Abstract
Sepsis is a serious clinical condition characterized by systemic inflammation. The long noncoding RNA (lncRNA) highly upregulated in liver cancer (HULC) was validated to partake in the development of sepsis. The present study aimed to investigate the potential mechanism of HULC in lipopolysaccharide (LPS)-induced sepsis. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blot analysis was employed to examine the expression of HULC, microRNA (miR)-128-3p, Rac family small GTPase 1 (RAC1) and pro-inflammatory factors [IL-6, TNF-α, intercellular adhesion molecule (ICAM1) and vascular cell adhesion molecule (VCAM1)] in the serum of patients with sepsis or LPS-induced human dermal microvascular endothelial cells (HMEC-1). Flow cytometry and western blot assays were performed to detect cell apoptosis. The targeted relationship among HULC, miR-128-3p and RAC1 was confirmed by a dual-luciferase reporter assay, RNA binding protein immunoprecipitation (RIP) assay and RNA pull-down assay. HULC and RAC1 were found to be upregulated, and miR-128-3p was downregulated in the serum of patients with sepsis and LPS-stimulated HMEC-1 cells. LPS promoted apoptosis and inflammation, which were decreased by silencing of HULC. HULC targeted miR-128-3p and negatively regulated its expression. HULC knockdown protected HMEC-1 cells from LPS-induced injury by upregulating miR-128-3p. RAC1 was a target of miR-128-3p, and gain of RAC1 also relieved the silencing of HULC-mediated suppressive effects on apoptosis and inflammation in LPS-stimulated HMEC-1 cells. In conclusion, HULC knockdown partially reversed LPS-induced sepsis via the regulation of miR-128-3p/RAC1 axis.
Keywords: lipopolysaccharide, sepsis, highly upregulated in liver cancer, microRNA-128-3p, Rac family small GTPase 1, apoptosis, inflammation
Introduction
Sepsis often occurs following infection or injury, and is one of the primary causes of morbidity and mortality worldwide (1). It was reported that sepsis was responsible for 2.8 million deaths in high-income countries every year (2). The pathophysiology of sepsis is complex, and there are a number of risk factors that can contribute to the development of sepsis (3). Therefore, it is necessary to understand the mechanistic pathways underlying sepsis in order to develop an effective treatment.
Long non-coding RNAs (lncRNAs) are a novel family of regulatory RNA molecules that are >200 nucleotides and have numerous diverse functions (4). lncRNAs have roles in the regulation of inflammatory responses in sepsis (5). For example, lncRNA Transcript Predicting Survival in AKI was demonstrated to facilitate HK-2 cell apoptosis and inflammatory responses in sepsis-induced kidney injury (6). lncRNA hox transcript antisense RNA could repress proliferation, and upregulate apoptosis and inflammatory responses in sepsis in vitro (7). Wang et al (8) reported that lncRNA nuclear paraspeckle assembly transcript 1 (NEAT1) knockdown could ameliorate sepsis-induced myocardial damage in mice, including the decrease of edema in myocardial tissues in mice, as well as the inhibition of apoptosis and inflammation. Previous literature has suggested that lncRNA highly upregulated in liver cancer (HULC) was involved in the decrease of pre-inflammatory mediators in lipopolysaccharide (LPS)-induced sepsis in vitro (9). Nevertheless, the mechanism of action of HULC in LPS-induced sepsis remains to be elucidated.
MicroRNAs (miRNAs/miRs) are a category of important non-coding RNAs associated with human diseases that participate in various biological processes (10). At present, an increasing number of researchers are focusing on miRNAs relevant to sepsis. For instance, miR-25 was demonstrated to attenuate apoptosis and increase expression of certain proinflammatory cytokines in cardiomyocytes treated with LPS (11). Wang et al (12) demonstrated that miR-21-3p affected sepsis-related cardiac dysfunction by targeting SH3 domain-containing protein 2 (12). It was revealed that the expression of miR-128-3p was downregulated in the podocytes of a patient with sepsis (13), which prompted us to investigate the effect of miR-128-3p on the progression of LPS-induced sepsis in the present study.
Rac family small GTPase 1 (RAC1) serves as a key signal transducer that has been demonstrated to control cellular inflammatory responses (14). It was reported that RAC1 could modulate the formation of platelet-derived microparticles and generation of thrombin in sepsis (15). In the present study, the role of RAC1 in LPS-induced sepsis was examined.
Materials and methods
Clinical sample collection
A total of 110 patients with sepsis (male/female: 72/38; Age range, 18–38 years; mean age, 56.84±10.17 years) and 100 healthy controls (male/female: 64/36; Age range, 24–81 years; mean age 59.33±11.24 years) were recruited at the Renmin Hospital of Wuhan University (Hubei, China) between November 2016 and January 2019. The patients with sepsis, admitted to intensive care units (ICUs) were diagnosed as septic based on the ‘Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis’ (16). A total of 110 patients with sepsis, including 36 patients with sepsis, 47 patients with severe sepsis and 27 patients with septic shock (17), were involved in the study. The exclusion criteria were as follows: Patients with malignancies, those receiving immunosuppressant treatment, pregnant or lactating women and patients with human immunodeficiency virus. All participants submitted written informed consent. The current study was approved by the Ethics Committee of the Renmin Hospital of Wuhan University.
Samples of blood were taken from patients with sepsis within 24 h of admission to the ICU. The blood samples of 100 healthy participants were acquired during their physical examination. Serum was obtained after centrifugation at 400 × g for 15 min at 4°C and stored at −80°C.
Cell culture and LPS treatment
Human dermal microvascular endothelial cells (HMEC-1; CRL-3243) were commercially procured from American Type Culture Collection and grown in MCDB 131 Medium, no glutamine (Gibco; Thermo Fisher Scientific, Inc.) containing 10 ng/ml epidermal growth factor (EGF; Gibco; Thermo Fisher Scientific, Inc.), 1 µg/ml hydrocortisone (Sigma-Aldrich; Merck KGaA), 10 mM glutamine (Gibco; Thermo Fisher Scientific, Inc.) and 10% (V/V) fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C.
To mimic sepsis in vitro, HMEC-1 cells (5×104/100 µl) maintained in 6-well plates were treated with 1 µg/ml LPS (Beijing Solarbio Science & Technology Co., Ltd.) for 24 h, which could trigger strong immune-inflammatory responses (18,19), whereas cells treated with dimethyl sulfoxide (DMSO; Beijing Solarbio Science & Technology Co., Ltd.) served as controls.
