Abstract
Objectives
Data derived from primary care electronic medical records (EMRs) are being used for research and surveillance. Case definitions are required to identify patients with specific conditions in EMR data with a degree of accuracy. The purpose of this study is to identify and provide a summary of case definitions that have been validated in primary care EMR data.
Materials and Methods
We searched MEDLINE and Embase (from inception to June 2016) to identify studies that describe case definitions for clinical conditions in EMR data and report on the performance metrics of these definitions.
Results
We identified 40 studies reporting on case definitions for 47 unique clinical conditions. The studies used combinations of International Classification of Disease version 9 (ICD-9) codes, Read codes, laboratory values, and medications in their algorithms. The most common validation metric reported was positive predictive value, with inconsistent reporting of sensitivity and specificity.
Discussion
This review describes validated case definitions derived in primary care EMR data, which can be used to understand disease patterns and prevalence among primary care populations. Limitations include incomplete reporting of performance metrics and uncertainty regarding performance of case definitions across different EMR databases and countries.
Conclusion
Our review found a significant number of validated case definitions with good performance for use in primary care EMR data. These could be applied to other EMR databases in similar contexts and may enable better disease surveillance when using clinical EMR data. Consistent reporting across validation studies using EMR data would facilitate comparison across studies.
Systematic review registration
PROSPERO CRD42016040020 (submitted June 8, 2016, and last revised June 14, 2016)
Keywords: systematic review, electronic medical record, case definitions, primary care
BACKGROUND AND SIGNIFICANCE
Rationale
The collection and storage of vast amounts of health data are growing rapidly.1 These “big data” include electronic medical record (EMR)2 data and traditional coded administrative health data, among others. Administrative health data are generated and collected from the administration of the healthcare system, such as hospital discharge abstracts and physician billing claims; these data are routinely used for research and surveillance, as most are population based, relatively inexpensive compared to primary data collection, and exist in a structured format.3
EMRs are commonly used in primary care settings to record patient information and facilitate patient care, and thus contain comprehensive demographic and clinical information about diagnoses, prescriptions, physical measurements, laboratory test results, medical procedures, referrals, and risk factors.4 The increased digitization of health information and novel techniques developed for extracting and standardizing data from EMR systems have resulted in many primary care EMR databases being established globally for the purposes of health research and public health surveillance.5 A few prominent examples include the Clinical Practice Research Datalink (CPRD)6 and The Health Improvement Network (THIN),7 both in the UK, as well as the Canadian Primary Care Sentinel Surveillance Network (CPCSSN)8 in Canada.
When using administrative or EMR data for secondary purposes, it is important to have the ability to reliably identify cohorts of patients with a specific disease or condition of interest. Case definitions, also referred to as phenotypes, can be constructed from combinations of diagnostic codes, text words, medications, and/or laboratory results found in the patient record.5 Ideally, case definitions should be validated against a reference standard for disease identification; in most cases, either a manual review of patient charts or physician confirmation is typically used.
As administrative data have been widely utilized for secondary purposes for many decades, numerous case definitions specific to this data source have been developed and validated in a variety of countries and populations.9 EMR data are still a relatively new contribution to disease surveillance and health research, and a full summary of available validated case definitions has not been previously published.
OBJECTIVE
The objective of this study was to identify all case definitions for specific conditions, which have been tested and validated in primary care EMR data.
MATERIALS AND METHODS
We conducted a systematic review of primary studies that reported on the development and validation of case definitions for use in primary care EMR data. We followed a pre-specified protocol,10 in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) reporting guidelines.11 All data were obtained from publically available materials and did not require ethics approval from our institutions.
Data sources and search strategy
We searched MEDLINE (Ovid) and Embase (Ovid) with no date, country, or language restrictions. We also searched the bibliographies of all identified studies. Further, the websites for EMR databases were searched for bibliographic lists (eg, CPRD,6www.cprd.com), and content experts were contacted for information about other potentially ongoing or unpublished studies. The search of online databases included three themes:
Electronic medical records
Case definition
Validation study
We used a comprehensive set of MeSH terms and keyword searches for each of the three themes to ensure we captured all relevant literature. For example, the term “EMR” may be synonymous with a number of relevant keywords (eg, computerized medical records, EHR, etc.). These three search themes were then combined using the Boolean term “AND.” Supplementary File S1 presents our MEDLINE search strategy.
Study selection
Two reviewers independently screened all abstracts. Articles that reported original data for the development and validation of disease case definitions in primary care EMR data were considered for further review. All citations for which either reviewer felt that further review was warranted were kept for full-text review. Bibliographic details from all stages of the review were managed within the Synthesis software package.12
Two reviewers then scanned full-text articles for the following inclusion criteria:
The database under study was a primary care EMR database.
There was a description of a computerized case definition for a specific disease or condition.
A clearly stated reference standard was used to validate the case definition.
Performance metrics were reported (ie, sensitivity, specificity, positive predictive value, negative predictive value, kappa, receiver operating characteristic, likelihood ratio, and their synonyms).
Non-human studies were excluded. Studies reporting on dental health or other non-primary care settings were excluded. We excluded studies in which EMR data were based on patient self-report. We also excluded studies that examined definitions in EMR data linked to administrative health data (though administrative health data used for the reference standard were acceptable). Studies that were not original research and conference abstracts without an adequately detailed description of study methods and results were excluded.
Data extraction
A data extraction form was used to collect information from each included study. The following data elements were extracted: first author, publication year, country, condition(s) under study, sample size and characteristics, cases identified as positive or negative, EMR database or data platform, description of case definition and techniques used to generate it along with fields accessed, reference standard, and performance metrics (eg, sensitivity, specificity, and positive and negative predictive values).
Risk of bias assessment
Included studies were assessed for quality using a component-based approach. We used relevant items from the Quality Assessment of Diagnostic Accuracy Studies (QUADAS) quality assessment tool for diagnostic accuracy studies.13 This tool includes an assessment of bias in several domains, including patient selection, the validation strategy, and reporting of outcomes. Two authors independently assessed risk of bias in each domain and reported the risk of bias as high, low, or unclear. Disagreements were resolved by discussion or with a third reviewer as needed.
Data synthesis
Included studies were described in detail, including setting, target population, and database accessed. Case definitions were grouped by ICD-9 disease category, and definitions were summarized together with their performance metrics. It was not possible to pool data for specific conditions, given study heterogeneity; however, a qualitative comparison of performance of case definitions across conditions was done and reported narratively. Within disease conditions for which there was more than one validated case definition, we documented performance metrics across case definitions and specifically examined the relative importance of using different data elements in creating case definitions for diabetes. In addition to summarizing case definitions and their performance metrics by disease condition, we also produced a detailed inventory of the combinations of variables used, the data fields accessed, and the computer programming methods used.
