Abstract
Background
The electronic medical record contains a wealth of information buried in free text. We created a natural language processing (NLP) algorithm to identify atrial fibrillation (AF) patients using text alone.
Methods
We created three data sets from patients with at least one AF billing code from 2010 to 2017: a training set (n=886), an internal validation set from Site #1 (n=285), and an external validation set from Site #2 (n=276). A team of clinicians reviewed and adjudicated patients as AF present or absent, which served as the reference standard. We trained 54 algorithms to classify each patient, varying the model, number of features, number of stop words, and the method used to create the feature set. The algorithm with the highest F-score (the harmonic mean of sensitivity and positive predictive value) in the training set was applied to the validation sets. F-scores and area under the receiver operating characteristic curves (AUC) were compared between Site #1 and Site #2 using bootstrapping.
Results
Adjudicated AF prevalence was 75.1% at Site #1 and 86.2% at Site #2. Among 54 algorithms, the best performing model was logistic regression, using 1000 features, 100 stop words, and term frequency-inverse document frequency (TF-IDF) method to create the feature set, with sensitivity 92.8%, specificity 93.9%, and an AUC of 0.93 in the training set. The performance at Site #1 was sensitivity 92.5%, specificity 88.7%, with an AUC of 0.91. The performance at Site #2 was sensitivity 89.5%, specificity 71.1%, with an AUC of 0.80. The F-score was lower at Site #2 compared to Site #1 (92.5% [SD 1.1%] versus 94.2% [SD 1.1%]; p<0.001).
Conclusions
We developed a NLP algorithm to identify AF patients using text alone, with >90% F-score at two separate sites. This approach allows better use of the clinical narrative, and creates an opportunity for precise, high throughput cohort identification.
Keywords: atrial fibrillation, electronic medical record, digital phenotype, natural language processing, epidemiology, artificial intelligence
Introduction
Appropriate patient selection is a critical step in clinical trials, real world evidence generation, cohort studies, and quality improvement efforts. For clinical trials, investigators traditionally identify patients one by one, during routine care. This process is time- and labor-intensive, requires familiarity with inclusion criteria, and creates a bottleneck for enrollment. Real world evidence and quality measures, including measures in the Centers for Medicare and Medicaid Systems (CMS) Quality Payment Program (QPP), often rely on billing codes to identify patient groups, such as International Classification of Diseases (ICD) and Common Procedural Terminology (CPT) codes.1–3 This structured data approach is appealing because it uses readily available, standardized terminologies, but it lacks accuracy and is subject to variation between institutions and over time.4–7 In this project, our goal was to create a new method, an “electronic cohort definition,” to identify atrial fibrillation (AF) patients using data from the electronic medical record (EMR).
EMRs contain a wealth of data that can be used for more precise and efficient cohort identification. Specifically, the clinical narrative includes detailed descriptions of patients and their associated conditions. In our prior work, we developed a relatively simple, rules-based natural language processing (NLP) approach to identify patients with atrial fibrillation (AF) using clinical notes.8 We were able to achieve good sensitivity (90%), but the algorithm lacked specificity (63%). In the current study, we sought to improve model specificity by using a supervised machine learning approach to NLP, rather than a rules-based approach. Compared to rules-based NLP, supervised machine learning requires less pre-specification from the developer. In the rules-based approach, one must specify modifiers (e.g. “denies” or “ruled out for”) to provide context and classification; in machine learning, the algorithm learns the relevant modifiers from the training data itself. Rules-based NLP can be thought of as an “expert system,” in which a person with clinical knowledge needs to specify the rules. Each time the note language style changes, the rules would have to be re-designed. In machine learning, one could re-train the model as note styles change, with less need to manually re-design the system with new rules and modifiers. The machine learning approach may be better suited to improve specificity because it is difficult to explicitly list all the modification terms used in the clinical narrative. The trade-off is that one must account for noise, or terms which are often present but do not add to context and classification. For example, the term “clinic” may appear in many clinical notes, but it is irrelevant to understanding the AF reference and, if included, adds to computational time.
We also sought to create a portable tool that maintains performance in different institutions. To this end, we used only the clinical narrative (unstructured data) as source data, and tested performance at a second site. Different institutions and health systems may vary in their use of structured data elements (e.g. some practices do not use the proprietary CPT coding system), but the narrative is readily available from clinical notes. The data format (text files) may be consistent and clinicians may reference AF similarly between institutions (e.g., “Patient presents with AF that was diagnosed two weeks ago”), which makes NLP appealing for portability. In addition, simple approaches (using just one type of data) are easier to interpret and troubleshoot. Thus, the goal of this project was to create a text-based, portable classifier to identify AF patients across different institutions.
