Abstract
Human cytomegalovirus (HCMV) is an opportunistic human herpesvirus that causes a sight-threatening retinitis in immunosuppressed patients, especially those with AIDS. Using an established model of experimental murine cytomegalovirus (MCMV) retinitis in mice with retrovirus-induced immunodeficiency (MAIDS), we have been attempting to define with greater clarity the immunologic mechanisms that contribute to the progression of AIDS-related HCMV retinitis in the unique immunosuppressive setting of HIV infection. Toward this end, we provide herein a comprehensive assessment of immune response gene expression during the onset and development of MAIDS-related MCMV retinitis employing NanoString nCounter. In so doing, we analyzed and compared the intraocular expressions of 561 immune response genes within MCMV-infected eyes of groups of healthy mice, MCMV-infected mice with MAIDS of 4 weeks’ (MAIDS-4) duration, and MCMV-infected eyes of mice with MAIDS of 10 weeks’ (MAIDS-10) duration. These animal groups show a progression of retinal disease from absolute resistance to retinitis development in healthy mice to the development of classic full-thickness retinal necrosis in MAIDS-10 mice but through an intermediate stage of retinal disease development in MAIDS-4 mice. Our findings showed that increased susceptibility to MCMV retinitis during the progression of MAIDS is associated with robust upregulation or downregulation of a surprisingly large number of immune response genes that operate within several immune response pathways often unique to each animal group. Analysis of 14 additional immune response genes associated with programmed cell death pathways suggested involvement of necroptosis and pyroptosis during MAIDS-related MCMV retinitis pathogenesis. Use of the NanoString nCounter technology provided new and unexpected information on the immunopathogenesis of retinitis within MCMV-infected eyes of mice with retrovirus-induced immunosuppression. Our findings may provide new insights into the immunologic events that operate during the pathogenesis of AIDS-related HCMV retinitis.
Author summary
The immunodeficiency caused by HIV during the progression of AIDS creates a unique systemic immunosuppressive environment that often leads to the development of opportunistic virus infections. Among these is a sight-threatening retinitis caused by human cytomegalovirus (HCMV). While the clinical and histopathologic features of AIDS-related HCMV retinitis have been well documented, the immunologic mechanisms that continue to operate during HIV-induced immunosuppression and contribute to the onset and development of retinal tissue destruction caused by ocular HCMV infection in patients with AIDS remain unexplored. Herein, we report the first comprehensive analysis of a total of 571 immune response genes that are expressed during the pathogenesis of experimental murine cytomegalovirus (MCMV) retinitis in mice at different stages of retrovirus-induced immunosuppression (MAIDS) who exhibit different susceptibilities to retinal disease development. Our finding that different patterns of immune response gene expression are observed at different stages of retrovirus immunosuppression during increased susceptibility to retinal necrosis development adds new knowledge to our understanding of the contributions of unique innate and adaptive immune pathways towards the pathogenesis of AIDS-related HCMV retinitis and may extend to other AIDS-related opportunistic virus infections.
Introduction
The unique immunosuppressive environment created by HIV infection during the development of AIDS resulted in the emergence of a significant number of diseases caused by opportunistic viruses that prior to AIDS were no more than rare medical curiosities. One such AIDS-related opportunistic disease that appeared as a consequence of HIV-induced immunosuppression is a sight-threatening retinitis caused by human cytomegalovirus (HCMV), a β herpesvirus [1,2]. Although HCMV retinitis had been documented occasionally in a patient immunosuppressed for solid-organ or bone-marrow allografts [3], the appearance of an AIDS-related HCMV retinitis was quickly observed in up to 42% of this patient population at the outset of AIDS in the United States [4]. Today the number of cases of AIDS-related HCMV retinitis in the United States has decreased significantly due to the use of combination antiretroviral therapy (ART) but nonetheless remains an ophthalmologic problem in some parts of the world in patients who do not have access to ART or who do not respond to ART [5–8].
The clinical and histopathologic features of AIDS-related HCMV retinitis have been well documented [9–12]. To extend this knowledge to include the virologic, immunologic, and pathogenic mechanisms that operate to allow the onset and progression of HCMV retinitis in patients with AIDS, we have been using an experimental animal model of murine cytomegalovirus (MCMV) retinitis that develops in mice with retrovirus-induced immunodeficiency (MAIDS) [13]. MAIDS-related MCMV retinitis mimics AIDS-related HCMV retinitis in many ways. These include the appearance of generalized lymphadenopathy, polyclonal B cell activation, hypergammaglobulinemia, and a Th1 to Th2 shift in cytokine profile accompanied by progressive dysfunction of cellular immunity that takes place over weeks following systemic infection with an immunosuppressive murine retrovirus mixture [14]. These events culminate in the development of MAIDS by 8 to 10 weeks after retrovirus infection that allows susceptibility to a retinitis in eyes inoculated with MCMV [15]. Importantly, MAIDS-related MCMV retinitis exhibits histopathologic features identical to those of AIDS-related HCMV retinitis. These include the emergence of a full-thickness retinal necrosis with prominent cytomegalic cells that develops within MCMV-infected retinal tissues at a frequency of 80 to 100% by 10 days after intraocular MCMV inoculation [13].
Previous work by us has used this animal model of MAIDS-related MCMV retinitis as an experimental platform to understand with greater clarity the pathogenesis of AIDS-related HCMV retinitis as dictated within the host by the unique immunologic milieu created by an immunosuppressive retrovirus. Past emphasis has been on various components of innate and adaptive immunity of the host with more recent focus being given to individual cytokines and a number of programmed cell death pathways [16]. While informative, these past studies have not been at the depth needed to identify and compare the extraordinary number of immune response genes expressed simultaneously within the ocular compartment at critical times during the evolution of MAIDS-related MCMV retinitis. In an attempt to overcome this obstacle, we employed NanoString nCounter technology which allows for the direct measurement of immune response gene expression levels without amplification during the onset and progression of retinitis within MCMV-infected eyes of mice with MAIDS. Herein, we report the intraocular expression of 575 immune response genes within MCMV-infected eyes of mice at different stages of MAIDS development and compared with MCMV-infected eyes of immunologically normal mice. As expected, our results show that intraocular MCMV infection of mice with MAIDS results in the upregulation or downregulation of immune response genes associated with several distinct immune response pathways during retinitis development. In addition, some immune response genes and pathways identified in this investigation have been surprising and not recognized by us previously, thereby extending our understanding of the role of various immune responses toward the pathogenesis of MAIDS-related MCMV retinitis. Furthermore, we have observed dramatic differences in the expression of immune response genes to intraocular virus infection depending on the resistance or degree of susceptibility of MCMV-infected eyes to retinitis development.
Results
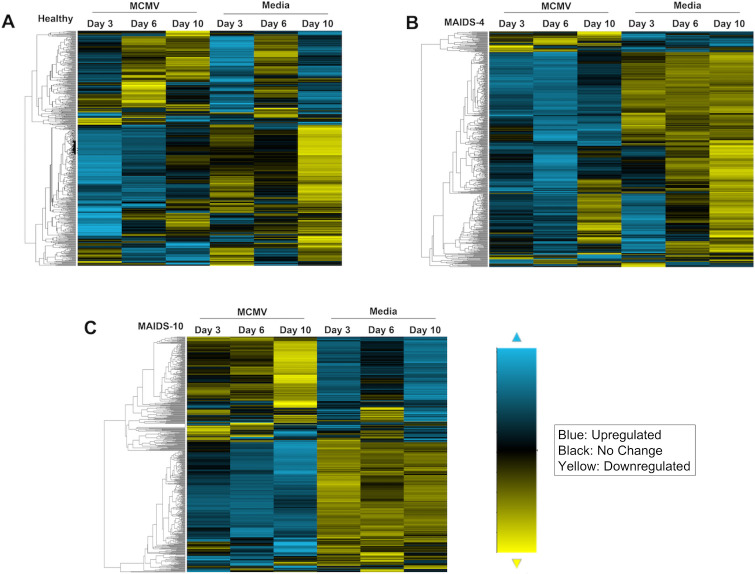
Hierarchical clustering analysis of 561 immune response gene transcripts within MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice
Several prior investigations by us have confirmed major differences in the susceptibility to development of full-thickness retinal necrosis when comparing the MCMV-infected eyes of healthy mice with the MCMV-infected eyes of mice at different stages of retrovirus-induced immunosuppression [13,17]. Whereas the retinal architecture of MCMV-infected eyes of healthy mice remain normal without histopathologic evidence of retinal disease, the MCMV-infected eyes of mice with MAIDS of 10-weeks duration (MAIDS-10 mice) show a progressive development of retinitis at 3, 6, and 10 days after intraocular MCMV inoculation that culminates in severe, full-thickness retinal necrosis at 10 days after inoculation [13]. Intermediate between the two histopathologic extremes of the eyes of healthy mice and MAIDS-10 mice inoculated with virus are the MCMV-infected eyes of mice with MAIDS of 4-weeks duration (MAIDS-4 mice) that show only mild retinal pigment epithelium (RPE) proliferation with preservation of the neurosensory retina but with an absolute absence of full-thickness retinal necrosis throughout the course of infection [17]. These diverse pathogenic outcomes among healthy mice, MAIDS-4 mice, and MAIDS-10 mice following intraocular MCMV infection therefore allowed us the unique opportunity to compare MCMV-infected eyes of these animal groups for simultaneous detection and quantification of transcript expression for a large number of immunologic pathways and related components throughout the present investigation.