RESULTS
Study identification
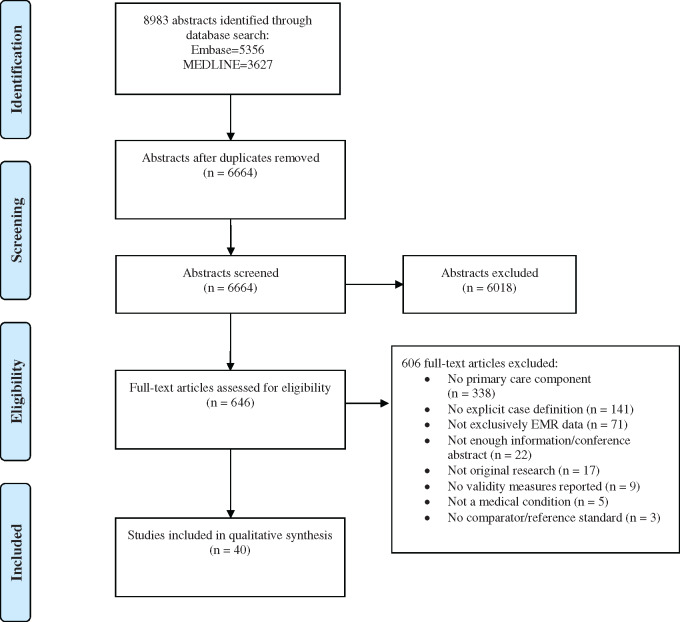
The initial search produced 8983 abstracts from the two databases; 6664 remained after removing duplicates (Figure 1). After the initial abstract screen, 646 articles went forward to full-text review, of which 40 met criteria for inclusion. Reviewer agreement was good in the full-text review stage, with a kappa value of 0.66. The most common reason for exclusion was setting, ie, not primary care (55.8%). Other reasons for exclusion included: not explicitly stating either the case definitions (23.3%) or validation results (1.5%), not using EMR data exclusively (11.7%), not focusing on a specific medical condition (0.8%), and not having a reference standard (0.5%).
Figure 1.
Study selection.
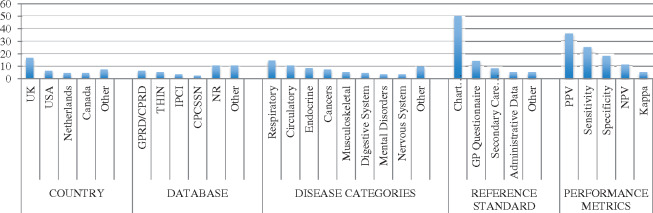
Table 1 describes the characteristics of the studies selected for inclusion (n = 40). Most studies were published between 2010 and 2016 (82.5%) and were conducted in Europe (n = 2562.5%). Twelve studies (30%) were conducted in North America, and the remaining three studies were from Australia and New Zealand (7.5%). Frequently used databases included the General Practice Research Database (GPRD) and its successor, the CPRD, THIN, Integrated Primary Care Information (IPCI), and CPCSSN. Most of the studies focused on a general clinical population, though some (7.5%) were specific to pediatric, adolescent, or senior groups. Sample sizes ranged between 75 and 190 000 patients, with a per-study median of just under 2000 patients. Figure 2 provides a summary of select study characteristics.
Table 1.
Study characteristics
| First Author | Year | Country | Condition | Patients | EMR Data Source | Sample size |
|---|---|---|---|---|---|---|
| Afzal14 | 2013 | Netherlands | Asthma | 5 to 18 years; registered in IPCI for at least 6 months | IPCI | 5032 |
| Afzal15 | 2013 | Netherlands |
|
All patients registered in IPCI | IPCI | 973; 3988 |
| Cea Soriano16 | 2016 | UK | Colorectal cancer | 40 to 89 years at diagnosis; no record of cancer prescription for low-dose aspirin prior to study entry | THIN | 3805 |
| Charlton17 | 2010 | UK | Major congenital malformations | Mother-baby pairs; mothers 14 to 49 years at the date of delivery | GPRD | 188 |
| Coleman18 | 2015 | Canada |
|
60 years and over; at least one of the five conditions of interest according to current CPCSSN algorithms | CPCSSN | 403 |
| Coloma19 | 2013 | Netherlands; Italy | Acute myocardial infarction | >1 year of continuous and valid data | IPCI; Health Search/CSD Patient DB | 400; 200 |
| Cowan20 | 2014 | USA | Asthma | Patients seen in the University of Wisconsin Department of Family Medicine Clinics between 2007-2009 | Health Plan Employer Data Information Set (HEDIS) | 190 000 |
| de Burgos-Lunar21 | 2011 | Spain | Diabetes mellitus; Hypertension | Over 18 years | Computerized Clinical Records of primary health care clinics in the Spanish National Health System | 423 |
| Dregan22 | 2012 | UK |
|
Patients with at least 12 months of follow-up prior to the start of observation and no alarm symptom or cancer diagnosis documented | GPRD | 42 556 |
| Dubreuil23 | 2016 | UK | Ankylosing spondylitis | 18 to 59 years | THIN | 85 |
| Faulconer24 | 2004 | UK | COPD | Over 45 years | EMIS | 10 975 |
| Gil Montalban25 | 2014 | Spain | Diabetes mellitus | 30 to 74 years | AP-Madrid | 2268 |
| Gray26 | 2000 | England | Ischaemic heart disease | 45 years or over | NR | 1680 |
| Gu27 | 2015 | New Zealand | Skin and Subcutaneous tissue infections | 20 years or under | Four New Zealand general practice EMRs | 307 |
| Hammad28 | 2013 | UK | Congenital cardiac malformations | Singleton live-birth babies | CPRD | 719 |
| Hammersley29 | 2011 | UK | Active seasonal allergic rhinitis | 15 to 45 years | Anonymized dataset | 1092 |
| Hirsch30 | 2014 | USA | Diabetes | 18 years or over; at least 2 outpatient encounters with any GHS provider in 2009 | EHR database from Geisinger Health System | 499 |
| Kadhim-Saleh31 | 2013 | Canada | Diabetes; Hypertension; Osteoarthritis; COPD; Depression | All patients who attended the Kingston (Ontario) PBRN and all 22 practices within the network | CPCSSN | 313 |
| Kang32 | 2015 | UK | Glaucoma; Cataract | 18 to 80 years | CPRD | 863; 986 |
| Krysko33 | 2015 | Canada | Multiple sclerosis | 20 years or over | EMRALD, EMR only | 943 |
| Levine34 | 2013 | USA | Skin and soft tissue infections | Primary care outpatients in an academic healthcare system | Oregon Health & Science University’s research data warehouse | 731 |
