Abstract
Tissue damage in diabetes is at least partly due to elevated reactive oxygen species production by the mitochondrial respiratory chain during hyperglycemia. Sustained hyperglycemia results in mitochondrial dysfunction and the abnormal expression of mitochondrial genes, such as NADH: Ubiquinone oxidoreductase subunit A13 (NDUFA13). Metformin, an AMP-activated protein kinase (AMPK) activator, protects cardiomyocytes from oxidative stress by improving mitochondrial function; however, the exact underlying mechanisms are not completely understood. The aim of the present study was to investigated the molecular changes and related regulatory mechanisms in the response of H9C2 cardiomyocytes to metformin under high glucose conditions. H9C2 cells were subjected to CCK-8 assay to assess cell viability. Reactive oxygen species generation was measured with DCFH-DA assay. Western blotting was used to analyze the expression levels of NDUFA13, AMPK, p-AMPK and GAPDH. Reverse transcription-quantitative PCR was used to evaluate the expression levels of mitochondrial genes and transcription factors. It was observed that metformin protected H9C2 cardiomyocytes by suppressing high glucose (HG)-induced elevated oxidative stress. In addition, metformin stimulated mitochondrial biogenesis, as indicated by increased expression levels of mitochondrial genes (NDUFA1, NDUFA2, NDUFA13 and manganese superoxide dismutase) and mitochondrial biogenesis-related transcription factors [peroxisome proliferator-activated receptor-gamma coactivator-1α, nuclear respiratory factor (NRF)-1, and NRF-2] in the metformin + HG group compared with the HG group. Moreover, metformin promoted mitochondrial NDUFA13 protein expression via the AMPK signaling pathway, which was abolished by pretreatment with the AMPK inhibitor, Compound C. The results suggested that metformin protected cardiomyocytes against HG-induced oxidative stress via a mechanism involving AMPK, NDUFA13 and mitochondrial biogenesis.
Keywords: metformin, cardiomyocyte, mitochondrial biogenesis, reactive oxygen species
Introduction
Diabetic patients have an notably increased incidence of heart failure according to the Framingham study (1); however, the molecular mechanisms underlying the pathogenesis of diabetic cardiomyopathy are not completely understood. Among the hypothesized mechanisms, hyperglycemia-induced oxidative stress is recognized as a critical participant in the pathogenesis and progression of diabetes (2). Increased levels of oxidative stress lead to harmful modifications of macromolecules, including DNA, proteins and lipids (3), which can result in cardiomyocyte apoptosis, hypertrophy and fibrosis, ultimately leading to cardiac remodeling and dysfunction (3). Therefore, developing an effective treatment to suppress increased oxidative stress levels and subsequent cardiomyocyte injury in patients with diabetes is important.
Metformin is widely used as a first-line hypoglycemic drug (4). Previous studies identified cardioprotective effects of metformin beyond its antihyperglycemic effects (5–7). The United Kingdom Prospective Diabetes Study demonstrated that early and intensive metformin intervention could reduce the incidence of myocardial infarction and increase the survival rate in patients with type 2 diabetes (5). Metformin exerts cardioprotective effects partly by suppressing high glucose (HG)-induced excessive reactive oxygen species production and inflammatory responses (8); however, the exact underlying mechanisms are not completely understood.
AMP-activated protein kinase (AMPK), a key mediator of the downstream effects of metformin, can be activated by cytokines, hormones and oral hypoglycemic agents that are commonly used to treat type 2 diabetes (9). Activated AMPK promotes the production of ATP by regulating glucose and fatty acid metabolism (9), indicating that AMPK may serve as an innate survival mechanism for the heart. For example, AMPK activation during myocardial ischemia reduces the infarct size in the hearts of diabetic model rats (10). In addition, upregulation of AMPK signaling in the vasculature improves microvascular function in the hearts of diabetic model mice (11).
Based on its role in regulating cellular energy status, AMPK is a major regulator of mitochondrial function. Hydroxytyrosol, a natural antioxidant, reduces oxidative stress and improves mitochondrial function, presumably by activating AMPK signaling in the brain of db/db mice (12). Therefore, it was hypothesized that AMPK may serve as a critical modulator of mitochondrial biogenesis and function, and enhance resistance to oxidative stress. However, the exact role and mechanisms underlying AMPK in the cardioprotective effects of metformin in response to oxidative stress are not completely understood.
Mitochondria are the primary energy generating organelles, but are also the main source of ROS (13). NADH: Ubiquinone oxidoreductase subunit A13 (NDUFA)13 is an indispensable assembly factor of complex I (14). The primary role of NDUFA13 in the antioxidant effect of resveratrol has been previously reported (15). Downregulated NDUFA13 expression increases mitochondrial ROS generation in H9C2 cardiomyocytes (16), and sustained high glucose decreases NDUFA13 expression levels (15).
Based on the aforementioned studies, it was hypothesized that NDUFA13 may be associated with alleviating oxidative stress in diabetic cardiomyopathy, and metformin may prevent diabetic myocardial injury by suppressing hyperglycemia-induced ROS overproduction. The present study aimed to investigate whether the protective effects of metformin were mediated by promoting mitochondrial biogenesis and NDUFA13 expression. Additionally, the exact role of the AMPK signaling pathway in metformin-induced effects was investigated.
Materials and methods
Cell culture and treatments
H9C2 cells (The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences) were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 mg/ml streptomycin at 37°C in a humidified incubator with 5% CO2 and 95% air.