Cell transfection
Small interfering RNA (siRNA) directed against HULC (si-HULC, 5′-CCUCCAGAACUGUGAUCCA-3′) and the negative control (si-NC, 5′-GGACUCUCGGAUUGUAAGAUU-3′) were acquired from Shanghai GeneChem Co., Ltd. To construct overexpression plasmids, the sequence of HULC or RAC1 was inserted into a pcDNA3.1 vector (Hanbio Biotechnology Co., Ltd.), generating pcDNA3.1-HULC (HULC) or pcDNA3.1-RAC1 (RAC1), with pcDNA3.1 (vector) as the control. In addition, miR-128-3p mimic (miR-128-3p, 5′-AAAGAGACCGGUUCACUGUGA-3′), miR-128-3p inhibitor (anti-miR-128-3p, 5′-UUUCUCUGGCCAAGUGACACU-3′) and their corresponding negative control (miR-NC, 5′-ACGUGACACGUUCGGAGAATT-3′ and anti-miR-NC, 5′-CUAACGCAUGCACAGUCGUACG-3′) were synthesized by GenePharma Co. Ltd. (Shanghai, China). When cell confluence reached 50%, LPS-treated HMEC-1 cells were transfected with 2 µg plasmids or 40 nM oligonucleotides using Lipofectamine® 3000 (Beijing Solarbio Science & Technology Co., Ltd.). The knockdown efficiency was measured in cells transfected without LPS treatment. After 48 h, cells were subjected to subsequent investigation.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Extraction of total serum RNA and RNA from HMEC-1 cells was performed with TRIzol® LS Reagent (Ambion; Thermo Fisher Scientific, Inc.). After determination of RNA concentration and purity using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.), 1 µg RNA was utilized to synthesize cDNA (42°C for 60 min and 80°C for 5 min) with the BeyoRT™ First Strand cDNA Synthesis kit (Beyotime Institute of Biotechnology) for HULC and RAC1, and miScript Reverse Transcription kit (Qiagen, Inc.) for miR-128-3p. Then, qPCR was performed with a SYBR-Green mix (Takara Bio, Inc.) with following thermocycling conditions: Initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 20 sec, annealing at 60°C for 30 sec and extension at 72°C for 20 sec. The relative expression of miR-128-3p, HULC, RAC1, IL-6, TNF-α, intercellular adhesion molecule (ICAM1) and vascular cell adhesion molecule (VCAM1) was evaluated by the 2−ΔΔCq method (20), miR-128-3p expression was normalized to U6, whereas HULC, RAC1, IL-6, TNF-α, ICAM1 and VCAM1 expression was normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Primers subjected for amplification were: miR-128-3p, forward (F): 5′-GGTCACAGTGAACCGGTC-3′ and reverse (R): 5′-GTGCAGGGTCCGAGGT-3′; U6, F: 5′-GCAGGAGGTCTTCACAGAGT-3′ and R: 5′-TCTAGAGGAGAAGCTGGGGT-3′; HULC, F: 5′-TCATGATGGAATTGGAGCCTT-3′ and R: 5′-CTCTTCCTGGCTTGCAGATTG-3′; RAC1, F: 5′-AAGAGAAAATGCCTGCTGTTGTAA-3′ and R: 5′-GCGTACAAAGGTTCCAAGGG-3′; IL-6, F: 5′-GGTACATCCTCGACGGCATCT-3′ and R: 5′-GTGCCTCTTTGCTGCTTTCAC-3′; TNF-α, F: 5′-CGAGTGACAAGCCTGTAGCC-3′ and R: 5′-GTTGACCTTGGTCTGGTAGG-3′; ICAM1, F: 5′-GGCCTCAGTCAGTGTGA-3′ and R: 5′-AACCCCATTCAGCGTCA-3′; VCAM1, F: 5′-CCGGATTGCTGCTCAGATTGGA-3′ and R: 5′-AGCGTGGAATTGGTCCCCTCA-3′; GAPDH, F: 5′-GCCAAAAGGGTCATCATCTC-3′ and R: 5′-GGCCATCCACAGTCTTCT-3′.
Cell apoptosis assay
Cell apoptosis was determined using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (BioVision, Inc.). In brief, HMEC-1 cells were collected and suspended in 200 µl binding buffer, and then double-stained with 5 µl Annexin V-FITC and 10 µl PI at room temperature for 20 min in the dark. Stained apoptotic cells (Annexin V+ and PI−) were identified by a flow cytometer (Accuri C6; BD Biosciences) and data were analyzed utilizing Cell Quest 6.0 software (BD Biosciences).
Western blot analysis
Whole proteins were extracted from HMEC-1 cells with a protein extraction kit (Beijing Solarbio Science & Technology Co., Ltd.). After quantification with a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology), 30 µg protein samples were separated via SDS-PAGE on a 10% gel, and then subsequently transferred onto a polyvinylidene fluoride membrane (Thermo Fisher Scientific, Inc.) using the wet electrophoretic transfer method for 2 h. The membranes were blocked in 5% skimmed milk at room temperature for 2 h, immersed in diluted primary antibodies at 4°C overnight, and then incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Goat Anti-Rabbit IgG H&L; cat. no. ab150077; 1:3,000; Abcam) at room temperature for 2 h. The primary antibodies were purchased from Abcam, including anti-cleaved-caspase-3 (anti-cleaved-cas3; cat. no. ab32042; 1:1,000), anti-cleaved-cas9 (cat. no. ab2324; 1:1500), anti-IL-6 (cat. no. ab233706; 1:1,500), anti-TNF-α (cat. no. ab183218; 1:1,000), anti-ICAM1 (cat. no. ab109361; 1:1,500), anti-VCAM1 (cat. no. ab134047; 1:1,500), anti-RAC1 (cat. no. ab155938; 1:1,000) and anti-β-actin (cat. no. ab115777; 1:2,000). An enhanced chemiluminescence kit (Amersham; Cytiva) was used to visualize the protein bands. The intensity of protein bands was determined using the Quantity One software (4.5.0 basic; Bio-Rad Laboratories, Inc.).
