Abstract
Renal cell carcinoma (RCC) is a common type of malignancy in the kidney, which accounts for ~80% of the cases within adult patients. The pathogenesis of RCC is complicated and involves alterations at both genetic and epigenetic levels. The aim of the present study was to investigate the roles of circRNAs in the pathogenesis of RCC. In the current study, exosomes were isolated via gradient centrifugation and identified using transmission electron microscope. The expression levels of circular RNA (circ)_400068, microRNA (miR)-210-5p and suppressor of cytokine signaling 1 (SOCS1) were examined using reverse transcription-quantitative PCR. Cell proliferation was evaluated using a Cell Counting Kit-8 assay, and the apoptotic rate was determined in transfected cells using flow cytometry. The protein expression levels of proliferation- and apoptosis-associated genes were assessed via western blot analysis. Upregulation of circ_400068 was detected in RCC plasma exosomes, tissue samples and cells. Additionally, treatment with exosomal circ_400068 promoted the proliferation and inhibited the apoptosis of healthy kidney cells, which were abrogated by short hairpin RNA-circ_400068. The results suggested that miR-210-5p was a potential downstream molecule of circ_400068, and SOCS1 was a novel target of miR-210-5p. Moreover, circ_400068 regulated the proliferation of HK-2 cells by targeting the miR-210-5p/SOCS1 axis, as the effects on cell proliferation caused by treatment using exosomes isolated from the culture media of RCC cells were abolished by miR-210-5p mimics. It was found that enhanced cell proliferation induced by miR-210-5p inhibitors was attenuated by the knockdown of SOCS1, while the influences triggered by miR-210-5p mimics were reversed by SOCS1 overexpression. Collectively, the present findings provided a novel insight into the crucial regulatory functions of circ_400068 in RCC, and the circ_400068/miR-210-5p/SOCS1 axis could be a candidate therapeutic target for the treatment of patients with RCC.
Keywords: renal cell carcinoma, circular RNA_400068, microRNA-210-5p, suppressor of cytokine signaling 1, proliferation, migration, invasion
Introduction
Renal cell carcinoma (RCC) is one of the most prevalent tumor types in the adult kidney, and the recurrence rate is high (1). In 2015, a ~66.8% increase in new RCC cases was reported, with a ~23.4% increase in mortality within patients with RCC in China (2). While there have been increased efforts for the development of diagnostic methods, including the utilization of ultrasound and CT technologies, it remains difficult to distinguish benign tumors from the malignancy (3). Thus, it is essential to identify novel non-invasive biomarkers and therapeutic candidates for RCC. The complex pathogenesis of RCC involves alterations at both the genetic and epigenetic levels, which can lead to tumorigenesis (4–8); however, the detailed mechanisms during the initiation and development of this disease are yet to be fully elucidated. Numerous pathways including PI3K/Akt/mTOR and Wnt/β-catenin could be involved in the progression of RCC (4,5,9).
Circular RNAs (circRNAs) are a novel group of non-coding RNAs that form a continuous loop and are more stable compared with their linear counterparts (10,11). Although certain circRNAs, such as CirS-7, have been reported as putative gene regulators (11), the detailed functions of most circRNAs are largely unknown. Due to the absence of free 5′- or 3′-ends, circRNAs are resistant to exonuclease-induced degradation (11). Previous studies have revealed that circRNAs could function as microRNA (miRNA/miR) ‘sponges’ that competitively inhibit the activity of miRNAs (11,12). In addition, circRNAs are involved in the pathogenesis of numerous types of diseases, such as nervous system disorders and cancer (13–15).
In the tumor microenvironment, crosstalk between malignant cells and adjacent healthy cells is crucial during the development of a tumor (16). Exosomes are a novel type of microparticles with a diameter of 40–100 nm, and these are produced by most cell types (17). Cell-secreted exosomes are detected in numerous types of eukaryotic fluids, such as blood, urine and cell culture media (18). Exosomes are crucial during cell-cell signaling. For instance, RNAs are able to shuttle from one cell to another, known as ‘exosomal shuttle RNAs’, and can affect protein production in healthy cells (19). Previous studies have also revealed that circRNAs are abundantly detected in exosomes, and function as miRNA sponges during gene regulation (20–22). circRNAs can be transferred into exosomes in colon cancer cells (23). In addition, mRNAs and miRNAs can be transferred by exosomes to facilitate genetic exchange between cells (24). However, the detailed functions of exosomal circRNAs in cancer remain largely unknown.
In the present study, the expression patterns of exosomal circRNAs in patients with RCC were examined, and the regulatory functions of circRNAs during the progression of RCC were elucidated. The effects of circ_400068-regulated miR-210-5p/suppressor of cytokine signaling 1 (SOCS1) signaling on the development of RCC were elucidated, which could provide novel insights into the therapeutic strategies of the treatment against this disease.
Materials and methods
Clinical samples
Human kidney tissue and plasma (10 ml) specimens were obtained from patients with RCC (n=28, aged 38–72 years old; 16 males and 12 females) at the Department of Urology of The First Affiliated Hospital of Jinzhou Medical University between May 2014 and April 2017. Para-carcinoma tissues were ≥5-cm from tumor margin. The expression levels of hsa_circ_400068 were categorized into low and high groups using the mean value. The biopsies were examined by two independent pathologists, and the clinicopathological features of enrolled patients were summarized in Table I. The study was approved by the Ethics Committee of The First Affiliated Hospital of Jinzhou Medical University, Written informed consents were signed by all patients.
Table I.