Methods
Training, Internal Validation, and External Validation
The candidate population for this study included patients with at least one ICD code for AF between 1/1/2010 and 1/1/2017 at site #1 and site #2. This study included a training set (n=886), internal validation set (n=285) from site #1, and an external validation set (n=276) from site #2. The presence of a single ICD code does not necessarily mean that the patient has AF; these codes are often used in “rule-out” testing. For example, patients who have strokes often receive event monitors to determine if undiagnosed AF is present, as a cause of the stroke. A single ICD code for AF appears for these patients, even if they are subsequently ruled out for AF. To train the NLP algorithm, we manually reviewed charts for 886 patients from the candidate population at site #1.
Among the candidate population, 786 were randomly selected and reviewed in our prior work, in which we achieved a specificity of 63% in identifying patients with AF.8 In order to improve specificity, we added 100 patients with a low probability of AF to the training set only. The internal and external validation sets included a random sample of patients, without over sampling of low probability patients. This yielded a total of 886 patients for training the algorithm, with an AF prevalence of 72.2% (n=640). Clinical notes for the training set were extracted from the University of Utah Health (Site #1) Enterprise Data Warehouse, an enterprise level data repository optimized for data analysis and reporting of healthcare data. It contains data extracted from many of the institution’s disparate source systems, which includes patient, visit, clinical, operational, financial, and research data. This provides the opportunities for data mining, outcomes, and decision support research. The notes were aggregated by medical record number (MRN) to create one text file for each MRN. We extracted phrases that included target terms (e.g. “AF,” “afib,” or “atrial fib”) consistent with AF from the text, plus one phrase before and after the phrase with the target terms. These 3-phrase text spans found in the text served as the data for each MRN. The purpose of using 3-phrase spans (rather than all the text), was to identify relevant modifiers (e.g., “not”), limit noise from other parts of the clinical notes, improve portability, and reduce processing time.
In order to train and identify the best performing algorithm, we varied the following parameters and calculated the sensitivity, specificity, positive predictive value, and negative predictive value for each algorithm:
Stop words: Stop words are common terms that do not provide context for the classification (e.g., is, and, the). Pre-specified stop words from existing Python packages are less applicable to healthcare settings. Therefore, we created a custom stop list. We used our entire corpus from the candidate population (n=1.8 million notes) to create a list of 500 stop words based on the frequency of single word tokens. In other words, words that appeared most frequently in the corpus (such as “reports” in medical text) were considered stop words. We varied the custom stop word list (100, 250, 500), plus added terms specific to Site #1 (e.g. “Utah”), numbers, and the English stop words from the scikit-learn Python package.9
Vectorizer: We used term counts and term frequency-inverse document frequency (TF-IDF) to generate the feature set from the training set.10,11
Model: We used logistic regression, extra trees, and naive Bayes classifiers.
Vectorizer refers to the method used to turn the words into computable tokens. In the training set, the entire corpus of text of all patients generates a vocabulary, all the words (i.e. tokens or features) available for analysis. The count method sums the frequency of each word in the vocabulary in a specific note and uses the values in the feature vector for that note. The TF-IDF method applies a weight to each of the words. TF is the term frequency, or how frequently a particular word appears in a specific note, and IDF is the inverse document frequency, or the total number of documents divided by the number of documents with the particular word. Conceptually, this weight approximates the importance of the word in the specific note and in the corpus overall.
We then applied the trained algorithms to an internal validation set, which included 285 randomly selected patients from the candidate population. Based on clinician chart review and adjudication, the prevalence of AF in the internal validation set was 75.1%. If the performance dropped substantially between the training and internal validation sets, we re-visited the training to find ways to improve algorithm performance. This iterative process was repeated until we reached a plateau or the performance was considered acceptable by the research team. The model with the highest F-score in the validation set was considered the best performing model.
External Validation
An important aspect of this work was to demonstrate portability through external validation at a different institution. We used a cohort of patients seen at Northwestern Memorial Healthcare in Chicago, IL (Site #2) with at least one billing ICD code for AF in the same time frame as the training and internal validations sets, between 1/1/2010 and 1/1/2017. From this candidate population, we randomly selected 276 patients for review by a team of four clinicians. This external review team classified patients using guidelines developed at Site #1, but team members from Site #1 had no part in the review. Based on clinician adjudication, the prevalence of AF in the validation set was 86.2%. As with the Site #1, the notes were aggregated to create one text file for each MRN; 3-phrase text spans including AF target terms were extracted. The trained algorithms from Site #1 were applied to Site #2 data. No modifications were made to the algorithm using Site #2 data.