| Lo Re35 | 2009 | UK | Hepatitis C virus infection; Viral hepatitis | Patients in the database identified with HCV diagnostic codes | THIN | 75 |
| MacRae36 | 2015 | New Zealand |
|
Under 18 years, enrolled in 36 primary care practices | Primary care EHR date | 1200 |
| Mamtani37 | 2015 | UK |
|
21 years or over; at least 6 months of follow-up preceding a first diagnostic code for bladder cancer | THIN | 87 |
| Margulis38 | 2009 | UK | Upper gastrointestinal complications; Peptic ulcer | 40 to 84 years | THIN | 44; 143 |
| Nielen39 | 2013 | Netherlands | Inflammatory arthritis | 30 years or over | LINH | 219 |
| Onofrei40 | 2004 | USA |
|
All patients with an active record | Providence Research Network | 1403; 793 |
| Quint41 | 2014 | UK | COPD | Over 35 years | CPRD-Gold | 704 |
| Rahimi42 | 2014 | Australia | Type 2 diabetes mellitus | At least 3 visits in a two-year period | ePBRN | 908 |
| Rakotz43 | 2014 | USA | Undiagnosed hypertension | 18 to 79 years | NR | 1586 |
| Rothnie44 | 2016 | UK | Acute exacerbation of COPD | Over 35 years | CPRD | 988 |
| Scott45 | 2015 | UK | Intra-abdominal surgery complications; small bowel obstruction; Lysis of adhesions | 18 years or over | THIN | 217 |
| Thiru46 | 2009 | UK | Coronary heart disease | 35 years or over | EMIS from the Northern Regional Research Network | 673 |
| Tian47 | 2013 | USA | Chronic pain | 18 years or over | ECW EHR system | 381 |
| Turchin48 | 2005 | USA |
|
18 years or over | EMR data from four primary care practices at the Brigham & Women’s Hospital, Boston MA | 150 |
| Valkhoff49 | 2014 | Netherlands; Italy | Upper gastrointestinal bleeding | 1- All ages; 2- 15 years or over | IPCI; Health Search/CSD Patient Database (HSD) | 400; 200 |
| Wang50 | 2012 | UK | Ovarian cancer diagnosis | 40 to 80 years | GPRD | 178 |
| Williamson5 | 2014 | Canada |
|
90% over 60 years | CPCSSN | 1920 |
| Xi51 | 2015 | Canada | Asthma | 16 years or over | Open-source Oscar EMR system | 150 |
| Zhou52 | 2016 | UK | Rheumatoid arthritis | Over 16 years | GP records contained in SAIL | 559 |
Abbreviations: IPCI = Integrated Primary Care Information; THIN = The Health Improvement Network; GPRD = General Practice Research Database; CPCSSN = Canadian Primary Care Sentinel Surveillance Network; COPD = Chronic obstructive pulmonary disease; EMIS = Egton Medical Information System; CPRD = Clinical Practice Research Datalink; HER = Electronic Health Records; EMRALD = Electronic Medical Record Administrative data Linked Database; PHO = Primary Health Organization; LINH = Netherlands Information Network of General Practice; LVEF = Left ventricular ejection fraction; CPRD-Gold = Clinical Practice Research Datalink-Gold; ePBRN = Electronic Practice Based Research Network; ECW = eClinicalWorks; SAIL = Secure Anonymised Information Linkage; NR= Not Reported.
Figure 2.
Summary of study characteristics.
Study quality assessment
Supplementary File S2 reports the study quality assessment. Most studies were of reasonably high quality with three studies meeting all quality criteria and 25 studies missing only one or two components of quality. Twelve studies were of questionable quality with three or more domains either not done or not reported. Most studies did not use blinding of the results of the case definition or did not report whether blinding was performed (85%). Further, over one-third (35%) of the studies did not report enough information about the case definition to allow for replication; nearly a quarter of the studies (22.5%) lacked adequate details about the reference standard used to validate the definition.
Medical conditions
Case definitions were found for a total of 47 medical conditions, which represented 13 chapters of the ICD-9,53 the most common being respiratory and circulatory conditions. Eight diseases had multiple case definitions: two for colorectal cancer, eight for diabetes, three for depression, six for hypertension, six for chronic obstructive pulmonary disease (COPD), three for asthma (one of which was pediatric asthma), two for skin and soft tissue infections, and five for arthritis (three osteoarthritis, one rheumatoid arthritis, one inflammatory arthritis).
Case definitions and validation
Most case definitions were constructed using diagnostic codes, such as ICD-953 or Read codes;54 these were sometimes supplemented with laboratory values (eg, glycated hemoglobin [HbA1c] for diabetes), medications (eg, metformin for diabetes), or physical measurements (eg, blood pressure, weight) (Table 2). Some studies tested machine learning programs that were also able to access unstructured data elements, such as free text clinical notes.14,15,50,55,56
Table 2.
