Abstract
Objective
Identifying patients who meet selection criteria for clinical trials is typically challenging and time-consuming. In this article, we describe our clinical natural language processing (NLP) system to automatically assess patients’ eligibility based on their longitudinal medical records. This work was part of the 2018 National NLP Clinical Challenges (n2c2) Shared-Task and Workshop on Cohort Selection for Clinical Trials.
Materials and Methods
The authors developed an integrated rule-based clinical NLP system which employs a generic rule-based framework plugged in with lexical-, syntactic- and meta-level, task-specific knowledge inputs. In addition, the authors also implemented and evaluated a general clinical NLP (cNLP) system which is built with the Unified Medical Language System and Unstructured Information Management Architecture.
Results and Discussion
The systems were evaluated as part of the 2018 n2c2-1 challenge, and authors’ rule-based system obtained an F-measure of 0.9028, ranking fourth at the challenge and had less than 1% difference from the best system. While the general cNLP system didn’t achieve performance as good as the rule-based system, it did establish its own advantages and potential in extracting clinical concepts.
Conclusion
Our results indicate that a well-designed rule-based clinical NLP system is capable of achieving good performance on cohort selection even with a small training data set. In addition, the investigation of a Unified Medical Language System-based general cNLP system suggests that a hybrid system combining these 2 approaches is promising to surpass the state-of-the-art performance.
Keywords: clinical natural language processing, cohort selection, clinical trial, rule-based system, UMLS
INTRODUCTION
Clinical trials are research studies to evaluate whether a new medical approach, such as a new medication or procedure, is safe and effective in people. They are also designed to answer scientific questions and assist researchers in developing new approaches to prevent, diagnose, or treat certain medical conditions.1 Typically, a successful clinical trial study requires large enough selected samples under certain criteria, such as defining case/control disease cohorts through the presence of certain medical conditions, to support it.2 Thus, accurate and robust cohort selection is critical to clinical trial studies.
Electronic health records (EHRs) contain a large amount of useful information of patients and can serve as a good platform for cohort selection in clinical trials.1,3 However, the structured data in EHRs are usually not sufficient to support the cohort selection, as much of the useful information lies buried in unstructured data, such as radiology reports, discharge summaries, medical history, laboratory results, and even email records.4–7 Typically these free text data are difficult and time-consuming for manual review or analysis. Therefore, natural language processing (NLP) systems which can automatically process these clinical narratives and assess patients’ qualification according to inclusion/exclusion criteria of clinical trials are highly desirable.
The 2018 National NLP Clinical Challenges (n2c2) Shared-Task and Workshop on Cohort Selection for Clinical Trials8 was organized to focus on this topic. The challenge was designed to encourage NLP systems which can automatically identify which patients meet selection criteria for clinical trials. The task requires NLP systems to process a set of longitudinal health records of each patient, compare them to a series of selection criteria, and determine if the patients meet or do not meet each criterion. The “met”/“not met” decision for each patient and different criteria will then be used for selecting patients for appropriate clinical trials.
In this article, we describe an integrated clinical NLP system as submitted to 2018 n2c2 task on cohort selection for clinical trials, which employs a generic rule-based framework plugged in with lexical-, syntactic-, and meta-level rules generated from the training data of the challenge. It was ranked fourth in the challenge. In addition, we developed and investigated a knowledge-based general clinical NLP (cNLP) system based on the Unified Medical Language System (UMLS)9 and Unstructured Information Management Architecture (UIMA).10 Evaluation and analysis were conducted upon different aspects between these systems with the n2c2 challenge data. We demonstrate that our systems are feasible and can be used for reliable clinical data mining.