H9C2 cells cultured to approximately 80% confluence were firstly incubated with various concentrations of glucose (5.5, 15, 25, 33.3 40 and 50 mM) for 36 h at 37°C. Cells treated with various concentrations of glucose and mannitol (Abcam) were then incubated for 12, 24, 36 and 48 h at 37°C. These included 5.5 and 33.3 mM glucose and 25 mM glucose plus 8.3 mM mannitol. According to changes in cell viability, 5.5 mM glucose was selected to mimic normal conditions and 33.3 mM glucose was selected to mimic the diabetic condition. Subsequently, cells were treated with low concentrations of metformin (0.5 or 1 mM; Sigma-Aldrich; Merck KGaA) for 36 h at 37°C. For subsequent experiments, 1 mM metformin was used, which was proven to be an effective in previous studies (17,18). The cardiomyocytes were divided into the following experimental groups: i) Control, medium containing 5.5 mM glucose; ii) HG, medium containing 33.3 mM glucose; iii) mannitol, medium containing 25 mM glucose + 8.3 mM mannitol; iv) metformin 0.5 mM, medium containing 33.3 mM glucose + 0.5 mM metformin; v) Met 1, medium containing 33.3 mM glucose + 1 mM metformin; and vi) Compound C, medium containing 33.3 mM glucose + 1 mM metformin + 10 µM Compound C (Sigma-Aldrich; Merck KGaA).
Cell Counting Kit-8 (CCK-8) assay for cell viability
Cell viability was assessed using a CCK-8 assay kit (Shanghai Yeasen Biotechnology Co., Ltd.) according to the manufacturer's protocol. H9C2 cells were seeded (5×104 cells/well) into 96-well plates. Subsequently, CCK-8 reagent was added to each well and incubated for 1 h at 37°C. The absorbance of each well was measured at a wavelength of 450 nm using a microplate reader (BioTek Instruments, Inc.).
Lactate dehydrogenase (LDH) release
Cell death was assessed using an LDH cytotoxicity assay kit (Shanghai Yeasen Biotechnology Co., Ltd.) according to the manufacturer's instructions. Following treatment for 36 h, the cell medium was collected. The absorbance of each sample was measured at a wavelength of 450 nm using a microplate reader.
Measurement of ROS, malondialdehyde (MDA) and superoxide dismutase (SOD)
Intracellular ROS levels were determined using a ROS assay kit (Shanghai Yeasen Biotechnology Co., Ltd.) according to the manufacturer's protocol. Dichlorofluorescein fluorescence was detected using an Olympus IX71 fluorescence microscope (Olympus Corporation; magnification, ×200). MDA levels were assessed using an MDA assay kit (Beyotime Institute of Biotechnology) according to the manufacturer's instructions. The absorbance of each sample was measured at a wavelength of 535 nm using a microplate reader. SOD activity was measured using a total SOD assay kit (Shanghai Yeasen Biotechnology Co., Ltd.) according to the manufacturer's protocol. The absorbance of each sample was measured at a wavelength of 450 nm using a microplate reader.
Western blotting
The H9c2 cells were lysed with RIPA buffer (Beyotime Institute of Biotechnology) for 45 min at 4°C. Total protein was quantified using a bicinchoninic acid assay kit (Beyotime Institute of Biotechnology). Equal amounts of protein were separated via 10–12% SDS-PAGE and transferred electrophoretically to PVDF membranes (Invitrogen; Thermo Fisher Scientific, Inc.). The membranes were blocked with Tris buffer (Beyotime Institute of Biotechnology) containing 0.1% Tween-20 (TBST) in 5% milk for 1 h at room temperature and then incubated overnight with primary antibodies targeted against: NDUFA13 (17 kD;1:1,000; cat. no. ab110240; Abcam), AMPK (62 KD;1:1,000; cat. no. 5831; Cell Signaling Technology, Inc.), phosphorylated (p)-AMPK (62 KD;1:1,000; cat. no. 50081; Cell Signaling Technology, Inc.) and GAPDH (37 KD;1:10,000; cat. no. 5174; Cell Signaling Technology, Inc.). Following primary incubation, the membranes were incubated with a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (1:10,000; cat. no. HA1001; HuaBio Inc.) for 2 h at room temperature. Protein bands were visualized using an enhanced chemiluminescence system (EMD Millipore). ImageJ software (version 1.41; National Institutes of Health) was used to quantify the bands of each protein and GAPDH was used as the loading control.
Immunofluorescence assay
Cells were fixed with 4% paraformaldehyde for 30 min at room temperature and permeabilized with 0.1% Triton X-100 for 10 min. Subsequently, cells were blocked with 3% bovine serum albumin (Beyotime Institute of Biotechnology) for 2 h at room temperature and incubated with an anti-NDUFA13 primary antibody (1:50; cat. no. ab110240; Abcam) overnight at 4°C. The slides were then washed with phosphate-buffered saline (PBS, Beyotime Institute of Biotechnology) and 0.1% Tween-20 (PBST) and incubated with a Texas Red/Alexa fluor-conjugated secondary antibody for 1 h at room temperature. The slides were mounted using mounting medium, counterstained with 6-diamidino-2-phenylindole (Invitrogen; Thermo Fisher Scientific, Inc.) for 10 min at room temperature and observed using an IX71 microscope (Olympus Corporation; magnification, ×200) and Image Pro Plus 3.0 software (Media Cybernetics, Inc.).
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol® reagent (Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA at 42°C for 1 h then 90°C for 5 min using a cDNA synthesis kit (Takara Biotechnology Co., Ltd.). Subsequently, qPCR was performed using SYBR green PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for the qPCR: Initial denaturation at 95°C for 2 min; and 40 cycles of 95°C for 15 sec and 60°C for 1 min. The sequences of the primers used for qPCR are presented in Table I. mRNA expression levels were quantified using the 2−∆∆Cq method (19) and normalized to the internal reference gene β-actin.
Table I.