Initial studies were performed to determine the overall change in expression of 561 immune defense genes during the pathogenesis of MCMV retinitis. This was accomplished using a commercial NanoString nCounter Murine Immunology Panel that included 15 additional internal reference and housekeeping genes [18]. The left eyes of groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3) were inoculated with MCMV; the contralateral right eyes of each animal group were mock-infected with maintenance medium only and served as internal controls. At 3, 6, and 10 days after intraocular inoculation, individual MCMV-infected and mock-infected eyes were collected from all animal groups, subjected to total RNA extraction, and the MCMV-infected or mock-infected eyes were pooled by groups and subjected to NanoString nCounter analysis. S1 Table details our findings for MCMV-infected eyes for each of the 561 immune defense gene analyzed whether upregulated or downregulated for each animal group of this investigation. Included are their overall fold-change expression profiles together with p values at 3, 6, and 10 days postinfection when compared with mock-infected eyes.
Hierarchical clustering analysis of the expression of 561 immune response gene transcripts analyzed for each group of MCMV-infected eyes or mock-infected eyes are presented in Fig 1. Inspection of these data indicates that patterns of gene expression differed greatly when comparing mock-infected and MCMV-infected eyes at each time point examined. More importantly, the patterns of immune response gene expression were remarkably distinct for each animal group, reflecting resistance or the degree of susceptibility to MCMV retinitis development. The MCMV-infected eyes of healthy mice without MAIDS that show absolute resistance to retinitis development [17] nonetheless exhibited active gene expression (Fig 1A). At least some of this transcriptional activity might be attributed to an intraocular trauma created in response to the needlestick that takes place during mock infection. In comparison, MCMV-infected eyes of MAIDS-4 mice and MAIDS-10 mice also showed active gene expression but with increased upregulation of genes when compared with MCMV-infected eyes of healthy mice. Of interest, MCMV-infected eyes of MAIDS-4 mice that fail to develop full-thickness retinal necrosis but nonetheless exhibit RPE proliferation showed an unexpected and extensive upregulation of a majority of the 561 immune response genes investigated at day 6 after virus inoculation (Fig 1B) when compared with retinitis-susceptible MCMV-infected eyes of MAIDS-10 mice at day 6 after virus inoculation (Fig 1C). This outcome was particularly surprising because of our previous observation that the MCMV-infected eyes of both animal groups harbor high but equivalent amounts of infectious virus [17]. Moreover, distinctly different expression patterns for individual immune response genes were observed within the MCMV-infected eyes of MAIDS-10 mice following development of full-thickness retinal necrosis but not within MCMV-infected eyes of MAIDS-4 mice resistant to full-thickness retinal necrosis development.
Fig 1. Hierarchical clustering analysis of 561 immune response gene transcripts within MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice.
Whole MCMV-infected eyes (MCMV) and mock-infected (Media) eyes were collected 3, 6, and 10 days after intraocular inoculation from groups of (A) healthy mice (n = 3), (B) MAIDS-4 mice (n = 3), and (C) MAIDS-10 mice (n = 3). Total RNA was extracted from individual MCMV-infected eyes or mock-infected eyes and pooled for each group. 100ng of RNA from each group were loaded onto a Murine Immunology Panel, specifically designed for the NanoString nCounter. Hierarchical clustering analysis for each of the 561 genes was performed using the nSolver software. The bar indicates range of transcriptional activity with blue indicating upregulation, yellow indicating downregulation, and black indicating no change in mRNA expression.
Comparison of MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice for the expression of genes associated with distinct immunologic pathways
We next processed the hierarchical clustering analysis of the 561 immune response gene transcripts from MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice at all time points examined for their involvement in 32 distinct immunologic pathways using the NanoString nSolver software. After determining the fold-change upregulation or downregulation of differentially expressed immune-response genes of MCMV-infected eyes when compared with mock-infected eyes for each animal group, a fold change of less than two was used to exclude that particular gene from further analysis. This approach revealed 17 genes were upregulated and 15 genes were downregulated within MCMV-infected eyes of healthy mice, 83 genes upregulated and 4 genes downregulated within MCMV-infected eyes of MAIDS-4 mice, and 92 genes upregulated and 54 genes downregulated within MCMV-infected eyes of MAIDS-10 mice (Fig 2). An increasing trend in immune response-related gene activity was observed when MCMV-infected eyes of healthy mice were compared to MCMV-infected eyes of MAIDS-4 mice and then compared to MCMV-infected eyes of MAIDS-10 mice. This trend is presumably due to an increased susceptibility of the MCMV-infected eyes of these animal groups toward development of full-thickness retinal necrosis as suggested in the resulting heatmaps (Fig 1).
Fig 2. Comparison of the number of upregulated and downregulated immune-response genes for MCMV-infected eyes of groups of healthy mice, MAIDS-4 mice, and MAIDS-10 mice.
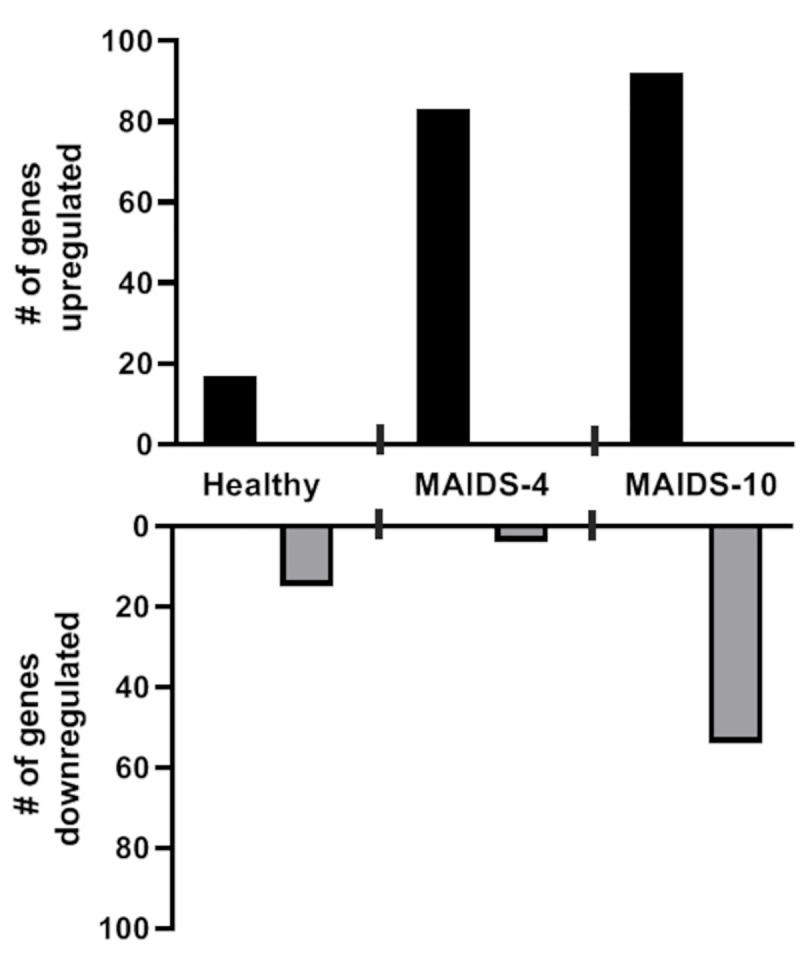
Comparison of the number of differentially expressed immune-response genes with a fold change of two-fold or greater for MCMV-infected eyes collected from groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3) when compared with mock-infected eyes at all time points examined.
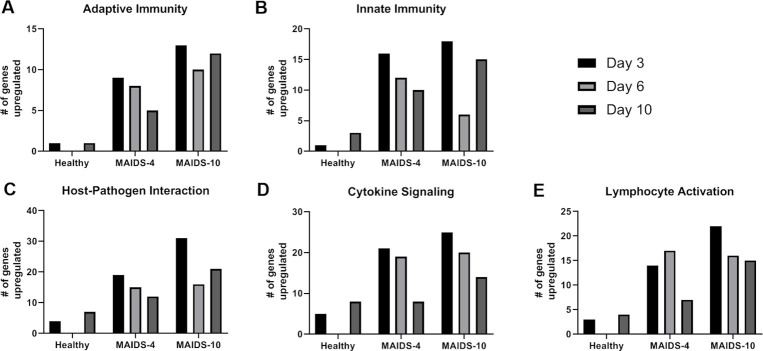
This trend also continued when the differentially expressed immune response genes showing upregulation of activity were organized into NanoString-defined immunologic pathways for each animal group with the understanding that each gene could be involved in multiple pathways. Categorization into immunologic pathways revealed that five of the 32 NanoString-defined pathways exhibited relatively substantial stimulation of gene activity within MCMV-infected eyes when compared with other pathways of MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice during progressive susceptibility to MCMV retinitis. Those pathways showing the most robust stimulation in gene activity included pathways associated with the broad categories of adaptive immunity, innate immunity, host-pathogen interactions, cytokine signaling, and lymphocyte activation (Fig 3). More functionally focused pathways such as those involved with NF-κB signaling, toll-like receptor signaling, NOD-like receptor signaling, chemokine signaling, type 1 interferon signaling, type 2 interferon signaling, tumor necrosis factor (TNF) family signaling, MHC class I antigen presentation, and phagocytosis and degradation also showed less robust but nonetheless heightened gene activity when comparing MCMV-infected eyes of MAIDS-4 and MAIDS-10 mice with MCMV-infected eyes of healthy mice (S1 Fig). Overall, these findings demonstrate that gene activity associated with immunologic pathways becomes progressively and dramatically more active in number and function as MCMV-infected eyes become progressively more susceptible to the onset and development of full-thickness retinal necrosis as retrovirus-induced immunosuppression ensues. Moreover, this progressive gene activity is far more complex than was originally thought and appears to involve a large number of immune pathways of which several were never considered by us to be involved in the evolution of cytomegalovirus retinitis in retrovirus-immunosuppressed hosts.