Clinicopathological parameters of patients with renal cell carcinoma recruited in the current study.
| hsa_circ_400068 expression | ||||
|---|---|---|---|---|
| Parameters | Total number of cases | Low | High | P-value |
| Sex | 0.392 | |||
| Male | 16 | 7 | 9 | |
| Female | 12 | 7 | 5 | |
| Age, years | 0.411 | |||
| >60 | 18 | 8 | 10 | |
| ≤60 | 10 | 6 | 4 | |
| Tumor size, cm | 0.409 | |||
| >5 | 14 | 6 | 8 | |
| ≤5 | 14 | 8 | 6 | |
| Histology gradea | 0.456 | |||
| I–II | 17 | 9 | 8 | |
| III–IV | 11 | 5 | 6 | |
Differences among variables were analyzed using Fisher's exact test.
The Fuhrman grading system was used. circRNA, circular RNA.
Extraction of exosomes in human plasma and RCC cells culture media
RCC cells were cultured using Dulbecco's modified Eagle's medium (DMEM) supplemented with streptomycin (100 µg/ml), penicillin (100 U/ml) and 10% (FBS; HyClone; Cytiva). Exosomes were isolated using gradient centrifugation as previously described (22). Circulating blood samples (~10 ml) were centrifugated at 2,000 × g at 4°C for 10 min and the plasma was obtained. After centrifugation at 3,000 × g for 30 min, debris and floating cells were aspirated. Subsequently, the supernatant was collected and centrifugated at 10,000 × g for 10 min to remove microvesicles, whose sizes are larger compared with exosomes. The supernatant was spun at 15,000 × g for 60 min. All centrifugation steps were performed at 4°C. Isolated exosomes pellets were rinsed using PBS and stored at −80°C until further use.
Confirmation of exosome using transmission electron microscope
Isolated exosomes pellets were resuspended in PBS and fixed using 2.5% of glutaraldehyde (pH 7.2) at 4°C overnight. A drop of 100 µl the suspension was placed on a parafilm sheet, and a copper grid coated by carbon was placed onto the drop for 10 sec and then removed. Uranyl acetate and phosphotungstic acid (0.5 µg, 2%) were added onto the grid at room temperature for 5 sec. Following the removal of excess liquid using filter paper, the grid was dried for 10 min at room temperature and the observed using a transmission electron microscope (H-7600; Hitachi, Ltd.). In total, 10 fields of view were randomly selected for each sample (magnification, ×200,000).
circRNA microarray
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and a RNeasy Mini kit (Qiagen GmbH). Samples were then grouped and Cy3-dUTP-labelled targets were generated using an Arraystar Super RNA Labeling Kit (Arraystar Inc.) for circRNA array. Human circRNA array v2 (CapitalBio Technology Co., Ltd.) was designed, ~5,000 genes were mounted onto the chip, the target sequences of these circRNAs were obtained from circBase (June, 2015 http://www.circbase.org/). Briefly, labeled targets were hybridized with the samples, which were scanned using Agilent Microarray scanner (Agilent Technologies, Inc.). Data were normalized according to the Quantile algorithm (https://www.fon.hum.uva.nl/praat/manual/quantile_algorithm.html). Arrays were performed using the protocol provided by Agilent Technologies, Inc. Genes whose fold change was >2 (P<0.05) were further analyzed. In total, the five most up- or downregulated circRNAs in plasma exosomes of patients with RCC are listed in Table II. circ_400068 was the most upregulated gene in RCC samples and was selected for further study.
Table II.
Top five up- and downregulated circRNAs in the array.
| A, Upregulated | |||||
|---|---|---|---|---|---|
| circRNA ID | Fold change | P-value | circRNA type | Genomic location | Gene symbol |
| hsa_circ_0092314 | 4.86 | 0.00772 | Exonic | chr22 | RANBP11 |
| hsa_circ_0000735 | 3.66 | 0.00664 | Exonic | chr17 | P2RX1 |
| hsa_circ_0023642 | 2.83 | 0.01453 | Exonic | chr11 | UVRAG |
| hsa_circ_0068610 | 2.33 | 0.03245 | Exonic | chr3 | TFRC |
| hsa_circ_0032821 | 2.15 | 0.04664 | Exonic | chr14 | CEP128 |
| B, Downregulated | |||||
| circRNA ID | Fold change | P-value | circRNA type | Genomic location | Gene symbol |
| hsa_circ_0005730 | 3.61 | 0.04235 | Exonic | chr5 | CDK7 |
| hsa_circ_0003645 | 3.22 | 0.02664 | Exonic | chr16 | C16orf62 |
| hsa_circ_0026134 | 2.76 | 0.01664 | Exonic | chr12 | TUBA1C |
| hsa_circ_0061274 | 2.52 | 0.01863 | Exonic | chr21 | NRIP1 |
| hsa_circ_0092368 | 2.19 | 0.00791 | Exonic | chr1 | HMGN2 |
circRNA, circular RNA; chr, chromosome.
Cell culture
The human clear cell RCC cell line Caki-1 and the papillary RCC cell line Caki-2, as well as a cell line of normal human kidney cells (HK-2) were obtained from the Shanghai Institute of Cell Biology, Chinese Academy of Science. Cells were maintained using DMEM supplemented with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin (all purchased from Gibco; Thermo Fisher Scientific, Inc.). Cells were incubated in a humidified incubator supplied with 5% CO2 at 37°C.