We compared algorithm performance between Site #1 and Site #2 by bootstrapping areas under the curves (AUC) for the receiver operating characteristic (ROC) curves and F-scores. For each site, we randomly selected 80% of the sample with replacement to create 100 samples. We ran the trained algorithm on each of these subsets to create 100 different AUCs and F-scores for the Site #1 and Site #2 validation sets. We compared the distribution of these values using a two-sided t-test and a p value <0.05 for significance. All analyses were performed using Python 3.6 and Stata Version 14. The machine learning and NLP tools were from the scikit-learn Python package.12 The data that support the findings of this study are available from the corresponding author upon reasonable request. Specifically, the code used to train and test the NLP algorithms will be available, but sensitive patient data will require appropriate data sharing agreements from qualified researchers trained in human subject confidentiality protocols.
For both sites, comorbid conditions were based on ICD billing codes present in the EMR for each patient. Patients were assigned a condition if a relevant code was present coincident with or before index AF diagnosis, looking back to 1/1/2010. Codes were aggregated into clinically meaningful groups using the Agency for Healthcare Research and Quality’s Clinical Classification software.13 Demographics and comorbid conditions were compared between AF and non-AF patients using chi-squared tests for categorical variables and t-test for continuous variables. These patient characteristics were used to describe the population, but were not used in the algorithms.
This study was reviewed and approved by the Institutional Review Board at the University of Utah, with waiver of informed consent.
Results
The validation set for Site #1 included 285 randomly selected patients, with mean age 67.7 years old (SD 15.0) and 42.8% female. The validation set for Site #2 included 276 randomly selected patients, with mean age 70.0 years old (SD 15.1) and 38.4% female. AF prevalence was 75.1% at Site #1 and 86.2% at Site #2 based on physician adjudication. At both sites, patients who were labelled as “AF present” by physician adjudication were older than patients labelled as “AF absent.” Cerebrovascular disease, or stroke, was more common among patients labelled as “AF absent” at Site #1, whereas comorbid conditions did not differ between “AF present” and “AF absent” patients at Site #2 (Table 1).
Table 1.
Characteristics of the internal validation (Site #1, n=285) and external validation (Site #2, n=276) populations, according to the presence or absence of atrial fibrillation
| Site #1 | Site #2 | |||||
|---|---|---|---|---|---|---|
| Characteristic* | AF Present (n=214) | AF Absent (n=71) | p-value | AF Present (n=238) | AF Absent (n=38) | p-value |
| Mean age (SD), years | 68.8 (14.1) | 64.4 (17.1) | 0.03 | 70.8 (13.9) | 65.1 (20.5) | 0.03 |
| Female | 91 (42.5%) | 31 (43.7%) | 0.87 | 91 (38.2%) | 15 (39.5%) | 0.88 |
| White race | 180 (84.1%) | 59 (83.1%) | 0.36 | 199 (83.6%) | 23 (60.5%) | 0.06 |
| Medicare insured | 139 (65.0%) | 38 (53.3%) | 0.26 | 121 (50.8%) | 14 (36.8%) | 0.27 |
| Comorbid conditions† | ||||||
| Acute myocardial infarction | 16 (7.5%) | 7 (9.9%) | 0.52 | 13 (5.5%) | 1 (2.6%) | 0.46 |
| Coronary artery disease | 75 (35.1%) | 24 (33.8%) | 0.85 | 66 (27.7%) | 8 (21.2%) | 0.39 |
| Congestive heart failure | 55 (25.7%) | 15 (21.1%) | 0.44 | 50 (21.0%) | 8 (21.1%) | 0.99 |
| Cerebrovascular disease | 35 (16.4%) | 33 (46.5%) | <0.01 | 30 (21.6%) | 6 (15.8%) | 0.59 |
| Diabetes | 76 (35.5%) | 25 (35.2%) | 0.96 | 56 (23.5%) | 12 (31.6%) | 0.29 |
| Chronic kidney disease | 31 (14.5%) | 14 (19.7%) | 0.30 | 24 (10.1%) | 5 (13.2%) | 0.57 |
| Hypertension | 139 (65.0%) | 50 (70.4%) | 0.40 | 123 (51.7%) | 16 (42.1%) | 0.27 |
n (%), unless otherwise specified
Comorbid conditions were identified from ICD billing codes present in the patients’ medical record
AF=atrial fibrillation; ICD=International Classification of Diseases
In total, we trained 54 different algorithms to identify AF patients from the clinical text extracted from the EMR. The performance results for all models can be found in the Online Supplement, Table S1. Based on the F-score, the best performing model was logistic regression, using 1000 features, 100 stop words, and TF-IDF to create the feature set. In the training set (Site #1), the test characteristics were: sensitivity 92.8%, specificity 93.9%, and F-score 95.1%.