Fig 3. Number of upregulated immune response genes within MCMV-infected eyes in groups of healthy mice, MAIDS-4 mice, and MAIDS-10 mice when categorized according to major immunologic pathways.
Transcriptional activity of immune response genes of MCMV-infected eyes in groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3) that registered a fold change of > two, when compared with mock-infected eyes at 3, 6, or 10 days postinfection. These genes were categorized into five NanoString-defined immunologic pathways that exhibited the most robust upregulation. Immunologic pathways included the broad categories of: (A) Adaptive Immunity, (B) Innate Immunity, (C) Host-Pathogen Interaction, (D) Cytokine Signaling, and (E) Lymphocyte Activation.
A comparison of the top 15 differentially expressed immunologic-associated genes within MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice
In an attempt to provide a more quantitative analysis of the progressive response(s) of the 561 immune response genes analyzed within MCMV-infected eyes as they relate to the progressive susceptibility to retinitis development, we focused on 15 differentially expressed genes which showed the greatest upregulation of gene expression at 3, 6, or 10 days after intraocular MCMV inoculation and compared these levels with those of mock-infected eyes for groups of healthy mice, MAIDS-4 mice, and MAIDS-10 mice. Table 1 summarizes our findings for MCMV-infected eyes for each animal group with respect to individual genes, their known function(s), their overall fold change at peak expression along with their p values when compared with mock-infected eyes [19–61]. In agreement with our previous observations, the fold-change for stimulation of the top 15 individual genes within MCMV-infected eyes appeared to increase markedly from healthy mice to MAIDS-4 mice and MAIDS-10 mice, seemingly as a reflection of increased susceptibility to the onset and development of MCMV retinitis among these animal groups, especially MAIDS-4 animals when compared with MAIDS-10 animals. While the average fold-increase for peak expression in activity of these 15 genes was 3.04 for MCMV-infected eyes of healthy mice, the average fold-increase for peak expression in activity increased to 20.14 and 20.74 for MCMV-infected eyes of MAIDS-4 mice and MAIDS-10 mice, respectively, suggesting a far more dynamic intraocular gene transcription activity in response to MCMV infection during progressive retrovirus-induced immunosuppression than during immunocompetence. It is also noteworthy that a subset of individual genes exhibited remarkable transcription activity such as ccl5, a gene encoding for a chemokine associated with inflammation [51], that showed an 83.32-fold increase in activity within MCMV-infected eyes of MAIDS-10 mice susceptible to full-thickness retinal necrosis development when compared to mock-infected eyes.
Table 1. Summary of the 15 differentially expressed immune response genes showing the greatest upregulation of activity within MCMV-infected eyes of groups of healthy mice, MAIDS-4 mice, and MAIDS-10.
| Top 15 Genes Upregulated in MCMV infected Eyes of Healthy Mice | |||
| Gene | Function [Ref.] | Peak Expression | p value |
| Bst1 | Facilitates pre-B-cell growth and induces cell migration [19] | 3.25 | 0.0298 |
| Casp3 | Activation plays a role in the execution -phase of apoptosis [20] | 2.36 | 0.0117 |
| Ccl3 | Associated with macrophage recruitment [21] | 2.28 | 0.0324 |
| Ccl9 | Induces chemotaxis of CD4+ T cells, CD8+ T cells, and monocytes [22] | 3.09 | 0.0107 |
| Cd2 | Regulates natural killer cell lytic activity and proinflammatory cytokine production [23] | 2.74 | 0.0365 |
| Clec5a | Involved in neutrophil extracellular trap formation and proinflammatory cytokine production [24] | 3.74 | 0.0244 |
| Emr1 | Murine marker of macrophages (F4/80) [25] | 2.48 | 0.0280 |
| H2-K1 | Bind to and present antigens derived from pathogens onto the cell surfaces for T cell recognition [26] | 4.37 | 0.0444 |
| Ifnar2 | Part of IFN-α and IFN-β receptor and critical for antiviral immunity [27] | 2.10 | 0.0377 |
| Itgb2 | Involved in extravasation into tissues during infection or injury [28] | 3.03 | 0.0016 |
| Jak2 | Involved in signal transduction of interferon and cytokine signaling [29] | 2.70 | 0.0131 |
| Ptafr | Involved in proinflammatory signaling [30] | 2.53 | 0.0230 |
| Ptgs2 | Involved in the production of prostacyclin, expressed in inflammation [31] | 3.80 | 0.0011 |
| Stat2 | Aids in the activation of the transcription of interferon stimulated genes [32] | 4.37 | 0.0350 |
| Tgfb1 | Inhibits the actions of T cells and the secretion of IFN-γ, TNF-α, and interleukins [33, 34, 35] | 2.75 | 0.0091 |
| Top 15 Genes Upregulated in MCMV infected Eyes of MAIDS-4 Mice | |||
| Gene | Function [Ref.] | Peak Expression | p value |
| Ccl12 | Attracts eosinophils, monocytes, and lymphocytes to the site of infection [36] | 10.88 | 0.0450 |
| Ccl2 | Involved in chemotaxis and regulating inflammation [37] | 51.23 | 0.0353 |
| Ccl7 | Promotes the recruitment of monocytes and neutrophils to the site infection [38] | 34.39 | 0.0350 |
| Ccr5 | Acts as a receptor for chemokines [39] | 11.10 | 0.0123 |
| Cfb | Regulates the alternative pathway of the complement system [40] | 27.79 | 0.0030 |
| Cxcl10 | Attracts CD8+ and CD4+ T cells to the site of inflammation [41] | 30.24 | 0.0374 |
| Cxcl9 | Attracts T cells to the site of inflammation [42] | 28.12 | 0.0413 |
| Icos | Involved in the induction and regulation of Th1, Th2, and Th17 immunity [43] | 10.01 | 0.0354 |
| Ifit2 | Plays a role in the stimulation of interferons as part of an anti-viral response [44] | 17.09 | 0.0008 |
| Irgm1 | Involved in the polarization of M1 [inflammatory driven] macrophages [45] | 12.16 | 0.0450 |
| Itgal | Involved in leukocyte cellular adhesion and costimulatory signaling [46] | 11.48 | 0.0185 |
| Lilrb3 | Functions as an inhibitory receptor to help balance the function of innate immune cells [47] | 9.59 | 0.0407 |
| Lilrb4 | Transduces a negative signal that inhibits stimulation of the immune response [48] | 16.53 | 0.0209 |
| Ptprc | Suppresses JAK kinases as a negative regulator of cytokine signaling [49] | 10.54 | 0.0490 |
| Slamf7 | Induces B cell proliferation [50] | 21.03 | 0.0411 |
| Top 15 Genes Upregulated in MCMV infected Eyes of MAIDS-10 Mice | |||
| Gene | Function [Ref.] | Peak Expression | p value |
| Ccl2 | Involved in chemotaxis and regulating inflammation [37] | 35.19 | 0.0323 |
| Ccl5 | Promotes the recruitment of leukocytes to the site of infection [51] | 83.32 | 0.0263 |
| Cd274 | Plays a major role in suppressing the adaptive arm of the immune system [52] | 7.36 | 0.0145 |
| Ctss | Degrades antigenic proteins for antigen presentation [53] | 12.69 | 0.0443 |
| Cybb | Involved in the formation of reactive oxygen species [54] | 9.83 | 0.0191 |
| Fcgr3 | Participates in signal transduction triggering lysis by natural killer cells [55] | 6.92 | 0.0425 |
| Fcgr4 | Promotes macrophage-mediated phagocytosis and antigen presentation to T cells [56] | 10.03 | 0.0307 |
| Ifi204 | Acts as a nuclear innate DNA sensor resulting in inflammasome activation [57] | 15.33 | 0.0202 |
| Il1rn | Binds non-productively to the interleukin-1 receptor preventing IL-1 from sending a signal [58] | 48.31 | 0.0017 |
| Irgm1 | Involved in the polarization of M1 [inflammatory driven] macrophages [45] | 8.36 | 0.0303 |
| Lilrb3 | Functions as an inhibitory receptor to help balance the function of innate immune cells [47] | 10.74 | 0.0062 |
| S100a9 | Controls macrophage accumulation and cytokine production [59] | 17.85 | 0.0179 |
| Slamf7 | Induces B cell proliferation [50] | 29.00 | 0.0425 |
| Socs1 | Involved in the negative feedback regulation of cytokine signaling [60] | 6.98 | 0.0208 |
| Tyrobp | Activates signal transduction and plays a role in inflammation [61] | 9.42 | 0.0468 |
The fold changes in transcriptional activities within MCMV-infected eyes of groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3) were determined for all 561 differentially expressed immune response genes. The 15 genes for the MCMV-infected eyes showing the greatest fold change increase when compared with mock-infected eyes are summarized, with known function(s), peak expression, and p values.
