Abstract
Objective
In this era of digitized health records, there has been a marked interest in using de-identified patient records for conducting various health related surveys. To assist in this research effort, we developed a novel clinical data representation model entitled medical knowledge-infused convolutional neural network (MKCNN), which is used for learning the clinical trial criteria eligibility status of patients to participate in cohort studies.
Materials and Methods
In this study, we propose a clinical text representation infused with medical knowledge (MK). First, we isolate the noise from the relevant data using a medically relevant description extractor; then we utilize log-likelihood ratio based weights from selected sentences to highlight “met” and “not-met” knowledge-infused representations in bichannel setting for each instance. The combined medical knowledge-infused representation (MK) from these modules helps identify significant clinical criteria semantics, which in turn renders effective learning when used with a convolutional neural network architecture.
Results
MKCNN outperforms other Medical Knowledge (MK) relevant learning architectures by approximately 3%; notably SVM and XGBoost implementations developed in this study. MKCNN scored 86.1% on F1metric, a gain of 6% above the average performance assessed from the submissions for n2c2 task. Although pattern/rule-based methods show a higher average performance for the n2c2 clinical data set, MKCNN significantly improves performance of machine learning implementations for clinical datasets.
Conclusion
MKCNN scored 86.1% on the F1 score metric. In contrast to many of the rule-based systems introduced during the n2c2 challenge workshop, our system presents a model that heavily draws on machine-based learning. In addition, the MK representations add more value to clinical comprehension and interpretation of natural texts.
Keywords: natural language processing, cohort selection, clinical trials, convolutional neural network, medical records
INTRODUCTION
Mining medically relevant information from clinical notes or pathology reports is important for building statistics on disease-specific mortality and symptomatic disease associations as well as for accessing knowledge of urgent and emerging medical issues in all therapeutic areas.1–6 To facilitate further research and to streamline hospital work, electronic health records (EHRs) were introduced in clinical informatics to digitally store, update, and maintain real-time clinical information.7 Much of the medical information in EHRs comes from clinical notes (eg, clinician’s case summaries), which are documented as free text, and thus require intensive use of natural language processing (NLP) techniques to extract structured information.8 Screening clinical narratives for the mention of diseases can help identify subjects of specific therapeutic areas (eg, neurology, cardiology), which, in turn, can contribute greatly to the analysis of a clinician’s diagnosis.9 Kocbek et al10 observed that metadata, like hospital admission data and radiology reports combined with pathology reports, are instrumental in detecting diseases like lung cancer and secondary malignant respiratory neoplasms at the time of admission.
Clinical narratives are complex and often require criteria-based elimination of information to catalog patients into study cohorts for disease or drug-associated research. Manual screening of narratives barring the human error of oversight is an effective method in cohort selection, but remains an enormously time-consuming process. An alternative to this approach is the automation of the recruitment and data gathering process via data optimization as described by Huang et al.11 While conducting a cohort identification study, Glicksberg et al12 used patient diagnostic summaries for assigning putative disease cohorts for attention deficit hyperactivity disorder, dementia, herpes zoster, sickle cell disease, and type 2 diabetes. They adapted International Classification of Disease (ICD)-9 code (https://www.cdc.gov/nchs/icd/icd9cm.htm) descriptors of key terms with word2vec embedding to produce a standardized representation for clinical topics like drug and diseases names.13 However, generic criteria, like cognitive capabilities, require clinical metadata or active text learning. Therefore, a superior clinical text representation using rules or alternate descriptor interpretation is important in building an effective clinical decision system.
Rule-based systems use real-time patient information and are subjected iteratively to validations to ensure stable clinical deployments.14–17 Despite the results, rule-based systems still suffer from constraints related to the cost of time and labor.18 For this reason, machine-learning models have also been explored in various clinical informatics studies. Yuan et al19 used support vector machines (SVMs) to extract simpler features like bag-of-words (BOW) and term-frequency-inverse document frequency (TF-IDF) from admission forms to identify autism in patients. Lexical and ontological features like BOW, TF-IDF, n-grams, Unified Medical Language Systems, ICD codes, and Medical Subject Heading (MeSH) or feature maps are used in previous studies to impart clinically relevant knowledge.8,10,12,19–21 In contrast, distributed representations generated by Deep Neural Networks (DNN) incorporate universality in token representations as shown in works by Hughes et al and Wang et al, where alternate word2vec and medical word embedding were respectively used for sentence-level classification.8,22,23
In this study, we employed an enhanced medical knowledge (MK)-infused representation with convolutional neural network (CNN) architecture (ie, MKCNN) to perform a cohort selection from clinical notes. We used data from Track 1: “cohort selection in clinical trials” of the 2018 National NLP Clinical Challenge (n2c2) shared task to develop our system and competed in the task. The cohort screening employed 13 criteria based on clinical and personal patient information, which is challenging in terms of use of a universal and clinically relevant feature descriptor. Therefore, we focused on developing a preprocessing module, which generates a medical criteria-relevant initial representation, henceforth known as representations (MKRs), both scalable and adaptable for medical and nonmedical criteria. These MK representations are then adapted in a bichannel CNN. CNN was adapted as it is capable of extracting local patterns from data regardless of their location, which is useful for identifying fixed-length medically relevant phrases.24 In addition, although CNN is a nonlinear model with minimal manual feature extraction effort, it still achieves better fit with data compared to linear models like conditional random field.25,26
MATERIALS AND METHODS
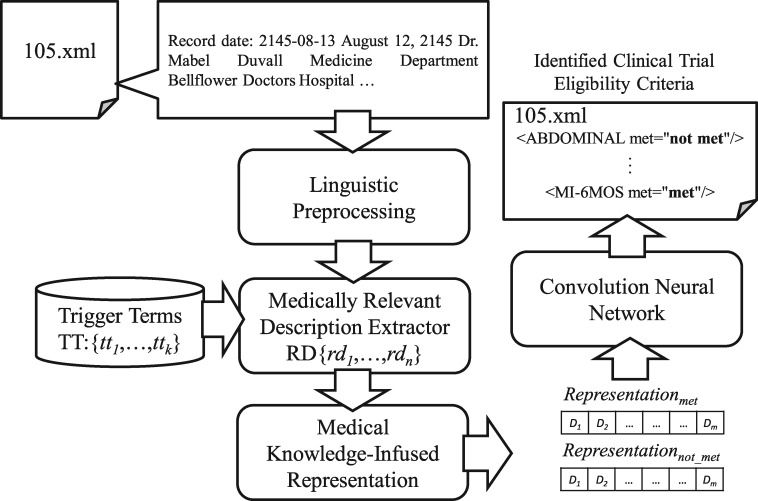
Figure 1 provides an overview of the proposed method, which consists of 3 key components: 1) a medically relevant description extractor (MDE), 2) a medical knowledge-infused representation, and 3) a convolutional neural network (CNN). The de-identified clinical records from the n2c2 training corpus are decomposed into candidate instances by MDE, which eliminates nonrelevant terms corresponding to the selection criteria. The candidate records representations are then reproduced separately for the “met” and “not-met” criteria via MKR for each criterion. This bichannel representation is employed by the CNN architecture for category-status classification.