Study results grouped by ICD chapter
| First Author | Condition | Reference Standard | Validation Results |
||||
|---|---|---|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Kappa | |||
| Certain infectious and parasitic diseases | |||||||
| Lo Re35 | Hepatitis C virus infection | GP questionnaire | . | . | 86 | . | . |
| Lo Re35 | Viral hepatitis | GP questionnaire | . | . | 76 | . | . |
| Neoplasms | |||||||
| Cea Soriano16 | Colorectal cancer | Chart review | . | . | 80 | . | . |
| Dregan22 | Colorectal cancer | Cancer registry data | 92 | 99 | 98 | 99* | . |
| Dregan22 | Lung cancer | Cancer registry data | 94 | 99 | 96 | 99* | . |
| Dregan22 | Gastro-esophageal cancer | Cancer registry data | 92 | 99 | 97 | 99* | . |
| Dregan22 | Urological cancer | Cancer registry data | 85 | 99 | 93 | 99* | . |
| Mamtani37 | Bladder cancer | GP questionnaire and medical reports | . | . | 99 | . | . |
| Mamtani37 | Muscle invasive bladder cancer | GP questionnaire and medical reports | . | . | 70 | . | . |
| Wang50 | Ovarian cancer | Chart review | 86 | . | 74 | . | . |
| Endocrine, nutritional, and metabolic diseases | |||||||
| Coleman18 | Diabetes | Chart review | 90 | 97 | 91 | 97* | 88 |
| de Burgos-Lunar21 | Diabetes mellitus | Chart review | 100 | 99 | 91 | 100 | 99 |
| Gil Montalban25 | Diabetes mellitus | Data from PREDIMERC | 74 | 99 | 88 | 97 | 78 |
| Hirsch30 | Diabetes | Chart review | 65-99 | 99-100* | 98-100 | 94-99* | . |
| Kadhim-Saleh31 | Diabetes | Primary audit of EMRs | 100 | 99 | 95 | 100 | . |
| Rahimi42 | Type 2 diabetes | Manual audit of EHR | 85* | 99* | 98 | 99 | . |
| Turchin48 | Overweight | Chart and billing code review | 74; 14 | 100 | . | . | 67 |
| Turchin48 | Diabetes | Chart and billing code review | 98; 98 | 98; 98 | . | . | 94 |
| Williamson5 | Diabetes | Chart review | 96 | 97 | 87 | 99 | . |
| Mental disorders | |||||||
| Coleman18 | Depression | Chart review | 73 | 96 | 87 | 90* | 72 |
| Kadhim-Saleh31 | Depression | Primary audit of EMRs | 39 | 97 | 79 | 86 | . |
| Williamson5 | Dementia | Chart review | 97 | 98 | 73 | 100 | . |
| Williamson5 | Depression | Chart review | 81 | 95 | 80 | 95 | . |
| Diseases of the nervous system | |||||||
| Krysko33 | Multiple sclerosis | Chart review | 92 | 100 | 99 | 100 | 95 |
| Tian47 | Chronic pain | Chart review | 85 | 98 | 91 | 96 | . |
| Williamson5 | Parkinsonism | Chart review | 99 | 99 | 82 | 100 | . |
| Williamson5 | Epilepsy | Chart review | 99 | 99 | 86 | 100 | . |
| Diseases of the eye and adnexa | |||||||
| Kang32 | Cataract | GP questionnaire | . | . | 92 | . | . |
| Kang32 | Glaucoma | GP questionnaire | . | . | 84 | . | . |
| MacRae36 | Otitis media | Chart review | 58 | 99 | 90 | 94 | . |
| Diseases of the circulatory system | |||||||
| Coleman18 | Hypertension | Chart review | 95 | 79 | 93 | 86* | 76 |
| Coloma19 | Acute myocardial infarction | Chart review/GP questionnaire | . | . | 60-97 | . | . |
| de Burgos-Lunar21 | Hypertension | Chart review | 85 | 97 | 85 | 97 | 77 |
| Gray26 | Ischemic heart disease | Chart review | 47-96 | . | 33-83 | . | . |
| Kadhim-Saleh31 | Hypertension | Primary audit of EMRs | 83 | 98 | 98 | 81 | . |
| Onofrei40 | Heart failure: LVEF ≤ 55% | Echocardiography database, echo in chart, chart review | 44 | 100 | 36 | 100 | . |
| Onofrei40 | Heart failure: LVEF ≤ 40% | Echocardiography database, echo in chart, chart review | 54 | 99 | 25 | 100 | . |
| Rakotz43 | Undiagnosed hypertension | AOBP measurement | . | . | 51-58 | . | . |
| Thiru46 | Coronary heart disease | Established EMR definitions | 65-98 | . | 40-74 | . | . |
| Turchin48 | Hypertension | Chart and billing code review | 91; 74 | 86; 92 | . | . | 77 |
| Williamson5 | Hypertension | Chart review | 85 | 94 | 93 | 86 | . |
| Diseases of the respiratory system | |||||||
| Afzal14 | Pediatric asthma | Chart review | 95-98 | 67-95 | 57-82 | . | . |
| Coleman18 | COPD | Chart review | 72 | 92 | 37 | 98* | 44 |
| Cowan20 | Asthma | Expert panel diagnosis | 82-92 | 95-98 | . | . | . |
| Faulconer24 | COPD | Chart review | 79 | 99* | 75 | 99* | . |
| Hammersley29 | Allergic rhinitis | Chart review | 17-85 | 86-100 | 15-100 | 96-99 | . |
| Kadhim-Saleh31 | COPD | Primary audit of EMRs | 41 | 99 | 80 | 94 | . |
| Quint41 | COPD | Chart review/GP questionnaire | . | . | 12-89 | . | . |
| Rothnie44 | Acute exacerbations of COPD | Chart review/GP questionnaire | 52-88 | . | 64-86 | . | . |
| Williamson5 | COPD | Chart review | 82 | 97 | 72 | 98 | . |
| Xi51 | Asthma | Chart review | 74-90 | 84-93 | 67-81* | 90-96* | . |
| MacRae36 | Upper respiratory tract infection | Chart review | 54 | 98 | 86 | 89 | . |
| MacRae36 | Lower respiratory tract infection | Chart review | 61 | 99 | 76 | 98 | . |
| MacRae36 | Wheeze illness | Chart review | 96 | 96 | 70 | 100 | . |
| MacRae36 | Throat infections | Chart review | 50 | 99 | 91 | 95 | . |
| MacRae36 | Other respiratory | Chart review | 66 | 99 | 68 | 99 | . |
| Diseases of the digestive system | |||||||
| Afzal15 | Hepatobiliary disease | Chart review | 89-92 | 68-79 | . | . | . |
| Margulis38 | Upper gastrointestinal complications | GP questionnaire | . | . | 95 | . | . |
| Margulis38 | Peptic ulcer | GP questionnaire | . | . | 94 | . | . |
| Valkhoff49 | Upper gastrointestinal bleeding | Expert panel review/chart review | . | . | 21; 78 | . | . |
| Scott45 | Intra-abdominal surgery complications, small bowel obstruction, and lysis of adhesions | GP questionnaire | . | . | 86-95 | . | . |
| Diseases of the skin and subcutaneous tissue | |||||||
| Gu27 | Skin and subcutaneous tissue infections | Chart review | 95 | 98 | 80 | 100 | . |
| Levine34 | Skin and soft tissue infections | Chart review | . | . | 53-92 | . | . |
| Diseases of the musculoskeletal system and connective tissue | |||||||
| Coleman18 | Osteoarthritis | Chart review | 63 | 94 | 96 | 51* | 46 |
| Dubreuil23 | Ankylosing spondylitis | GP questionnaire | 30-98 | . | 71-89 | . | . |
| Kadhim-Saleh31 | Osteoarthritis | Primary audit of EMRs | 45 | 100 | 100 | 68 | . |
| Nielen39 | Inflammatory arthritis | Chart review | . | . | 78 | . | . |
| Williamson5 | Osteoarthritis | Chart review | 78 | 95 | 88 | 90 | . |
| Zhou52 | Rheumatoid arthritis | Secondary care electronic patient records | 86-89 | 91-95 | 79-86 | . | . |
| Diseases of the genitourinary system | |||||||
| Afzal15 | Acute renal failure | Chart review | 62-71 | 88-92 | . | . | . |
| Congenital malformations, deformations, and chromosomal abnormalities | |||||||
| Charlton17 | Major congenital malformations | Chart review | . | . | 85 | . | . |
| Hammad28 | Congenital cardiac malformations | GP questionnaire | . | . | 0-100 | . | . |
Abbreviations: ICD = International Classification of Diseases; ICD-9-CM = International Classification of Diseases, 9th revision, clinical modification; NPV = negative predictive value; PPV = positive predictive value.
indicates calculated values based upon published data.