A detailed comparison of the top 15 immune response genes activated within MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice also revealed that some upregulated gene activities were exclusive to each animal group whereas other upregulated gene activities were shared between and among groups. This is depicted in the Venn diagram shown in Fig 4. Whereas the top 15 upregulated genes of the MCMV-infected eyes of healthy mice that are absolutely resistant to MCMV retinitis development were found to be exclusive to this animal group, while 4 of the top 15 upregulated genes of the eyes of MCMV-infected mice of MAIDS-4 and MAIDS-10 groups were shared between and among the animal groups. This observation, however, should not diminish the observation that 11 of the top 15 upregulated genes of the MCMV-infected eyes of these animal groups were exclusive to MAIDS-4 mice and MAIDS-10 mice, animals that exhibited remarkably distinct patterns of MCMV-induced retinal disease [17].
Fig 4. Venn diagram comparing the expression of 15 differentially expressed immune response genes showing the greatest upregulation of activity within MCMV-infected eyes of groups of healthy mice, MAIDS-4 mice, and MAIDS-10.
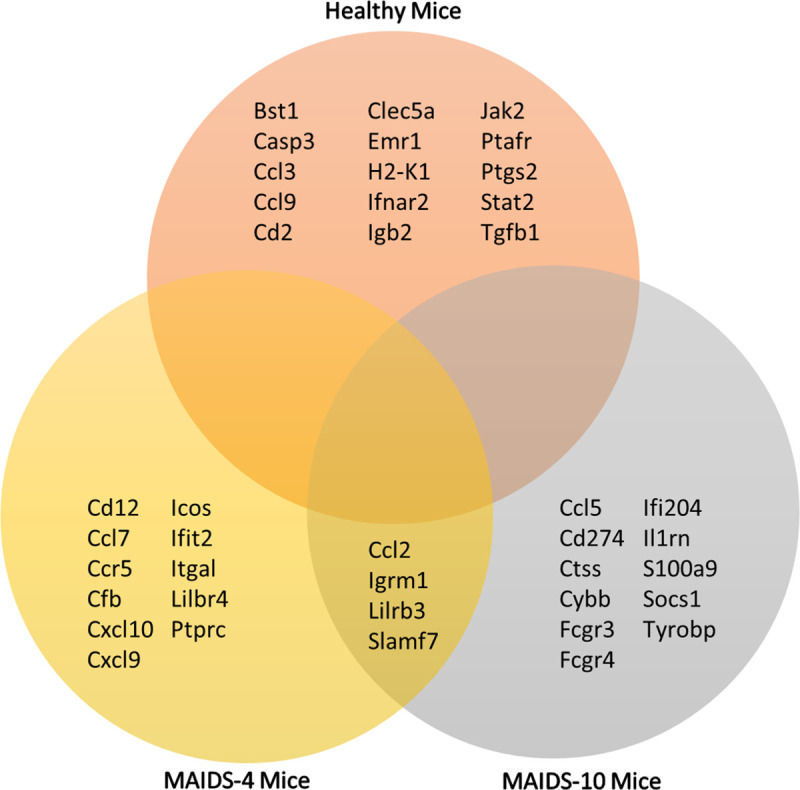
Following analysis of the differentially expressed transcriptional activity for 561 immune response genes within MCMV-infected eyes of groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3), those 15 genes showing the greatest upregulation of activity at all times examined postinfection (see Table 1) were compared for possible, shared activities among the three animal groups. While none of the genes presented shared activities among all three groups, of the 15 most active genes, 4 genes were found to be commonly expressed within the MCMV-infected eyes of MAIDS-4 mice and MAIDS-10 mice.
Analysis of 14 additional immune response gene transcripts of MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice associated with programmed cell death pathways
Use of the commercially available NanoString nCounter Murine Immunology Panel provided a wealth of new and, at times, unexpected changes in the expression of 561 immune response genes during the onset and development of MAIDS-related MCMV retinitis. Given our present interest in programmed cell death pathways and their relative roles in the pathogenesis of MCMV-induced full-thickness retinal necrosis in MAIDS-10 mice [62,63], we created a custom panel consisting of 14 genes. This panel included 3 genes associated with autophagy, 3 genes associated with necroptosis, 2 genes associated with parthanatos, and 6 genes associated with pyroptosis and inflammasomes. Importantly, gene transcription analysis using this custom gene panel for cell death pathways was performed using the same samples collected from MCMV-infected eyes of healthy mice, MAIDS-4 mice, and MAIDS-10 mice that were earlier used to generate data using the commercially available murine immunology gene panel. Genes associated with apoptosis were excluded from this custom panel because we have already determined previously, using mice with MAIDS deficient in key apoptosis-associated genes, that this programmed cell death pathway contributes only minimally to the pathogenesis of MAIDS-related MCMV retinitis [62].
A summary of the upregulation or downregulation for each of the 14 immune response genes within MCMV-infected eyes for each animal group at 3, 6, and 10 days after intraocular MCMV inoculation is shown in Table 2. The overall positive (upregulated) or negative (downregulated) fold changes at peak expression including their p values of these response genes when compared with mock-infected eyes is also shown. Owing to the relatively small number of 14 genes being analyzed in this experiment, a two-fold change in gene activity was not used to exclude some genes for analysis as was done for the 561 immune response genes analyzed above. The MCMV-infected eyes of healthy mice which are absolutely resistant to the development of MCMV retinitis [17] exhibited a pattern of cell death pathway-associated gene activities that suggested significant quiescence of activity for each pathway at all days postinfection examined. Indeed, downregulation of gene activity was consistently observed for all 3 necroptosis genes and all 6 pyroptosis and associated inflammasome genes at 3, 6, and 10 days postinfection. Gene activities for autophagy and parthanatos were also found to be either downregulated or only minimally upregulated (< 2-fold increase) within MCMV-infected eyes of healthy mice at all days postinfection examined. As MCMV-infected eyes of animals at different stages of MAIDS development became more susceptible to the development of retinal disease at different stages of MAIDS development, however, genes associated with some, but not all, cell death pathways under investigation became increasingly active. This was apparent within the MCMV-infected eyes of MAIDS-4 mice and MAIDS-10 mice for necroptosis and pyroptosis and pyroptosis-associated inflammasomes but not for autophagy and parthanatos.
Table 2. Summary of differentially expressed immune response genes associated with programmed cell death pathways within MCMV-infected eyes of groups of healthy mice, MAIDS-4 mice, and MAIDS-10 mice.
| Fold Change of MCMV-infected Eyes of Healthy Mice | |||||||
| Cell Death Pathway | Genes | Day 3 | Day 6 | Day 10 | |||
| Δ | p value | Δ | p value | Δ | p value | ||
| Autophagy | Atg12 | 1.07 | 0.3855 | 1.24 | 0.0792 | 1.14 | 0.1344 |
| Autophagy | Atg5 | -1.05 | 0.0155 | -1.13 | 0.0269 | -1.02 | 0.0047 |
| Autophagy | Becn1 | 0.05 | 0.5346 | -0.05 | 0.5090 | 1.07 | 0.3855 |
| Necroptosis | Mlkl | -3.53 | 0.1688 | -1.59 | 0.0012 | -4.34 | 0.0500 |
| Necroptosis | Ripk1 | -1.46 | 0.0491 | -1.31 | 0.0248 | -1.63 | 0.0048 |
| Necroptosis | Ripk3 | -2.34 | 0.0983 | -2.01 | 0.0632 | -2.87 | 0.0016 |
| Parthanatos | Parg | 1.07 | 0.1772 | 1.24 | 0.2209 | 0.02 | 0.5158 |
| Parthanatos | Parp1 | 0.07 | 0.5445 | 1.33 | 0.0385 | 1.11 | 0.1695 |
| Inflammasome/Pyroptosis | Aim2 | -2.27 | 0.0457 | -1.45 | 0.0013 | -2.07 | 0.0073 |
| Inflammasome/Pyroptosis | Casp11 | -6.83 | 0.2365 | -2.86 | 0.0683 | -7.90 | 0.1394 |
| Inflammasome/Pyroptosis | Gsdmd | -2.64 | 0.0437 | -1.95 | 0.0636 | -2.91 | 0.0033 |
| Inflammasome/Pyroptosis | Nlrc4 | -3.78 | 0.1043 | -1.81 | 0.1011 | -4.14 | 0.0538 |
| Inflammasome/Pyroptosis | Nlrp1b | -0.01 | 0.5154 | -0.07 | 0.5284 | -1.24 | 0.0510 |
| Inflammasome/Pyroptosis | Nlrp3 | -2.42 | 0.0483 | -0.01 | 0.5328 | -2.25 | 0.0837 |
| Fold Change of MCMV-infected Eyes of MAIDS-4 Mice | |||||||
| Cell Death Pathway | Genes | Day 3 | Day 6 | Day 10 | |||
| Δ | p value | Δ | p value | Δ | p value | ||
| Autophagy | Atg12 | 0.05 | 0.5378 | -1.13 | 0.0314 | -0.44 | 0.5109 |
| Autophagy | Atg5 | 1.52 | 0.2518 | 1.19 | 0.0172 | 0.10 | 0.6342 |
| Autophagy | Becn1 | 1.25 | 0.3402 | 1.19 | 0.0855 | 0.05 | 0.5810 |
| Necroptosis | Mlkl | 4.73 | 0.1322 | 6.69 | 0.0625 | 6.01 | 0.2067 |
| Necroptosis | Ripk1 | 1.98 | 0.1282 | 2.25 | 0.1306 | 2.34 | 0.0547 |
| Necroptosis | Ripk3 | 3.18 | 0.2738 | 4.49 | 0.2026 | 5.29 | 0.0467 |
| Parthanatos | Parg | -0.03 | 0.5322 | -0.13 | 0.5056 | -1.28 | 0.0612 |
| Parthanatos | Parp1 | 1.06 | 0.5000 | 1.21 | 0.1669 | -1.49 | 0.1188 |
| Inflammasome/Pyroptosis | Aim2 | 2.56 | 0.0470 | 2.72 | 0.1099 | 3.03 | 0.0995 |
| Inflammasome/Pyroptosis | Casp11 | 8.74 | 0.1416 | 11.25 | 0.0666 | 8.81 | 0.0533 |
| Inflammasome/Pyroptosis | Gsdmd | 3.59 | 0.0783 | 5.05 | 0.1068 | 3.59 | 0.1095 |
| Inflammasome/Pyroptosis | Nlrc4 | 7.67 | 0.1999 | 7.40 | 0.0953 | 4.61 | 0.0291 |
| Inflammasome/Pyroptosis | Nlrp1b | 1.58 | 0.2863 | 1.74 | 0.1732 | 4.54 | 0.2836 |
| Inflammasome/Pyroptosis | Nlrp3 | 4.23 | 0.1047 | 3.41 | 0.0672 | 3.64 | 0.1983 |
| Fold Change of MCMV-infected Eyes of MAIDS-10 Mice | |||||||
| Cell Death Pathway | Genes | Day 3 | Day 6 | Day 10 | |||
| Δ | p value | Δ | p value | Δ | p value | ||
| Autophagy | Atg12 | -0.07 | 0.5202 | 1.19 | 0.3286 | 1.08 | 0.0424 |
| Autophagy | Atg5 | 1.32 | 0.0101 | 1.36 | 0.1665 | 1.69 | 0.0092 |