Figure 1.
Overview of the cohort selection method.
Medically relevant description extractor
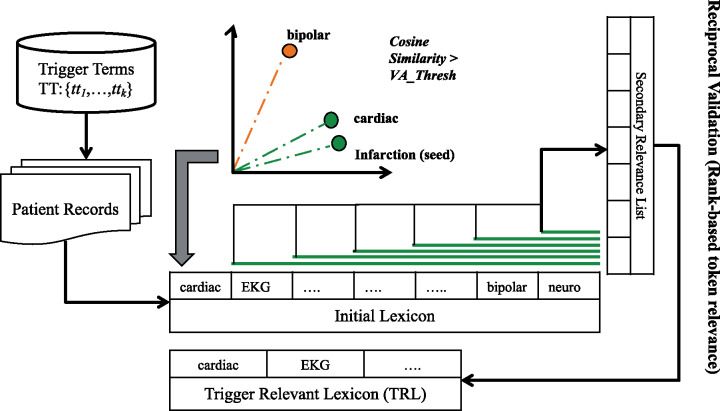
Free text descriptions are often too elaborate and noisy; therefore, preprocessing raw text to generate targeted topic summaries can help boost the effectiveness of classifiers. Our initial linguistic preprocessing includes sentence segmentation and punctuation and stop-word elimination using NLTK (https://pythonspot.com/nltk-stop-words/). Noises from multiple unrelated reports like pathology or radiology accumulate during separate hospital visits, which are sorted by the MDE module. Our approach focuses on using a criteria-relevant trigger term to initialize and expand trigger relevant lexicon (TRL)—a criterion-associated dictionary containing terms assimilated from the corpus that are inferentially related to the criterion. TRL operates by finding tokens that have similar scores on a vector-based cosine similarity index with the seed token based on word embeddings. As shown in Figure 2, for criterion MI-6MOS (6-months myocardial infarction), a corresponding trigger word, “infarction,” is used as the seed to expand the vocabulary for the criterion description. The sentences from the clinical records are tokenized and their respective embeddings are evaluated with the trigger term using cosine similarity. Selective threshold values are defined for each criterion to filter out unrelated tokens (noise). The first pass generates an initial TRL list. Reciprocal validation is used to establish strict relevance association between lexicon terms, while the rest are purged from the list. Each nth token (current secondary seed) in the initial list is reciprocally evaluated using the cosine metric with other terms in the list. These tokens are reranked according to the similarity index to generate a secondary relevance list. Tokens prior to n in the initial list, and maintaining top “n-1” ranks in the secondary list, are declared as reciprocally validated. Upon successful validation, the corresponding secondary seed terms are either included in the final TRL list or otherwise dropped from it. The MDE algorithm is illustrated in Box 1. Based on the overlapping terms in trigger relevance lexicon, medically relevant description (M) candidate instances are then mined from the clinical notes.
Figure 2.
Process of medically relevant description extractor.
Box 1: Algorithm: Medically Relevant Description Extractor
INPUT:
T: {T1, T2,……Tm} – Tokenized words from Clinical Record Ri
S: {S1, S2,……S13} – Seed Term for each of the select criteria for cohort.
BEGIN
1: FORi = 1 to N = 13
2: seed = Si
3: finalTRL = list()
4: inTRL = rankedVectorCosineSim(seed, T)
5: FOR EACHrelToken, posininTRL
6: buffTRL =rankedVectorCosineSim (relToken, T)
7: FOR EACHindexin range(0, pos-1)
8: IF (if_Present(inTRL [index], buffTRL[0, pos-1],))
9: finalTRL.add(inTRL [index])
10: END IF
11: END FOR
12: END FOR
OUTPUT: finalTRL
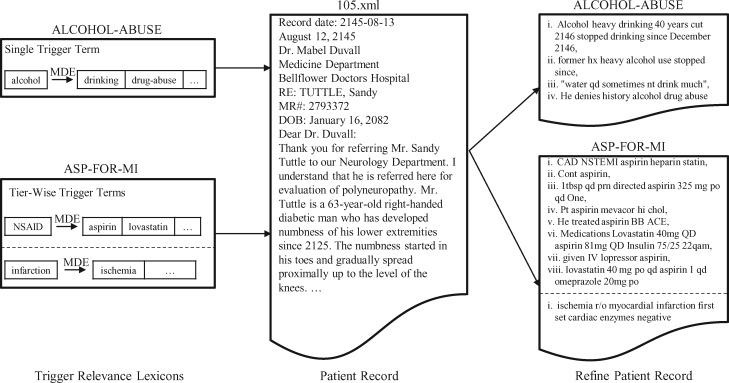
For example, criteria “ALCOHOL-ABUSE” with a seed trigger term (eg, “alcohol”) uses MDE to build the relevant lexicon by picking tokens, like “drinking, drug-abuse,” from the training set. Afterwards, this token dictionary is used to exclude sentences with no overlapping tokens reducing candidate instance size as shown in Figure 3. Single trigger-based processing is more challenging for complex or descriptive criteria such as “ADVANCED-CAD,” “ASP-FOR-MI,” “CREATININE,” and “MAJOR-DIABETES.” Therefore, tier-wise triggers are used to build separate lexicons. For example, “ASP-FOR-MI” is supplemented with 2 trigger terms “NSAID” and “infarction” to build relevant lexicons. Tokens like “aspirin, lovastatin” which have higher cosine similarities with “NSAID” are used to screen for candidate instances.