The reporting of performance metrics was variable across studies, with some describing several metrics and others reporting just one. The most frequently used validation measure was the positive predictive value (PPV); sensitivity and specificity were also frequently measured. Seven studies reported true positives and false positives.14,21,33,34,39,42,51 Only one study reported likelihood ratios40 (Figure 2 and Table 2). The most common reference standard used was manual chart review, though others included a physician questionnaire, registries based on other data sources, and other diagnostic tests.
Case definitions for malignancies (n = 8) performed well overall, with mostly high sensitivities, specificities, and PPVs. With respect to chronic illnesses, definitions for diabetes (n = 8) also performed well across the three metrics. Definitions for hypertension (n = 5) and ischemic heart disease (n = 3) had moderate performance, while definitions for heart failure (n = 2) were highly specific but not very sensitive. Similarly, definitions for COPD (n = 6), overweight (n = 1), osteoarthritis (n = 3), and depression (n = 3) also had high specificities but low sensitivities. Asthma definitions performed moderately well across sensitivity and specificity. Some less common diseases, ie, dementia, Parkinson’s disease, epilepsy, and multiple sclerosis, had definitions with good sensitivities and specificities, but PPVs tended to be moderate. Case definitions for acute infections (otitis media and respiratory infections) had excellent specificities but low sensitivities. Supplementary File S3 contains further detail on the published case definitions.
In the case of diabetes, 19 separate tests of validation were reported across eight studies, three of which were performed at different times in the same database (CPCSSN). These definitions used various elements of EMR data alone or in combination, including diagnostic codes, reason for visit, medications, laboratory data, problems lists, and in one case, free text (Table 3). There are no consistent trends indicating that one or more elements increases performance. However, in the case of Hirsch et al, case definition performance improved slightly with the addition of other data elements to diagnostic codes. In most cases in which diagnostic codes were not used, performance was markedly lower. Also noted is that PPV decreased slightly when case definition sensitivity increased. Free text was used only in one instance; however, it did not have superior performance to diagnostic codes.
Table 3.
Diabetes case definition elements and performance
| First Author | Database | Diagnostic Codes | Reason for Visit | Medications | Lab results | Problem List | Classification System | Free Text | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coleman18 | CPCSSN | X | X | X | 90 | 97 | 91 | . | ||||
| de Burgos-Lunar21 | EMRs in Spanish National Health System | X | 100 | 99 | 91 | 100 | ||||||
| Gil Montalban25 | AP-Madrid | X | X | 74 | 99 | 88 | 97 | |||||
| Hirsch30 | Geisinger Health System | X | 96 | . | 98 | . | ||||||
| Hirsch30 | Geisinger Health System | X | 75 | . | 100 | . | ||||||
| Hirsch30 | Geisinger Health System | X | 65 | . | 100 | . | ||||||
| Hirsch30 | Geisinger Health System | X | 84 | . | 99 | . | ||||||
| Hirsch30 | Geisinger Health System | X | X | 96 | . | 98 | . | |||||
| Hirsch30 | Geisinger Health System | X | X | X | 99 | . | 98 | . | ||||
| Hirsch30 | Geisinger Health System | X | X | X | 98 | . | 98 | . | ||||
| Hirsch30 | Geisinger Health System | X | X | X | X | 99 | . | 98 | . | |||
| Kadhim-Saleh31 | CPCSSN | X | X | X | X | 100 | 99 | 95 | 100 | |||
| Rahimi42 | ePBRN | X | . | . | 100 | 100 | ||||||
| Rahimi42 | ePBRN | X | . | . | 97 | 99 | ||||||
| Rahimi42 | ePBRN | X | . | . | 16 | 99 | ||||||
| Rahimi42 | ePBRN | X | X | X | . | . | 98 | 99 | ||||
| Turchin48 | EMR from four practices | X | 98 | 98 | . | . | ||||||
| Turchin48 | EMR from four practices | X | 98 | 98 | . | . | ||||||
| Williamson5 | CPCSSN | X | X | X | 96 | 97 | 87 | 99 |
DISCUSSION
Summary of findings
We undertook this project to summarize all studies that have developed and validated case definitions using primary care EMR data. Validated case definitions are important tools, as they can be adapted and applied to different EMR databases to conduct research or surveillance. Our review identified 40 studies that validated case definitions for 47 conditions. The most common conditions were diabetes (eight definitions), hypertension (six definitions), and COPD (six definitions), though multiple definitions have also been developed for depression (three definitions), osteoarthritis (three definitions), and asthma and respiratory infections (four definitions). The majority of other conditions was limited to a single case definition.
The case definitions we identified may be useful for research or surveillance efforts that require identification of one of the 47 conditions in primary care EMR data. While not all will be easily transferred for use with other data sources, these definitions can serve as an important starting point. Further, for conditions for which we have not identified a case definition, there is an opportunity to develop, test, and publish case definitions so they are available to others. For instance, Barnett et al conducted a literature review, followed by a consensus exercise to identify 40 conditions likely to be chronic and have significant impact on patients’ treatment needs, function, quality of life, morbidity, and mortality.57 Tonelli et al identified case definitions with moderate to high validity for use in administrative health data for 30 of these conditions.9 Our review identified EMR case definitions for only 16 of these conditions. While it appears that case definitions are created opportunistically for research that focuses on one or more specific conditions, creating case definitions requires considerable effort, and having these available for use and adaptation may facilitate future research.
This review also summarized the methods and data elements used for developing the case definitions. Diagnostic codes were the most common feature used to define the conditions, and also the most simplistic method, as many definitions relied solely on diagnostic codes. While diagnostic codes were highly sensitive and specific for some conditions (eg, cancer), they were much less sensitive for others (eg, heart failure, depression), perhaps due to less specific diagnostic codes for these types of conditions. For instance, heart failure could present as shortness of breath, or together with another condition such as arrhythmia or diabetes. Similarly, depression could present symptomatically as insomnia, fatigue, malaise, etc., and be given a diagnostic code specific to the symptomatology. In several instances, diagnostic codes were augmented with a combination of medications, laboratory data, problem lists, and/or free text searches. In the case of diabetes, these additional data elements were useful in improving the sensitivity and specificity of the case definition.5,30 Of note, diabetes case definitions tended to perform better overall than those for COPD, another common chronic condition. While this may be due to better diagnostic coding on the part of physicians, it is also possible to identify diabetes based on laboratory values. Having additional data elements available for disease identification may improve case definition performance.