Sequences of primers used for reverse transcription-quantitative PCR.
| Gene | Sequence (5′→3′) | Product size (bp) |
|---|---|---|
| β-actin | F: GCGTCCACCCGCGAGTACAA | 118 |
| R: ACATGCCGGAGCCGTTGTCG | ||
| NDUFA1 | F: TGCTGCCGGAAGAGCGGTGA | 189 |
| R: TCCTTGCCCCCGTTGGTGAACT | ||
| NDUFA2 | F: ACTGAGGACTGAACAAGCCCACCA | 223 |
| R: GCGACATCCCAGCGGGTAGC | ||
| NDUFA13 | F: CTACTGGAGAATAATGAGGTGGAAC | 175 |
| R: CCAGTTGGGCACATCTTTCA | ||
| Mn-SOD | F: GTGTCTGTGGGAGTCCAAGG | 149 |
| R: TGCTCCCACACATCAATCCC | ||
| PGC-1α | F: GGGGCACATCTGTTCTTCCA | 156 |
| R: GCTTGACTGGGATGACCGAA | ||
| NRF1 | F: ACACAGCATAGCCCATCTCG | 226 |
| R: GGTCATTTCACCGCCCTGTA | ||
| NRF2 | F: AGCAAGACTTGGGCCACTTA | 112 |
| R: TCTGGCTTCTTGCTCTTGGG |
NDUF, NADH: Ubiquinone oxidoreductase subunit; Mn-SOD, manganese superoxide dismutase; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1α; NRF, nuclear respiratory factor.
Statistical analysis
Data are expressed as the mean ± standard deviation of at least three independent experiments. Comparisons among multiple groups were analyzed using one-way ANOVA followed by Tukey's post hoc test. Statistical analyses were performed using GraphPad Prism software (version 7; GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
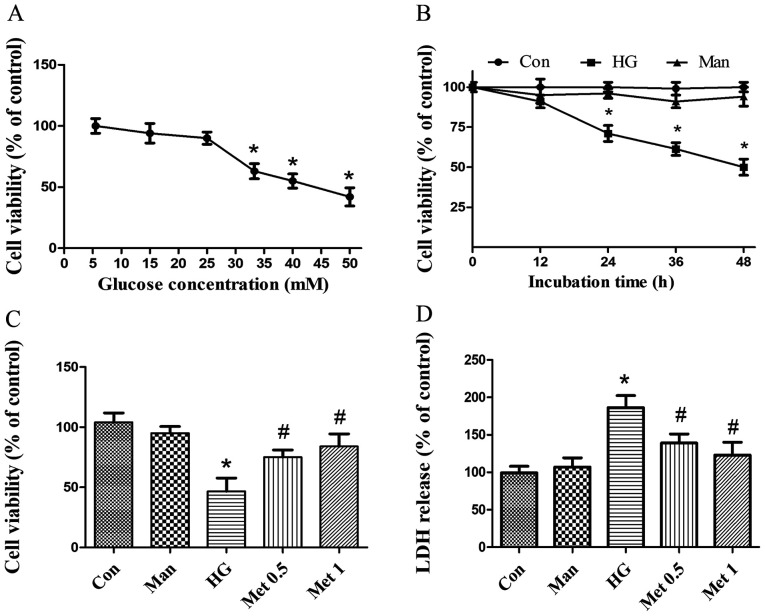
Metformin protects H9C2 cells from HG-induced cytotoxicity
To investigate the effect of metformin on cardiomyocyte survival in various concentrations of glucose (5.5–50 mM), H9C2 cell viability was assessed. Cell viability was significantly decreased following incubation with 33.3 mM glucose (Fig. 1A) for at least 24 h (Fig. 1B) compared with the control group. Subsequently, cardiomyocytes were pretreated with metformin and then incubated with 33.3 mM glucose for 36 h. Metformin (0.5 and 1 mM) significantly increased cell viability (Fig. 1C) and decreased LDH release (Fig. 1D) under HG conditions compared with the HG group. The results indicated that metformin rescued cardiomyocytes from HG-induced injury. Mannitol was used as an osmotic control and did not mimic the effects of 33.3 mM glucose (Fig. 1). For subsequent experiments, 33.3 mM glucose was used to induce a HG situation and 1 mM metformin was applied, unless stated otherwise.
Figure 1.
Effect of metformin on cardiomyocyte survival under high glucose conditions. (A) Effect of different concentrations of glucose on H9C2 cell viability. *P<0.05 vs. Con. (B) H9C2 cell viability following incubation with 5.5 glucose, 33.3 mM glucose or 33.3 mM Man for up to 48 h. *P<0.05 vs. Con. (C) H9C2 cell viability following incubation with 33.3 mM glucose and 0.5 or 1 mM Met. *P<0.05 vs. Con; #P<0.05 vs. HG. (D) H9C2 cell LDH release following incubation with 33.3 mM glucose and 0.5 or 1 mM Met. Man (33.3 mM) was used as an osmotic control. *P<0.05 vs. Con; #P<0.05 vs. HG. LDH, lactate dehydrogenase; Con, control; HG, high glucose; Man, mannitol; Met, metformin.
Clinically, the median plasma concentration of metformin is 330 µM (20); however, the concentration of metformin is several times higher in tissues compared with in the blood (20). Therefore, intracellular metformin concentrations are 10–15% of the drug present in the medium (21), meaning the dose used in the present study was higher than the therapeutic dose.
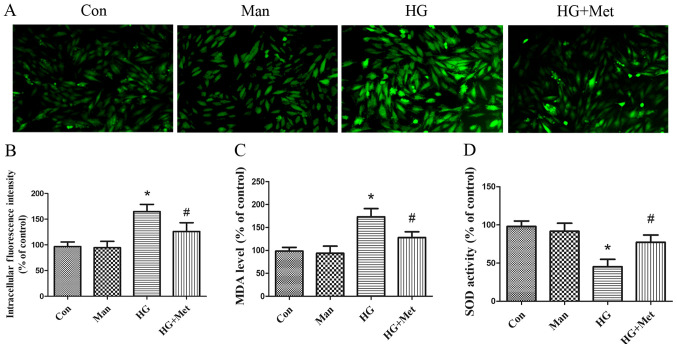
Metformin suppresses HG-induced oxidative stress in H9C2 cells
Markers of oxidative stress, including ROS, MDA and SOD, were detected. Compared with the control and mannitol groups, ROS and MDA levels were significantly increased in the high glucose group. However, metformin pretreatment significantly decreased ROS (Fig. 2A and B) and MDA (Fig. 2C) levels compared with the HG group. By contrast, HG significantly inhibited SOD activity compared with the control and mannitol groups, whereas metformin reversed HG-mediated inhibition of SOD activity in H9C2 cells (Fig. 2D).