Figure 3.
An example of a refined clinical record using medically relevant description extractor.
Medical knowledge-infused representation
In the next phase, we use class knowledge-infused (K) discriminative lexicons on the medically relevant (M) candidate instances to generate candidate record embeddings. These criteria specific class knowledge-infused (K) lexicons are tokens with significant association to patient’s met (positive) and not-met (negative) class knowledge, as evaluated by the log likelihood ratio (LLR).27 The following equation is used to calculate the likelihood of the assumption that the occurrence of critical terms in the relevant descriptors is not random:
| (1) |
where I denotes the set of positive relevant descriptors in the training data; N(M) and N(¬M) are the numbers of relevant positive and negative descriptors, respectively; and N(t∧M) is the number of positive relevant descriptors containing the term t. A maximum likelihood estimation is performed to obtain probabilities p(t), p(t|M), and p(t|¬M). Tokens are ranked based on respective LLR scores; those with higher scores are ranked higher and, thus, are strongly related to the respective descriptor. For instance, for the criterion “ABDOMINAL,” “cholecystectomy: 31.73” is significantly associated with abdominal surgery when compared to “patient: 0.012.”
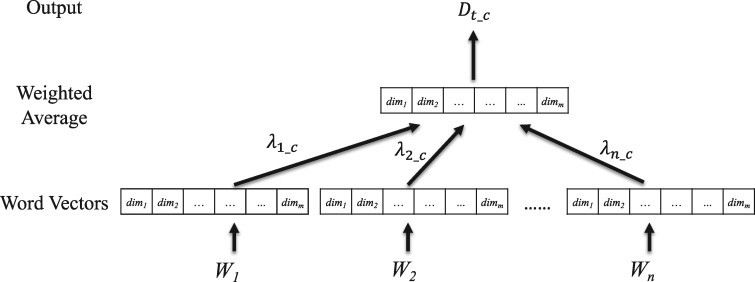
Next, we generate a knowledge-infused (K) weight based distributed vector representation for medically-relevant (M) candidate records. Between the continuous bag-of-word (CBOW) and skip-gram model,22 we adopted the former model to learn a 300-dimension embedding vector, which is commonly used in many DNN-based text representation tasks.28,29 The CBOW method learns the embedding of a word from the neighboring context.
As shown in Figure 4, a weighted average embedding is generated by integrating word embeddings with respective word LLR values to produce MKR for the clinical record. The document is represented as a weighted average of the word embeddings for category c, and the weight for a word is determined by its LLR value for category c. In essence, each clinical record is projected onto a point in the latent feature space using a distributed representation, which can then be evaluated using any classifier. More specifically, a clinical record is represented by 2 vectors, and , as the input of the CNN. In comparison with the BOW model, this distributed representation can incorporate more contextual information from the entire record. Moreover, semantic relations of various surface words can also be captured by the vector space projection of these distributed representations, which would otherwise consume a sizeable n-gram dictionary storage in the BOW approach.
Figure 4.
The procedure of integrating word embeddings using their respective LLR values to produce an MK representation.
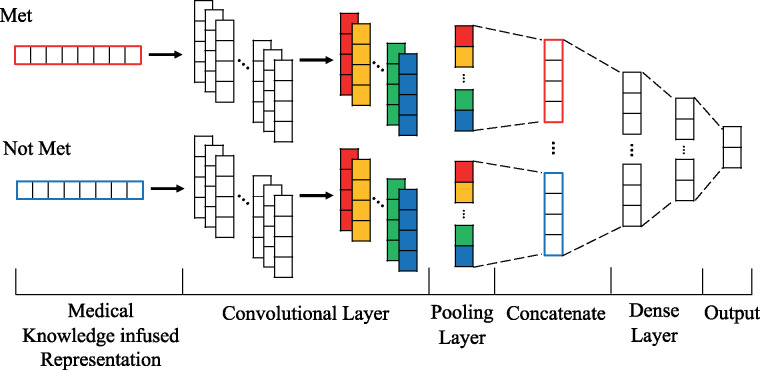
Convolutional neural network for identifying the status of selection criteria
Figure 5 illustrates the integration of MKR into the CNN. Bichannel “met” and “not-met” MK representations are used as initial embedding in the CNN architecture, which contains 256 filters of a window size of 4. The initial filters perform convolutions and generate a new feature map C = [C1, C2, …, Cm], which is further refined by another 128 filters with the same region size. Subsequent 1D-max pooling layer captures the maximum value over the convolved features. Features from both channels are concatenated into a single output with 256 dimensions. Finally, a dense layer with a softmax activation considers the concatenated feature vector as input and outputs met or not-met status for each record.
Figure 5.
Illustration of an integrated multiple convolutional neural networks architecture for recognizing clinical trial eligibility criteria.