Finally, while less common than traditional expert committee created definitions, machine learning programs have been used in an attempt to efficiently identify the best case definitions using multiple data elements.5,9 It is difficult to comment on whether these programs perform better, as the only condition for which both methods have been used is diabetes, and both performed similarly, albeit in different databases.5,18,21,25,30,31,42,48 That said, machine learning techniques are likely a more efficient way to generate candidate case definitions, and may therefore play an important role in increasing both the number of conditions for which case definitions are available, and their performance, as many more candidate definitions could be tested and validated quickly. The database used is also likely to be an important factor in the performance of case definitions, as data quality influences the predictive accuracy of any one data element for a specific disease. Therefore, while advanced techniques such as machine learning and free text mining may lead to higher performing case definitions, improving the quality and completeness of data within a database is also an important consideration for moving this field forward.
Strengths
Research and surveillance using primary care EMR databases are increasing, as vast amounts of clinical data are becoming available for secondary purposes. To our knowledge, this is the first systematic review of its kind that describes validated case definitions used in primary care EMR data. These results may improve our ability to more efficiently define cohorts with specific conditions without requiring a full validation exercise, which is resource and time intensive. In addition, this review summarizes the disease conditions for which validated case definitions have been developed and encourages further research to develop and validate case definitions for other disease conditions, for which such definitions do not exist or are lacking.
Limitations
Although this review was thorough in its methods, a lack of detailed reporting in many papers may have led to their exclusion. For instance, 141 papers did not explicitly describe their case finding algorithms, and 22 papers were missing requisite data. Also, 12 studies did not report validity measures (ie, specificity, PPV, etc.) or did not describe a reference standard. Among the studies included in our review, not all metrics of interest were reported. For instance, some studies reported only PPV, which limits our ability to comment on the sensitivity of the case definition, or its performance in a population with a different prevalence of disease. The generalizability of each case definition is also unknown, as they were conducted in a unique variety of populations, settings, healthcare systems, and EMR systems/databases.
Recommendations for reporting of case definition validation studies
Given the variability in reporting, adherence to reporting guidelines, such as those described in the Standards for Reporting of Diagnostic Studies (STARD) statement,58 may strengthen this growing field of research. STARD lists the essential elements to be included in a report of a diagnostic accuracy study. STARD contains 30 elements, most of which apply to reporting validation studies of case definitions. However, as most diagnostic accuracy studies are undertaken in a clinical context, with the test under study being one that diagnoses disease in an individual, certain elements should be modified to reflect the unique aspects of case definition validation studies. The following are specific recommendations for reporting of case definition validation studies:
Identify the study as a case definition validation study, including the condition(s) in question.
Specify the intended use of the case definition (eg, patient identification for clinical purposes, surveillance, research).
Describe the database that the case definition was applied to and how the elements populating the database were collected (eg, for clinical care, health care administration, other).
Describe how the sample used for validation was selected.
Describe the clinical and demographic characteristics of the population whose information is included in the database.
Describe the process by which the case definition(s) was/were derived (eg, using statistical methods or machine learning, expert opinion, other).
Clearly describe the case definition(s) under study in sufficient detail to allow others to replicate their implementation.
Clearly describe the reference standard used to verify the performance of the case definition(s) and rationale for its use.
State whether the reference standard was applied independently of the case definition.
In studies in which the case definition(s) is/are derived from data using machine learning or other statistical methods, the testing dataset and validation dataset should be clearly described.
Clearly describe the methods for assessing performance and the specific metrics used (eg, sensitivity, specificity, PPV, negative predictive value [NPV], other), including how missing or indeterminate data were handled.
Presentation of results should include cross tabulation of the case definition(s) results by the reference standard results as well as the performance metrics and their 95% confidence intervals.
CONCLUSION
Data collected in primary care electronic medical records are becoming an important resource for conducting research and understanding disease patterns and prevalence. This review provides a summary of validated case definitions for a number of clinical conditions in primary care EMR data, most of which identify conditions with relatively good accuracy. The case definitions identified through this review may be used as a starting point for research and disease surveillance that require the identification of medical conditions in primary care EMR data. However, there are a number of conditions for which no case definition has been reported in the peer-reviewed literature. To improve the utility of future studies, authors publishing on case definitions with validity outcomes should adhere to detailed reporting standards.
List of Abbreviations
COPD: Chronic Obstructive Pulmonary Disease
CPCSSN: Canadian Primary Care Sentinel Surveillance Network
CPRD: Clinical Practice Research Datalink
EHR: Electronic Health Record
EMR: Electronic Medical Record
GPRD: General Practice Research Database
ICD-9: International Classification of Disease version 9
IPCI: Integrated Primary Care Information
NPV: Negative Predictive Value
PPV: Positive Predictive Value
PRISMA: Preferred Reporting Items for Systematic Reviews and Meta-analyses
QUADAS: Quality Assessment of Diagnostic Accuracy Studies
STARD: Standards for Reporting of Diagnostic Accuracy Studies
THIN: The Health Improvement Network
Supplementary Material
ACKNOWLEDGMENTS
Not applicable.
FUNDING
This was an investigator-initiated project. No sources of funding are related to the research reported.
AUTHORS’ CONTRIBUTIONS
This review was conceived by PER, TSW, GEF, and KAM, and the protocol was designed with input by SS, NES, AR, BCL, SG, RB, and HQ. NES, AR, BCL, SG, and KAM designed the search strategy. RB and HQ contributed as knowledge users. SS, NES, AR, BCL, PER, and KAM drafted the manuscript, and all authors critically revised it and approved the final version. KAM will act as the guarantor for this review.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Conflict of interest statement. The authors declare that they have no competing interests.