Dual-luciferase reporter assay
The miRcode (http://www.mircode.org/) was used to search which miRNAs directly interacted with HULC. The possible target genes of miR-128-3p were also predicted by the starBase database (http://starbase.sysu.edu.cn/agoClipRNA.php?source=circRNA). A dual-luciferase reporter assay was performed to confirm the interaction between miR-128-3p and HULC or RAC1. The segmental sequence of HULC or 3′untranslated region (3′UTR) of RAC1 at the predicted binding sites were amplified, followed by insertion into a pGL3 basic vector (Promega Corporation) to construct HULC wild-type (WT) or RAC1 WT. The binding sites were mutated by Q5 Site Directed Mutagenesis kit (New England Biolabs, Inc.), and HULC mutant (MUT) and RAC1 MUT were constructed in a similar manner. 1×105 HMEC-1 cells were co-transfected with 2 µg HULC WT, HULC MUT, RAC1 WT or RAC1 MUT and 40 nM miR-128-3p or miR-NC with Lipofectamine 3000 (Beijing Solarbio Science & Technology Co., Ltd.) at 37°C. After 48 h, the luciferase activity was measured using the Dual-Lucy Assay kit (Beijing Solarbio Science & Technology Co., Ltd.). The relative luciferase activity indicated the ratio of firefly luciferase activity to Renilla luciferase activity.
RNA binding protein immunoprecipitation (RIP)
RIP assays were performed using an EZ-Magna RIP kit (EMD Millipore), following the manufacturer's instructions to further confirm the target relationship among HULC, miR-128-3p and RAC1. A total of 1×107 HMEC-1 cells were lysed in RIP lysis buffer, then the cell extract was incubated with magnetic beads and an antibody against argonaute 2 (Ago2; cat. no. ab32381; 1:50; Abcam) or immunoglobin G (IgG; cat. no. ab109761; 1:50; Abcam) at 4°C overnight. After protein digestion, immunoprecipitated RNA was subjected to RNA isolation and RT-qPCR for detection of HULC, miR-128-3p and RAC1 expression.
RNA pull-down assays
For the RNA pull-down assay, the Magnetic RNA-Protein Pull-Down kit (Thermo Fisher Scientific, Inc.) was used. Biotin-labeled miR-128-3p (Bio-miR-128-3p) and its corresponding negative control (Bio-miR-NC) were constructed by Guangzhou RiboBio Co., Ltd. Then, cells were lysed and incubated with beads binding with Bio-miR-128-3p or Bio-miR-NC. Finally, beads were washed with wash buffer and subjected for examination of HULC expression via RT-qPCR.
Statistical analysis
Statistical analyses of data derived from 3 parallel repeat experiments were performed using SPSS 21.0 software (IBM Corp.) and GraphPad Prism 7 (GraphPad Software Inc.). Data are presented as the mean ± standard deviation. Comparisons were conducted with a Student's t test (for 2 groups) or one-way ANOVA followed by a Tukey's post hoc test (for ≥3 groups). Pearson correlation analysis was performed to identify the correlation between the expression of miR-128-3p and HULC or RAC1 in the serum of patients with sepsis. P<0.05 was considered to indicate a statistically significant difference.
Results
Dysregulation of HULC and miR-128-3p in serum of patients with sepsis
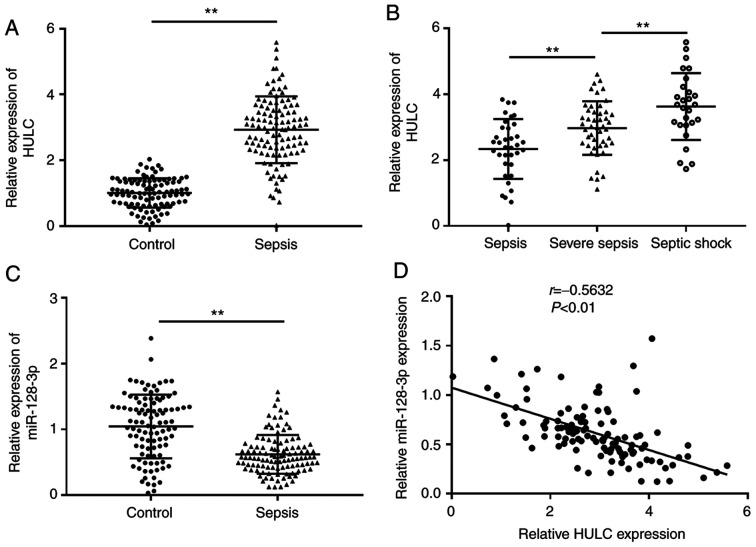
Initially, the expression of HULC and miR-128-3p was evaluated in the serum of patients with sepsis and healthy participants (control) via RT-qPCR. As depicted in Fig. 1A, the expression of HULC in the serum of patients with sepsis was significantly higher than that in the control. Among the 110 patients with sepsis, the highest level of HULC was discovered in patients with septic shock (n=27), the second-highest level of HULC expression was observed in patients with severe sepsis (n=47) compared with patients with sepsis (n=36) (Fig. 1B). RT-qPCR analysis suggested that miR-128-3p expression was significantly downregulated in the serum of patients with sepsis compared with those in the control group (Fig. 1C). In addition, there was an inverse correlation between expression levels of HULC and miR-128-3p in the serum of patients with sepsis (Fig. 1D). Thus, HULC and miR-128-3p may be related to the development of sepsis.
Figure 1.
Dysregulation of HULC and miR-128-3p in serum of patients with sepsis. (A and C) Expression of (A) HULC and (C) miR-128-3p in the serum of 110 patients with sepsis and 100 healthy participants. (B) HULC expression level in the serum of patients with sepsis (n=36), patients with severe sepsis (n=47) and patients with septic shock (n=27). (D) Pearson correlation analysis for HULC and miR-128-3p expression levels in the serum of 110 patients with sepsis (r=−0.5632, P<0.01). **P<0.01. HULC, highly upregulated in liver cancer; miR, microRNA.
Silencing of HULC partially reverses LPS-induced apoptosis and inflammation in a cell model of sepsis
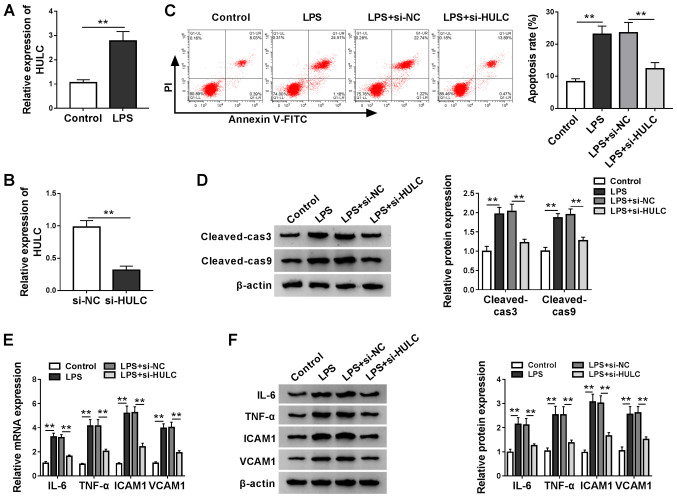
To construct a cell model of sepsis, HMEC-1 cells were treated with LPS or DMSO as a carrier control. Utilizing RT-qPCR analysis, it was found that LPS treatment significantly elevated HULC expression when compared with the control (Fig. 2A). In addition, knockdown efficiency of si-HULC transfection was determined by RT-qPCR (Fig. 2B). Then, functional analyses for the role of HULC in LPS-induced HMEC-1 cells were performed. Flow cytometry indicated that HULC knockdown decreased the LPS-induced upregulation of apoptosis (Fig. 2C). Subsequent western blotting and semi-quantitative analysis showed similar results. In Fig. 2D, protein expression levels of cleaved-cas3 and cleaved-cas9 were significantly upregulated by LPS, which was reversed in the LPS + si-HULC group. RT-qPCR and western blot assays were employed to determine the expression levels of IL-6, TNF-α, ICAM1 and VCAM1, which indicated that LPS stimulated expression of the four pro-inflammatory factors, and HULC knockdown largely attenuated the aforementioned promotion (Fig. 2E and F). Collectively, silencing of HULC could partially reverse LPS-induced apoptosis and inflammation in HMEC-1 cells.