After applying the best performing model to the validation sets, the test characteristics for Site #1 were sensitivity 92.5%, specificity 88.7%, and F-score 94.3%. The test characteristics at Site #2 were sensitivity 89.5%, specificity 71.1%, and F-score 92.2%. Table 2 shows the test characteristics for the top 10 performing algorithms, based on F-score.
Table 2.
Test characteristics for top 10 performing algorithms to identify AF patients, sorted based on F-score
| Parameters | Site #1 Validation | Site #2 Validation | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model Type | No. features | No. stop words | Vectorizer | Acc | PPV | NPV | Sens | Spec | F Score | Acc | PPV | NPV | Sens | Spec | F Score |
| Logistic | 1000 | 100 | TF-IDF | 0.916 | 0.961 | 0.797 | 0.925 | 0.887 | 0.943 | 0.870 | 0.951 | 0.519 | 0.895 | 0.711 | 0.922 |
| Logistic | 500 | 100 | TF-IDF | 0.902 | 0.951 | 0.772 | 0.916 | 0.859 | 0.933 | 0.862 | 0.939 | 0.500 | 0.899 | 0.632 | 0.918 |
| Logistic | 1500 | 100 | TF-IDF | 0.898 | 0.956 | 0.756 | 0.907 | 0.873 | 0.930 | 0.870 | 0.947 | 0.520 | 0.899 | 0.684 | 0.922 |
| Logistic | 1000 | 100 | Count | 0.877 | 0.950 | 0.709 | 0.883 | 0.859 | 0.915 | 0.841 | 0.937 | 0.444 | 0.874 | 0.632 | 0.904 |
| Logistic | 500 | 100 | Count | 0.877 | 0.954 | 0.705 | 0.879 | 0.873 | 0.915 | 0.855 | 0.950 | 0.482 | 0.878 | 0.711 | 0.913 |
| Logistic | 1500 | 100 | Count | 0.877 | 0.954 | 0.705 | 0.879 | 0.873 | 0.915 | 0.841 | 0.937 | 0.444 | 0.874 | 0.632 | 0.904 |
| Extra trees | 1500 | 100 | TF-IDF | 0.863 | 0.903 | 0.735 | 0.916 | 0.704 | 0.910 | 0.859 | 0.916 | 0.486 | 0.920 | 0.474 | 0.918 |
| Extra trees | 1000 | 100 | TF-IDF | 0.860 | 0.903 | 0.725 | 0.911 | 0.704 | 0.907 | 0.862 | 0.920 | 0.500 | 0.920 | 0.500 | 0.920 |
| Logistic | 1000 | 250 | TF-IDF | 0.863 | 0.931 | 0.695 | 0.883 | 0.803 | 0.906 | 0.830 | 0.928 | 0.415 | 0.870 | 0.579 | 0.898 |
| Extra trees | 500 | 100 | TF-IDF | 0.853 | 0.894 | 0.716 | 0.911 | 0.676 | 0.903 | 0.870 | 0.928 | 0.525 | 0.920 | 0.553 | 0.924 |
“Parameters” refers to the model specification. “Model” is which model was used, “No. features” is the number of features, “No. stop words” number of stop words, “Vectorizer” is the method used to create the feature set. The table is sorted according to the F-score in Site #1, the internal validation data set.
Acc=accuracy; PPV=positive predictive value; NPV=negative predictive value; Sens=sensitivity; Spec=specificity; TF-IDF=term frequency-inverse document frequency
The ROC AUC was 0.93 for the training set, 0.91 for the Site #1 validation set, and 0.80 for Site #2 validation set (Figure). Using bootstrapping with 100 randomly selected subsets in the validation sets, the mean AUC was less for Site #2 compared to Site #1 (0.91 [SD 0.02] versus 0.80 [SD 0.03]; p<0.001). Similarly, using bootstrapping with 100 randomly selected subsets in the validation sets, the mean F-score was 94.2% (SD 1.1%) for Site #1 compared to 92.5% (SD 1.1%) for Site #2 (p<0.001).
Figure 1. Receiver operator characteristic (ROC) curves for Training, Site #1, and Site #2 data sets.