Comparative analysis models
We also developed baselines and other MKR models to estimate the significance of our approach and performance variation in different classification platforms. Our first baseline employs a tokenized representation evaluated on the Naïve Bayes classifier, smoothed using the Laplace correction.27 It learns the statistical relevance of each token in a clinical record within different syntactic and semantic context. We also adopt a whole text multikernel CNN model using static word embeddings of instances as another baseline, dubbed here as Convolutional Neural Network for text (TextCNN) (https://github.com/bhaveshoswal/CNN-text-classification-keras).30 In addition to these 2 baselines, we also experiment with other models using MKRs and other classifiers. To this end, we first incorporate our MKR with an SVM classifier, denoted as MKSVM. This model uses the radial basis function kernel-based SVM with the enhanced embedding representation of MK to determine the category of each record.
Another model that we use to evaluate the effectiveness of MKRs is the extreme gradient boosting (XGBoost) classifier, denoted as MKXGBoost. XGBoost is a gradient-boosting decision tree that integrates multiple learners for classification problems.31 We include XGBoost (https://xgboost.readthedocs.io/en/latest/python/python_intro.html ) in this study as it has been validated on real-life, large-scale, and imbalanced data sets and solves many data science-related problems in a fast and accurate way.32,33 The last model that we include in the comparative analysis is a variant of our MKR that employs a multifilter, max-pooling CNN architecture called Knowledge-infused Convolutional Neural Network (KCNN). This model transforms the entire medical record into knowledge-infused (K) LLR-weighted embeddings for text representation.
Evaluation data set and experiment settings
Our evaluations were conducted on de-identified data from the 2014 i2b2/UTHealth Shared Task developed by Partners HealthCare (https://dbmi.hms.harvard.edu/programs/healthcare-data-science-program/clinical-nlp-research-data-sets), which was re-released as part of the 2018 n2c2 Shared Task for modeling criteria status prediction in cohort selection for clinical trials. It consists of 288 longitudinal patient records annotated by medical experts for 13 selection criteria. The selection criteria were based on patient’s personal and clinical history as detailed in Table 1.
Table 1.
Statistics of the n2c2 shared task data set
| No. of records | Train: 202 |
Test: 86 |
Criteria | ||
|---|---|---|---|---|---|
| met | not met | met | not met | ||
| ABDOMINAL | 77 | 125 | 30 | 56 | History of intra abdominal surgery, small or large intestine resection or small bowel obstruction |
| ADVANCED-CAD | 125 | 77 | 45 | 41 | Advanced cardiovascular disease, where “advanced” means having 2 or more of the following:
|
| ALCOHOL-ABUSE | 7 | 195 | 3 | 83 | Current alcohol use over weekly recommended limits |
| ASP-FOR-MI | 162 | 40 | 68 | 18 | Use of aspirin to prevent myocardial infarction |
| CREATININE | 82 | 120 | 24 | 62 | Serum creatinine > upper limit of normal |
| DIETSUPP-2MOS | 105 | 97 | 44 | 42 | Use of a dietary supplement (excluding Vitamin D) in the past 2 months |
| DRUG-ABUSE | 12 | 190 | 3 | 83 | Drug abuse, current or past |
| ENGLISH | 192 | 10 | 73 | 13 | Patient ability to speak English |
| HBA1C | 67 | 135 | 35 | 51 | Any HbA1c value between 6.5% and 9.5% |
| KETO-1YR | 1 | 201 | 0 | 86 | Diagnosis of ketoacidosis in the past year |
| MAJOR-DIABETES | 113 | 89 | 43 | 43 | Major diabetes-related complications (major complication is defined as any of the following that are a result of uncontrolled diabetes: amputation, kidney damage, skin conditions, retinopathy, nephropathy, and neuropathy) |
| MAKES-DECISIONS | 194 | 8 | 83 | 3 | Patient ability to make their own medical decisions |
| MI-6MOS | 18 | 184 | 8 | 78 | Myocardial infarction in the past 6 months |
For the initial MDE module, pretrained GloVe embeddings are used for cosine similarity evaluation.34 In order to generate MKRs, the model learned embeddings from 202 records in the training corpus using the Gensim word2vec (https://radimrehurek.com/gensim/models/word2vec.html) toolkit. Based on the statistical significance learned from our experiments using McNemar’s test on CBOW and skip-gram at 95% confidence level, we selected the CBOW method to learn 300-dimension word embeddings as the basis of MKRs. Our CNN architecture is implemented with Keras (https://keras.io/) under the following configurations: we set a Dropout probability of 0.25 after each layer, ReLU activation on the Dense Layer, and train for 20 epochs.
The results are evaluated with precision, recall, and F1-measure,35 as well as micro-averaged metrics to compare the overall performance. These measures are defined based on a contingency table of predictions for a target criteria Ci. The precision P(Ci), recall R(Ci), F1-measure F1(Ci), macro-average Macro(Ci), and micro-average Micro(Ci) are defined as follows:
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
where TP(Ci) denotes the number of true positives, the number of positive instances that are correctly classified. The FP(Ci) denotes the number of false positives, which are negative instances that are erroneously classified as positives. Analogously, TN(Ci) and FN(Ci) stand for the number of true negatives and false negatives, respectively. The F1 value is used to determine the relative effectiveness of the compared methods. The Fμ value is averaged over eligibility criteria status classes (ie, “met” and “not met”).
RESULTS
The experimental results are displayed in Table 2. Our 2 baselines, Naïve Bayes (NB) and TextCNN, perform consistently lower for each criterion when compared to others. Interestingly, TextCNN scored approximately 10% lower than NB and was the lowest among all the CNN-related runs executed in this study. Table 2 also contains MKR and other variant models including MKSVM, MKXGBoost, and KCNN. MKSVM and MKXGBoost show an approximately 7% and 6% increase in performance, respectively, over the NB implementation. KCNN, a variant of our bichannel, MKR scores 82.3% on the F1-measure metric—lower than both MKSVM and MKXGBoost runs, signifying that combined MKRs add further medical relevance to the representation than less sophisticated class knowledge-based infusion. MKCNN scores higher than any other listed CNN or kernel-based classifiers averaging 86.1% on F1-measure. It registers a 4% performance leap over the KCNN implementation.