Figure 2.
Silencing of HULC partially reverses LPS-induced apoptosis and inflammation in a cell model of sepsis. (A) HULC expression levels in HMEC-1 cells treated with LPS or DMSO (control). (B) HULC expression in HMEC-1 cells transfected with si-HULC or si-NC. (C-F) HMEC-1 cells treated with LPS or DMSO (control) were transfected with si-HULC or si-NC. (C) Apoptotic rate of transfected HMEC-1 cells. (D) Protein expression levels of cleaved-cas3 and cleaved-cas9 in transfected HMEC-1 cells. The (E) mRNA and (F) protein expression levels of IL-6, TNF-α, ICAM1 and VCAM1 in transfected HMEC-1 cells. **P<0.01. HULC, highly upregulated in liver cancer; ICAM1, intercellular adhesion molecule; VCAM1, vascular cell adhesion molecule; siRNA, small interfering RNA; NC, negative control; LPS, lipopolysaccharide; HMEC-1, human dermal microvascular endothelial cells; DMSO, dimethyl sulfoxide; cas, caspase.
HULC was a sponge of miR-128-3p
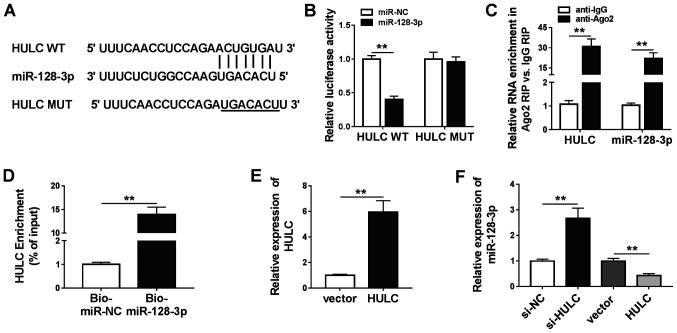
Analysis utilizing miRcode revealed that miR-128-3p, miR-9, miR-150, miR-203, miR-27a-3p and miR-218-5p were predicted to have possible binding positions for HULC. miR-128-3p (Fig. 3A) was selected for subsequent investigations as it exhibited the most significant downregulation in LPS-stimulated HMEC-1 cells transfected with HULC in a preliminary study among the six miRNAs (data not shown). To validate this prediction, a dual-luciferase reporter assay was performed. As shown in Fig. 3B, co-transfection with miR-128-3p significantly inhibited the luciferase activity of HULC WT in HMEC-1 cells compared with the miR-NC group. There was no significant difference in luciferase activity in HULC MUT. Besides, the RIP assay revealed that both HULC and miR-128-3p were abundant in Ago2 RIP of HMEC-1 cells compared with IgG RIP (Fig. 3C). A large amount of HULC was found to be pulled down by Bio-miR-128-3p rather than Bio-miR-NC in HMEC-1 cells (Fig. 3D). The aforementioned data demonstrated that HULC could sponge miR-128-3p. To ascertain the regulatory effect of HULC on miR-128-3p expression, the present study upregulated HULC expression levels by transfection, which was confirmed in HMEC-1 cells (Fig. 3E). Following this, it was found that silencing of HULC increased miR-128-3p expression, and overexpression of HULC inhibited miR-128-3p expression (Fig. 3F). Overall, the data showed that HULC targeted miR-128-3p and negatively modulated its expression.
Figure 3.
HULC is a sponge for miR-128-3p. (A) The putative binding sites between HULC and miR-128-3p, as well as the MUT. (B) The luciferase activities of HULC WT and HULC MUT in HMEC-1 cells. (C) The enrichment of HULC and miR-128-3p in the samples bound to anti-Ago2 or anti-IgG. (D) The enrichment of HULC pulled down by Bio-miR-128-3p or Bio-miR-NC in HMEC-1 cells. (E) The expression of HULC in HMEC-1 cells transfected with an empty vector or HULC-overexpression vector. (F) The expression level of miR-128-3p in HMEC-1 cells transfected with si-NC, si-HULC, an empty vector or a HULC-overexpression vector. **P<0.01. WT, wild-type; MUT, mutant; miR, microRNA; NC, negative control; IgG, immunoglobin G; HULC, highly upregulated in liver cancer; HMEC-1, human dermal microvascular endothelial cells; Ago2, argonaute 2; Bio, biotin; siRNA, small interfering RNA.