Figure 2.
Effects of metformin (1 mM) on high glucose-induced oxidative stress in H9C2 cells. Cells were divided into four groups: i) Con, normal medium; ii) Man, 33.3 mM Man; iii) HG, 33.3 mM glucose; and iv) HG + Met, 33.3 mM glucose pretreated with 1 mM Met. ROS levels were detected by (A) dichlorofluorescein fluorescence intensity and (B) quantified. Magnification, ×200, Scale bars=50 µm. (C) MDA and (D) SOD levels. *P<0.05 vs. Con; #P<0.05 vs. HG. Man, mannitol; HG, high glucose; Met, metformin; ROS, reactive oxygen species; MDA, malondialdehyde; SOD, superoxide dismutase; Con, control.
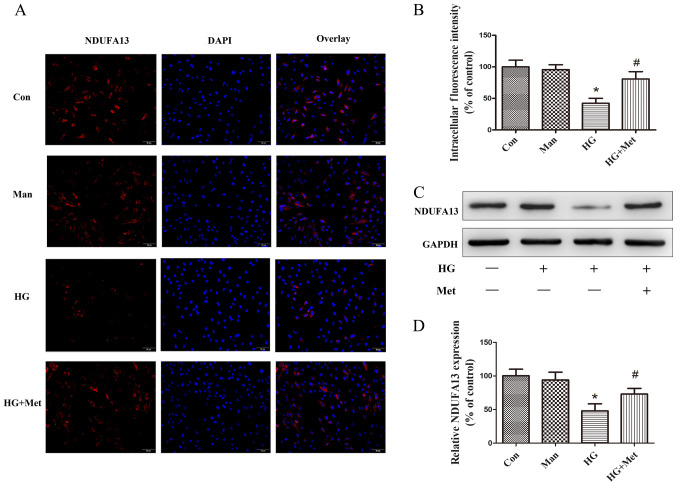
Metformin prevents HG-mediated downregulation of NDUFA13
To examine the effects of metformin on NDUFA13 expression, immunofluorescence and western blotting assays were performed. The immunofluorescence results suggested that the expression of NDUFA13 was significantly decreased in the HG group compared with the control and mannitol groups, but this effect was partly reversed by pretreatment with metformin (Fig. 3A and B). The western blotting results were consistent with the immunofluorescence results (Fig. 3C and D). Mannitol was used as the osmotic control and did not mimic the effects of 33.3 mM glucose (Fig. 3).
Figure 3.
Effects of metformin (1 mM) on NDUFA13 expression in H9C2 cells under high glucose conditions. Cells were divided into four groups: i) Con, normal medium; ii) Man, 33.3 mM Man; iii) HG, 33.3 mM glucose; and iv) HG + Met, 33.3 mM glucose pretreated with 1 mM Met. NDUFA13 protein expression was (A) evaluated by immunofluorescence and (B) quantified. Magnification, ×200, Scale bars=50 µm. (C) NDUFA13 protein expression levels were (C) determined by western blotting and (D) semi-quantified. *P<0.05 vs. Con; #P<0.05 vs. HG. NDUFA13, NADH: Ubiquinone oxidoreductase subunit A13; Man, mannitol; HG, high glucose; Met, metformin; Con, control.
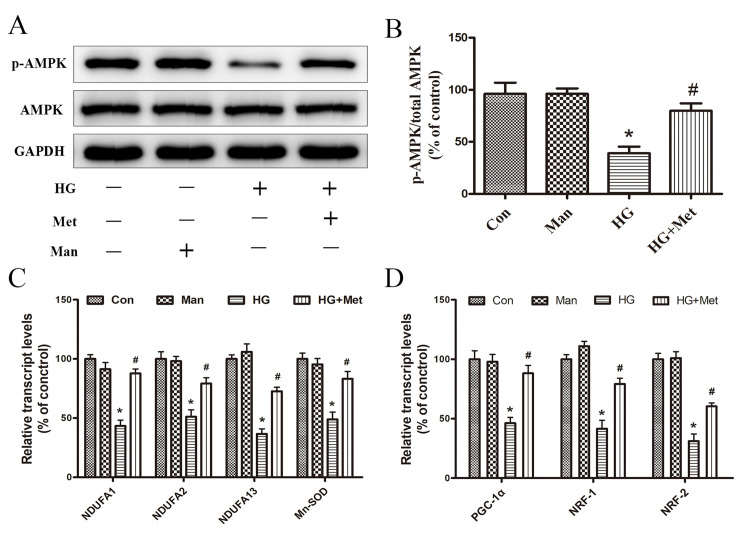
Metformin activates AMPK and induces mitochondrial biogenesis
The activity of AMPK was evaluated by detecting the phosphorylation of Thr-172 (p-AMPK) via western blotting. p-AMPK expression levels were significantly decreased in the HG group compared with the control group, but metformin reversed HG-mediated effects (Fig. 4A and B).
Figure 4.