REFERENCES
- 1. Murdoch TB, Detsky AS.. The inevitable application of big data to health care. JAMA 2013; 30913: 1351–2. [DOI] [PubMed] [Google Scholar]
- 2. Deniz S, Şengül A, Aydemir Y, Çeldir Emre J, Özhan MH.. Clinical factors and comorbidities affecting the cost of hospital-treated COPD. Int J Chron Obstruct Pulmon Dis 2016; 111: 3023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Quan H, Smith M, Bartlett-Esquilant G, Johansen H, Tu K, Lix L.. Mining administrative health databases to advance medical science: geographical considerations and untapped potential in Canada. Can J Cardiol 2012; 282: 152–4. [DOI] [PubMed] [Google Scholar]
- 4. Biro SC, Barber DT, Kotecha JA.. Trends in the use of electronic medical records. Can Fam Physician 2012; 581: e21.. [PMC free article] [PubMed] [Google Scholar]
- 5. Williamson T, Green ME, Birtwhistle R, et al. Validating the 8 CPCSSN case definitions for chronic disease surveillance in a primary care database of electronic health records. Ann Fam Med 2014; 124: 367–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herrett E, Gallagher AM, Bhaskaran K, et al. Data resource profile: Clinical Practice Research Datalink (CPRD). Int J Epidemiol 2015; 443: 827–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blak B, Thompson M, Dattani H, Bourke A.. Generalisability of The Health Improvement Network (THIN) database: demographics, chronic disease prevalence and mortality rates. Inform Prim Care 2011; 194: 251–5. [DOI] [PubMed] [Google Scholar]
- 8. Garies S, Birtwhistle R, Drummond N, Queenan J, Williamson T.. Data resource profile: national electronic medical record data from the Canadian Primary Care Sentinel Surveillance Network (CPCSSN). Int J Epidemiol 2017; 464: 1091–2f. [DOI] [PubMed] [Google Scholar]
- 9. Tonelli M, Wiebe N, Fortin M.. Methods for identifying 30 chronic conditions: application to administrative data. BMC Med Inform Decis Mak 2015; 15: 31.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Souri S, Symonds NE, Rouhi A, et al. Identification of validated case definitions for chronic disease using electronic medical records: a systematic review protocol. Syst Rev 2017; 61: 38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P.. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med 2009; 1514: 264–9 W64. [DOI] [PubMed] [Google Scholar]
- 12. Yergens D. Synthesis v2.4 and v3.0. 2015. http://www.synthesis.info. Accessed June 1, 2015.
- 13. Whiting P, Rutjes AW, Reitsma JB, Bossuyt PM, Kleijnen J.. The development of QUADAS: a tool for the quality assessment of studies of diagnostic accuracy included in systematic reviews. BMC Med Res Methodol 2003; 31: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Afzal Z, Engelkes M, Verhamme KM, et al. Automatic generation of case-detection algorithms to identify children with asthma from large electronic health record databases. Pharmacoepidemiol Drug Saf 2013; 228: 826–33. [DOI] [PubMed] [Google Scholar]
- 15. Afzal Z, Schuemie MJ, van Blijderveen JC, Sen EF, Sturkenboom MC, Kors JA.. Improving sensitivity of machine learning methods for automated case identification from free-text electronic medical records. BMC Med Inform Decis Mak 2013; 131: 30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cea Soriano L, Soriano-Gabarro M, Garcia Rodriguez LA.. Validity and completeness of colorectal cancer diagnoses in a primary care database in the United Kingdom. Pharmacoepidemiol Drug Saf 2016; 254: 385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Charlton RA, Weil JG, Cunnington MC, de Vries CS.. Identifying major congenital malformations in the UK General Practice Research Database (GPRD): a study reporting on the sensitivity and added value of photocopied medical records and free text in the GPRD. Drug Saf 2010; 339: 741–50. [DOI] [PubMed] [Google Scholar]
- 18. Coleman KJ, Lutsky MA, Yau V, et al. Validation of autism spectrum disorder diagnoses in large healthcare systems with electronic medical records. J Autism Dev Disord 2015; 457: 1989–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coloma PM, Valkhoff VE, Mazzaglia G, et al. Identification of acute myocardial infarction from electronic healthcare records using different disease coding systems: a validation study in three European countries. BMJ Open 2013; 36: e002862.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cowan K, Tandias A, Arndt B, Hanrahan L, Mundt M, Guilbert T.. Defining asthma: validating automated electronic health record algorithm with expert panel diagnosis. Am J Respir Clin Care Med 2014.; 189: A2297. [Google Scholar]
- 21. de Burgos-Lunar C, Salinero-Fort MA, Cardenas-Valladolid J, et al. Validation of diabetes mellitus and hypertension diagnosis in computerized medical records in primary health care. BMC Med Res Methodol 2011; 111: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dregan A, Moller H, Murray-Thomas T, Gulliford MC.. Validity of cancer diagnosis in a primary care database compared with linked cancer registrations in England. Population-based cohort study. Cancer Epidemiol 2012; 365: 425–9. [DOI] [PubMed] [Google Scholar]
- 23. Dubreuil M, Peloquin C, Zhang Y, Choi HK, Inman RD, Neogi T.. Validity of ankylosing spondylitis diagnoses in The Health Improvement Network. Pharmacoepidemiol Drug Saf 2016; 254: 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Faulconer E, DeLusignan S.. An eight-step method for assessing diagnostic data quality in practice: chronic obstructive pulmonary disease as an exemplar. Inform Prim Care 2004; 124: 243–54. [PubMed] [Google Scholar]
- 25. Gil Montalbán E, Ortiz Marrón H, López-Gay Lucio-Villegas D, Zorrilla Torrás B, Arrieta Blanco F, Nogales Aguado P.. [Validity and concordance of electronic health records in primary care (AP-Madrid) for surveillance of diabetes mellitus. PREDIMERC study]. Gac Sanit 2014; 285: 393–6. [DOI] [PubMed] [Google Scholar]
- 26. Gray J, Majeed A, Kerry S, Rowlands G.. Identifying patients with ischaemic heart disease in general practice: cross sectional study of paper and computerised medical records. BMJ 2000; 3217260: 548–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gu Y, Kennelly J, Warren J, Nathani P, Boyce T.. Automatic detection of skin and subcutaneous tissue infections from primary care electronic medical records. Stud Health Technol Inform 2015; 214: 74–80. [PubMed] [Google Scholar]
- 28. Hammad TA, Margulis AV, Ding Y, Strazzeri MM, Epperly H.. Determining the predictive value of Read codes to identify congenital cardiac malformations in the UK Clinical Practice Research Datalink. Pharmacoepidemiol Drug Saf 2013; 2211: 1233–8. [DOI] [PubMed] [Google Scholar]
- 29. Hammersley V, Flint R, Pinnock H, Sheikh A.. Developing and testing search strategies to identify patients with active seasonal allergic rhinitis in general practice. Prim Care Respir J 2010; 201: 71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hirsch AG, Scheck McAlearney A.. Measuring diabetes care performance using electronic health record data: the impact of diabetes definitions on performance measure outcomes. Am J Med Qual 2014; 294: 292–9. [DOI] [PubMed] [Google Scholar]