Cell transfection
To generate circ_400068 and SOCS1 knockdown HK-2 cells, SureSilencing short hairpin (sh)RNA plasmids (Qiagen GmbH) targeting circ_400068 (sh-circ_400068) and the scrambled negative control (sh-NC; sense, 5′-CACCGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACACGTTCGGAGAATTTTTTG-3′ and antisense, 5′-AGCTCAAAAAATTCTCCGAACGTGTCACGTAATCTCTTGACGTGACACGTTCGGAGAAC-3′), as well as small interfering (si)RNA against SOCS1 (si-SOCS1; 5′-CCUGCACGGAGCAUUAACUTT-3′) and the NC (si-NC; 5′-ACGUGACACGUUCGGAGAATT-3′) were obtained from Shanghai GenePharma Co., Ltd. Subsequent experiments were performed 48 h post-transfection.
For miR-210-5p interference, miR-210-5p mimics (5′-CUGUGCGUGUGACAGCGGCUGA-3′), miR-210-5p inhibitors (5′-UCAGCCGCUGUCACACGCACAG-3′) and miR-NC (sense, 5′-UUCUUUUCGAACGUGUCACGUTT-3′ and antisense, ACGUGACACGUUCGGAGAATT-3′) with the non-targeting sequence were purchased from Shanghai GenePharma Co., Ltd and transfected into HK-2 cells (ATCC).
To produce HK-2 cells overexpressing circ_400068 (LV-circ_400068) or SOCS1(LV-SOCS1), wild-type (WT) and scrambled (LV-NC) sequences were integrated into PLCDH-cir vectors (Guangzhou RiboBio Co., Ltd.).
In total, 25 nM plasmids were used for transfection together with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Fresh DMEM containing 10% FBS was added into cells 12 h after transfection. Transfected cells were further selected using puromycin (0.5 µg/ml; Sigma-Aldrich; Merck KGaA) for an additional 2 weeks. Non-transfected cells or cells transfected with sh-circ_400068 and miR-210-5p mimics were further treated using exosomes (108 particles/ml) isolated from the culture media of RCC cells (RCC-Exo) 37°C for 24 h.
Reverse transcription-quantitative PCR (RT-qPCR)
miRNA was extracted using a miRNeasy Mini kit (Qiagen China Co., Ltd.). A TaqMan miRNA assay (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to examine the expression of miR-210-5p, and the reaction was completed on Applied Biosystem 7500 system (Thermo Fisher Scientific, Inc.). For the detection of mRNAs and circRNAs, total RNA was isolated using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocols. The concentration of extracted RNA was evaluated using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.). Then, cDNA was produced using PrimeScript™ RT kit (Takara Biotechnology Co., Ltd.), and PCR program used was: 42°C for 45 min, 99°C for 5 min and 5°C for 5 min in a PCR cycler. qPCR was conducted using SYBR Green PCR Master mix (Takara Biotechnology Co., Ltd.). U6 snRNA was used as internal control for miRNAs. Endogenous GAPDH was used as a reference for circRNA and mRNA. The forward and reverse primer pairs used were as follows: circ_400068, forward: 5′-TGATCTCACCCTAAGTTCGC-3′ and reverse: 5′-CATACCAATCCTCAATCCTC-3′; miR-210-5p, forward: 5′-CCTGCAATATTTGCATGTCG-3′ and reverse: 5′-GTCCCTATTGGCGTTACTATGG-3′; SOCS1, forward: 5′-GCAUCCGCGUGCACUUUCAUU-3′ and reverse: 5′-AAUGAAAGUGCACGCGGAUGC-3′; GAPDH, forward: 5′-ATGTCGTGGAGTCTACTGGC-3′ and reverse: 5′-TGACCTTGCCCACAGCCTTG-3′; and U6, forward: 5′-CTCGCTTCGGCAGCACA-3′ and reverse: 5′-AACGCTTCACGAATTTGCGT-3′. The thermocycling program included: Initial denaturation at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 20 sec and 72°C for 10 sec and final extension at 72°C for 5 min. Relative gene expression was analyzed using the 2−∆∆Cq method (25).
Cell proliferation assay
At 24 h post-transfection, cells were harvested and seeded at 5×103 per well onto a 96-well plate. Cell proliferation was determined at day 1, 2, 3 and 4. According to the manufacturer's protocol, 10 µl Cell Counting Kit (CCK)-8 solution (Beyotime Institute of Biotechnology) was used for each coloring reaction. After a further incubation at 37°C for 2 h, the absorbance at 450 nm was quantified using a plate reader (Bio-Rad Laboratories, Inc.). Each experiment was performed in triplicate independently.
Cell apoptosis analysis
Cell apoptotic rate was calculated as the percentage of early apoptotic cells, the percentage of late apoptotic cells, or the percentage of early + late apoptotic cells. Transfected cells were placed onto a 6-well plate at 4×104 per well and then centrifugated at 5,000 × g at room temperature for 5 min. Cell pellets were washed and re-suspended using PBS. Cells were stained with PI at room temperature for 5 min. To evaluate cell apoptotic activity, cell suspension were incubated at 4°C in dark for 30 min and staining was performed using 5 µl Annexin V-FITC (Sigma-Aldrich; Merck KGaA). Cell apoptotic rate was determined using a flow cytometer FACSAria III (BD Biosciences) and the data were interpreted using FlowJo software (version 7.6; FlowJo LLC).