BACKGROUND
Cohort selection based on patients’ narrative records is challenging for clinical NLP as the selection criteria are usually complex and require combining multiple clinical NLP components such as clinical concept extraction (eg, disease, symptom, and medication), assertion detection (eg, negation, uncertainty), laboratory results extraction (eg, creatine measurement), temporal information extraction (eg, date of diagnosis, frequency and duration of taking medication), patients’ meta-information extraction (eg, gender, age) and so on. The challenges lie in both the performance of individual clinical NLP components and also the integration among them. Misinformation in any component could result in inaccurate decisions on patients’ qualification of certain criteria, leading to contamination of the study cohorts. For instance, 1 of the criteria in the n2c2 challenge is to decide whether the patients had advanced cardiovascular disease or not. In order to certify this criterion, the NLP system needs to detect whether the patients were taking 2 or more medications for CAD; had the history of myocardial infarction; were currently experiencing angina; and had ischemia currently or in the past. Thus, multiple NLP components, such as concepts detection, assertion, and temporal information detection are involved. Table 1 establishes the definition, examples, required NLP components, and data size of each criterion used in the 2018 n2c2 challenge.
Table 1.
Definition and basic information of the 13 selection criteria as used in the n2c2 challenge
| Criterion Name | Criteria | NLP components |
Examples | Number of records |
|||||
|---|---|---|---|---|---|---|---|---|---|
| CE | AD | Time | Lab | Meta | Met | Not met | |||
| DRUG-ABUSE | Drug abuse, current or past | Y | Y | “Drugs- According to the pt he has used cocaine and crack as recent 5-6 years ago.” | 15 | 273 | |||
| ALCOHOL-ABUSE | Current alcohol use over weekly recommended limits | Y | Y | Y | Y | “He does admit to heavy drinking, approximately 3–4 drinks per day.” | 10 | 278 | |
| ENGLISH | Patient must speak English | Y | Y | “HPI: 76 year old Spanish speaking male with numerous medical problems” | 265 | 23 | |||
| MAKES-DECISIONS | Patient must make their own medical decisions | Y | Y | “Pt. is minimally responsive at baseline, cared for at home by her husband with the assistance of VNA.” | 277 | 11 | |||
| ABDOMINAL | History of intra-abdominal surgery, small or large intestine resection or small bowel obstruction | Y | Y | “bowel surgery 15 yrs ago- g.a.- no complications” | 107 | 181 | |||
| MAJOR-DIABETES | Major diabetes-related complication, defined as any of the following that are a result of (or strongly correlated with) uncontrolled diabetes:
|
Y | Y |
|
156 | 132 | |||
| ADVANCED-CAD | Advanced cardiovascular disease, defined as having 2 or more of the following:
|
Y | Y | Y | Y | “Lasix, enalapril, and amlodipine”“prior transient ischemic attack”“presents with NSTEMI.”“describes intermittent chest pain, which he has had for a number of months without significant change.” | 170 | 118 | |
| MI-6MOS | Myocardial infarction in the past 6 months | Y | Y | Y | Y | “NSTEMI on Oct 8, 2111” | 26 | 262 | |
| KETO-1YR | Diagnosis of ketoacidosis in the past year | Y | Y | Y | Y | “evidence of DKA on urine or labs” | 1 | 287 | |
| DIETSUPP-2MOS | Taken a dietary supplement (excluding Vitamin D) in the past 2 months | Y | Y | Y | Y | “calcium carbonate 1250mg po tid” | 149 | 139 | |
| ASP-FOR-MI | Use of aspirin to prevent myocardial infarction | Y | Y | “Aspirin (ACETYLSALICYLIC Acid) 325MG TABLET PO QD x 30 days” | 230 | 58 | |||
| HBA1C | Any HbA1c value between 6.5 and 9.5% | Y | Y | Y | “Hgb A1c 7.30 6/28/96.” | 102 | 186 | ||
| CREATININE | Serum creatinine > upper limit of normal | Y | Y | Y | Y | “creatinine of 1.69” | 106 | 182 | |
The table also shows the examples, number of records “met” or “not met,” each criterion in the challenge released data sets, and NLP components used by each criterion. For NLP components, “CE” stands for concept extraction, “AD” stands for assertion detection, “Time” stands for time-related information extraction, “Lab” stands for laboratory results extraction, and “Meta” stands for meta-information extraction, such as patient’s gender and the most recent timestamp in all notes.