HULC knockdown ameliorates LPS-induced apoptosis and inflammation in HMEC-1 cells by targeting miR-128-3p
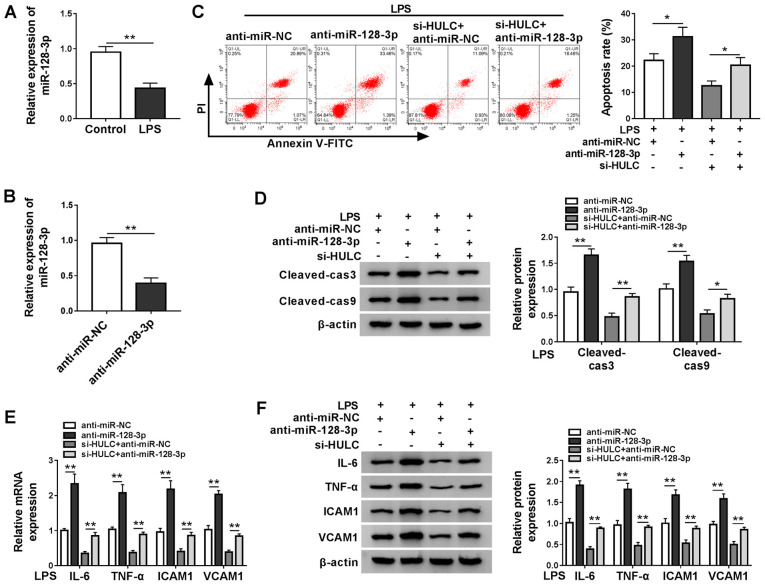
miR-128-3p expression was measured in LPS-treated HMEC-1 cells and the control group, RT-qPCR demonstrated that LPS treatment downregulated miR-128-3p expression (Fig. 4A). Subsequently, the ability of anti-miR-128-3p to interfere with miR-128-3p expression was confirmed by RT-qPCR (Fig. 4B). To clarify the underlying mechanism of HULC in LPS-induced apoptosis and inflammation of HMEC-1 cells, LPS-treated HMEC-1 cells were transfected with anti-miR-NC, anti-miR-128-3p, si-HULC + anti-miR-NC or si-HULC + anti-miR-128-3p. As demonstrated in Fig. 4C, miR-128-3p interference promoted the apoptosis of HMEC-1 cells treated with LPS, and also elevated cell apoptosis of LPS-treated HULC-knockdown HMEC-1 cells. Western blot analysis indicated that miR-128-3p knockdown elevated the expression levels of cleaved-cas3 and cleaved-cas9 in LPS-treated HMEC-1 cells in the untransfected group and the si-HULC group (Fig. 4D). Furthermore, silencing of miR-128-3p also significantly promoted the inflammatory response in LPS-treated HMEC-1 cells in both the si-HULC group and the untransfected group (Fig. 4E and F). Taken together, silencing of HULC weakened LPS-induced apoptosis and the inflammatory response in HMEC-1 cells by targeting miR-128-3p.
Figure 4.
HULC knockdown ameliorates LPS-induced apoptosis and inflammation in HMEC-1 cells by targeting miR-128-3p. (A) miR-128-3p expression in HMEC-1 cells treated with LPS or DMSO (control). (B) miR-128-3p expression levels in HMEC-1 cells transfected with anti-miR-NC or anti-miR-128-3p. (C-F) HMEC-1 cells treated with LPS were transfected with anti-miR-NC, anti-miR-128-3p, si-HULC + anti-miR-NC or si-HULC + anti-miR-128-3p. (C) Apoptotic rate of transfected HMEC-1 cells. (D) Protein expression levels of cleaved-cas3 and cleaved-cas9 in transfected HMEC-1 cells. The (E) mRNA and (F) protein expression levels of IL-6, TNF-α, ICAM1 and VCAM1 in transfected HMEC-1 cells. *P<0.05. **P<0.01. HULC, highly upregulated in liver cancer; LPS, lipopolysaccharide; miR, microRNA; NC, negative control; siRNA, small interfering RNA; ICAM1, intercellular adhesion molecule; VCAM1, vascular cell adhesion molecule; DMSO, dimethyl sulfoxide; HMEC-1, human dermal microvascular endothelial cells; cas, caspase.
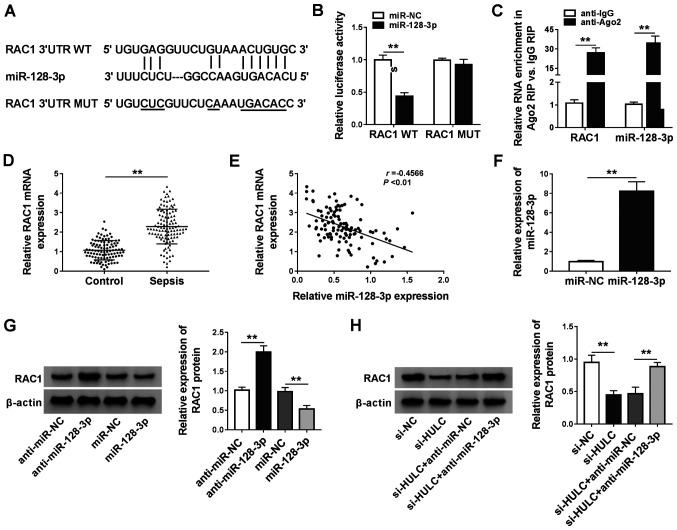
RAC1 is a target of miR-128-3p
The starBase software was applied for searching the target genes of miR-128-3p, RAC1, kruppel-like factor 4 (KLF4), disintegrin and metalloproteinase domain-containing protein 10 and forkhead box protein O4 were identified as candidates. In a preliminary study, among these four predicted genes, RAC1 expression levels decreased the most in LPS-induced HMEC-1 cells transfected with miR-128-3p (data not shown). The binding sequence between miR-128-3p and the 3′UTR of RAC1 is exhibited in Fig. 5A, along with a mutant version. Dual-luciferase reporter assays and RIP assays demonstrated that there was a relationship between miR-128-3p and RAC1 (Fig. 5B and C). Furthermore, the present study found that RAC1 was significantly upregulated in the serum of patients with sepsis compared with that in the control group (Fig. 5D). Furthermore, expression levels of RAC1 mRNA was inversely correlated with miR-128-3p expression in the serum of patients with sepsis (Fig. 5E). The overexpression efficiency of miR-128-3p is presented in Fig. 5F. Introduction of miR-128-3p reduced protein expression levels of RAC1, but transfection with anti-miR-128–3 resulted in the opposite effect (Fig. 5G). Additionally, silencing HULC expression led to the reduction of RAC1 expression, which was reversed by anti-miR-128-3p (Fig. 5H). These results suggested that RAC1 was a downstream target of miR-128-3p in HMEC-1 cells.
Figure 5.
RAC1 is a target of miR-128-3p. (A) The predicted binding position between miR-128-3p, the 3′UTR of RAC1 and the MUT. (B) The luciferase activities of RAC1 WT and RAC1 MUT in HMEC-1 cells. (C) The enrichment of RAC1 and miR-128-3p in the samples bound to the anti-Ago2 or anti-IgG. (D) The mRNA expression of RAC1 in the serum of 110 patients with sepsis and 100 healthy participants. (E) Pearson correlation analysis for expression levels of miR-128-3p and RAC1 mRNA in the serum of 110 patients with sepsis (r=−0.4566, P<0.01). (F) miR-128-3p expression in HMEC-1 cells transfected with miR-NC or miR-128-3p. (G) The protein expression of RAC1 in HMEC-1 cells transfected with anti-miR-NC, anti-miR-128-3p, miR-NC or miR-128-3p. (H) The protein expression of RAC1 in HMEC-1 cells transfected with si-NC, si-HULC, si-HULC + anti-miR-NC or si-HULC + anti-miR-128-3p. **P<0.01. RAC1, Rac family small GTPase 1; miR, microRNA; 3′UTR, 3′untranslated region; WT, wild-type; MUT, mutant; NC, negative control; IgG, immunoglobin G; Ago2, argonaute 2; HULC, highly upregulated in liver cancer; siRNA, small interfering RNA; HMEC-1, human dermal microvascular endothelial cells.