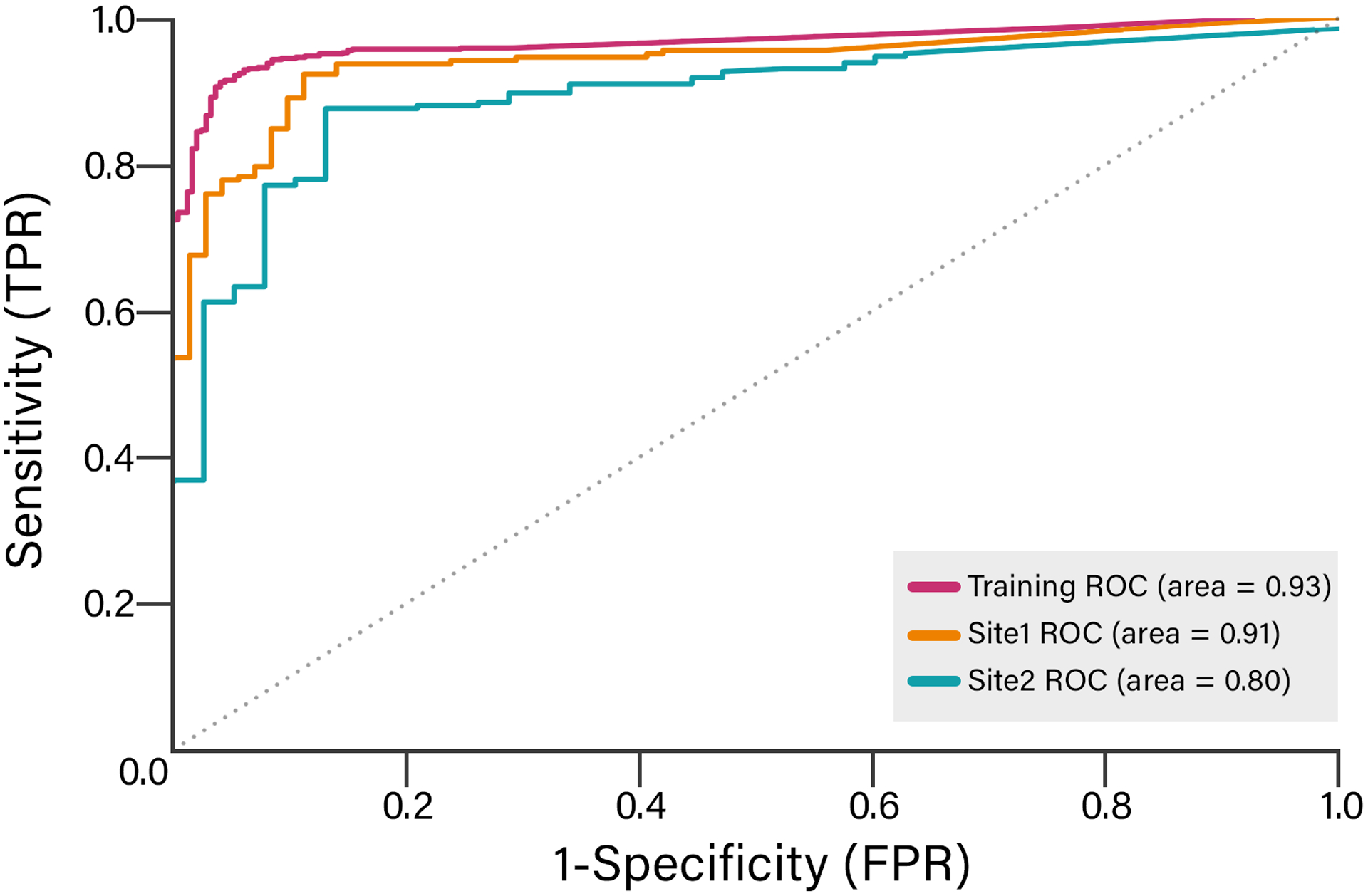
This plot compares the ROC curves for a natural language processing algorithm to identify AF patients at different institutions. The algorithm used a logistic regression model, 1000 features, 100 stops words, and the TF-IDF method to create the feature set.
ROC=receiver operator characteristic; TF-IDF=term frequency-inverse document frequency
Discussion
In this project, we created a sensitive and specific algorithm to identify AF patients using data from the EMR. Our approach differs from prior work in that we relied only on clinical text, which includes the narrative written by clinicians. In our development site, Site #1, the algorithm achieved 92.5% sensitivity and 88.7% specificity, an improvement from our prior work. In the external validation site, Site #2, algorithm performance was somewhat lower, with sensitivity 89.5% and specificity 71.1%.