Effects of metformin (1 mM) on the expression of AMPK, p-AMPK, and mitochondrial biogenesis genes under high glucose conditions. Cells were divided into four groups: i) Con, normal medium; ii) Man, 33.3 mM Man; iii) HG, 33.3 mM glucose; and iv) HG + Met, 33.3 mM glucose pretreated with 1 mM Met. (A) Protein expression levels of p-AMPK and AMPK were (A) determined by western blotting and (B) the ratio of p-AMPK/AMPK was semi-quantified. mRNA expression levels of (C) NDUFA1, NDUFA2, NDUFA13, Mn-SOD, (D) PGC-1α, NRF1 and NRF2 were detected via reverse transcription-quantitative PCR. *P<0.05 vs. Con; #P<0.05 vs. HG. AMPK, AMP-activated protein kinase; p, phosphorylated; Man, mannitol; HG, high glucose; Met, metformin; NDUF, NADH: Ubiquinone oxidoreductase subunit; Mn-SOD, manganese superoxide dismutase; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1α; NRF, nuclear respiratory factor; Con, control.
RT-qPCR was performed to evaluate the effects of metformin on the mRNA expression levels of mitochondrial genes (NDUFA1, NDUFA2, NDUFA13 and Mn-SOD) and transcription factors (PGC-1α, NRF-1 and NRF-2). The results suggested that, compared with the control group, the expression levels of mitochondrial genes related to intracellular ROS production were significantly decreased in the HG group, which was reversed by metformin (Fig. 4C). Therefore, the results provided a potential explanation for the ability of metformin to reduce ROS production. Additionally, the inhibitory effect of HG on mitochondria biogenesis-related transcription factor expression was reversed by metformin pretreatment (Fig. 4D), which may be associated with metformin-mediated promotion of cardiomyocyte survival under high glucose conditions. Mannitol was used as an osmotic control and did not mimic the effects of 33.3 mM glucose (Fig. 4).
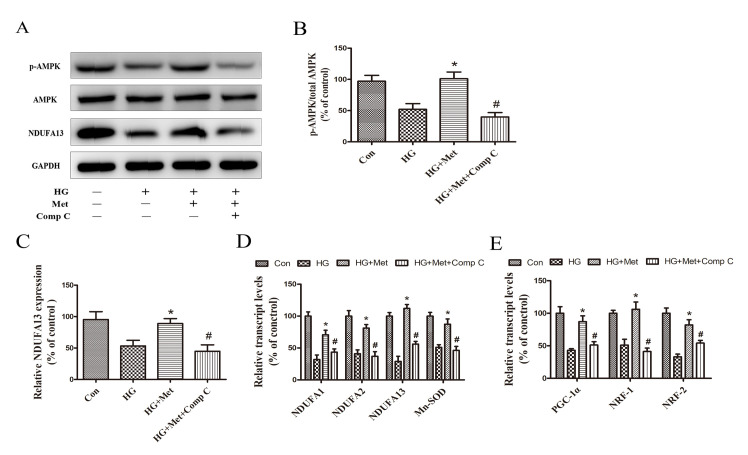
Compound C reduced the expression of NDUFA13 and mitochondrial biogenesis via suppressing AMPK pathway
To examine the dependency of AMPK in metformin-induced mitochondrial biogenesis and NDUFA13 expression, a selective AMPK chemical inhibitor (Compound C) was used. The results indicated that metformin-induced AMPK activation, as evidenced by an increase in p-AMPK expression levels, was blocked by pretreatment with Compound C (Fig. 5A and B). Compared with the HG + Met group, mitochondrial gene and mitochondrial biogenesis-related transcription factor expression levels were also significantly decreased in the HG + Met + Comp C group (Fig. 5D and E). In addition, metformin-induced NDUFA13 upregulation reversed by Compound C under HG conditions (Fig. 5A and C). The results suggested that the roles of metformin in NDUFA13 expression and mitochondrial biogenesis were AMPK signaling pathway-dependent.
Figure 5.
Role of the AMPK signaling pathway in the expression NDUFA13 and mitochondrial biogenesis. Cells were divided into four groups: i) Con, normal medium; ii) HG, 33.3 mM glucose; iii) HG + Met, 33.3 mM glucose pretreated with 1 mM Met; and iv) HG + Met + Comp C, 33.3 mM glucose pretreated with 1 mM Met and Comp C. Protein expression levels were (A) determined by western blotting and (B) semi-quantified for the ratio of p-AMPK/AMPK and (C) NDUFA13. mRNA expression levels of (D) NDUFA1, NDUFA2, NDUFA13, Mn-SOD, (E) PGC-1α, NRF1 and NRF2 were measured via reverse transcription-quantitative PCR. *P<0.05 vs. HG; #P<0.05 vs. HG + Met. AMPK, AMP-activated protein kinase; NDUF, NADH: Ubiquinone oxidoreductase subunit; HG, high glucose; Met, metformin; Comp C, Compound C; p, phosphorylated; Mn-SOD, manganese superoxide dismutase; PGC-1α, peroxisome proliferator-activated receptor-γ coactivator-1α; NRF, nuclear respiratory factor; Con, control.
Discussion
In the present study, metformin exerted protective effects on H9C2 cardiomyocytes by suppressing HG-induced oxidative stress, as evidenced by ameliorating HG-induced decreases in cell viability and SOD levels, and increases in ROS and MDA levels. mRNA expression levels of the mitochondrial genes (NDUFA1, NDUFA2, NDUFA13 and Mn-SOD) were upregulated by metformin under HG conditions compared with the HG group. Furthermore, under HG conditions, the expression levels of the mitochondrial protein NDUFA13 were increased by metformin compared with the HG group. The results also indicated that mitochondrial biogenesis-related transcription factors (PGC-1α, NRF1 and NRF2) were targets of metformin. The present study identified the role of the AMPK signaling pathway in the mechanism underlying metformin-mediated regulation of NDUFA13 and mitochondrial biogenesis based on the effects of the AMPK inhibitor, Compound C. The results indicated that metformin protected cardiac cells against HG-induced oxidative stress via a mechanism involving p-AMPK, NDUFA13 and mitochondrial biogenesis.