Bioinformatics and dual-luciferase reporter assay
TargetScan (version 6.2; www.targetscan.org/) and miRanda (version 5.0; www.microrna.org/microrna/) were used to predict the putative downstream targets of circ_400068 and miR-210-5p. Wild-type (WT) fragment of the 3′untranslated region (UTR) on circ_400068 or SOCS1 with potential complementary binding sites of miR-210-5p were obtained from Shanghai GenePharma Co., Ltd. The sequences were integrated onto pmirGLO Dual-Luciferase miRNA Target Expression vector (Promega Corporation) according to the manufacturer's protocols. A circ_400068 or SOCS1 3′UTR-mutant (MUT) vector containing the mutant binding site of miR-210-5p was produced using a QuikChange Multi Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc.). Subsequently, the plasmids were used to co-transfect 293 cells (ATCC) with miR-210-5p mimics or mock control (2 µg/µl) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Luciferase activity was examined 48 h after transfection using a Dual Luciferase Reporter Assay system (Promega Corporation), and firefly luciferase activity was normalized using Renilla luciferase.
Western blotting
Total protein was extracted from tissues and cells using RIPA buffer (Beyotime Institute of Biotechnology). The concentration of isolated protein was evaluated using BCA assay (Beyotime Institute of Biotechnology). Equal amount (30 µg) of protein samples were loaded onto 10% SDS-PAGE and were subsequently transferred to PVDF membrane (EMD Millipore). Then, the membranes were blocked in TBS containing 5% non-fat milk at room temperature for 1 h, followed by incubation using primary antibodies: Ki-67 (1:5,000; cat. no. ab16667; Abcam), caspase-7 (1:500; cat. no. ab32522; Abcam), cleaved caspase-7 (1:500; cat. no. ab256474; Abcam), SOCS1 (1:100; cat. no. ab62584; Abcam), E-cadherin (E-cad; 1:5,000; cat. no. ab15148; Abcam), p53 (1:1,000; cat. no. ab131442; Abcam), GAPDH (1:10,000; cat. no. ab8245; Abcam) and STAT1 (1:5,000; cat. no. ab47425; Abcam) at 4°C overnight. The following day, membranes were rinsed and incubated in horseradish peroxidase-conjugated anti-rabbit (1:10,000; cat. no. sc-2357; Santa Cruz Biotechnology Inc.) or anti-mouse IgG (1:10,000; cat. no. sc-2371; Santa Cruz Biotechnology Inc.) at room temperature for 1 h. Protein bands were visualized using an ECL detection kit (Pierce Biotechnology; Thermo Fisher Scientific, Inc.), and blots were analyzed using ImageJ (version 1.3; National Institutes of Health). GAPDH was used as internal standard.
Statistical analysis
Data are presented as the mean ± standard deviation, and were analyzed using JMP 9.0 software (SAS Institute, Inc.). All the experiments were performed three times. The significance of differences were interpreted using paired or unpaired Student's t-test or one-way ANOVA followed by a post hoc Tukey's test. The association between circ_400068 expression in RCC tissues and exosomes was evaluated using Pearson's correlation test. P<0.05 was considered to indicate a statistically significant difference.
Results
circ_400068 is upregulated in RCC samples and cells
According to the data from circRNA microarray (Table II), circ_400068 was upregulated in plasma exosomes of patients with RCC and selected for further functional study. The results indicated that the expression levels of plasma exosomal and tissue circ_400068 in patients with RCC were moderately positively correlated (Fig. 1A). Therefore, the expression of circ_400068 in RCC tissues was evaluated. Upregulation of circ_400068 was observed in RCC tissues compared with adjacent healthy controls (Fig. 1B). Additionally, the expression of circ_400068 was elevated in RCC cells lines in comparison with healthy kidney cells (Fig. 1C). However, no significant association was observed between circ_400068 in RCC tissues and clinicopathological characteristics of the patients (Table II). These findings suggested that the expression of circ_400068 was upregulated in RCC, which could be associated with the development of this disease.
Figure 1.
circ_400068 is upregulated in RCC samples and cells. (A) Expression levels of circ_400068 in RCC plasma exosome and tissues were positively correlated. (B) circ_400068 expression was determined in tissue samples from patients with RCC. (C) Expression of circ_400068 was significantly upregulated in Caki-1 and Caki-2 cells compared with HK-2 cells. *P<0.05 vs. HK-2 cells. RCC, renal cell carcinoma; circ, circular RNA.
Treatment with exosomal circ_400068 promotes the proliferation and inhibits the apoptosis of kidney cells, which were abrogated by sh-circ_400068
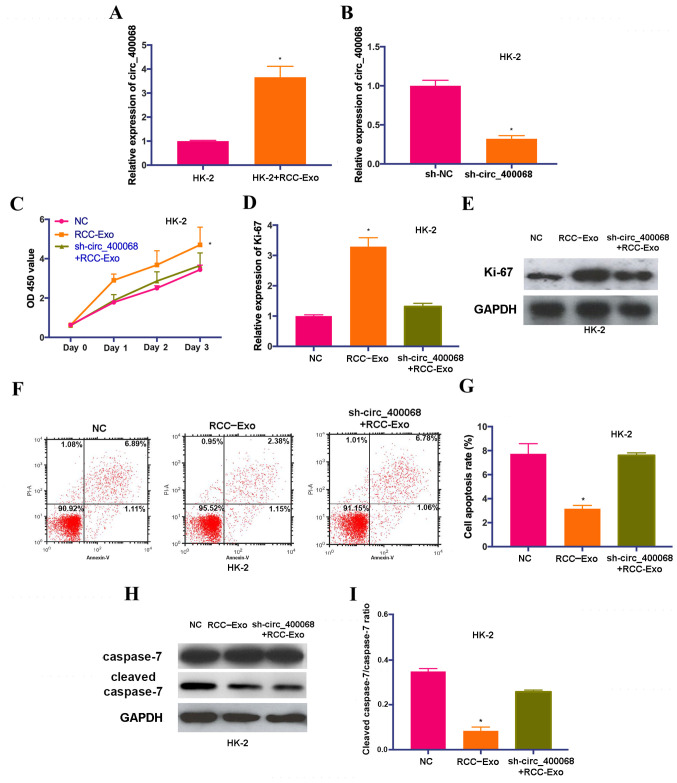
In order to elucidate the roles of circ_400068 on cellular functions, proliferation and apoptosis were examined in HK-2 cells treated with exosomal circ_400068 extracted from RCC-Exo. The results demonstrated that the expression of circ_400068 in HK-2 cells was increased following this treatment (Fig. 2A). In order to perform further functional experiments, the cells were also treated with sh-circ_400068, and the transfection efficiencies were confirmed using RT-qPCR (Fig. 2B).