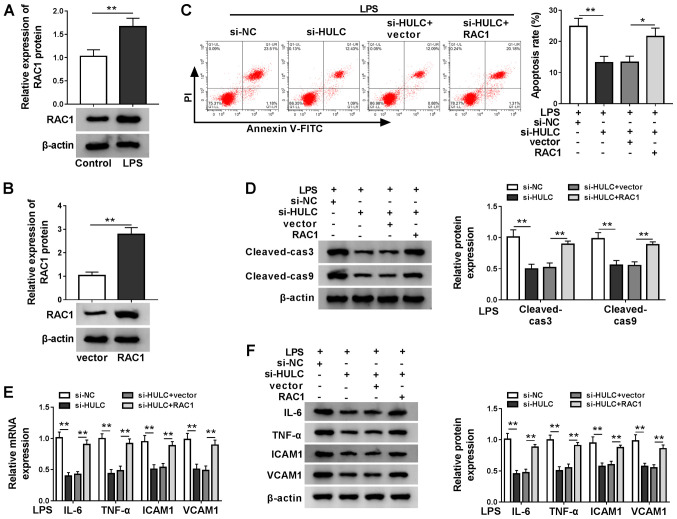
Upregulation of RAC1 could reverse the effect of HULC depletion on apoptosis and inflammation in HMEC-1 cells
As indicated in Fig. 6A, LPS treatment also upregulated RAC1 expression levels in HMEC-1 cells compared with the control group. RAC1 expression levels were successfully upregulated by transfection with RAC1 (Fig. 6B). Then, to validate the anti-apoptotic and anti-inflammatory roles of RAC1 in the HULC-knockdown cells, LPS-induced HMEC-1 cells were transfected with si-NC, si-HULC, si-HULC + vector or si-HULC + RAC1. Furthermore, the aforementioned HULC knockdown-induced inhibition of apoptosis, reduction in cleaved-cas3 and cleaved-cas9 expression levels and decrease in the inflammatory response were all attenuated by the co-transfection of RAC1 (Fig. 6C-F). Taken together, HULC knockdown had anti-apoptotic and anti-inflammatory effects in LPS-induced HMEC-1 cells via the downregulation of RAC1.
Figure 6.
Upregulation of RAC1 reverses the anti-apoptotic and anti-inflammatory effects of HULC knockdown in HMEC-1 cells. (A) The protein expression of RAC1 in HMEC-1 cells treated with LPS or DMSO (control). (B) RAC1 protein expression in HMEC-1 cells transfected with an empty vector or a RAC1-overexpression vector. (C-F) HMEC-1 cells treated with LPS were transfected with si-NC, si-HULC, si-HULC + vector or si-HULC + RAC1. (C) Apoptotic rate of transfected HMEC-1 cells. (D) Protein expression levels of cleaved-cas3 and cleaved-cas9 in transfected HMEC-1 cells. The (E) mRNA and (F) protein expression levels of IL-6, TNF-α, ICAM1 and VCAM1 in transfected HMEC-1 cells. *P<0.05, **P<0.01. HULC, highly upregulated in liver cancer; siRNA, small interfering RNA; NC, negative control; LPS, lipopolysaccharide; RAC1, Rac family small GTPase 1; ICAM1, intercellular adhesion molecule; VCAM1, vascular cell adhesion molecule; HMEC-1, human dermal microvascular endothelial cells; DMSO, dimethyl sulfoxide.
Discussion
The primary feature of sepsis is the systemic dysregulation of the inflammatory response after infection (1), which can result in multiple organ failure and even death (21). Hence, there is an urgent need to develop a deeper understanding about sepsis initiation and progression. The present study identified HULC/miR-128-3p/RAC1 as a novel potential regulatory axis in a sepsis cell model using LPS-treated HMEC-1 cells.
LPS is an endotoxin that is able to regulate the development of myocardial injury caused by sepsis (22), and has been widely used to induce sepsis models in vitro (23,24). In the present study, 1 µg/ml LPS was used to treat HMEC-1 cells. Initially, the effect of LPS on HMEC-1 cells was measured. The data from RT-qPCR, western blot analysis and flow cytometry suggested that LPS treatment reinforced apoptosis and inflammatory responses in treated HMEC-1 cells, which was in line with previous studies (7,24), thus indicating the successful establishment of a sepsis model in vitro.
RT-qPCR in the present study revealed that HULC was highly expressed in the serum of patients with sepsis, especially the patients with septic shock; as well as in LPS-treated HMEC-1 cells. The HULC gene is 16 kb long and is located at chromosome 6p24.3; it is the first recognized non-coding RNA that is upregulated in liver cancer tissues, harboring the potential to be a biomarker of hepatocellular carcinoma (25,26). Additionally, HULC knockdown has been demonstrated to inhibit gastric cancer progression via the mediation of the miR-9-5p/myosin heavy chain 9 axis (27). Chu et al (28) found that HULC aggravated ovarian carcinoma progression through the activation of the PI3K/AKT/mTOR signaling pathway via the downregulation of miR-125a-3p. The aforementioned reports implied the oncogenic role of HULC in human cancer types. In the present study, HULC was knocked down by transfection with specific siRNAs to explore its role in sepsis in vitro. Functional experiments showed that HULC knockdown reduced apoptosis and expression levels of proinflammatory cytokines (IL-6, TNF-α, ICAM1 and VCAM1) in HMEC-1 cells treated with LPS. In other words, silencing of HULC ameliorated the LPS-mediated injury in HMEC-1 cells, which was consistent with a previous report (9). Multiple reports have proposed that lncRNAs play pivotal roles in sepsis by targeting miRNAs (6,7). For example, NEAT1 upregulated Toll-like receptor 4 to promote sepsis-induced liver injury by sponging let-7a (29). In the present study, miR-128-3p was predicted to target HULC, using the online software microRNA.org. The target relationship between HULC and miR-128-3p was confirmed by dual-luciferase reporter, RIP and RNA pull-down assays. From the present data, miR-128-3p was downregulated in the serum of patients with sepsis and LPS-induced HMEC-1 cells. A significant inverse correlation between HULC and miR-128-3p was discovered in serum of patients with sepsis.