Although the association between mitochondrial biogenesis and ROS production is not completely understood, it has been reported that hyperglycemia-induced ROS impairs mitochondrial biogenesis and mitochondrial function (12,22). Metformin is an orally administered biguanide that is widely used to treat type 2 diabetes (4). The cardioprotective effects of metformin have received increasing attention and may be related to the ability of metformin to regulate oxidative stress; however, the exact mechanism is not completely understood (23).
A previous study identified the role of metformin in mitochondrial biogenesis, which is an endogenous protective response to stress (8). Mitochondrial biogenesis is significantly impaired in cardiomyocytes from db/db mice, as indicated by reduced mitochondrial DNA content and mitochondrial respiratory chain activity (24). Metformin administration promotes mitochondrial biogenesis and improves mitochondrial function in the skeletal muscle and human umbilical vein endothelial cells by activating PGC-1α (25). Activated PGC-1α is a powerful inducer of NRF-1, NRF-2 and mitochondrial transcription factor A, which initiates the expression of nuclear and mitochondrial genes encoding mitochondrial respiratory chain proteins (26). Although the role of metformin in mitochondrial biogenesis has been investigated, the effects of metformin in cardiomyocytes have not been fully elucidated.
The results of the current study suggested that pretreatment with metformin increased the expression of mitochondrial complex I subcomplexes, including NDUFA1, NDUFA2 and NDUFA13, under HG conditions. Additionally, the expression levels of mitochondrial biogenesis-related transcription factors, such as PGC-1α, NRF-1 and NRF-2, and the antioxidant enzyme, Mn-SOD, were regulated by metformin under HG conditions. The results suggested that metformin stimulated cardiomyocyte mitochondrial biogenesis, and may serve a critical role in cellular defense and cell survival responses to HG-induced oxidative stress.
The present study investigated the role of AMPK activation in metformin-mediated effects in cardiomyocytes. AMPK is a highly conserved sensor of cellular energy homeostasis and is the most recognized factor that mediates multiple effects of metformin (27). AMPK activation causes mitochondrial biogenesis (28). Mice expressing a dominant-negative form of AMPK in skeletal muscle are unable to increase mitochondrial biogenesis in response to energy deprivation (29). Activated AMPK stimulates PGC-1α, NRF1 and NRF-2, which in turn promote mitochondrial biogenesis (30). In the present study, metformin activated AMPK and enhanced the expression of PGC-1α, NRF1 and NRF-2 under HG conditions compared with the HG group. The expression of mitochondrial genes NDUFA1, NDUFA2, NDUFA13 and Mn-SOD was also regulated by metformin. The effects of metformin on expression were reversed by Compound C; therefore, the results suggested that AMPK served an important role in promoting mitochondrial biogenesis and the expression of mitochondrial genes in response to metformin. Additionally, alternative signaling pathways involving AMPK, Mn-SOD and PGC-1α, and those independent of mitochondrial genes should be investigated. Tumor suppressor p53-mediated PGC-1α upregulation suppresses increased levels of oxidative stress via nuclear factor (erythroid-derived 2)-like2-mediated expression of Mn-SOD and γ-glutamyl cysteine ligase without modulating mitochondrial biogenesis (31).
The present study suggested that the expression of the mitochondrial protein NDUFA13, a newly identified accessory subunit of mitochondrial complex I, was increased by metformin under HG conditions. Furthermore, the results indicated that NDUFA13 was regulated by metformin via an AMPK signal-dependent pathway, as evidenced by the inhibition of NDUFA13 induction in HG conditions by Compound C.
NDUFA13 was originally identified as a death activator in tumor cells and was recognized as an indispensable subunit of mitochondrial complex I (32). Elimination of NDUFA13 prevents the assembly and electron transfer activity of complex I, and also influences other complexes in the mitochondrial respiratory chain (14). The role of NDUFA13 in H9C2 cardiomyocytes and the relationship among NDUFA13, metformin and AMPK are not completely understood. However, mutations in the genes encoding subunits of complex I and complex I increase ROS generation (33). NDUFA13 downregulation increases basal ROS generation, which may serve as a survival signal by activating the STAT3/Bcl-2 signaling pathway (34). In the present study, NDUFA13 expression levels were decreased in HG-induced H9C2 cardiomyocytes compared with control H9C2 cardiomyocytes. Therefore, it was hypothesized that reduced expression levels of NDUFA13 in H9C2 cardiomyocytes were responsible for elevated ROS production under HG conditions, which may explain the antioxidant effects of metformin.
Moreover, it was observed that mitochondrial Mn-SOD was upregulated by metformin under HG conditions. Mn-SOD is a major antioxidant mitochondrial enzyme that serves as a primary ROS regulator. Mn-SOD-deficient mitochondria are more susceptible to oxidative stress and exhibit ultrastructural damage (35). The present study demonstrated that increased Mn-SOD expression might be involved in the mechanism underlying metformin-mediated amelioration of oxidative stress.
The present study had several limitations. AMPK kinase inhibitor Compound C can also inhibit vascular endothelial growth factor and bone morphogenetic proteins receptors (36,37). Therefore, further investigations into the effects of AMPK deficiency, for example, are required to verify the results of the present study. In addition, only the H9c2 cell line was used in the present study; therefore, in vivo studies or in vitro studies involving additional cell lines are required to verify the conclusions of the present study.
