-6. [DOI] [PubMed] [Google Scholar]
- 3.Xiao Z, Zhang M, Liu Y, Ren L. Botulinum toxin type a inhibits connective tissue growth factor expression in fibroblasts derived from hypertrophic scar. Aesthetic Plast Surg. 2011;35:802–807. doi: 10.1007/s00266-011-9690-3. [DOI] [PubMed] [Google Scholar]
- 4.Son D, Harijan A. Overview of surgical scar prevention and management. J Korean Med Sci. 2014;29:751–757. doi: 10.3346/jkms.2014.29.6.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowin AJ, Holmes TM, Brosnan P, Ferguson MW. Expression of TGF-beta and its receptors in murine fetal and adult dermal wounds. Eur J Dermatol. 2001;11:424–431. [PubMed] [Google Scholar]
- 6.Tredget EE, Yang L, Delehanty M, Shankowsky H, Scott PG. Polarized Th2 cytokine production in patients with hypertrophic scar following thermal injury. J Interferon Cytokine Res. 2006;26:179–189. doi: 10.1089/jir.2006.26.179. [DOI] [PubMed] [Google Scholar]
- 7.Macias MJ, Martin-Malpartida P, Massague J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem Sci. 2015;40:296–308. doi: 10.1016/j.tibs.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YE. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 10.Meng XM, Chung AC, Lan HY. Role of the TGF-β/BMP-7/Smad pathways in renal diseases. Clin Sci (Lond) 2013;124:243–254. doi: 10.1042/CS20120252. [DOI] [PubMed] [Google Scholar]
- 11.Chin GS, Liu W, Peled Z, Lee TY, Steinbrech DS, Hsu M, Longaker MT. Differential expression of transforming growth factor-beta receptors I and II and activation of Smad 3 in keloid fibroblasts. Plast Reconstr Surg. 2001;108:423–429. doi: 10.1097/00006534-200108000-00022. [DOI] [PubMed] [Google Scholar]
- 12.Walraven M, Gouverneur M, Middelkoop E, Beelen RH, Ulrich MM. Altered TGF-β signaling in fetal fibroblasts: What is known about the underlying mechanisms? Wound Repair Regen. 2014;22:3–13. doi: 10.1111/wrr.12098. [DOI] [PubMed] [Google Scholar]
- 13.Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/S1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 14.Rheinwald JG, Green H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/S0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 15.Andaloussi SE, Lehto T, Mager I, Rosenthal-Aizman K, Oprea II, Simonson OE, Sork H, Ezzat K, Copolovici DM, Kurrikoff K, et al. Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo. Nucleic Acids Res. 2011;39:3972–3987. doi: 10.1093/nar/gkq1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ezzat K, Andaloussi SE, Zaghloul EM, Lehto T, Lindberg S, Moreno PM, Viola JR, Magdy T, Abdo R, Guterstam P, et al. PepFect 14, a novel cell-penetrating peptide for oligonucleotide delivery in solution and as solid formulation. Nucleic Acids Res. 2011;39:5284–5298. doi: 10.1093/nar/gkr072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan C, Dong Y, Xie Y, Su Y, Zhang X, Leavesley D, Upton Z. Shikonin reduces TGF-β1-induced collagen production and contraction in hypertrophic scar-derived human skin fibroblasts. Int J Mol Med. 2015;36:985–991. doi: 10.3892/ijmm.2015.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 21.Applied Biosystems. Guide to performing relative quantitation of gene expression using real-time quantitative PCR. 2004. https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_042380.pdf
- 22.Verrecchia F, Chu ML, Mauviel A. Identification of novel TGF-beta/Smad gene targets in dermal fibroblasts using a combined cDNA microarray/promoter transactivation approach. J Biol Chem. 2001;276:17058–17062. doi: 10.1074/jbc.M100754200. [DOI] [PubMed] [Google Scholar]
- 23.Fan C, Xie Y, Dong Y, Su Y, Upton Z. Investigating the potential of Shikonin as a novel hypertrophic scar treatment. J Biomed Sci. 2015;22:70. doi: 10.1186/s12929-015-0172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S, Liu J, Yang M, Lai W, Ye L, Chen J, Hou X, Ding H, Zhang W, Wu Y, et al. Fn14, a downstream target of the TGF-β signaling pathway, regulates fibroblast activation. PLoS One. 2015;10:e0143802. doi: 10.1371/journal.pone.0143802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins K, Davey RB, Wallis KA. Silicone gel: A new treatment for burn scars and contractures. Burns Incl Therm Inj. 1983;9:201–204. doi: 10.1016/0305-4179(83)90039-6. [DOI] [PubMed] [Google Scholar]