Other researchers have developed EMR-based methods to identify AF patients, mostly using structured data in the form of billing codes.1 Our work differs because we focused entirely on unstructured data in the form of clinical text. Text analysis is appealing because the critical and detailed information is often in the clinical narrative, and this approach is more aligned with the clinician workflow. While the current system seeks to generate structured data by forcing clinicians to click checkboxes and toggle through dialogue boxes, many clinicians prefer to dictate a rich, clinical narrative. We envision a future system wherein the EMR will adapt to the clinician workflow, not the other way around: the clinician narrates the history and algorithms like the one described here will extract clinically meaningful information.14–16
Prior work in AF cohort identification has generally relied on ICD billing codes and reported positive predictive value (PPV), rather than sensitivity and specificity. The unknown impact of false negatives (1-sensitivity; or the proportion of AF patients who would be missed by an algorithm), therefore limits many prior studies. Khurshid et al., for example, demonstrated the performance of seven different algorithms to detect AF using structured data from the EMR.17 Along with reporting only PPV, the algorithm included outcome variables (order for an oral anticoagulant), which could bias the results when applied to subsequent studies or quality improvement efforts.
Our work is an example of digital, or electronic, phenotyping, a method which has specific applications in genotype-phenotype association studies. The Electronic Medical Records and Genomics (eMERGE) Network, for example, has made major progress in electronic phenotyping in order to enable “high-throughput genomics research.”18,19 The network includes an AF definition, but the definition is highly dependent on electrocardiograms that demonstrate AF.20 As we demonstrated in our prior work, electrocardiograms with AF were present in only 37% of the AF patients, which may enhance true positives but limits overall detection rate.21 In addition, the eMERGE definitions are defined to explicitly identify patients without AF, which enables case-control study designs. This approach may be suitable for genomic studies, but is not ideal for cohort studies, quality improvement, or real world evidence generation.
Compared to our prior work using rules-based NLP,8 this study used machine learning approaches to develop the algorithm. We were able to substantially improve the specificity for our development site, Site #1; the specificity was somewhat lower when the algorithm was applied to Site #2, but still improved compared with prior efforts. This drop in performance between institutions is expected and has implications for implementation. When models are trained at one institution, “overfitting” occurs. In other words, the model has learned the patterns and habits of one healthcare system, which may differ from other systems. If NLP were used to, for example, identify patients for a clinical trial, the models have to be trained with a combined dataset or individually at each site. Alternatively, one could use NLP for screening, such that the number of charts reviewed for the classification could be decreased, while still broadening the net for clinical trial inclusion. We observed that the prevalence of AF in the candidate population at Site #2 was higher than Site #1, and that cerebrovascular disease was more common among patients without AF at Site #1. In other words, the AF-specific coding practices differ between institutions. Upon review, this pattern appears to be due to the practice of ordering event monitors at Site #1 to evaluate for AF among patients who experience a stroke. These patients appear in the Site #1 candidate population, but subsequently are “ruled out” for AF. One possibility is that similar patients at Site #2 do not receive an AF billing code until they “rule in” for AF. The feature vector, developed at Site #1, includes the term “event,” as in “event monitor,” which could result in portability issues at Site #2. One solution is to train the model on data from multiple sites, which will probably be needed for future implementation.
Moving forward, one could consider combining structured and unstructured data in a more complex classification model. In combination with larger datasets from more institutions, this approach could raise the option to create AF sub-classifications (e.g. valvular or perioperative AF). The trade-off is that broader data sets, including different types of data, require harmonization across institutions to ensure portability, an added layer of work. An NLP limited approach could theoretically facilitate portability by requiring only clinical notes to create and run the algorithm. Furthermore, neural network approaches may improve the performance of future NLP models, with some tradeoffs. In our prior work to identify bleeding, for example, we found that a neural network approach did not perform as well as other models.22 In our current approach, we were able to address a very specific task with limited computational demand, simple implementation at an external site, and perhaps in a way that is more understandable by clinicians. In addition, state-of-the-art NLP approaches like Google’s BERT (Bidirectional Encoder Representations from Transformers) have not yet been trained on clinical language, which differs markedly from other types of text.23
Limitations
Our study used a binary classification for AF -- present or not present. In clinical reality, the AF phenotype is more complex. For example, a patient who experiences AF in the setting of hyperthyroidism differs from an older patient in permanent AF. Multiclass algorithms require large, labelled data sets, which is a labor- and time-intensive roadblock that requires domain experts.24 In addition, our candidate population included patients with at least one code for AF, so we may have missed some patients who have AF in the absence of a billing code. Internal and external validation at different institutions is a strength, but both institutions are academic medical centers, which may limit more general portability. EMR-based cohort definitions could be biased by the patient’s contact with the healthcare system; the more contact the patient has, the more data are available for analysis, and the more likely the patient will be included in the cohort. Models that include a broad set of features (e.g. all prior diagnoses codes) will be more prone to this bias. In our case, we limited the analyses to a small feature set, specifically the target terms and surrounding text span. This approach probably does not eliminate the bias related to data density, but may limit its effect. For example, one could imagine a scenario in which an AF patient’s first encounter in the healthcare system is for a catastrophic intracranial hemorrhage, with death in the first 24 hours. This patient would have very little data in the EMR, but notes would say “Patient has a known history of AF and is treated with apixaban.” Our algorithm would include that patient despite the sparse data in the EMR.