In conclusion, the present study suggested that metformin protected cardiomyocytes against HG-induced oxidative stress. The results indicated that metformin-mediated protection occurred at least in part via promoting mitochondrial biogenesis and NDUFA13 expression. In addition, the present study indicated that the AMPK signaling pathway was associated with the mechanisms underlying metformin-mediated regulation of NDUFA13 and mitochondrial biogenesis. The results suggested that AMPK may serve as a potential therapeutic target for normalizing mitochondrial function in diabetes, and NDUFA13 may serve as a useful target for designing novel pharmacological approaches to prevent diabetic complications.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science Foundation of China (grant no. 81771496), Natural Science Foundation of China (grant no. 881800331), School of Medicine, Shanghai Jiao Tong University (grant no. 16XJ21006), Shanghai Municipal Health Commission (grant no. 201940079) and Interdisciplinary Program of Shanghai Jiao Tong (grant no. YG2017QN23).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
QYZ, LLW and XDL designed the study. YGL, GYW, YZ, MLY and YGB performed the experiments. XDL and YGL drafted the manuscript. MW and XBL helped with the statistical analysis. QYZ, MW and XBL revised the paper. All authors reviewed the manuscript and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hayat SA, Patel B, Khattar RS, Malik RA. Diabetic cardiomyopathy: Mechanisms, diagnosis and treatment. Clin Sci (Lond) 2004;107:539–557. doi: 10.1042/CS20040057. [DOI] [PubMed] [Google Scholar]
- 2.Cai L, Kang YJ. Oxidative stress and diabetic cardiomyopathy: A brief review. Cardiovasc Toxicol. 2001;1:181–193. doi: 10.1385/ct:1:3:181. [DOI] [PubMed] [Google Scholar]
- 3.Khullar M, Al-Shudiefat AA, Ludke A, Binepal G, Singal PK. Oxidative stress: A key contributor to diabetic cardiomyopathy. Can J Physiol Pharmacol. 2010;88:233–240. doi: 10.1139/Y10-016. [DOI] [PubMed] [Google Scholar]
- 4.Sanchez-rangel E, Inzucchi SE. Metformin: Clinical use in type 2 diabetes. Diabetologia. 2017;60:1586–1593. doi: 10.1007/s00125-017-4336-x. [DOI] [PubMed] [Google Scholar]
- 5.Intensive blood-glucose control with sulphonylureas or insulin compared with conventional treatment and risk of complications in patients with type 2 diabetes (UKPDS 33). UK prospective diabetes study (UKPDS) group. Lancet. 1998;352:837–853. [PubMed] [Google Scholar]
- 6.Varjabedian L, Bourji M, Pourafkari L, Nader ND. Cardioprotection by metformin: Beneficial effects beyond glucose reduction. Am J Cardiovasc Drugs. 2018;18:181–193. doi: 10.1007/s40256-018-0266-3. [DOI] [PubMed] [Google Scholar]
- 7.Calvert JW, Gundewar S, Jha S, Greer JJM, Bestermann WH, Tian R, Lefer DJ. Acute metformin therapy confers cardioprotection against myocardial infarction via AMPK-eNOS-mediated signaling. Diabetes. 2008;57:696–705. doi: 10.2337/db07-1098. [DOI] [PubMed] [Google Scholar]
- 8.Hu M, Ye P, Liao H, Chen M, Yang F. Metformin protects H9C2 cardiomyocytes from high-glucose and hypoxia/reoxygenation injury via inhibition of reactive oxygen species generation and inflammatory responses: Role of AMPK and JNK. J Diabetes Res. 2016;2016:2961954. doi: 10.1155/2016/2961954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim AS, Miller EJ, Young LH. AMP-activated protein kinase: A core signalling pathway in the heart. Acta Physiol (Oxf) 2009;196:37–53. doi: 10.1111/j.1748-1716.2009.01978.x. [DOI] [PubMed] [Google Scholar]
- 10.Paiva MA, Rutter-Locher Z, Gonçalves LM, Providência LA, Davidson SM, Yellon DM, Mocanu MM. Enhancing AMPK activation during ischemia protects the diabetic heart against reperfusion injury. Am J Physiol Heart Circ Physiol. 2011;300:H2123–H2134. doi: 10.1152/ajpheart.00707.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusmic C, L'abbate A, Sambuceti G, Drummond G, Barsanti C, Matteucci M, Cao J, Piccolomini F, Cheng J, Abraham NG. Improved myocardial perfusion in chronic diabetic mice by the up-regulation of pLKB1 and AMPK signaling. J Cell Biochem. 2010;109:1033–1044. doi: 10.1002/jcb.22486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng A, Li H, Xu J, Cao K, Li H, Pu W, Yang Z, Peng Y, Long J, Liu J, Feng Z. Hydroxytyrosol improves mitochondrial function and reduces oxidative stress in the brain of db/db mice: Role of AMP-activated protein kinase activation. Br J Nutr. 2015;113:1667–1676. doi: 10.1017/S0007114515000884. [DOI] [PubMed] [Google Scholar]
- 13.Vakifahmetoglu-norberg H, Ouchida AT, Norberg E. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun. 2017;482:426–431. doi: 10.1016/j.bbrc.2016.11.088. [DOI] [PubMed] [Google Scholar]