- 31. Kadhim-Saleh A, Green M, Williamson T, Hunter D, Birtwhistle R.. Validation of the diagnostic algorithms for 5 chronic conditions in the Canadian Primary Care Sentinel Surveillance Network (CPCSSN): a Kingston Practice-based Research Network (PBRN) report. J Am Board Fam Med 2013; 262: 159–67. [DOI] [PubMed] [Google Scholar]
- 32. Kang EM, Pinheiro SP, Hammad TA, Abou-Ali A.. Evaluating the validity of clinical codes to identify cataract and glaucoma in the UK Clinical Practice Research Datalink. Pharmacoepidemiol Drug Saf 2015; 241: 38–44. [DOI] [PubMed] [Google Scholar]
- 33. Krysko KM, Ivers NM, Young J, O’Connor P, Tu K.. Identifying individuals with multiple sclerosis in an electronic medical record. Mult Scler 2015; 212: 217–24. [DOI] [PubMed] [Google Scholar]
- 34. Levine PJ, Elman MR, Kullar R, et al. Use of electronic health record data to identify skin and soft tissue infections in primary care settings: a validation study. BMC Infect Dis 2013; 131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lo Re V 3rd, Haynes K, Forde KA, Localio AR, Schinnar R, Lewis JD.. Validity of The Health Improvement Network (THIN) for epidemiologic studies of hepatitis C virus infection. Pharmacoepidemiol Drug Saf 2009; 189: 807–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MacRae J, Darlow B, McBain L, et al. Accessing primary care Big Data: the development of a software algorithm to explore the rich content of consultation records. BMJ Open 2015; 58: e008160.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mamtani R, Haynes K, Boursi B, et al. Validation of a coding algorithm to identify bladder cancer and distinguish stage in an electronic medical records database. Cancer Epidemiol Biomarkers Prev 2015; 241: 303–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Margulis AV, Garcia Rodriguez LA, Hernandez-Diaz S.. Positive predictive value of computerized medical records for uncomplicated and complicated upper gastrointestinal ulcer. Pharmacoepidemiol Drug Saf 2009; 1810: 900–9. [DOI] [PubMed] [Google Scholar]
- 39. Nielen MM, Ursum J, Schellevis FG, Korevaar JC.. The validity of the diagnosis of inflammatory arthritis in a large population-based primary care database. BMC Fam Pract 2013; 141: 79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Onofrei M, Hunt J, Siemienczuk J, Touchette D, Middleton B.. A first step towards translating evidence into practice: heart failure in a community practice-based research network. Inform Prim Care 2004; 123: 139–45. [DOI] [PubMed] [Google Scholar]
- 41. Quint JK, Mullerova H, DiSantostefano RL, et al. Validation of chronic obstructive pulmonary disease recording in the Clinical Practice Research Datalink (CPRD-GOLD). BMJ Open 2014; 47: e005540.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rahimi A, Liaw ST, Taggart J, Ray P, Yu H.. Validating an ontology-based algorithm to identify patients with type 2 diabetes mellitus in electronic health records. Int J Med Inform 2014; 8310: 768–78. [DOI] [PubMed] [Google Scholar]
- 43. Rakotz MK, Ewigman BG, Sarav M, et al. A technology-based quality innovation to identify undiagnosed hypertension among active primary care patients. Ann Fam Med 2014; 124: 352–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rothnie KJ, Müllerová H, Hurst JR, et al. Validation of the recording of acute exacerbations of copd in uk primary care electronic healthcare records. PloS One 2016; 113: e0151357.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scott FI, Mamtani R, Haynes K, Goldberg DS, Mahmoud NN, Lewis JD.. Validation of a coding algorithm for intra-abdominal surgeries and adhesion-related complications in an electronic medical records database. Pharmacoepidemiol Drug Saf 2016; 254: 405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thiru K, Donnan P, Weller P, Sullivan F.. Identifying the optimal search strategy for coronary heart disease patients in primary care electronic patient record systems. Inform Prim Care 2009; 174: 215–24. [DOI] [PubMed] [Google Scholar]
- 47. Tian TY, Zlateva I, Anderson DR.. Using electronic health records data to identify patients with chronic pain in a primary care setting. J Am Med Inform Assoc 2013; 20 (e2): e275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turchin A, Pendergrass ML, Kohane IS.. DITTO- a tool for identification of patient cohorts from the text of physician notes in the electronic medical record. AMIA Annu Symp Proc. 2005; 2005: 744–8. [PMC free article] [PubMed] [Google Scholar]
- 49. Valkhoff VE, Coloma PM, Masclee GMC, et al. Validation study in four health-care databases: upper gastrointestinal bleeding misclassification affects precision but not magnitude of drug-related upper gastrointestinal bleeding risk. J Clin Epidemiol 2014; 678: 921–31. [DOI] [PubMed] [Google Scholar]
- 50. Wang Z, Shah AD, Tate AR, Denaxas S, Shawe-Taylor J, Hemingway H.. Extracting diagnoses and investigation results from unstructured text in electronic health records by semi-supervised machine learning. PloS One 2012; 71: e30412.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xi NW, Agarwal G, Chan D, Gershon A, Gupta S.. Identifying patients with asthma in primary care electronic medical record systems chart analysis-based electronic algorithm validation study. Can Fam Physician 2015; 610: 474–83. [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou S-M, Fernandez-Gutierrez F, Kennedy J, et al. Defining disease phenotypes in primary care electronic health records by a machine learning approach: a case study in identifying rheumatoid arthritis. PloS One 2016; 115: e0154515.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. World Health Organization. International Classification of Disease. Geneva: World Health Organization; 1979. [Google Scholar]
- 54. O'Neil M, Payne C, Read J.. Read Codes Version 3: a user led terminology. Methods In Med 1995; 34 (1–2): 187–92. [PubMed] [Google Scholar]
- 55. Hripcsak G, Bakken S, Stetson PD, Patel VL.. Mining complex clinical data for patient safety research: a framework for event discovery. J Biomed Inform 2003;36 (1–2): 120–30. [DOI] [PubMed] [Google Scholar]
- 56. Denny JC, Miller RA, Johnson KB, Spickard A.. Development and evaluation of a clinical note section header terminology. AMIA Annu Symp Proc 2008; 2008: 156–60. [PMC free article] [PubMed] [Google Scholar]
- 57. Barnett K, Mercer SW, Norbury M, Watt G, Wyke S, Guthrie B.. Epidemiology of multimorbidity and implications for health care, research, and medical education: a cross-sectional study. Lancet 2012; 3809836: 37–43. [DOI] [PubMed] [Google Scholar]
- 58. Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016; 611: e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.