Figure 2.
Exosomal circ_400068 enhances the proliferation and suppresses the apoptosis of kidney cells, and the effects are reversed by sh-circ_400068. (A) Expression of circ_400068 was elevated in HK-2 cells treated with RCC-Exo. (B) Transfection efficiency of sh-circ_400068 in HK-2 cells was assessed using reverse transcription quantitative PCR compared with the sh-NC. (C) Proliferative activity in HK-2 cells were enhanced following the treatment with RCC-Exo, which was abrogated by sh-circ_400068. (D) The RNA levels of Ki-67 were increased in cells treated with RCC-exo and reduced by sh-circ_400068. (E) The protein levels of Ki-67 were enhanced in cells treated with RCC-exo, which was reversed by sh-circ_400068. (F) Flow cytometry results demonstrated that the (G) cell apoptotic rate was significantly reduced in RCC-Exo-treated HK-2 cells, and the effects were rescued by the treatment with sh-circ_400068. (H) Western blotting results indicated that the (I) expression of cleaved caspase-7 was decreased in HK-2 cells treated with RCC-Exo, which was abrogated by sh-circ_400068. *P<0.05 vs. NC. NC, negative control; sh-, short hairpin RNA; RCC-Exo, exosomes isolated from the culture media of RCC cells; circ, circular RNA; RCC, RCC, renal cell carcinoma; OD, optical density.
HK-2 cells were also co-treated with sh-circ_400068 and RCC-Exo. The results of CCK-8 assay suggested that the proliferation of HK-2 cells was significantly promoted after the treatment with RCC-Exo, and the effects were abolished by sh-circ_400068 (Fig. 2C). In line with these findings, the expression of Ki-67 was upregulated in HK-2 cells treated with RCC-Exo, which was markedly reversed by the treatment with sh-circ_400068 (Fig. 2D and E). Moreover, flow cytometry results indicated that the apoptotic rate of RCC-Exo-treated cells was significantly decreased compared with non-treated cells (Fig. 2F and G). The expression of cleaved caspase-7 was significantly downregulated in HK-2 cells treated with RCC-Exo compared with the NC, and the effects were reversed by sh-circ_400068 (Fig. 2H and I). These data indicated that exosomal circ_400068 produced by RCC cells was able to enhance cell proliferation and suppress the apoptosis of healthy kidney cells, which could contribute to the progression of RCC.
miR-210-5p is a putative downstream target of circ_400068 in RCC
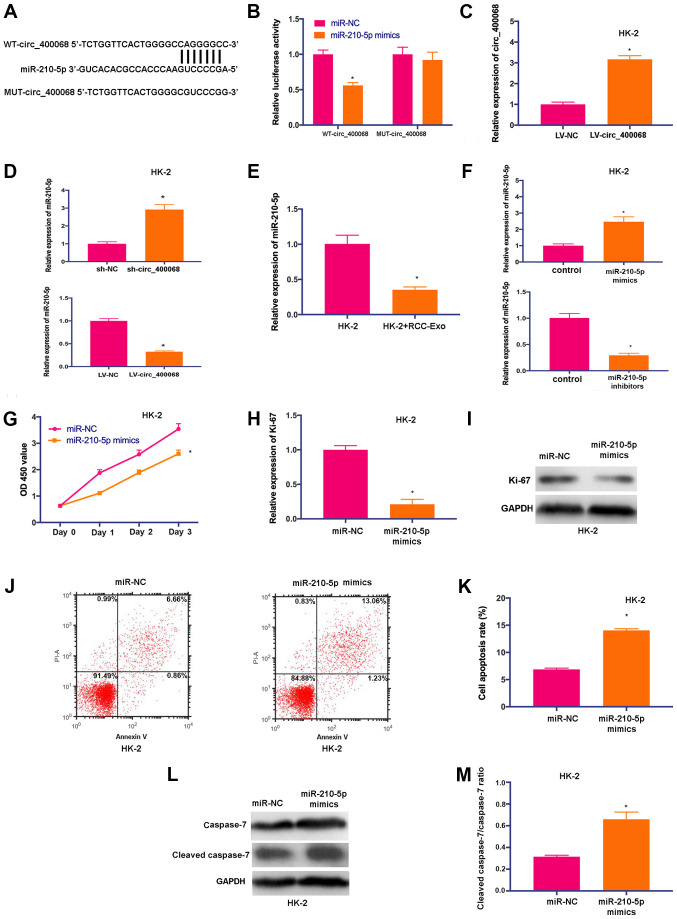
In order to investigate whether circ_400068 exerts its regulatory functions by targeting corresponding miRNAs in RCC, the complementary binding sites of miR-210-5p on circ_400068 transcripts were predicted using miRanda (Fig. 3A). Luciferase reporters carrying WT (WT-circ_400068) and MUT (MUT-circ_400068) fragments of the predicted miR-210-5p binding sites were produced. The data indicated that miR-210-5p mimics significantly abolished the luciferase activity of vectors containing WT binding sequences, but not the MUT control (Fig. 3B). In order to perform further functional experiments, the cells were also treated with LV-circ_400068, and the transfection efficiencies were confirmed using RT-qPCR (Fig. 3C).