- 14.Huang G, Lu H, Hao A, Ng DC, Ponniah S, Guo K, Lufei C, Zeng Q, Cao X. GRIM-19, a cell death regulatory protein, is essential for assembly and function of mitochondrial complex I. Mol Cell Biol. 2004;24:8447–8456. doi: 10.1128/MCB.24.19.8447-8456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li YG, Han BB, Li F, Yu JW, Dong ZF, Niu GM, Qing YW, Li JB, Wei M, Zhu W. High glucose induces down-regulated GRIM-19 expression to activate STAT3 signaling and promote cell proliferation in cell culture. PLoS One. 2016;11:e0153659. doi: 10.1371/journal.pone.0153659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sazanov LA. Respiratory complex I: Mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry. 2007;46:2275–2288. doi: 10.1021/bi602508x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu X, Zhang L, Li X, Zhou Z, Jiao L, Shao Y, Li M, Leng B, Zhou Y, et al. Metformin protects against H2O2-induced cardiomyocyte injury by inhibiting the miR-1a-3p/GRP94 pathway. Mol Ther Nucleic Acids. 2018;13:189–197. doi: 10.1016/j.omtn.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang J, Jung HH, Yang JY, Lee S, Choi J, Im GJ, Chae SW. Protective effect of metformin against cisplatin-induced ototoxicity in an auditory cell line. J Assoc Res Otolaryngol. 2014;15:149–158. doi: 10.1007/s10162-013-0431-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Method. 2001;408:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Foretz M, Guigas B, Bertrand L, Pollak M, Viollet B. Metformin: From mechanisms of action to therapies. Cell Metab. 2014;20:953–966. doi: 10.1016/j.cmet.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Dowling RJO, Lam S, Bassi C, Mouaaz S, Aman A, Kiyota T, Al-Awar R, Goodwin PJ, Stambolic V. Metformin pharmacokinetics in mouse tumors: Implications for human therapy. Cell Metab. 2016;23:567–568. doi: 10.1016/j.cmet.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 22.Xie L, Zhu X, Hu Y, Li T, Gao Y, Shi Y, Tang S. Mitochondrial DNA oxidative damage triggering mitochondrial dysfunction and apoptosis in high glucose-induced HRECs. Invest Ophthalmol Vis Sci. 2008;49:4203–4209. doi: 10.1167/iovs.07-1364. [DOI] [PubMed] [Google Scholar]
- 23.Kukidome D, Nishikawa T, Sonoda K, Imoto K, Fujisawa K, Yano M, Motoshima H, Taguchi T, Matsumura T, Araki E. Activation of AMP-activated protein kinase reduces hyperglycemia-induced mitochondrial reactive oxygen species production and promotes mitochondrial biogenesis in human umbilical vein endothelial cells. Diabetes. 2006;55:120–127. [PubMed] [Google Scholar]
- 24.Yan W, Zhang H, Liu P, Wang H, Liu J, Gao C, Liu Y, Lian K, Yang L, Sun L, et al. Impaired mitochondrial biogenesis due to dysfunctional adiponectin-AMPK-PGC-1α signaling contributing to increased vulnerability in diabetic heart. Basic Res Cardiol. 2013;108:329. doi: 10.1007/s00395-013-0329-1. [DOI] [PubMed] [Google Scholar]
- 25.Karnewar S, Neeli PK, Panuganti D, Kotagiri S, Mallappa S, Jain N, Jerald MK, Kotamraju S. Metformin regulates mitochondrial biogenesis and senescence through AMPK mediated H3K79 methylation: Relevance in age-associated vascular dysfunction. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1115–1128. doi: 10.1016/j.bbadis.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 26.Chen SD, Yang DI, Lin TK, Shaw FZ, Liou CW, Chuang YC. Roles of oxidative stress, apoptosis, PGC-1α and mitochondrial biogenesis in cerebral ischemia. Int J Mol Sci. 2011;12:7199–7215. doi: 10.3390/ijms12107199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adeghate E, Singh J. Structural changes in the myocardium during diabetes-induced cardiomyopathy. Heart Fail Rev. 2014;19:15–23. doi: 10.1007/s10741-013-9388-5. [DOI] [PubMed] [Google Scholar]
- 28.Nanjaiah H, Vallikannan B. Enhanced phosphorylation of AMPK by lutein and oxidised lutein that lead to mitochondrial biogenesis in hyperglycemic HepG2 cells. J Cell Biochem. 2019;120:15255–15267. doi: 10.1002/jcb.28793. [DOI] [PubMed] [Google Scholar]
- 29.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci USA. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song P, Kwon Y, Yea K, Moon HY, Yoon JH, Ghim J, Hyun H, Kim D, Koh A, Berggren PO, et al. Apolipoprotein a1 increases mitochondrial biogenesis through AMP-activated protein kinase. Cell Signal. 2015;27:1873–1881. doi: 10.1016/j.cellsig.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 31.Aquilano K, Baldelli S, Pagliei B, Cannata SM, Rotilio G, Ciriolo MR. p53 orchestrates the PGC-1α-mediated antioxidant response upon mild redox and metabolic imbalance. Antioxid Redox Signal. 2013;18:386–399. doi: 10.1089/ars.2012.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreira S, Correia M, Soares P, Máximo V. GRIM-19 function in cancer development. Mitochondrion. 2011;11:693–699. doi: 10.1016/j.mito.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 33.Fato R, Bergamini C, Leoni S, Strocchi P, Lenaz G. Generation of reactive oxygen species by mitochondrial complex I: Implications in neurodegeneration. Neurochem Res. 2008;33:2487–2501. doi: 10.1007/s11064-008-9747-0. [DOI] [PubMed] [Google Scholar]
- 34.Hu H, Nan J, Sun Y, Zhu D, Xiao C, Wang Y, Zhu L, Wu Y, Zhao J, Wu R, et al. Electron leak from NDUFA13 within mitochondrial complex I attenuates ischemia-reperfusion injury via dimerized STAT3. Proc Natl Acad Sci USA. 2017;114:11908–11913. doi: 10.1073/pnas.1704723114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams MD, Van Remmen H, Conrad CC, Huang TT, Epstein CJ, Richardson A. Increased oxidative damage is correlated to altered mitochondrial function in heterozygous manganese superoxide dismutase knockout mice. J Biol Chem. 1998;273:28510–28515. doi: 10.1074/jbc.273.43.28510. [DOI] [PubMed] [Google Scholar]
- 36.Yu PB, Hong CC, Sachidanandan C, Babitt JL, Deng DY, Hoyng SA, Lin HY, Bloch KD, Peterson RT. Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat Chem Biol. 2008;4:33–41. doi: 10.1038/nchembio.2007.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao J, Ho JN, Lewis JA, Karim KA, Daniels RN, Gentry PR, Hopkins CR, Lindsley CW, Hong CC. In vivo structure-activity relationship study of dorsomorphin analogues identifies selective VEGF and BMP inhibitors. ACS Chem Biol. 2010;5:245–253. doi: 10.1021/cb9002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.