| Autophagy | Becn1 | 1.41 | 0.0465 | 1.28 | 0.2578 | 1.59 | 0.1850 |
| Necroptosis | Mlkl | 5.75 | 0.1482 | 4.16 | 0.1853 | 3.43 | 0.1166 |
| Necroptosis | Ripk1 | 2.20 | 0.0978 | 1.96 | 0.0565 | 2.17 | 0.0272 |
| Necroptosis | Ripk3 | 3.80 | 0.0057 | 3.93 | 0.0228 | 4.14 | 0.0091 |
| Parthanatos | Parg | -1.14 | 0.0119 | 1.21 | 0.2048 | -1.26 | 0.0085 |
| Parthanatos | Parp1 | -0.13 | 0.5111 | 1.19 | 0.1190 | -1.63 | 0.0736 |
| Inflammasome/Pyroptosis | Aim2 | 2.08 | 0.1594 | 2.44 | 0.0244 | 2.04 | 0.1032 |
| Inflammasome/Pyroptosis | Casp11 | 12.65 | 0.0358 | 5.07 | 0.1811 | 5.83 | 0.0631 |
| Inflammasome/Pyroptosis | Gsdmd | 4.41 | 0.0707 | 2.68 | 0.1036 | 3.63 | 0.0073 |
| Inflammasome/Pyroptosis | Nlrc4 | 7.20 | 0.2324 | 6.05 | 0.3290 | 4.23 | 0.0659 |
| Inflammasome/Pyroptosis | Nlrp1b | -0.10 | 0.5529 | 1.80 | 0.3369 | 1.12 | 0.3362 |
| Inflammasome/Pyroptosis | Nlrp3 | 3.08 | 0.0882 | 3.36 | 0.1534 | 1.40 | 0.2788 |
The fold changes in transcriptional activities within MCMV-infected eyes when compared with mock-infected eyes of groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3) were determined for genes associated with autophagy, necroptosis, parthanatos, and pyroptosis including inflammasomes at 3, 6, and 10 days postinfection.
Discussion
Herein we have confirmed and extended our understanding of some of the immunologic events that take place during the onset and development of cytomegalovirus retinitis in the unique setting of retrovirus-induced immunosuppression. This was accomplished by performing a comprehensive transcriptional analysis of immune response genes during the pathogenesis of MAIDS-related MCMV retinitis, a well-characterized, reproducible, and clinically relevant mouse model of AIDS-related HCMV retinitis [13]. Our findings show that (i) the pathogenesis of retinal disease in MCMV-infected eyes of MAIDS-10 mice susceptible to full-thickness retinal necrosis development is associated with the robust, differential expression of extensive number of immune response genes that operate in several distinct immune response pathways; (ii) the temporal development of MCMV retinitis within the eyes of MAIDS-10 mice is a dynamic process that involves both the upregulation and downregulation of several immune response genes at different times after intraocular MCMV infection; and (iii) the pattern of immune response gene activation differs remarkably within MCMV-infected eyes of healthy mice resistant to retinitis development when compared with MCMV-infected eyes of mice at different stages of MAIDS that exhibit a profound difference in their susceptibility to full-thickness retinitis development. A more focused companion investigation also provided compelling evidence for the stimulation of the transcription of multiple genes associated with the necroptosis and pyroptosis programmed cell death pathways during the development MAIDS-related MCMV retinitis.
For our investigation of immune response gene expression during the pathogenesis of MAIDS-related MCMV retinitis, we used the NanoString nCounter assay. This recently developed technology platform is capable of making without amplification a direct multiplexed measurement of gene expression within the ocular compartment following intraocular MCMV inoculation of mice for comparisons during immunocompetence and MAIDS without recourse to any need for sequence amplication. This amplification-free microarray technology is a powerful tool that allows for the simultaneous quantification of the expression of hundreds of genes at several distinct time points during disease pathogenesis. Even so, we initiated this investigation mindful of several limitations that this experimental approach enjoys. Firstly, the amount of data obtained after the performance of a single experiment is overwhelming and requires thoughtful use of statistical analysis to help provide meaningful conclusions [64]. Secondly, subsequent performance of quantitative RT-PCR and/or western blot assays is needed to confirm the upregulation or downregulation of an individual gene under investigation. Thirdly, the data generated are descriptive and often do not provide mechanistic insights into the function of an individual gene or its functional gene product during a pathogenic event. Finally, it has not escaped our attention that the NanoString nCounter assay provides only fold-change differences in transcription activity that may or may not be biologically significant to disease pathogenesis in that the quantitative fold-change in activity of an individual gene may not be a true reflection of the unique kinetics of that particular gene’s mRNA or its gene product and consequently pertinent changes in its functional ability. For example, a two-fold increase in transcriptional activity may be biologically significant for one gene, but another gene may require a far greater transcriptional increase for a greater duration and at a particular time to be biologically significant.
Despite these perceived limitations, the NanoString nCounter assay has proven to be a useful and powerful tool to point us quickly in new directions of investigation and thereby expand our knowledge base of a series of immune functions that contribute (or do not contribute) to the onset of MCMV-induced retinal disease and progression to full-thickness necrosis during MAIDS. Use of the NanoString nCounter assay has also provided the opportunity to confirm previous findings by us and others on the role of various immune responses toward the pathogenesis of experimental MCMV retinitis during immunosuppression. Most of the findings presented herein are in agreement with findings documented by us in past publications that have focused on various immune-mediated pathways and/or individual molecules associated with innate or adaptive immunity vis-a-vis the pathogenesis of MAIDS-related MCMV retinitis. These include roles for humoral immunity [65], cellular immunity [66,67], suppressor of cytokine signaling (SOCS) pathways [60,68], TNF-α [17], interferon-γ [17], and a number of proinflammatory cytokines associated with innate immunity [62,69] as summarized by us in a recent review [16]. The use of a custom panel consisting of a smaller subset of genes associated with necroptosis and pyroptosis as well as pyroptosis-related inflammasomes in this report also served to confirm our previous determination that these programmed cell death pathways appear to be involved in the progression of MCMV retinitis during MAIDS [62] as evidenced by this earlier report of significant intraocular stimulation of necroptosis-associated receptor interacting protein kinase 3 (RIPK3), pyroptosis-asssociated caspase-1, interleukin-1β, interleukin-18, and the AIM2 inflammasome.
Careful inspection of the data obtained in the present investigation also exposed an unexpected difference in findings reported herein using the NanoString technology and a publication by us for another programmed cell death pathway, parthanatos, and MAIDS-related MCMV retinitis [63]. Whereas the transcription levels of parg and parp1, two genes associated with the parthanatos pathway [70–75], show negligible activity within MCMV-infected eyes of MAIDS-10 at either 3, 6, or 10 days after intraocular MCMV inoculation in the present investigation, we have documented previously a significant increase in parp1 mRNA and protein production as well as parg mRNA and protein synthesis by quantitative RT-PCR and western blot assays, respectively, at the same times examined after intraocular MCMV inoculation of MAIDS-10 mice [63]. The reason for this major discrepancy in findings remains unclear but may be due to differences in the regional specificity of probes used by the NanoString nCounter assays [18]. That these parthanatos-associated genes are indeed upregulated is undeniable given the marked intraocular stimulation of protein production previously shown by us for their respective gene products [63]. These apparently conflicting observations, however, only serve to underscore the need to confirm all NanoString nCounter data using further quantitative RT-PCR and/or western blot analysis for each gene of interest.