Figure 3.
miR-210-5p is a novel target of circ_400068 in RCC. (A) Potential binding sites of miR-210-5p on transcript of circ_400068 were predicted. (B) miR-210-5p mimics significantly decreased the luciferase activity of WT-circ_400068 compared with miR-NC, but not MUT-circ_400068. (C) Transfection efficiency of LV-circ_400068 in HK-2 cells. (D) miR-210-5p expression was determined in HK-2 cells treated with sh-circ_400068 or LV-circ_400068, as well as (E) RCC-Exo using RT-qPCR. (F) Transfection efficiencies of miR-210-5p mimics or inhibitors in HK-2 cells were measured via RT-qPCR compared with miR-NC. (G) Proliferation of HK-2 cells was suppressed following the treatment with miR-210-5p mimics. (H) Ki-67 expression was decreased by miR-210-5p mimic in HK-2 cells compared with miR-NC, as determined by (I) western blotting. (J) Flow cytometry analysis demonstrated that (K) the apoptotic activity was promoted in HK-2 cells treated with miR-210-5p mimics compared with miR-NC. (L) Western blotting results indicated that (M) cleaved caspase-7 expression was significantly elevated by the transfection with miR-210-5p mimics in HK-2 cells compared with miR-NC. *P<0.05 vs. respective control. miR, microRNA; NC, negative control; MUT, mutant; WT, wild-type; RCC, renal cell carcinoma; RT-qPCR, reverse transcription quantitative PCR; sh-, short hairpin RNA; RCC-Exo, exosomes isolated from the culture media of RCC cells; circ, circular RNA.
To further study the influences of circ_400068 on miR-210-5p expression, the expression of miR-210-5p was detected in HK-2 cells transfected with sh-circ_400068 or LV-circ_400068. The expression of miR-210-5p was upregulated in cells following the transfection with sh-circ_400068, but downregulated after LV-circ_400068 transfection (Fig. 3D). Furthermore, the expression of miR-210-5p was significantly decreased in RCC-Exo-treated cells (Fig. 3E).
In order to perform further functional studies, HK-2 cells were transfected with miR-210-5p mimics or inhibitors, and the transfection efficiencies were confirmed via RT-qPCR (Fig. 3F). The proliferative activity of HK-2 cells was significantly inhibited after the treatment with miR-210-5p mimics (Fig. 3G). In addition, the expression of Ki-67 was downregulated in HK-2 cells transfected with miR-210-5p mimics (Fig. 3H and I). It was identified that cell apoptosis was enhanced following the treatment with miR-210-5p mimics (Fig. 3J and K). Similarly, the expression of cleaved caspase-7 was significantly increased in HK-2 cells treated with miR-210-5p mimics (Fig. 3L and M). Thus, the results suggested that miR-210-5p could be a promising target of circ_400068 in RCC.
SOCS1 is a novel target of miR-210-5p
Using the TargetScan database, complementary binding sequences between SOCS1 and miR-210-5p were predicted (Fig. 4A). In order to elucidate whether SOCS1 was a putative target of miR-210-5p, WT and MUT sequences of SOCS1 were inserted after a firefly luciferase coding domain. It was found that the overexpression of miR-210-5p significantly decreased the luciferase activity of the SOCS1-WT reporter but not SOCS1-MUT control (Fig. 4B).
Figure 4.
SOCS1 is a putative target of miR-210-5p. (A) Novel binding sites of SOCS1 on the miR-210-5p transcript. (B) Transfection with miR-210-5p mimics resulted in significant decrease in the luciferase activity of WT-SOCS1, while no decrease was detected in MUT-SOCS1 group. (C) SOCS1 expression was evaluated in HK-2 cells transfected with miR-210-5p mimics and inhibitors using reverse transcription-quantitative PCR compared with miR-NC. (D) Protein expression levels of SOCS1 and its downstream molecules in transfected HK-2 cells were examined using western blotting compared with miR-NC. *P<0.05 vs. miR-NC. RCC, renal cell carcinoma; miR, microRNA; NC, negative control; MUT, mutant; WT, wild-type; SOCS1, suppressor of cytokine signaling 1.
Additional experiments were performed to investigate whether miR-210-5p affects the expression levels of SOCS1 and its downstream molecules. RT-qPCR results demonstrated that the expression of SOCS1 was downregulated by miR-210-5p mimics, but upregulated by miR-210-5p inhibitors compared with miR-NC (Fig. 4C). IT was found that the protein expression levels of SOCS1, E-cad and p53 were notably decreased in HK-2 cells treated with miR-210-5p mimics, while their expression levels were elevated by treatment with miR-210-5p inhibitors. However, the expression of STAT1 was significantly upregulated by miR-210-5p mimics and downregulated by miR-210-5p inhibitors (Fig. 4D). These findings indicated that SOCS1 signaling could be downstream target of miR-210-5p.