Conclusions
The EMR is an untapped resource for clinical information, but much of the data is buried in the clinical narrative. Disease specific patient cohort identification is one important aspect of digital phenotyping using the EMR. In this project, we developed a high performing NLP algorithm using the clinical narrative alone, with >90% F-score at two separate sites, to identify AF patients. From a clinician’s standpoint, simply describing the patients’ condition using narrative language is preferable to clicking checkboxes and assigning billing codes. Precise NLP algorithms, as we have demonstrated here, could allow physicians to move toward dictation and away from “death by a thousand clicks,”25 while also improving cohort identification for clinical research and quality improvement efforts.
Supplementary Material
What is known
Accurate cohort identification is critically important, as we move into an era of real world evidence using electronic medical records (EMR) for data.
Prior methods rely on structured data (e.g. International Classification of Diseases billing codes or oral anticoagulation orders) to identify atrial fibrillation (AF) patients from large databases, with limited performance.
What the study adds
In this project, we demonstrate that natural language processing (NLP) of clinical text can identify AF patients with 92.5% sensitivity and 88.7% specificity; the algorithm performance was 89.5% sensitivity and 71.1% specificity in an external health system’s data.
These results show promise for NLP as a means to extract clinically relevant information from the EMR.
We envision a future system wherein the EMR will adapt to the clinician workflow, not the other way around: the clinician narrates the history and algorithms like the one described here will extract clinically meaningful information for downstream use.
Sources of funding:
Research reported in this publication was supported, in part, by the National Institutes of Health’s National Center for Advancing Translational Sciences, Grant Number UL1TR001422. This work was also supported by grants from the National Heart Lung and Blood Institute, K08HL136850 (to RUS).
Disclosures/conflicts of interest:
BAS is supported by a grant from the National Heart Lung and Blood Institute (K23HL143156). RUS is supported by a donation from Women As One. BAS receives research support from Boston Scientific and Janssen; consulting to Janssen, Bayer, and Merit Medical; speaking for North American Center for Continuing Medical Education (funded by Sanofi).
References
- 1.Jensen PN, Johnson K, Floyd J, Heckbert SR, Carnahan R, Dublin S. A systematic review of validated methods for identifying atrial fibrillation using administrative data. Pharmacoepidemiol Drug Saf. 2012;21 Suppl 1:141–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherman RE, Anderson SA, Dal Pan GJ, Gray GW, Gross T, Hunter NL, LaVange L, Marinac-Dabic D, Marks PW, Robb MA, Shuren J, Temple R, Woodcock J, Yue LQ, Califf RM. Real-World Evidence — What Is It and What Can It Tell Us? N Engl J Med. 2016;375:2293–2297. [DOI] [PubMed] [Google Scholar]
- 3.Quality ID #326 (NQF 1525): Atrial Fibrillation and Atrial Flutter: Chronic Anticoagulation Therapy – National Quality Strategy Domain: Effective Clinical Care – Meaningful Measure Area: Management of Chronic Conditions. [Last accessed 05/05/020];Available from: https://qpp.cms.gov/docs/QPP_quality_measure_specifications/Claims-Registry-Measures/2019_Measure_326_MedicarePartBClaims.pdf
- 4.Nordstrom BL, Whyte JL, Stolar M, Mercaldi C, Kallich JD. Identification of metastatic cancer in claims data. Pharmacoepidemiol Drug Saf. 2012;21 Suppl 2:21–28. [DOI] [PubMed] [Google Scholar]
- 5.Bernstam EV, Herskovic JR, Reeder P, Meric-Bernstam F. Oncology research using electronic medical record data. J Clin Orthod. 2010;28:e16501–e16501. [Google Scholar]
- 6.Hersh WR, Weiner MG, Embi PJ, Logan JR, Payne PRO, Bernstam EV, Lehmann HP, Hripcsak G, Hartzog TH, Cimino JJ, Saltz JH. Caveats for the use of operational electronic health record data in comparative effectiveness research. Med Care. 2013;51:S30–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy C, Murphy S, Cohen JA, Rehman S, Jones-O’Connor M, Olshan DS, Singh A, Vaduganathan M, Januzzi JL Jr, Wasfy JH. Misclassification of Myocardial Injury as Myocardial Infarction: Implications for Assessing Outcomes in Value-Based Programs. JAMA Cardiol. 2019;4:460–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shah RU, Mukherjee R, Zhang Y, Jones AE, Springer J, Hackett I, Steinberg BA, Lloyd-Jones DM, Chapman WW. Impact of Different Electronic Cohort Definitions to Identify Patients With Atrial Fibrillation From the Electronic Medical Record. J Am Heart Assoc. 2020;9:e014527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.scikit-learn. Github; [Last accessed 05/05/020]. Available from: https://github.com/scikit-learn/scikit-learn
- 10. [May 5, 2020];sklearn.feature_extraction.text.CountVectorizer — scikit-learn 0.21.3 documentation. Available from: https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html. Accessed.