In summary, our experience using the NanoString nCounter assay to provide a novel transcriptional analysis of 575 immune response genes within MCMV-infected eyes of mice at different stages of MAIDS development compared with MCMV-infected eyes of immunologically normal mice has provided new, and at times, unexpected information on the pathogenesis of MAIDS-related MCMV retinitis. By extension, our findings may also improve our understanding of the pathogenesis AIDS-related HCMV retinitis as well as other AIDS-related opportunistic virus infections. While helpful in many ways, we have also identified areas of caution when using this powerful research tool. Future use of this technology by us will be directed toward the identification of host genes expressed by different cell populations of retinal tissues during the onset and progression of MAIDS-related MCMV retinitis. Moreover, because host RNA constitutes an overwhelming portion of the total RNA recovered from infected tissue samples when compared with pathogen RNA which usually comprises a vanishingly small portion of total RNA [76], use of this technology will also allow us to investigate with greater precision the pattern of mRNA synthesis for individual MCMV genes that are expressed within retinal tissues and retina-related cell populations during the course of disease development following intraocular MCMV infection of retrovirus-immunosuppressed mice.
Materials and methods
Viruses
Stocks of MCMV (Smith) were propagated through salivary glands of groups of BALB/c mice (Harlan Laboratories, USA) as described previously [13]. Briefly, fourteen days following intraperitoneal injection of 102 to 103 plaque forming units (PFU) of MCMV contained within 0.2 ml volume, salivary glands were harvested aseptically, pooled, and homogenized (10% [wt/mol]) in Dulbecco’s modified eagle media (DMEM, Corning Life Sciences, Manassas, VA, #10–013). Virus stocks were clarified by centrifugation, aliquoted, and stored in liquid nitrogen prior to quantification by standard plaque assay on monolayers of mouse embryonic fibroblasts as described previously [13]. A fresh aloquot of MCMV stock was thawed and used for each experiment.
Stocks of murine retrovirus (LP-BM5 murine leukemia virus [MuLV]) were prepared in monolayers of SC-1 fibroblasts (ATCC #CRL-1404) and SC-1/MuLV LP-BM5 cells [77] kindly provided by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (Germantown, MD). Six days following seeding at a 1:1 ratio, the cells were scraped into SC-1 media and stored at -80°C. Prior to use, LP-BM5 stocks were thawed and clarified by centrifugation to remove cellular debris.
Animals
Adult female wildtype BALB/c mice used for the preparation of MCMV stocks were purchased from Harlan Laboratories (Indianapolis, IN, USA). Adult female wildtype C57BL/6 mice used for all MAIDS studies were purchased from Jackson Laboratory (Bar Harbor, ME, USA). All mice were maintained on alternative 12-hr light/dark cycles and allowed unrestricted access to food and water. All procedures were conducted with strict compliance to National Institutes of Health and the Association for Research in Vision and Ophthalmology (ARVO) statement for Use of Animals in Ophthalmic and Vision Research guidelines and in accordance with Georgia State University Institutional Animal Care and Use Committee (IACUC) approved protocols.
Induction of MAIDS
MAIDS was induced by injecting 1.0 ml of inoculum containing approximately 5 × 103 to 5 × 104 infectious LP-BM5 murine leukemia retrovirus into the peritoneum of C57BL/6 mice. Mice with MAIDS of 4 weeks’ duration (MAIDS-14 mice) and 10 weeks’ duration (MAIDS-10 mice) were used throughout the investigation and compared with age-matched healthy C57BL/6 mice.
Experimental mouse model of MCMV retinitis
Details for the MAIDS model of MCMV retinitis used throughout the investigation have been described by us previously [13,16,62]. Briefly, the left eyes of groups of healthy mice, MAIDS-4 mice, and MAIDS-10 mice were subjected to intraocular (subretinal) inoculation with approximately 104 PFU of MCMV contained within a 2-ul volume of DMEM. The right contralateral eyes of all mice were inoculated intraocularly with DMEM and served as controls.
NanoString nCounter assay
Whole MCMV-infected and control eyes were collected from all animal groups at 3, 6, and/or 10 days after intraocular inoculation and stored at 4C in RNAlater solution (Ambion, Austin, TX) prior to NanoString nCounter analysis. At time of analysis, eyes were individually homogenized in 1.0 ml of TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA) using a 2-ml Ten Broeck tissue grinder (Wheaton, Millville, NJ). Total RNA was extracted in chloroform and purified using the PureLink RNA Mini Kit according to the manufacturer’s instructions (Ambion/ThermoFisher, Grand Island, NY). The extracted RNA from each group and their respective time points were pooled and the RNA concentrations were determined using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Pittsburgh, PA). Approximately 100 ng of the purified total RNA from each time point of each animal group were analyzed using the nCounter Analysis System (NanoString Technologies, Seattle, WA) according to the manufacturer’s instructions in combination with the Murine Immunology Panel which contained 561 unique RNA barcodes. Probes for 15 internal and housekeeping genes such as ribosomal protein L10, beta-actin, beta-2-microglobuin, glyceraldehyde 3-phosphate dehydrogenase, and ribosomal protein L19 were incorporated into the NanoString codesets of this panel. A second custom-made panel that included 14 unique genes not included in the Murine Immunology Panel but associated with programmed cell death pathways as well as three housekeeping genes was used in another set of studies. Analysis of raw mRNA data was performed using the NanoString nSolver™ analysis software version 4.0.
Statistical analysis
Two independent experiments were performed for each study and were run independently through the NanoString nSolver™ analysis software. All p values were determined on fold changes of raw mRNA counts as determined by the NanoString nSolver™ software and performed with a significance level (α) set to 0.05; p values of < 0.05 were considered statistically significant. Statistical analysis were performed by comparing MCMV-infected eyes with mock-infected eyes (controls) by unpaired, two-sided Student’s t-test.
Supporting information
Transcriptional activity of immune response genes of MCMV-infected eyes of groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3) with a fold change of greater than two when compared with mock-infected eyes at 3, 6, or 10 days postinfection were categorized into more focused NanoString-defined immunologic pathways that exhibited the robust upregulation. Immunologic pathways included those associated with (A) TLR Signaling (B)Phagocytosis and Degradation, (C) NOD-like Receptor (NLR) Signaling, (D) Type I Interferon (IFN) Signaling, and (E) Type II Interferon (IFN) Signaling, (F) Tumor Necrosis Factor (TNF) Signaling, (G) Chemokine Signaling, (H) NF-kB Signaling, and (I) MHC Class I Antigen Presentation.
(TIF)
Variability between individual eyes per group is not shown due to pooling of individual eyes.
(PDF)
Acknowledgments
We thank the Biology Core Facilities, Department of Biology, Georgia State University, for excellent technical assistance.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This study was funded by NIH NEI RO1 EY024630 (RDD), NIH NEI P30 EY006360 (RDD), and NIH NEI T32 EY007092 (JJC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dix RD, Cousins SW. AIDS-related cytomegalovirus retinitis: lessons from the laboratory. Curr Eye Res. 2004;29(2–3):91–101. 10.1080/02713680490504641 [DOI] [PubMed] [Google Scholar]
- 2.Tufail A, Holland G. Ocular infection and immunity: Mosby Incorporated; 1996. [Google Scholar]
- 3.Ho M. Advances in understanding cytomegalovirus infection after transplantation. Transplant Proc. 1994;26(5 Suppl 1):7–11. [PubMed] [Google Scholar]
- 4.Hoover DR, Saah AJ, Bacellar H, Phair J, Detels R, Anderson R, et al. Clinical manifestations of AIDS in the era of Pneumocystis prophylaxis. 1993;329(26):1922–6. [DOI] [PubMed] [Google Scholar]
- 5.Heiden D, Ford N, Wilson D, Rodriguez WR, Margolis T, Janssens B, et al. Cytomegalovirus retinitis: the neglected disease of the AIDS pandemic. PLoS Med. 2007;4(12):e334 10.1371/journal.pmed.0040334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stewart MW. Optimal management of cytomegalovirus retinitis in patients with AIDS. Clin Ophthalmol. 2010;4:285–99. 10.2147/opth.s6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugar EA, Jabs DA, Ahuja A, Thorne JE, Danis RP, Meinert CL, et al. Incidence of cytomegalovirus retinitis in the era of highly active antiretroviral therapy. American journal of ophthalmology. 2012;153(6):1016–24.e5. 10.1016/j.ajo.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jabs DA, Ahuja A, Van Natta ML, Lyon AT, Yeh S, Danis R. Long-term Outcomes of Cytomegalovirus Retinitis in the Era of Modern Antiretroviral Therapy: Results from a United States Cohort. Ophthalmology. 2015;122(7):1452–63. 10.1016/j.ophtha.2015.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofman FM, Hinton DR. Tumor necrosis factor-alpha in the retina in acquired immune deficiency syndrome. Investigative ophthalmology & visual science. 1992;33(6):1829–35. [PubMed] [Google Scholar]