Many previous works and NLP systems contribute to addressing this issue in aspects of different clinical NLP components required by this task. Different NLP systems have been developed for clinical concepts extraction, such as MetaMap,11 cTAKES,12 HiTEX,13 and MedTagger.14 Series of systems have been developed for negation and assertion detection including NegEx,15 ConText,16 and DEEPEN.17 NLP challenges on clinical text mining have also been organized to assess the state of the art on different aspects, including concept extraction,18 assertion detection,18,19 medication information extraction,20 temporal information extraction,21 heart disease risk factor identification,22 smoking status identification,23 etc. Among these systems and works, various approaches have been used, and most of them can be classified as rule-based, machine learning, or hybrid. Though machine learning approaches are widely used for individual focused clinical NLP tasks, rule-based systems still hold their superiority in integration and interpretation and are still the dominant methods in commercial products.24Successful stories in applying rule-based systems have been reported widely in medical information extraction6,25 and clinical decision support.26,27
In this article, we describe an integrated clinical NLP system for clinical cohort selection by combining individual NLP components (enabled by a generic rule-based framework) and task-specific multi-level rules, which achieved good performance in the 2018 n2c2 challenge. We also investigated a UMLS-based general cNLP system for this task.
MATERIALS AND METHODS
Task and data
In the 2018 n2c2 challenge, there were 13 criteria for NLP systems to evaluate based on patients’ longitudinal health records. The names of these 13 criteria are: “DRUG-ABUSE,” “ALCOHOL-ABUSE,” “ENGLISH,” “MAKES-DECISIONS,” “ABDOMINAL,” “MAJOR-DIABETES,” “ADVANCED-CAD,” “MI-6MOS,” “KETO-1YR,” “DIETSUPP-2MOS,” “ASP-FOR-MI,” “HBA1C,” and “CREATININE”. Table 1 establishes the definition and examples of each criterion. As shown in Table 1, several selection criteria consist of multiple sub-criteria and require intra-criterion rules/logic to organize and integrate sub-criteria-level evidence into the final decision. The task required NLP systems to extract and evaluate annotation evidence for every sub-criterion in the 13 main criteria and provide patient-level decisions on whether the patient met or did not meet these 13 criteria. Regarding the complexity of the criteria logic, almost all criteria demanded the collaboration of various NLP components including clinical concept extraction, assertion detection, laboratory results extraction, temporal information extraction, and patients’ meta-information extraction. Table 1 also provides a high-level summary of the clinical NLP components employed by each criterion.
The data used by 2018 n2c2 shared task were from the 2014 Informatics for Integrating Biology & the Bedside (i2b2)/UTHealth Shared-Tasks on de-identification and heart disease risk factors,22 which was provided by Partners HealthCare. The data set contains longitudinal records of 288 patients, with about 2 to 5 records for each patient. During the challenge, these 288 patient records were split into training and testing data sets with a population of 202 and 86, respectively. All the files had been de-identified and annotated in patient level to determine the patients’ qualification against the 13 selection criteria as previously mentioned. In addition, 10 records from the training data set were further annotated with textual–span-level annotations which provided detailed evidence to support the patient-level decision. Only the training data set with the annotations was released for participants to develop their NLP systems, and the final evaluation of the submitted systems was conducted by the organizer based on the held-out test data set.
Systems overview
We developed 2 NLP systems to address the issues mentioned previously. A challenge-oriented rule-based system which mainly uses the challenge released training data for constructing rules and domain knowledge was our main system for this task. In addition, a hybrid general cNLP system which was initially designed for general medical information extraction and computer-assistant coding 28 was retested with n2c2 data. This general cNLP system is a rule/machine learning hybrid system which is built with the UMLS knowledge base and UIMA framework and employs various machine learning models pretrained with a much larger medical data set. We intentionally did minimal tuning of the general cNLP system with n2c2 data and only applied it to 3 highly medical-related criteria: “ABDOMINAL,” “MAJOR-DIABETES,” and “ADVANCED-CAD”.