Table 2.
Comparison of different models in their ability to recognize the status of the selection criteria
| Precision/Recall/F1 score (%) |
||||||
|---|---|---|---|---|---|---|
| Criteria/Models | NB | TextCNN | MKSVM | MKXGBoost | KCNN | MKCNN |
| ABDOMINAL | 60.5/59.3/59.5 | 44.6/48.0/42.6 | 70.5/69.4/69.8 | 71.8/71.8/71.8 | 83.3/53.3/46.3 | 76.3/78.4/76.8 |
| ADVANCED-CAD | 63.8/62.0/61.1 | 26.2/50.0/34.4 | 67.4/67.3/67.3 | 74.2/72.7/72.6 | 88.0/87.6/87.2 | 71.9/68.9/68.4 |
| ALCOHOL-ABUSE | 48.3/50.0/49.1 | 48.2/49.4/48.8 | 48.2/47.6/47.9 | 48.2/48.2/48.2 | 48.3/50.0/49.1 | 98.8/66.7/74.4 |
| ASP-FOR-MI | 90.0/52.8/49.7 | 38.0/41.3/22.9 | 92.5/66.7/70.9 | 69.6/67.1/68.1 | 39.5/50.0/44.2 | 93.0/69.4/74.3 |
| CREATININE | 58.7/58.3/58.5 | 52.9/50.9/47.5 | 80.9/67.1/69.5 | 80.3/70.5/73.0 | 80.9/67.1/69.5 | 77.5/69.7/71.8 |
| DIETSUPP-2MOS | 52.2/52.1/51.8 | 52.2/52.2/52.2 | 77.0/76.8/76.7 | 77.0/76.6/76.6 | 63.5/61.1/59.7 | 75.6/75.5/75.6 |
| DRUG-ABUSE | 48.3/50.0/49.1 | 48.2/49.4/48.8 | 65.5/65.5/65.5 | 58.8/64.3/60.7 | 98.8/66.7/74.4 | 48.3/50.0/49.1 |
| ENGLISH | 42.4/50.0/45.9 | 52.6/51.1/50.4 | 81.9/81.9/81.9 | 84.8/82.6/83.6 | 42.4/50.0/45.9 | 85.0/75.6/79.2 |
| HBA1C | 45.7/47.9/42.9 | 46.0/48.9/40.8 | 85.4/76.2/77.3 | 77.9/61.9/59.4 | 59.6/56.0/54.0 | 85.4/76.2/77.3 |
| KETO-1YR | 50.0/50.0/50.0 | 50.0/50.0/50.0 | 50.0/50.0/50.0 | 50.0/50.0/50.0 | 50.0/50.0/50.0 | 50.0/50.0/50.0 |
| MAJOR-DIABETES | 76.8/69.8/67.7 | 46.0/46.5/44.6 | 80.6/80.2/80.2 | 82.6/82.6/82.6 | 76.8/76.7/76.7 | 81.5/81.4/81.4 |
| MAKES-DECISIONS | 48.3/50.0/49.1 | 48.2/48.8/48.5 | 48.3/50.0/49.1 | 48.1/46.4/47.2 | 48.3/50.0/49.1 | 48.3/50.0/49.1 |
| MI-6MOS | 45.3/50.0/47.6 | 50.0/50.0/40.1 | 45.3/50.0/47.6 | 62.4/55.0/56.3 | 45.3/50.0/47.6 | 45.3/50.0/47.6 |
| Macro-average | 56.2/54.0/52.5 | 46.4/49.0/44.0 | 68.7/65.3/65.7 | 68.1/65.4/65.4 | 63.4/59.1/58.0 | 72.1/66.3/67.3 |
| Micro-average | 77.6/78.1/77.8 | 66.4/66.0/66.2 | 85.0/84.5/84.7 | 83.9/83.2/83.5 | 82.8/82.0/82.3 | 86.0/86.1/86.1 |
The bolded numbers indicate the best performance in the criteria with comparisons.Abbreviations: KCNN, Knowledge-infused Convolutional Neural Network; MKCNN, medical knowledge-infused convolutional neural network; MKSVM, medical knowledge-infused support vector machine; MKXGBoost, medical knowledge-infused extreme gradient boosting; NB, Naïve Bayes; TextCNN, convolutional neural network for text.
We also list the results from the n2c2 shared task to which we submitted 3 separate runs (Table 3). Submission 1 focused on criteria-based instance denoising using medically relevant descriptors (M) trained on multiwidth filters CNN model. For submissions 2 and 3, we experimented with knowledge-infused (K) weighted embeddings for the representation of the entire text trained on an SVM for classification. In addition, rules were also added in the initial screening of selected categories for submission 3. Heuristic rules were added to improve the prediction performance in skewed data sets for criteria (eg, English, Alcohol-abuse, Drug-abuse, MI6MOS, and Make-Decision). We have also added results from 1 of the top-performing systems in the n2c2 shared task Track 1, “MediUniGraz,” a rule-based model, to showcase a comparative assessment of our model.36 Performances of various systems that participated in this task were assessed as per the following F1 score range: average score: 0.80; minimum: 0.21; median: 0.82; and maximum: 0.91.36
Table 3.