miR-128-3p regulates the inflammatory responses triggered by TNF-α by targeting sirtuin 1 (Sirt1) in bone marrow mesenchymal stem cells (30). Selenium could mitigate LPS-induced myocardial inflammation by modulating the miR-128-3p/p38MAPK-NF-κB pathway (31). miR-128-3p also functions as a tumor suppressor in human breast cancer (32), glioma (33) and hepatocellular carcinoma (34). In the present study, miR-128-3p interference aggravated LPS-mediated damage in HMEC-1 cells, and abolished the HULC knockdown-mediated reduction of apoptosis and expression levels of proinflammatory cytokines in LPS-treated HMEC-1 cells. miR-128-3p can modulate inflammatory responses by binding to the 3′UTR of Sirt1 (30) and KLF4 (35). The present study hypothesized that miR-128-3p took part in the LPS-induced proinflammatory response by targeting specific genes. Consequently, the binding position between miR-128-3p and RAC1 were searched using starBase, following which they were validated using dual-luciferase reporter and RIP assays. RT-qPCR analysis demonstrated the high expression of RAC1 in the serum of patients with sepsis and LPS-treated HMEC-1 cells. In addition, RAC1 mRNA expression was negatively correlated with miR-128-3p in the serum of patients with sepsis.
RAC1 activity is associated with mitogen-activated protein kinases involved in proinflammatory activities, therefore repressing RAC1 activity is hypothesized to be a mechanism to alleviate the coagulation dysfunction in abdominal sepsis (15). Jiang et al (36) proposed that RAC1 signaling affected inflammation caused by cigarette smoke in vitro and in vivo via Erk1/2 MAPK and STAT3 pathways. Inactivation of RAC1 reversed PM2.5-induced inflammation in mouse airways and human bronchial epithelial cells via the AKT signaling pathway (37). In a preliminary study, miR-128-3p upregulation suppressed the inflammatory response in LPS-stimulated HMEC-1 cells, which could be reversed by the introduction of RAC1. Functional analyses in the current study demonstrated that accumulation of RAC1 reversed the downregulated apoptosis and proinflammatory response induced by HULC knockdown in LPS-treated HMEC-1 cells, which also indicated the participation of RAC1 in the proinflammatory response induced by LPS.
A few limitations exist in the present study. The specific signaling pathways involved in the HULC/miR-128-3p/RAC1 axis in the inflammatory response during LPS-induced sepsis in HMEC-1 cells have not yet been investigated. Furthermore, mouse models could help to further study the role of HULC in vivo.
In conclusion, HULC and RAC1 expression were elevated, and miR-128-3p expression declined in patients with sepsis and LPS-induced HMEC-1 cells. HULC knockdown could protect HMEC-1 cells from LPS-triggered injury, which was reversed by miR-128-3p knockdown or RAC1 overexpression. HULC sponged miR-128-3p to upregulate RAC1 in LPS-induced HMEC-1 cells, therefore inhibiting HULC expression could be a potential strategy in the treatment of sepsis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XL and YuL conceptualized the study and developed the methodology. Data analysis and interpretation were performed by JX, HX and YaL. Validation and investigation were conducted by HX, YaL and WY. The original draft of the manuscript, along with the review and editing was conducted by WY, XL and YuL. All authors read and approved the final version of the manuscript.
Ethics approval and consent to participate
All participants submitted written informed consent. The present study was approved by the ethical review committee of the Renmin Hospital of Wuhan University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Deutschman CS, Tracey KJ. Sepsis: Current dogma and new perspectives. Immunity. 2014;40:463–475. doi: 10.1016/j.immuni.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Adhikari NK, Fowler RA, Bhagwanjee S, Rubenfeld GD. Critical care and the global burden of critical illness in adults. Lancet. 2010;376:1339–1346. doi: 10.1016/S0140-6736(10)60446-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cecconi M, Evans L, Levy M, Rhodes A. Sepsis and septic shock. Lancet. 2018;392:75–87. doi: 10.1016/S0140-6736(18)30696-2. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482:339–346. doi: 10.1038/nature10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho J, Chan H, Wong SH, Wang MH, Yu J, Xiao Z, Liu X, Choi G, Leung CC, Wong WT, et al. The involvement of regulatory non-coding RNAs in sepsis: A systematic review. Crit Care. 2016;20:383. doi: 10.1186/s13054-016-1555-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen J, Liu L, Zhang F, Gu J, Pan G. LncRNA TapSAKI promotes inflammation injury in HK-2 cells and urine derived sepsis-induced kidney injury. J Pharm Pharmacol. 2019;71:839–848. doi: 10.1111/jphp.13049. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Gu X, Zhou L, Wang S, Zhu L, Huang Y, Cao F. Long non-coding RNA-HOTAIR promotes the progression of sepsis by acting as a sponge of miR-211 to induce IL-6R expression. Exp Ther Med. 2019;18:3959–3967. doi: 10.3892/etm.2019.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang SM, Liu GQ, Xian HB, Si JL, Qi SX, Yu YP. LncRNA NEAT1 alleviates sepsis-induced myocardial injury by regulating the TLR2/NF-kappaB signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:4898–4907. doi: 10.26355/eurrev_201906_18078. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Fu Y, Song YF, Li N. Increased expression of lncRNA UCA1 and HULC is required for pro-inflammatory response during LPS induced sepsis in endothelial cells. Front Physiol. 2019;10:608. doi: 10.3389/fphys.2019.00608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rivera-Barahona A, Perez B, Richard E, Desviat LR. Role of miRNAs in human disease and inborn errors of metabolism. J Inherit Metab Dis. 2017;40:471–480. doi: 10.1007/s10545-017-0018-6. [DOI] [PubMed] [Google Scholar]