- 10.Pepose JS, Holland GN, Nestor MS, Cochran AJ, Foos RY. Acquired immune deficiency syndrome. Pathogenic mechanisms of ocular disease. Ophthalmology. 1985;92(4):472–84. 10.1016/s0161-6420(85)34008-3 [DOI] [PubMed] [Google Scholar]
- 11.Jabs DA. Cytomegalovirus retinitis and the acquired immunodeficiency syndrome—bench to bedside: LXVII Edward Jackson Memorial Lecture. Am J Ophthalmol. 2011;151(2):198–216.e1. 10.1016/j.ajo.2010.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alston CI, Carter J.J, Dix R.D. Cytomegalovirus and the-Eye: AIDS-Related Retinitis and Beyond. Herpesviridae. 2017:42. [Google Scholar]
- 13.Dix RD, Cray C, Cousins SW. Mice immunosuppressed by murine retrovirus infection (MAIDS) are susceptible to cytomegalovirus retinitis. Curr Eye Res. 1994;13(8):587–95. 10.3109/02713689408999892 [DOI] [PubMed] [Google Scholar]
- 14.Gazzinelli RT, Makino M, Chattopadhyay SK, Snapper CM, Sher A, Hügin AW, et al. CD4+ subset regulation in viral infection. Preferential activation of Th2 cells during progression of retrovirus-induced immunodeficiency in mice. The Journal of Immunology. 1992;148(1):182 [PubMed] [Google Scholar]
- 15.Jolicoeur P. Murine acquired immunodeficiency syndrome (MAIDS): an animal model to study the AIDS pathogenesis. FASEB J. 1991;5(10):2398–405. 10.1096/fasebj.5.10.2065888 [DOI] [PubMed] [Google Scholar]
- 16.Carter J, Alston CI, Oh J, Duncan L-A, Esquibel Nemeno JG, Byfield SN, et al. Mechanisms of AIDS-related cytomegalovirus retinitis. Future Virology. 2019;14(8):545–60. [Google Scholar]
- 17.Dix RD, Cousins SW. Susceptibility to murine cytomegalovirus retinitis during progression of MAIDS: correlation with intraocular levels of tumor necrosis factor-alpha and interferon-gamma. Curr Eye Res. 2004;29(2–3):173–80. 10.1080/02713680490504876 [DOI] [PubMed] [Google Scholar]
- 18.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26(3):317–25. 10.1038/nbt1385 [DOI] [PubMed] [Google Scholar]
- 19.Funaro A, Ortolan E, Bovino P, Lo Buono N, Nacci G, Parrotta R, et al. Ectoenzymes and innate immunity: the role of human CD157 in leukocyte trafficking. Frontiers in bioscience (Landmark edition). 2009;14:929–43. 10.2741/3287 [DOI] [PubMed] [Google Scholar]
- 20.Choudhary GS, Al-harbi S, Almasan A. Caspase-3 Activation Is a Critical Determinant of Genotoxic Stress-Induced Apoptosis In: Mor G, Alvero AB, editors. Apoptosis and Cancer: Methods and Protocols. New York, NY: Springer New York; 2015. p. 1–9. [DOI] [PubMed] [Google Scholar]
- 21.Gibaldi D, Vilar-Pereira G, Pereira IR, Silva AA, Barrios LC, Ramos IP, et al. CCL3/Macrophage Inflammatory Protein-1α Is Dually Involved in Parasite Persistence and Induction of a TNF- and IFNγ-Enriched Inflammatory Milieu in Trypanosoma cruzi-Induced Chronic Cardiomyopathy. Frontiers in immunology. 2020;11:306 10.3389/fimmu.2020.00306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hara T, Bacon KB, Cho LC, Yoshimura A, Morikawa Y, Copeland NG, et al. Molecular cloning and functional characterization of a novel member of the C-C chemokine family. The Journal of Immunology. 1995;155(11):5352 [PubMed] [Google Scholar]
- 23.McNerney ME, Kumar V. The CD2 Family of Natural Killer Cell Receptors In: Compans RW, Cooper MD, Honjo T, Koprowski H, Melchers F, Oldstone MBA, et al. , editors. Immunobiology of Natural Killer Cell Receptors. Berlin, Heidelberg: Springer Berlin Heidelberg; 2006. p. 91–120. [Google Scholar]
- 24.Sung PS, Hsieh SL. CLEC2 and CLEC5A: Pathogenic Host Factors in Acute Viral Infections. Frontiers in immunology. 2019;10:2867 10.3389/fimmu.2019.02867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamann J, Koning N, Pouwels W, Ulfman LH, van Eijk M, Stacey M, et al. EMR1, the human homolog of F4/80, is an eosinophil-specific receptor. 2007;37(10):2797–802. [DOI] [PubMed] [Google Scholar]
- 26.Shiina T, Blancher A, Inoko H, Kulski JK. Comparative genomics of the human, macaque and mouse major histocompatibility complex. Immunology. 2017;150(2):127–38. 10.1111/imm.12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shepardson KM, Larson K, Johns LL, Stanek K, Cho H, Wellham J, et al. IFNAR2 Is Required for Anti-influenza Immunity and Alters Susceptibility to Post-influenza Bacterial Superinfections. Frontiers in immunology. 2018;9:2589–. 10.3389/fimmu.2018.02589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shukla AK, McIntyre LL, Marsh SE, Schneider CA, Hoover EM, Walsh CM, et al. CD11a expression distinguishes infiltrating myeloid cells from plaque-associated microglia in Alzheimer's disease. 2019;67(5):844–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocrine reviews. 1998;19(3):225–68. 10.1210/edrv.19.3.0334 [DOI] [PubMed] [Google Scholar]
- 30.Bhosle VK, Rivera JC, Zhou TE, Omri S, Sanchez M, Hamel D, et al. Nuclear localization of platelet-activating factor receptor controls retinal neovascularization. Cell discovery. 2016;2:16017 10.1038/celldisc.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunzmann AT, Murray LJ, Cardwell CR, McShane CM, McMenamin ÚC, Cantwell MM. PTGS2 (Cyclooxygenase-2) Expression and Survival among Colorectal Cancer Patients: A Systematic Review. Cancer Epidemiology Biomarkers & Prevention. 2013;22(9):1490 10.1158/1055-9965.EPI-13-0263 [DOI] [PubMed] [Google Scholar]
- 32.Blaszczyk K, Nowicka H, Kostyrko K, Antonczyk A, Wesoly J, Bluyssen HAR. The unique role of STAT2 in constitutive and IFN-induced transcription and antiviral responses. Cytokine & Growth Factor Reviews. 2016;29:71–81. 10.1016/j.cytogfr.2016.02.010 [DOI] [PubMed] [Google Scholar]
- 33.Wahl SM, Hunt DA, Wong HL, Dougherty S, McCartney-Francis N, Wahl LM, et al. Transforming growth factor-beta is a potent immunosuppressive agent that inhibits IL-1-dependent lymphocyte proliferation. J Immunol. 1988;140(9):3026–32. [PubMed] [Google Scholar]
- 34.Letterio JJ, Roberts AB. Regulation of immune responses by TGF-beta. Annual review of immunology. 1998;16:137–61. 10.1146/annurev.immunol.16.1.137 [DOI] [PubMed] [Google Scholar]
- 35.Gilbert KM, Thoman M, Bauche K, Pham T, Weigle WO. Transforming growth factor-beta 1 induces antigen-specific unresponsiveness in naive T cells. Immunological investigations. 1997;26(4):459–72. 10.3109/08820139709022702 [DOI] [PubMed] [Google Scholar]
- 36.Jia GQ, Gonzalo JA, Lloyd C, Kremer L, Lu L, Martinez-A C, et al. Distinct expression and function of the novel mouse chemokine monocyte chemotactic protein-5 in lung allergic inflammation. The Journal of experimental medicine. 1996;184(5):1939–51. 10.1084/jem.184.5.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697–710. 10.18632/oncotarget.7376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ford J, Hughson A, Lim K, Bardina SV, Lu W, Charo IF, et al. CCL7 Is a Negative Regulator of Cutaneous Inflammation Following Leishmania major Infection. Frontiers in immunology. 2018;9:3063 10.3389/fimmu.2018.03063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annual review of immunology. 1999;17:657–700. 10.1146/annurev.immunol.17.1.657 [DOI] [PubMed] [Google Scholar]
- 40.Yang M, Fan J-j, Wang J, Zhao Y, Teng Y, Liu P. Association of the C2–CFB locus with non-infectious uveitis, specifically predisposed to Vogt–Koyanagi–Harada disease. Immunologic Research. 2016;64(2):610–8. 10.1007/s12026-015-8762-x [DOI] [PubMed] [Google Scholar]
- 41.Karin N, Razon H. Chemokines beyond chemo-attraction: CXCL10 and its significant role in cancer and autoimmunity. Cytokine. 2018;109:24–8. 10.1016/j.cyto.2018.02.012 [DOI] [PubMed] [Google Scholar]
- 42.Tokunaga R, Zhang W, Naseem M, Puccini A, Berger MD, Soni S, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—A target for novel cancer therapy. Cancer Treat Rev. 2018;63:40–7. 10.1016/j.ctrv.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wikenheiser DJ, Stumhofer JS. ICOS Co-Stimulation: Friend or Foe? Frontiers in immunology. 2016;7:304 10.3389/fimmu.2016.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su W, Xiao W, Chen L, Zhou Q, Zheng X, Ju J, et al. Decreased IFIT2 Expression In Human Non-Small-Cell Lung Cancer Tissues Is Associated With Cancer Progression And Poor Survival Of The Patients. OncoTargets and therapy. 2019;12:8139–49. 10.2147/OTT.S220698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y-H, He M, Wang Y, Liao A-H. Modulators of the Balance between M1 and M2 Macrophages during Pregnancy. 2017;8(120). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naeim F. Chapter 2—Principles of Immunophenotyping In: Naeim F, Rao PN, Grody WW, editors. Hematopathology. Oxford: Academic Press; 2008. p. 27–55. [Google Scholar]