circ_400068 regulates the proliferation of HK-2 cells by targeting the miR-210-5p/SOCS1 axis
To investigate the involvement of miR-210-5p and SOCS1 in exosomal circ_400068-modulated biological feature changes in HK-2 cells, the cells were co-treated with miR-210-5p mimics and RCC-Exo, miR-210-5p mimics and LV-SOCS1 or miR-210-5p inhibitors and si-SOCS1. The transfection efficiencies of si-SOCS1 and LV-SOCS1 were confirmed via RT-qPCR (Fig. 5A and B). The data suggested that the effects on cell proliferation and apoptosis caused by RCC-Exo were significantly abolished by miR-210-5p mimics (Fig. 5C-E). Additionally, promoted cell proliferation caused by miR-210-5p inhibitors was significantly attenuated by the knockdown of SOCS1, whereas the influences on cell proliferation and apoptosis triggered by miR-210-5p mimics were significantly reversed by SOCS1 overexpression (Fig. 5F-K). Collectively, these findings suggested that circ_400068 may regulate the proliferation of HK-2 cells via the miR-210-5p/SOCS1 signaling pathway. It was also indicated that exosomal circ_400068 originated from the tumor has potential as a putative non-invasive biomarker for the prognosis of RCC, and it may affect the proliferation of healthy kidney cells by targeting the miR-210/SOCS1 axis.
Figure 5.
circ_400068 modulates the proliferation of kidney cells via miR-210-5p/SOCS1 signaling. Transfection efficiencies of (A) si-SOCS1 and (B) LV-SOCS1 were evaluated using reverse transcription-quantitative PCR. (C) Effects on HK-2 cell proliferation induced by the treatment with RCC-Exo were significantly abolished by miR-210-5p mimics. (D) Influences on HK-2 cell apoptosis caused by RCC-Exo were significantly reversed by miR-210-5p mimics. (E) Effects on HK-2 cell apoptotic rate induced by the treatment with RCC-Exo were significantly abolished by miR-210-5p mimics. (F) Enhanced HK-2 cell proliferation caused by miR-210-5p inhibitors were markedly attenuated by the knockdown of SOCS1. (G) Reduced HK-2 cell apoptosis caused by miR-210-5p inhibitors were markedly abolished by the knockdown of SOCS1. (H) Decreased HK-2 cell apoptotic rate induced by miR-210-5p inhibitors were markedly abolished by the knockdown of SOCS1. (I) Influences on HK-2 cell proliferation induced by miR-210-5p mimics were notably reversed by overexpression of SOCS1. (J) Effects on HK-2 cell apoptosis caused by miR-210-5p mimics were notably abrogated by overexpressed SOCS1. (K) Influences on HK-2 cell apoptotic rate induced by miR-210-5p mimics were notably abolished by overexpression of SOCS1. *P<0.05 vs. negative control group. RCC, renal cell carcinoma; miR, microRNA; NC, negative control; SOCS1, suppressor of cytokine signaling 1; sh-, short hairpin RNA; RCC-Exo, exosomes isolated from the culture media of RCC cells; circ, circular RNA; OD, optical density.
Discussion
RCC is the most prevalent kidney cancer type in adults (1). Although the diagnostic and therapeutic methods have been improved for RCC, it remains difficult to distinguish benign and malignant tumors (2). The pathogenesis of RCC is complex, and the detailed mechanisms are largely unknown.
circRNAs are potential gene regulators, which can act as miRNA ‘sponges’ (12). Impaired circRNAs expression levels are associated with the onset and development of nervous system disorders and cancer (13–15). During the progression of a tumor, crosstalk between cancer cells and adjacent healthy cells is crucial, and exosomes produced by malignant cells serve essential roles during this process (16,19). Previous studies have reported that exosomal circRNAs are novel regulators of gene expression (20–22). For instance, circRNAs are transferred into exosomes and secreted by colon cancer cells (23).
In the present study, the circRNAs expression array data suggested that circ_400068 was significantly upregulated in RCC exosomes. Further functional studies were performed to investigate the effects of exosomal circ_400068 produced by RCC cells on the proliferation of healthy kidney cells. Treatment with RCC-Exo promoted the proliferation of healthy kidney cells, while cell apoptosis was significantly inhibited. In order to examine the detailed function of exosomal circ_400068 in tumorigenesis, its downstream molecules were predicted, and it was identified that a novel signaling pathway involving a circ_400068/miR-210-5p/SOCS1 axis may regulate the proliferation of kidney cells. miR-210 is involved in numerous biological processes during the development of cancer, such as apoptosis, angiogenesis and DNA damage response, and it is considered as potential biomarker and therapeutic target in numerous types of tumors, including RCC (26–30). In line with the present findings, the tumor-promoting role of SOCS1 and its detailed regulatory functions in colorectal cancer cells via the downstream molecules E-cad, p53 and STAT1 have been revealed in a previous report (31).
However, there are limitations to the present study. For instance, the effects of sh-circ_400068 on the proliferation and apoptosis of RCC cells in vivo could be evaluated in future studies. Furthermore, whether circ_400068 regulates SOCS1 in a miR-210-5p-dependent manner could be investigated.