The general cNLP system was initially expected to serve as a competitor of our rule-based system to achieve better performance, especially since the rule-based system was constructed based on a relatively small data set. However, the comparison and analysis between them provided interesting insights about the capability and generalizability of a highly task-specific rule-based system versus a general cNLP system when dealing with small data sets.
Both of the systems contain 3 main modules: (1) Evidence Extraction; (2) Assertion; (3) Criteria Logic. The Evidence Extraction module serves to extract all the useful information such as diseases, medications, and time mentions which could be used in the final criteria qualification evaluation. The Assertion module extracts negation, history, family, evaluation, and uncertainty information to support the decision-making. Finally, the Criteria Logic module provides the decision according to the criteria qualification requirements. The 2 systems share the same criteria logic but differ in Evidence Extraction and Assertion modules regarding algorithm and architecture.
Rule-based system
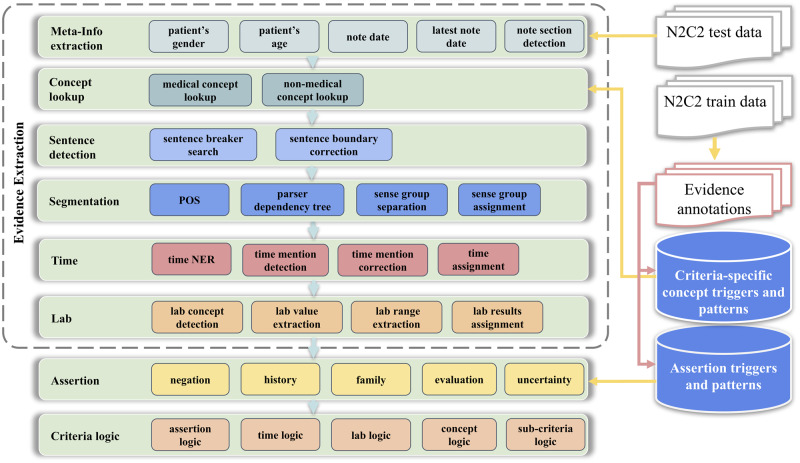
The challenge-oriented rule-based system consists of multiple functional modules, which all share the same generic rule-based framework. In each module, the rule-based framework uses a “bottom-up” design and directly starts with lexical-level rules, such as key word triggers or regular expressions lookup, in the note. Then, the lexical-level evidence/mentions will be used by syntactic-level rules for validation or relation assignment. For example, a lexical-level extracted mention (eg, time mention) will be assigned to other mentions (eg, the corresponding medical concept) in syntactic-level. In high-level architecture, the pipeline starts with patient-level/note-level meta-information extraction and Concept Lookup in the notes. Once a certain concept such as disease or medication is found, the Sentence Boundary Detection module extracts the corresponding sentence. Then the Segmentation module extracts the sense group which relates to the core concept based on part of sentence (POS) tag and parser dependency tree. The following Time and Lab modules extract possible time mentions and lab results, and decide whether to assign them to the evidence or not. In the Assertion module, key words to address the assertion condition are used as triggers, and assertion logics in syntactic-level are applied to evaluate whether the core concept should be assigned with assertion attributes. In this system, the criteria-specific medical/nonmedical concepts and assertion triggers are the key inputs of the framework and most of them are extracted from the n2c2 training data set. Figure 1 shows the high-level architecture of the rule-based system.
Figure 1.
Architecture of the rule-based system. This system utilizes task-specific multilevel rules generated from the 2018 n2c2 challenge released training data.
More specifically, the rules used in our individual functional modules can be classified into 3 levels: lexical, syntactic, and meta-information level.