Comparison of the proposed method with previous submissions for the n2c2 Shared Task
| Precision/Recall/F1 score (%) |
||||||
|---|---|---|---|---|---|---|
| Criteria/Models | Submission 1 (CNN) | Submission 2 (SVM) | Submission 3 (SVM+Rule) | MKCNN | MedUniGraz | |
| ABDOMINAL | 76.0/77.6/76.5 | 32.6/50.0/39.4 | 32.6/50.0/39.4 | 76.3/78.4/76.8 | 87.2/87.2/87.2 | |
| ADVANCED-CAD | 76.1/67.3/65.2 | 80.2/70.8/69.3 | 26.2/50.0/34.4 | 71.9/68.9/68.4 | 79.0/79.0/79.0 | |
| ALCOHOL-ABUSE | 48.2/49.4/48.8 | 48.3/50.0/49.1 | 48.3/50.0/49.1 | 98.8/66.7/74.4 | 48.2/49.4/48.8 | |
| ASP-FOR-MI | 77.8/67.2/70.1 | 39.5/50.0/44.2 | 39.5/50.0/44.2 | 93.0/69.4/74.3 | 92.5/66.7/70.9 | |
| CREATININE | 73.8/78.8/74.4 | 65.0/68.6/62.5 | 14.0/50.0/21.8 | 77.5/69.7/71.8 | 79.6/82.3/80.7 | |
| DIETSUPP-2MOS | 58.4/58.3/58.1 | 63.6/58.6/54.9 | 25.6/50.0/33.8 | 75.6/75.5/75.6 | 91.9/91.8/91.9 | |
| DRUG-ABUSE | 58.8/64.3/60.7 | 48.3/50.0/49.1 | 48.3/50.0/49.1 | 48.3/50.0/49.1 | 73.8/66.1/69.1 | |
| ENGLISH | 58.0/60.3/58.7 | 96.2/76.9/83.0 | 96.2/76.9/83.0 | 85.0/75.6/79.2 | 96.8/80.8/86.4 | |
| HBA1C | 78.3/78.3/78.3 | 54.8/50.5/39.7 | 29.7/50.0/37.2 | 85.4/76.2/77.3 | 95.5/92.9/93.8 | |
| KETO-1YR | 50.0/50.0/50.0 | 50.0/50.0/50.0 | 50.0/50.0/50.0 | 50.0/50.0/50.0 | 50.0/50.0/50.0 | |
| MAJOR-DIABETES | 79.6/79.1/79.0 | 68.2/58.1/51.4 | 25.0/50.0/33.3 | 81.5/81.4/81.4 | 84.0/83.7/83.7 | |
| MAKES-DECISIONS | 48.2/48.8/48.5 | 48.2/48.8/48.5 | 48.2/48.8/48.5 | 48.3/50.0/49.1 | 48.3/50.0/49.1 | |
| MI-6MOS | 58.2/54.3/55.2 | 45.2/48.7/46.9 | 45.2/48.7/46.9 | 45.3/50.0/47.6 | 98.2/81.3/87.5 | |
| Macro-average | 64.7/64.1/63.3 | 56.9/56.2/52.9 | 40.7/51.9/43.9 | 72.1/66.3/67.3 | 78.8/73.9/75.2 | |
| Micro-average | 81.7/82.4/81.9 | 78.3/79.0/78.5 | 74.0/74.7/73.3 | 86.0/86.1/86.1 | 90.8/91.2/91.0 | |
The bolded numbers indicate the best performance in the criteria with comparisons. Abbreviations: CNN, convolutional neural network; MKCNN, medical knowledge-infused convolutional neural network; SVM, support vector machine.
DISCUSSION
As a part of this clinical topic detection study, we developed a medical knowledge-infused clinical representation model to eliminate additional noise introduced by nondescriptive text. The comparative analysis of MKSVM, MKXGBoost and MKCNN with NB, TextCNN, and KCNN runs in Table 2 shows a gradual performance improvement using the clinically relevant representation—particularly with CNN, when compared with whole text representation. The latter fails to bring feature distinction owing to the high recurrence of irrelevant phrases in noisy data sets. For instance, n-grams like “Record date,” “Labs/Studies,” and “attending physician” appear repeatedly throughout the records and are repetitively captured as significant tokens or features in the convolutional filters. However, such n-grams have little medical relevance with criteria like cardiovascular health or drug-abuse history. In contrast, the construction of bichannel embeddings involves learning independently from the criteria relevant instances “M,” and differentially weighted according to infused-knowledge “K.” They enable us to explore the latent lexical MK features from an otherwise noisy data set.
We find in Table 3 that submission 1 had a comparatively stable performance over the other 2, and maintained F1 scores of 60%–79% for 7 out of 13 criteria. Our initial analysis indicated that the utilization of medically relevant representations can improve the overall performance. Therefore, we built on our previous work to develop an adaptable representation (ie, the “MKR”). A subsequent case study example for “ALOCHOL-ABUSE” from the current work shows that MKRs reduce the candidate instance size (eg, “intermittently binge drinking, occasional smoking” via MDE and identify differentiable class weights in instances (eg, “binge” [score: 16.09] and “drinking” [score: 8.63] for “met” and “not-met” classes, respectively). This relevance-based scoring helps in criteria-relevant detection of the record. MKRs are also useful in assessing nonrelevant instances as shown in another example where only “not-met” category lexicons are identified: “alcohol” [score: 7.33] and “occasional” [score: 1.81] for reduced instances (“denies smoking, occasional alcohol”). This signifies that the knowledge generated via MKR is statistically and semantically criteria-relevant.
Table 4 showcases a list of top 10 words from clinical records with selective “met” criteria relevance. The MKR module using LLR exploits the semantic co-occurrence of the tokens with a class/category to elevate word significance; for example, “binge” [score: 16.09], “dependence” [score: 16.09], and “help” [score: 10.66] were found to be significantly correlated with “met” status for ALCOHOL-ABUSE. Although these words do not have a direct inferential association with “alcohol,” they have associative relevance in describing alcohol-related abuse.