- 26.de Oliveira GV, Nunes TA, Magna LA. Silicone versus nonsilicone gel dressings: A controlled trial. Dermatol Surg. 2001;27:721–726. doi: 10.1097/00042728-200108000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Niessen FB, Spauwen PH, Schalkwijk J, Kon M. On the nature of hypertrophic scars and keloids: A review. Plast Reconstr Surg. 1999;104:1435–1458. doi: 10.1097/00006534-199910000-00031. [DOI] [PubMed] [Google Scholar]
- 28.Casi G, Neri D. Antibody-drug conjugates and small molecule-drug conjugates: Opportunities and challenges for the development of selective anticancer cytotoxic agents. J Med Chem. 2015;58:8751–8761. doi: 10.1021/acs.jmedchem.5b00457. [DOI] [PubMed] [Google Scholar]
- 29.An S, Fu L. Small-molecule PROTACs: An emerging and promising approach for the development of targeted therapy drugs. EBioMedicine. 2018;36:553–562. doi: 10.1016/j.ebiom.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Yang J, Ran B, Yang X, Zheng W, Long Y, Jiang X. Small Molecular TGF-β1-inhibitor-loaded electrospun fibrous scaffolds for preventing hypertrophic scars. ACS Appl Mater Interfaces. 2017;9:32545–32553. doi: 10.1021/acsami.7b09796. [DOI] [PubMed] [Google Scholar]
- 31.Cardarelli F, Digiacomo L, Marchini C, Amici A, Salomone F, Fiume G, Rossetta A, Gratton E, Pozzi D, Caracciolo G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci Rep. 2016;6:25879. doi: 10.1038/srep25879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Lullo GA, Sweeney SM, Korkko J, Ala-Kokko L, San Antonio JD. Mapping the ligand-binding sites and disease-associated mutations on the most abundant protein in the human, type I collagen. J Biol Chem. 2002;277:4223–4231. doi: 10.1074/jbc.M110709200. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y, Tao H, Jin C, Liu Y, Lu X, Hu X, Wang X. Transforming growth factor-β1 induces type II collagen and aggrecan expression via activation of extracellular signal-regulated kinase 1/2 and Smad2/3 signaling pathways. Mol Med Rep. 2015;12:5573–5579. doi: 10.3892/mmr.2015.4068. [DOI] [PubMed] [Google Scholar]
- 34.Santoro MM, Gaudino G. Cellular and molecular facets of keratinocyte reepithelization during wound healing. Exp Cell Res. 2005;304:274–286. doi: 10.1016/j.yexcr.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 35.Brown NJ, Kimble RM, Gramotnev G, Rodger S, Cuttle L. Predictors of re-epithelialization in pediatric burn. Burns. 2014;40:751–758. doi: 10.1016/j.burns.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 36.Das S, Mandal M, Chakraborti T, Mandal A, Chakraborti S. Structure and evolutionary aspects of matrix metalloproteinases: A brief overview. Mol Cell Biochem. 2003;253:31–40. doi: 10.1023/A:1026024829294. [DOI] [PubMed] [Google Scholar]
- 37.Eto H, Suga H, Aoi N, Kato H, Doi K, Kuno S, Tabata Y, Yoshimura K. Therapeutic potential of fibroblast growth factor-2 for hypertrophic scars: Upregulation of MMP-1 and HGF expression. Lab Invest. 2012;92:214–223. doi: 10.1038/labinvest.2011.127. [DOI] [PubMed] [Google Scholar]
- 38.Liu J, Shen JX, Wu HT, Li XL, Wen XF, Du CW, Zhang GJ. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med. 2018;25:211–223. [PubMed] [Google Scholar]
- 39.Ma L, Gan C, Huang Y, Wang Y, Luo G, Wu J. Comparative proteomic analysis of extracellular matrix proteins secreted by hypertrophic scar with normal skin fibroblasts. Burns Trauma. 2014;2:76–83. doi: 10.4103/2321-3868.130191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seifert O, Bayat A, Geffers R, Dienus K, Buer J, Löfgren S, Matussek A. Identification of unique gene expression patterns within different lesional sites of keloids. Wound Repair Regen. 2008;16:254–265. doi: 10.1111/j.1524-475X.2007.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Amadeu TP, Braune AS, Porto LC, Desmouliere A, Costa AM. Fibrillin-1 and elastin are differentially expressed in hypertrophic scars and keloids. Wound Repair Regen. 2004;12:169–174. doi: 10.1111/j.1067-1927.2004.012209.x. [DOI] [PubMed] [Google Scholar]