In conclusion, the present study demonstrated that exosomal circ_400068 was upregulated in the culture media of RCC cells, and circ_400068 could be a novel oncogenic factor. The crosstalk of RCC cells and adjacent healthy kidney cells may be crucial during tumor progression, and exosomal circ_400048 secreted by RCC cells could serve essential roles during this process. Thus, this novel circ_400068/miR-210-5p/SOCS1 axis could be a therapeutic candidate for the treatment of RCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Fund Program of Liaoning Province (grant no. 20180530058).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JS designed this study. HX and JS performed the experiments and conducted the data analysis. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of The First Affiliated Hospital of Jinzhou Medical University. Written informed consents were signed by all patient.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Dabestani S, Beisland C, Stewart GD, Bensalah K, Gudmundsson E, Lam TB, Gietzmann W, Zakikhani P, Marconi L, Fernandéz-Pello S, et al. Intensive Imaging-based follow-up of surgically treated localised renal cell carcinoma does not improve Post-recurrence survival: Results from a European multicentre database (RECUR) Eur Urol. 2019;75:261–264. doi: 10.1016/j.eururo.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Sheth S, Scatarige JC, Horton KM, Corl FM, Fishman EK. Current concepts in the diagnosis and management of renal cell carcinoma: Role of multidetector ct and Three-dimensional CT. Radiographics. 2001;21(Suppl):S237–S254. doi: 10.1148/radiographics.21.suppl_1.g01oc18s237. [DOI] [PubMed] [Google Scholar]
- 4.Chappell WH, Steelman LS, Long JM, Kempf RC, Abrams SL, Franklin RA, Bäsecke J, Stivala F, Donia M, Fagone P, et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: Rationale and importance to inhibiting these pathways in human health. Oncotarget. 2011;2:135–1365. doi: 10.18632/oncotarget.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elfiky AA, Aziz SA, Conrad PJ, Siddiqui S, Hackl W, Maira M, Robert CL, Kluger HM. Characterization and targeting of phosphatidylinositol-3 kinase (PI3K) and mammalian target of rapamycin (mTOR) in renal cell cancer. J Transl Med. 2011;9:133. doi: 10.1186/1479-5876-9-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Cristofano C, Minervini A, Menicagli M, Salinitri G, Bertacca G, Pefanis G, Masieri L, Lessi F, Collecchi P, Minervini R, et al. Nuclear expression of hypoxia-inducible factor-1alpha in clear cell renal cell carcinoma is involved in tumor progression. Am J Surg Pathol. 2007;31:1875–1881. doi: 10.1097/PAS.0b013e318094fed8. [DOI] [PubMed] [Google Scholar]
- 7.Li M, Wang Y, Song Y, Bu R, Yin B, Fei X, Guo Q, Wu B. Expression profiling and clinicopathological significance of DNA methyltransferase 1, 3A and 3B in sporadic human renal cell carcinoma. Int J Clin Exp Pathol. 2014;7:7597–609. [PMC free article] [PubMed] [Google Scholar]
- 8.Shang D, Liu Y, Ito N, Kamoto T, Ogawa O. Defective Jak-Stat activation in renal cell carcinoma is associated with interferon-alpha resistance. Cancer Sci. 2007;98:1259–1264. doi: 10.1111/j.1349-7006.2007.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Von Schulz-Hausmann S, Schmeel L, Schmeel F, Schmidt-Wolf I. Targeting the Wnt/beta-catenin pathway in renal cell carcinoma. Anticancer Res. 2014;34:4101–4108. [PubMed] [Google Scholar]
- 10.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 11.Vicens Q, Westhof E. Biogenesis of circular RNAs. Cell. 2014;159:13–14. doi: 10.1016/j.cell.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Conn S, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 14.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao Z, Luo J, Hu K, Lin J, Huang H, Wang Q, Zhang P, Xiong Z, He C, Huang Z, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11:422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tlsty T, Coussens L. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 17.Théry C, Zitvogel L, Amigorena S. Exosomes: Composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–759. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 18.Keller S, Sanderson M, Stoeck A, Altevogt P. Exosomes: From biogenesis and secretion to biological function. Immunol. Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Luo F, Wang B, Li H, Xu Y, Liu X, Shi L, Lu X, Xu W, Lu L, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370:125–135. doi: 10.1016/j.canlet.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilusz J, Sharp P. Molecular biology. A circuitous route tononcoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang PF, Wei CY, Huang XY, Peng R, Yang X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, et al. Circular RNA circTRIM33-12 acts as the sponge of microRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18:105. doi: 10.1186/s12943-019-1031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dou Y, Cha DJ, Franklin JL, Higginbotham JN, Jeppesen DK, Weaver AM, Prasad N, Levy S, Coffey RJ, Patton JG, Zhang B. Circular RNAs are Down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6:37982. doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee J, Lötvall J. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using Real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Zhao A, Li G, Péoc'h M, Genin C, Gigante M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol. 2013;94:115–120. doi: 10.1016/j.yexmp.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Han Y, Zhang H, Nie L, Jiang Z, Fa P, Gui Y. Synthetic miRNA-mowers targeting miR-183-96-182 cluster or miR-210 inhibit growth and migration and induce apoptosis in bladder cancer cells. PLoS One. 2012;7:e52280. doi: 10.1371/journal.pone.0052280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M, Moriyama M. Overexpression of miR-210, a downstream target of HIF1a, causes centrosome amplification in renal carcinoma cells. J Pathol. 2011;224:280–288. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- 29.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and Krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen WY, Liu WJ, Zhao YP, Zhou L, Zhang TP, Chen G, Shu H. Induction, modulation and potential targets of miR-210 in pancreatic cancer cells. Hepatobiliary Pancreat Dis Int. 2012;11:319–324. doi: 10.1016/S1499-3872(12)60168-4. [DOI] [PubMed] [Google Scholar]
- 31.Tobelaim W, Beaurivage C, Champagne A, Pomerleau V, Simoneau A, Chababi W, Yeganeh M, Thibault P, Klinck R, Carrier JC, et al. Tumour-promoting role of SOCS1 in colorectal cancer cells. Sci Rep. 2015;5:14301. doi: 10.1038/srep14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.