- 11. [May 5, 2020];sklearn.feature_extraction.text.TfidfVectorizer — scikit-learn 0.21.3 documentation. Available from: https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.TfidfVectorizer.html. Accessed.
- 12. [May 5, 2020];scikit-learn: machine learning in Python — scikit-learn 0.21.3 documentation. Available from: https://scikit-learn.org/stable/. Accessed.
- 13.Agency for Healthcare Research and Quality. Clinical Classifications Software (CCS) for ICD-9-CM Fact Sheet. Available from: https://www.hcup-us.ahrq.gov/toolssoftware/ccs/ccsfactsheet.jsp. Accessed May 5, 2020
- 14.Friedberg MW, Chen PG, Van Busum KR, Pham C, Aunon FM. Factors Affecting Physician Professional Satisfaction and Their Implications for Patient Care, Health Systems, and Health Policy. Rand Corporation; 2013. [PMC free article] [PubMed] [Google Scholar]
- 15.Collier R Electronic health records contributing to physician burnout. CMAJ. 2017;189:E1405–E1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collier R Rethinking EHR interfaces to reduce click fatigue and physician burnout. CMAJ. 2018;190:E994–E995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurshid S, Keaney J, Ellinor PT, Lubitz SA. A Simple and Portable Algorithm for Identifying Atrial Fibrillation in the Electronic Medical Record. Am J Cardiol. 2016;117:221–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Electronic Medical Records and Genomics (eMERGE) Network. Genome.gov Available from: https://www.genome.gov/Funded-Programs-Projects/Electronic-Medical-Records-and-Genomics-Network-eMERGE. Accessed May 5, 2020
- 19.Pacheco JA, Rasmussen LV, Kiefer RC, Campion TR, Speltz P, Carroll RJ, Stallings SC, Mo H, Ahuja M, Jiang G, LaRose ER, Peissig PL, Shang N, Benoit B, Gainer VS, Borthwick K, Jackson KL, Sharma A, Wu AY, Kho AN, Roden DM, Pathak J, Denny JC, Thompson WK. A case study evaluating the portability of an executable computable phenotype algorithm across multiple institutions and electronic health record environments. J Am Med Inform Assoc. 2018;25:1540–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atrial Fibrillation - Demonstration Project | PheKB [Internet]. [cited 2019 Sep 23];Available from: https://phekb.org/phenotype/atrial-fibrillation-demonstration-project
- 21.Shah RU, Mukherjee R, Zhang Y, Jones AE, Springer J, Hackett I, Steinberg BA, Lloyd-Jones DM, Chapman WW. Impact of Different Electronic Cohort Definitions to Identify Patients With Atrial Fibrillation From the Electronic Medical Record. J Am Heart Assoc. 2020;9:e014527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taggart M, Chapman WW, Steinberg BA, Ruckel S, Pregenzer-Wenzler A, Du Y, Ferraro J, Bucher BT, Lloyd-Jones DM, Rondina MT, Shah RU. Comparison of 2 Natural Language Processing Methods for Identification of Bleeding Among Critically Ill Patients. JAMA Netw Open. 2018;1:e183451–e183451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devlin J, Chang M-W, Lee K, Toutanova K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv.org 2018. Available from: http://arxiv.org/abs/1810.04805 [Google Scholar]
- 24.The real-world potential and limitations of artificial intelligence. Available from: https://www.mckinsey.com/featured-insights/artificial-intelligence/the-real-world-potential-and-limitations-of-artificial-intelligence. Accessed October 15, 2019.
- 25.Fry E, Schulte F. Death by a Thousand Clicks: Where Electronic Health Records Went Wrong. 2019. Available from: https://fortune.com/longform/medical-records/. Accessed May 5, 2020
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