- 47.Jones DC, Hewitt CRA, López-Álvarez MR, Jahnke M, Russell AI, Radjabova V, et al. Allele-specific recognition by LILRB3 and LILRA6 of a cytokeratin 8-associated ligand on necrotic glandular epithelial cells. Oncotarget. 2016;7(13):15618–31. 10.18632/oncotarget.6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.John S, Chen H, Deng M, Gui X, Wu G, Chen W, et al. A Novel Anti-LILRB4 CAR-T Cell for the Treatment of Monocytic AML. Molecular therapy: the journal of the American Society of Gene Therapy. 2018;26(10):2487–95. 10.1016/j.ymthe.2018.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rheinländer A, Schraven B, Bommhardt U. CD45 in human physiology and clinical medicine. Immunology Letters. 2018;196:22–32. 10.1016/j.imlet.2018.01.009 [DOI] [PubMed] [Google Scholar]
- 50.Kim JR, Mathew SO, Patel RK, Pertusi RM, Mathew PA. Altered expression of signalling lymphocyte activation molecule (SLAM) family receptors CS1 (CD319) and 2B4 (CD244) in patients with systemic lupus erythematosus. Clinical and experimental immunology. 2010;160(3):348–58. 10.1111/j.1365-2249.2010.04116.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aldinucci D, Colombatti A. The inflammatory chemokine CCL5 and cancer progression. Mediators of inflammation. 2014;2014:292376 10.1155/2014/292376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Cai Y, Zhang D, Sun J, Xu C, Zhao W, et al. miR-195/miR-497 Regulate CD274 Expression of Immune Regulatory Ligands in Triple-Negative Breast Cancer. J Breast Cancer. 2018;21(4):371–81. 10.4048/jbc.2018.21.e60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao P, Lieu T, Barlow N, Metcalf M, Veldhuis NA, Jensen DD, et al. Cathepsin S causes inflammatory pain via biased agonism of PAR2 and TRPV4. The Journal of biological chemistry. 2014;289(39):27215–34. 10.1074/jbc.M114.599712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gul-Kahraman K, Yilmaz-Bozoglan M, Sahna E. Physiological and pharmacological effects of melatonin on remote ischemic perconditioning after myocardial ischemia-reperfusion injury in rats: Role of Cybb, Fas, NfκB, Irisin signaling pathway. 2019;67(2):e12589 10.1111/jpi.12589 [DOI] [PubMed] [Google Scholar]
- 55.Vivier E, Morin P, O'Brien C, Druker B, Schlossman SF, Anderson P. Tyrosine phosphorylation of the Fc gamma RIII(CD16): zeta complex in human natural killer cells. Induction by antibody-dependent cytotoxicity but not by natural killing. J Immunol. 1991;146(1):206–10. [PubMed] [Google Scholar]
- 56.Jakus Z, Németh T, Verbeek JS, Mócsai A. Critical but overlapping role of FcgammaRIII and FcgammaRIV in activation of murine neutrophils by immobilized immune complexes. J Immunol. 2008;180(1):618–29. 10.4049/jimmunol.180.1.618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell host & microbe. 2011;9(5):363–75. 10.1016/j.chom.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perrier S, Darakhshan F, Hajduch E. IL-1 receptor antagonist in metabolic diseases: Dr Jekyll or Mr Hyde? FEBS letters. 2006;580(27):6289–94. 10.1016/j.febslet.2006.10.061 [DOI] [PubMed] [Google Scholar]
- 59.Shabani F, Farasat A, Mahdavi M, Gheibi N. Calprotectin (S100A8/S100A9): a key protein between inflammation and cancer. Inflammation Research. 2018;67(10):801–12. 10.1007/s00011-018-1173-4 [DOI] [PubMed] [Google Scholar]
- 60.Alston CI, Dix RD. Reduced frequency of murine cytomegalovirus retinitis in C57BL/6 mice correlates with low levels of suppressor of cytokine signaling (SOCS)1 and SOCS3 expression within the eye during corticosteroid-induced immunosuppression. Cytokine. 2017;97:38–41. 10.1016/j.cyto.2017.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haure-Mirande J-V, Audrain M, Fanutza T, Kim SH, Klein WL, Glabe C, et al. Deficiency of TYROBP, an adapter protein for TREM2 and CR3 receptors, is neuroprotective in a mouse model of early Alzheimer's pathology. Acta Neuropathol. 2017;134(5):769–88. 10.1007/s00401-017-1737-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chien H, Dix RD. Evidence for multiple cell death pathways during development of experimental cytomegalovirus retinitis in mice with retrovirus-induced immunosuppression: apoptosis, necroptosis, and pyroptosis. J Virol. 2012;86(20):10961–78. 10.1128/JVI.01275-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oh JJ, Carter JJ, Nemeno JGE, Dix RD. Parthanatos-associated proteins are stimulated intraocularly during development of experimental murine cytomegalovirus retinitis in mice with retrovirus-induced immunosuppression. Journal of medical virology. 2020;92(3):394–8. 10.1002/jmv.25619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang H, Horbinski C, Wu H, Liu Y, Sheng S, Liu J, et al. NanoStringDiff: a novel statistical method for differential expression analysis based on NanoString nCounter data. Nucleic acids research. 2016;44(20):e151 10.1093/nar/gkw677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dix RD, Cray C, Cousins SW. Antibody Alone Does Not Prevent Experimental Cytomegalovirus Retinitis in Mice with Retrovirus-lnduced Immunodeficiency (MAIDS). Ophthalmic Research. 1997;29(6):381–92. 10.1159/000268039 [DOI] [PubMed] [Google Scholar]
- 66.Dix RD, Giedlin M, Cousins SW. Systemic cytokine immunotherapy for experimental cytomegalovirus retinitis in mice with retrovirus-induced immunodeficiency. Investigative ophthalmology & visual science. 1997;38(7):1411–7. [PubMed] [Google Scholar]
- 67.Dix RD, Cousins SW. Interleukin-2 immunotherapy of murine cytomegalovirus retinitis during MAIDS correlates with increased intraocular CD8+ T-cell infiltration. Ophthalmic Res. 2003;35(3):154–9. 10.1159/000070051 [DOI] [PubMed] [Google Scholar]
- 68.Blalock EL, Chien H, Dix RD. Systemic reduction of interleukin-4 or interleukin-10 fails to reduce the frequency or severity of experimental cytomegalovirus retinitis in mice with retrovirus-induced immunosuppression. Ophthalmology and eye diseases. 2012;4:79–90. 10.4137/OED.S10294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chien H, Alston CI, Dix RD. Suppressor of Cytokine Signaling 1 (SOCS1) and SOCS3 Are Stimulated within the Eye during Experimental Murine Cytomegalovirus Retinitis in Mice with Retrovirus-Induced Immunosuppression. Journal of Virology. 2018;92(18):e00526–18. 10.1128/JVI.00526-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Experimental neurology. 2009;218(2):193–202. 10.1016/j.expneurol.2009.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Y, Kim NS, Haince JF, Kang HC, David KK, Andrabi SA, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos). Science signaling. 2011;4(167):ra20 10.1126/scisignal.2000902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fatokun AA, Dawson VL, Dawson TM. Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. British journal of pharmacology. 2014;171(8):2000–16. 10.1111/bph.12416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, Feng X, Koh DW. Activation of cell death mediated by apoptosis-inducing factor due to the absence of poly(ADP-ribose) glycohydrolase. Biochemistry. 2011;50(14):2850–9. 10.1021/bi101829r [DOI] [PubMed] [Google Scholar]
- 74.Blenn C, Althaus FR, Malanga M. Poly(ADP-ribose) glycohydrolase silencing protects against H2O2-induced cell death. The Biochemical journal. 2006;396(3):419–29. 10.1042/BJ20051696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davidovic L, Vodenicharov M, Affar EB, Poirier GG. Importance of poly(ADP-ribose) glycohydrolase in the control of poly(ADP-ribose) metabolism. Experimental cell research. 2001;268(1):7–13. 10.1006/excr.2001.5263 [DOI] [PubMed] [Google Scholar]
- 76.Xu W, Solis NV, Filler SG, Mitchell AP. Pathogen Gene Expression Profiling During Infection Using a Nanostring nCounter Platform. Methods in molecular biology (Clifton, NJ). 2016;1361:57–65. 10.1007/978-1-4939-3079-1_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Atherton SS, Newell CK, Kanter MY, Cousins SW. Retinitis in euthymic mice following inoculation of murine cytomegalovirus (MCMV) via the supraciliary route. Curr Eye Res. 1991;10(7):667–77. 10.3109/02713689109013858 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptional activity of immune response genes of MCMV-infected eyes of groups of healthy mice (n = 3), MAIDS-4 mice (n = 3), and MAIDS-10 mice (n = 3) with a fold change of greater than two when compared with mock-infected eyes at 3, 6, or 10 days postinfection were categorized into more focused NanoString-defined immunologic pathways that exhibited the robust upregulation. Immunologic pathways included those associated with (A) TLR Signaling (B)Phagocytosis and Degradation, (C) NOD-like Receptor (NLR) Signaling, (D) Type I Interferon (IFN) Signaling, and (E) Type II Interferon (IFN) Signaling, (F) Tumor Necrosis Factor (TNF) Signaling, (G) Chemokine Signaling, (H) NF-kB Signaling, and (I) MHC Class I Antigen Presentation.
(TIF)
Variability between individual eyes per group is not shown due to pooling of individual eyes.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.