Table 4.
Top 10 keywords of each criterion extracted by relevant descriptor extraction
| Criteria | Keywords |
|---|---|
| ABDOMINAL | {cholecystectomy, nephrectomy, colostomy, history, appendectomy, bowel, renal, partial, splenectomy, HTN} |
| ADVANCED-CAD | {myocardial, infarction, inferior, test, stress, ischemic, ischemia, anterior, CAD, left} |
| ALCOHOL-ABUSE | {binge, dependence, help, therapy, CT, night, seizures, withdrawal, says, days} |
| ASP-FOR-MI | {mg, 81, 325, stroke, Capsule, pain, aspirin, qd, 81mg, today} |
| CREATININE | {value, 1.6, 1.8, 2.5, 2.4, 1.7, 2.2, 2.1, 2.3, 2.9} |
| DIETSUPP-2MOS | {D, TAB, PO, vitamin, QD, 1000, MULTIVITAMINS, Calcium, Vitamin, po} |
| DRUG-ABUSE | {cocaine, H, heroin, O, past, ago, There, previous, Admits, 95} |
| ENGLISH | {english, fluently, cultures, right, history, speaking, language, speak, tongue, moderate} |
| HBA1C | {6.6, 6.8, 6.9, 6.7, 7.5, 7.2, 7.3, 7, 8.5, 7.4} |
| MAJOR-DIABETES | {diabetic, nephropathy, neuropathy, peripheral, diabetes, disease, hypertension, retinopathy, p, chronic} |
| MAKES-DECISIONS | {SH, can, manage, independently, two, Lives, Father, history, sons, found} |
| MI-6MOS | {ASA, limited, 20, BB, RBBB, heparin, NSR, following, improves, 2091} |
One of the limitations of the proposed method is the prediction for ketoacidosis criteria, with only a single training record which makes it difficult to evaluate using machine-learning models. In addition, criteria like “ADVANCED-CAD” also show subpar performances due to lower precision scores. Subsequent error analysis reveal that the instances picked by MDE in such cases are not evaluated under multiword trigger context as required by the criteria. For instance, MKR identified 45 “met” significant words (eg, “myocardial,” “infarction,” “inferior”) with the highest category score of 35.89, and only 3 “not-met” significant words (eg, “no,” “hemorrhage,” “territorial”) with the highest category score of 20.66. This imbalance in weighted token size and higher score for “met” lead to a false positive prediction for this record. Therefore, for complex selection criteria, we need to develop a multitrigger context-based MDE screening. Semantic features like symptom co-occurrence can also improve the performance for some criteria (eg, co-occurrence of alcohol and drug-abuse history in patient records).
CONCLUSION
Rule/guideline-based systems have been helpful in text classification of clinical literature in the past. However, in order to more effectively and expediently achieve this goal, we developed an automated module to represent criteria-specific clinical text for the purpose of model learning, which is scalable and flexible for clinical applications. This system can reduce the dependency on the human component required in rule-based systems. The novelty of our method lies in its enhanced feature representation framework, which combines the discriminative factors of medical criteria-based denoising (MDE) and class–category knowledge-infused representation (MKR). Our system performs well over the generic representations learned from DNN as showcased in our experiments with TextCNN, KCNN, and MKSVM. However, despite the promising performance on a machine-learning platform, our model was less effective for a few criteria, such as “MI-6MOS” and “DRUG-ABUSE.”
For our future work, we intend to integrate expert clinician knowledge with the current model. We also plan to develop another study from this work that will focus on identifying deeper semantic elements for an enhanced MKR. The current task of cohort selection criteria is a text classification problem. Therefore, the proposed method has the capacity to be extended to various NLP tasks, such as sentiment analysis, question answering systems, and topic detection. Moreover, our method is adaptable to different languages. Therefore, we also intend to 1) implement our model on Chinese clinical notes-based studies using Chinese character embeddings, 2) generate Trigger Relevant Lexicon, and 3) use Chinese word segmentation toolkit (eg, Jieba) for linguistic preprocessing.
FUNDING
This research was supported by the Ministry of Science and Technology of Taiwan under grants MOST 106-2218-E-038-004-MY2, MOST 107-2410-H-038-017-MY3, and MOST 107-2634-F-001-005.
AUTHOR CONTRIBUTIONS
YCC, JHC, and WLH designed the study and conceived the research question. CJC and NW are Joint First Authors. CJC and NW conducted the experiments, statistical analyses, and reviewed and interpreted the findings. YCC, NW, and CJC wrote the manuscript, and reviewed and noted points of revision for the manuscript.
ACKNOWLEDGMENT
The authors are grateful to The Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan for providing financial support for this project.
Conflict of interest statement
None declared.