- 42.Liu R, Hossain MM, Chen X, Jin JP. Mechanoregulation of SM22α/Transgelin. Biochemistry. 2017;56:5526–5538. doi: 10.1021/acs.biochem.7b00794. [DOI] [PubMed] [Google Scholar]
- 43.Assinder SJ, Stanton JA, Prasad PD. Transgelin: An actin-binding protein and tumour suppressor. Int J Biochem Cell Biol. 2009;41:482–486. doi: 10.1016/j.biocel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 44.Ulrich D, Ulrich F, Unglaub F, Piatkowski A, Pallua N. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in patients with different types of scars and keloids. J Plast Reconstr Aesthet Surg. 2010;63:1015–1021. doi: 10.1016/j.bjps.2009.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Poormasjedi-Meibod MS, Hartwell R, Kilani R, Ghahary A. Anti-scarring properties of different tryptophan derivatives. PLoS One. 2014;9:e91955. doi: 10.1371/journal.pone.0091955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin-Malpartida P, Batet M, Kaczmarska Z, Freier R, Gomes T, Aragón E, Zou Y, Wang Q, Xi Q, Ruiz L, et al. Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors. Nat Commun. 2017;8:2070. doi: 10.1038/s41467-017-02054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walton KL, Johnson KE, Harrison CA. Targeting TGF-β Mediated SMAD signaling for the prevention of fibrosis. Front Pharmacol. 2017;8:461. doi: 10.3389/fphar.2017.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu A, Kligman LH, Samulewicz SJ, Howe CC. Impaired wound healing in mice deficient in a matricellular protein SPARC (osteonectin, BM-40) BMC Cell Biol. 2001;2:15. doi: 10.1186/1471-2121-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chavez-Muñoz C, Hartwell R, Jalili RB, Jafarnejad SM, Lai A, Nabai L, Ghaffari A, Hojabrpour P, Kanaan N, Duronio V, et al. SPARC/SFN interaction, suppresses type I collagen in dermal fibroblasts. J Cell Biochem. 2012;113:2622–2632. doi: 10.1002/jcb.24137. [DOI] [PubMed] [Google Scholar]
- 50.Dougherty KM, Pearson JM, Yang AY, Westrick RJ, Baker MS, Ginsburg D. The plasminogen activator inhibitor-2 gene is not required for normal murine development or survival. Proc Natl Acad Sci USA. 1999;96:686–691. doi: 10.1073/pnas.96.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rohani MG, Parks WC. Matrix remodeling by MMPs during wound repair. Matrix Biol. 2015;44-46:113–121. doi: 10.1016/j.matbio.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Li Y, Li N, Teng W, Wang M, Zhang Y, Xiao Z. TGF-β1 promotes scar fibroblasts proliferation and transdifferentiation via up-regulating MicroRNA-21. Sci Rep. 2016;6:32231. doi: 10.1038/srep32231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai X, He T, Liu J, Wang Y, Fan L, Tao K, Shi J, Tang C, Su L, Hu D. Loureirin B inhibits fibroblast proliferation and extracellular matrix deposition in hypertrophic scar via TGF-β/Smad pathway. Exp Dermatol. 2015;24:355–360. doi: 10.1111/exd.12665. [DOI] [PubMed] [Google Scholar]
- 54.Kim ES, Kim MS, Moon A. TGF-beta-induced upregulation of MMP-2 and MMP-9 depends on p38 MAPK, but not ERK signaling in MCF10A human breast epithelial cells. Int J Oncol. 2004;25:1375–1382. [PubMed] [Google Scholar]
- 55.Gill SE, Parks WC. Metalloproteinases and their inhibitors: Regulators of wound healing. Int J Biochem Cell Biol. 2008;40:1334–1347. doi: 10.1016/j.biocel.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu SS, Dotor J, Hontanilla B. Effect of P144® (Anti-TGF-β) in an ‘in vivo’ human hypertrophic scar model in nude mice. PLoS One. 2015;10:e0144489. doi: 10.1371/journal.pone.0144489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tredget EE, Wang R, Shen Q, Scott PG, Ghahary A. Transforming growth factor-beta mRNA and protein in hypertrophic scar tissues and fibroblasts: Antagonism by IFN-alpha and IFN-gamma in vitro and in vivo. J Interferon Cytokine Res. 2000;20:143–151. doi: 10.1089/107999000312540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the present study are available from the corresponding author upon reasonable request.