- 11.Yao Y, Sun F, Lei M. miR-25 inhibits sepsis-induced cardiomyocyte apoptosis by targetting PTEN. Biosci Rep. 2018;38:BSR20171511. doi: 10.1042/BSR20171511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Bei Y, Shen S, Huang P, Shi J, Zhang J, Sun Q, Chen Y, Yang Y, Xu T, et al. miR-21-3p controls sepsis-associated cardiac dysfunction via regulating SORBS2. J Mol Cell Cardiol. 2016;94:43–53. doi: 10.1016/j.yjmcc.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 13.Wang S, Wang J, Zhang Z, Miao H. Decreased miR-128 and increased miR-21 synergistically cause podocyte injury in sepsis. J Nephrol. 2017;30:543–550. doi: 10.1007/s40620-017-0405-y. [DOI] [PubMed] [Google Scholar]
- 14.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 15.Wang Y, Luo L, Morgelin M, Thorlacius H. Rac1 regulates sepsis-induced formation of platelet-derived microparticles and thrombin generation. Biochem Biophys Res Commun. 2017;487:887–891. doi: 10.1016/j.bbrc.2017.09.126. [DOI] [PubMed] [Google Scholar]
- 16.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Shi K, Chen M, Xu L, Hong J, Hu B, Yang X, Sun R. Elevated miR-155 expression induces immunosuppression via CD39(+) regulatory T-cells in sepsis patient. Int J Infect Dis. 2015;40:135–141. doi: 10.1016/j.ijid.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Li C, Wu J, Li Y, Xing G. Cytoprotective effect of heat shock protein 27 against lipopolysaccharide-induced apoptosis of renal epithelial HK-2 cells. Cell Physiol Biochem. 2017;41:2211–2220. doi: 10.1159/000475636. [DOI] [PubMed] [Google Scholar]
- 19.Zhang A, Lu H, Wen D, Sun J, Du J, Wang X, Gu W, Jiang J. The potential roles of long non-coding RNAs in lipopolysaccharide-induced human peripheral blood mononuclear cells as determined by microarray analysis. FEBS Open Bio. 2019;9:148–158. doi: 10.1002/2211-5463.12556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, Kumar A, Sevransky JE, Sprung CL, Nunnally ME, et al. Surviving sepsis campaign: International Guidelines for Management of Sepsis and Septic Shock: 2016. Crit Care Med. 2017;45:486–552. doi: 10.1097/CCM.0000000000002255. [DOI] [PubMed] [Google Scholar]
- 22.Virzi GM, Clementi A, Brocca A, Ronco C. Endotoxin effects on cardiac and renal functions and cardiorenal syndromes. Blood Purif. 2017;44:314–326. doi: 10.1159/000480424. [DOI] [PubMed] [Google Scholar]
- 23.Quoilin C, Mouithys-Mickalad A, Lécart S, Fontaine-Aupart MP, Hoebeke M. Evidence of oxidative stress and mitochondrial respiratory chain dysfunction in an in vitro model of sepsis-induced kidney injury. Biochim Biophys Acta. 2014;1837:1790–1800. doi: 10.1016/j.bbabio.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Zhang W, Lu F, Xie Y, Lin Y, Zhao T, Tao S, Lai Z, Wei N, Yang R, Shao Y, He J. miR-23b negatively regulates sepsis-induced inflammatory responses by targeting ADAM10 in human THP-1 monocytes. Mediators Inflamm. 2019;2019:5306541. doi: 10.1155/2019/5306541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Li Z, Zhang Y, Zhong Q, Chen Q, Zhang L. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int J Clin Exp Med. 2015;8:12956–12962. [PMC free article] [PubMed] [Google Scholar]
- 26.Panzitt K, Tschernatsch M M, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, Zatloukal K. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 27.Liu T, Liu Y, Wei C, Yang Z, Chang W, Zhang X. LncRNA HULC promotes the progression of gastric cancer by regulating miR-9-5p/MYH9 axis. Biomed Pharmacother. 2019;121:109607. doi: 10.1016/j.biopha.2019.109607. [DOI] [PubMed] [Google Scholar]
- 28.Chu P, Xu L, Su H. HULC functions as an oncogene in ovarian carcinoma cells by negatively modulating miR-125a-3p. J Physiol Biochem. 2019;75:163–171. doi: 10.1007/s13105-019-00669-5. [DOI] [PubMed] [Google Scholar]
- 29.Zhang CC, Niu F. LncRNA NEAT1 promotes inflammatory response in sepsis-induced liver injury via the Let-7a/TLR4 axis. Int Immunopharmacol. 2019;75:105731. doi: 10.1016/j.intimp.2019.105731. [DOI] [PubMed] [Google Scholar]
- 30.Wu L, Zhang G, Guo C, Zhao X, Shen D, Yang N. MiR-128-3p mediates TNF-α-induced inflammatory responses by regulating Sirt1 expression in bone marrow mesenchymal stem cells. Biochem Biophys Res Commun. 2020;521:98–105. doi: 10.1016/j.bbrc.2019.10.083. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Wang S, Zhang Q, Li X, Xu S. Selenomethionine alleviates LPS-induced chicken myocardial inflammation by regulating the miR-128-3p-p38 MAPK axis and oxidative stress. Metallomics. 2020;12:54–64. doi: 10.1039/C9MT00216B. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Li D, Fang L. MiR-128-3p suppresses breast cancer cellular progression via targeting LIMK1. Biomed Pharmacother. 2019;115:108947. doi: 10.1016/j.biopha.2019.108947. [DOI] [PubMed] [Google Scholar]
- 33.Huo L, Wang B, Zheng M, Zhang Y, Xu J, Yang G, Guan Q. miR-128-3p inhibits glioma cell proliferation and differentiation by targeting NPTX1 through IRS-1/PI3K/AKT signaling pathway. Exp Ther Med. 2019;17:2921–2930. doi: 10.3892/etm.2019.7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang CY, Huang XP, Zhu JY, Chen ZG, Li XJ, Zhang XH, Huang S, He JB, Lian F, Zhao YN, Wu GB. miR-128-3p suppresses hepatocellular carcinoma proliferation by regulating PIK3R1 and is correlated with the prognosis of HCC patients. Oncol Rep. 2015;33:2889–2898. doi: 10.3892/or.2015.3936. [DOI] [PubMed] [Google Scholar]
- 35.Lu Q, Meng Q, Qi M, Li F, Liu B. Shear-sensitive lncRNA AF131217.1 inhibits inflammation in HUVECs via regulation of KLF4. Hypertension. 2019;73:e25–e34. doi: 10.1161/HYPERTENSIONAHA.118.12476. [DOI] [PubMed] [Google Scholar]
- 36.Jiang JX, Zhang SJ, Shen HJ, Guan Y, Liu Q, Zhao W, Jia YL, Shen J, Yan XF, Xie QM. Rac1 signaling regulates cigarette smoke-induced inflammation in the lung via the Erk1/2 MAPK and STAT3 pathways. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1778–1788. doi: 10.1016/j.bbadis.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 37.Zhang S, Zhang W, Zeng X, Zhao W, Wang Z, Dong X, Jia Y, Shen J, Chen R, Lin X. Inhibition of Rac1 activity alleviates PM2.5-induced pulmonary inflammation via the AKT signaling pathway. Toxicol Lett. 2019;310:61–69. doi: 10.1016/j.toxlet.2019.04.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.