REFERENCES
- 1. Liao KP, Ananthakrishnan AN, Kumar V, et al. Methods to develop an electronic medical record phenotype algorithm to compare the risk of coronary artery disease across 3 chronic disease cohorts. PLoS ONE 2015; 108: e0136651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McCoy TH, Castro VM, Cagan A, et al. Sentiment measured in hospital discharge notes is associated with readmission and mortality risk: an electronic health record study. PLoS ONE 2015; 108: e0136341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uzuner Ö. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc 2009; 164: 561–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Uzuner Ö, Solti I, Cadag E.. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 175: 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Uzuner Ö, Luo Y, Szolovits P.. Evaluating the state-of-the-art in automatic de-identification. J Am Med Inform Assoc 2007; 145: 550–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uzuner Ö, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008; 151: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quintana Y, Safran C, Global health informatics—an overview In de Fátima Marin H, Massad E, Gutierrez MA, et al. (Eds.). Global Health Informatics. London: Academic Press; 2017: 1–13. [Google Scholar]
- 8. Weng W-H, Wagholikar KB, McCray AT, et al. Medical subdomain classification of clinical notes using a machine learning-based natural language processing approach. BMC Med Inform Decis Mak 2017; 171: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bernhardt PJ, Humphrey SM, Rindflesch TC. Determining prominent subdomains in medicine. In: AMIA Annual Symposium Proceedings; 2005: 46–50. [PMC free article] [PubMed]
- 10. Kocbek S, Cavedon L, Martinez D, et al. Text mining electronic hospital records to automatically classify admissions against disease: measuring the impact of linking data sources. J Biomed Inform 2016; 64: 158–67. [DOI] [PubMed] [Google Scholar]
- 11. Huang GD, Bull J, Johnston McKee K, et al. Clinical trials recruitment planning: a proposed framework from the clinical trials transformation initiative. Contemp Clin Trials 2018; 66: 74–9. [DOI] [PubMed] [Google Scholar]
- 12. Glicksberg BS, Miotto R, Johnson KW, et al. Automated disease cohort selection using word embeddings from electronic health records. Pac Symp Biocomput 2018; 23: 145–56. [PMC free article] [PubMed] [Google Scholar]
- 13. Mikolov T, Sutskever I, Chen K, et al. Distributed Representations of Words and Phrases and their Compositionality. arXiv: 1310.4546 [cs. CL] Published Online First: October 16, 2013. http://arxiv.org/abs/1310.4546.
- 14. Wagholikar KB, MacLaughlin KL, Kastner TM, et al. Formative evaluation of the accuracy of a clinical decision support system for cervical cancer screening. J Am Med Inform Assoc 2013; 204: 749–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mann CJ. Observational research methods. Research design II: cohort, cross sectional, and case-control studies. Emerg Med J 2003; 201: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Trafton JA, Martins SB, Michel MC, et al. Designing an automated clinical decision support system to match clinical practice guidelines for opioid therapy for chronic pain. Implement Sci 2010; 5: 26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lobach DF, Johns EB, Halpenny B, et al. Increasing complexity in rule-based clinical decision support: the symptom assessment and management intervention. JMIR Med Inform 2016; 44: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sebastiani F. Machine learning in automated text categorization. ACM Comput Surv 2002; 341: 1–47. [Google Scholar]
- 19. Yuan J, Holtz C, Smith T, et al. Autism spectrum disorder detection from semi-structured and unstructured medical data. EURASIP J Bioinform Syst Biol 2017; 2017: 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marafino BJ, Davies JM, Bardach NS, et al. N-gram support vector machines for scalable procedure and diagnosis classification, with applications to clinical free text data from the intensive care unit. J Am Med Inform Assoc 2014; 215: 871–5. doi: 10.1136/amiajnl-2014-002694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goodwin TR, Harabagiu SM.. Learning relevance models for patient cohort retrieval. JAMIA Open 2018; 12: 265–75. doi: 10.1093/jamiaopen/ooy010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mikolov T, Chen K, Corrado G, et al. Efficient Estimation of Word Representations in Vector Space. arXiv: 1301.3781 [cs] Published Online First: January 16, 2013http://arxiv.org/abs/1301.3781.
- 23. Hughes M, Li I, Kotoulas S, et al. Medical text classification using convolutional neural networks. Stud Health Technol Inform 2017; 235: 246–50. [PubMed] [Google Scholar]
- 24. Goldberg Y. Neural network methods for natural language processing. San Rafael, CA: Morgan & Claypool; 2017. doi: 10.2200/S00762ED1V01Y201703HLT037 [Google Scholar]
- 25. Poria S, Cambria E, Gelbukh A.. Aspect extraction for opinion mining with a deep convolutional neural network. Know-Based Syst 2016; 108: 42–9. [Google Scholar]
- 26. Do HH, Prasad P, Maag A, et al. Deep learning for aspect-based sentiment analysis: a comparative review. Expert Syst Appl 2019; 118: 272–99. [Google Scholar]
- 27. Manning CD, Raghavan P, Schutze H. Introduction to Information Retrieval. https://nlp.stanford.edu/IR-book/information-retrieval-book.html. Accessed February 8, 2019.
- 28. Melamud O, McClosky D, Patwardhan S, et al. The role of context types and dimensionality in learning word embeddings. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; June 12–17, 2016: 1030–40; San Diego, CA.
- 29. Yadav S, Ekbal A, Saha S, et al. Deep learning architecture for patient data de-identification in clinical records. ClinicalNLP@COLING 2016; 2016: 32–41. https://www.aclweb.org/anthology/W16-4206. Accessed January 25, 2019.
- 30. Kim Y. Convolutional Neural Networks for Sentence Classification. arXiv: 1408.5882 [cs] Published Online First: 25 August 2014.http://arxiv.org/abs/1408.5882.
- 31. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining-KDD ’16; August 13–17, 2016: 785–94; San Francisco, CA.
- 32. Wei X, Jiang F, Wei F, et al. An ensemble model for diabetes diagnosis in large-scale and imbalanced dataset In: Proceedings of the Computing Frontiers Conference on ZZZ-CF’17; May 15–17, 2017: 71–8; Siena, Italy. [Google Scholar]
- 33. Nagata M, Takai K, Yasuda K, et al. Prediction models for risk of type-2 diabetes using health claims. In: Proceedings of the BioNLP 2018 Workshop.; July 19, 2018: 172 –6; Melbourne, Australia. http://www.aclweb.org/anthology/W18-2322.
- 34. Pennington J, Socher R, Manning C. Glove: Global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP); October 25–29, 2014: 1532–43; Doha, Qatar.
- 35. Manning CD, Schütze H.. Foundations of Statistical Natural Language Processing. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 36. Stubbs A, Filannino M, Uzuner O. n2c2 shared task Track 1-wrap-up slides, 2018. https://portal.dbmi.hms.harvard.edu/projects/n2c2-t1/. Accessed November 13, 2018.