Lexical level
Lexical rules contain positive or negative lexical patterns that indicate the presence of the target concepts. For instance, “alcohol,” “EtOH,” and “drunk” are the triggers indicating the patients’ alcohol use status. For each concept, a semantic group with the vocabulary and patterns was generated. Those semantic groups were initially generated by analyzing the evidence-level annotations of the training data. Later on, an active learning process was performed to tailor the lexical triggers and patterns in the semantic group. This process contained 6 steps: (1) Create or update the vocabularies/patterns for the target semantic group; (2) Apply triggers/patterns for concept extractions; (3) Integrate the evidence-level results to patient-level; (4) Compare to gold standard results and conduct error analysis; (5) Generate new triggers/patterns or remove some from the semantic group according to the error analysis; (6) Back to step 1 and repeat the process until expected performance has been achieved. In addition, several open access resources were borrowed to add more terms of interest. For instance, a list of dietary supplements29 was used to generate the vocabularies for dietary supplements. A list of countries, nationalities, and their languages30 was used to generate rules indicating whether the patient spoke English or not. Some terms of interest were also added from UMLS, such as different expressions of myocardial infarction and intraabdominal surgery. This process utilized the rich expressions of concepts and CUI–CUI relations in UMLS. For instance, we first identified the corresponding concept unique identifier (CUI) for the target semantic group (eg, “C0198482” for abdomen surgery), and then utilized the CUI–CUI relations in UMLS to extract the children of the target CUI as the candidates (eg, “C0008320” as cholecystectomy). After this step, we manually reviewed these CUI candidates as well as their expressions and then conducted the aforementioned active learning process to refine the patterns. Table 2 shows the semantic groups and examples of the triggers.
Table 2.
Semantic groups and example terms for extracting concepts and other types of mentions
| Semantic groups | Example terms | Size |
|---|---|---|
| Drug abuse | drug abuse, IVDA, substance abuse, illicit | 14 |
| Alcohol abuse | alcohol, drunk, EtOH, beer | 11 |
| Language | Spanish, Cantonese, French | 171 |
| Cannot make decisions | unresponsive, unconscious, confusing | 16 |
| Myocardial ischemia | myocardial ischemia, NSTEMI, MI, myocardial infarction | 19 |
| Ketoacidosis | ketoacidosis, DKA | 2 |
| Dietary supplements | Vitamin, Ca, Folic acid, MVI | 259 |
| Aspirin | aspirin, ASA, Ecotrin, Bufferin | 8 |
| HbA1C | Hemoglobin A1c, HgbA1c, HgA1c | 6 |
| Creatinine | Creatinine, CRE, Crt | 3 |
| Intra-abdominal surgery | bowel obstruction, small bowel resection, hysterectomy, cholecystectomy | 31 |
| diabetic complications | amputation, erythema, renal insufficiency, retinopathy, neuropathy, nephropathy | 40 |
| CAD medications | Zocor, Lasix, hydrochlorothiazide | 104 |
| Angina | angina, chest pain, CP | 4 |
| Ischemia | ischemia, ischemic | 4 |
| Negation | denies, no, negative, never, rule out | 27 |
| History | past, s/p, PMH, HX | 8 |
| Family | mother, father, wife, husband | 10 |
| Evaluation | consult, prevent, screen | 7 |
| Uncertainty | ?, check, unsure, possible | 9 |
| Gender | male, female, he, she | 10 |
Abbreviations: ASA, acetylsalicylic acid (aspirin); DKA, diabetic ketoacidosis; EtOH, ethyl alcohol; IVDA, intravenous drug abuse; MI, myocardial infarction; MVI, multivitamin; NSTEMI, non-ST-elevation myocardial infarction.
Syntactic level
Rules in this level utilize POS tags and the parser dependency tree of the sentence, and were mainly used to validate the relations between the core concepts and their modification attributes, such as assertions, time mentions, and lab results. There could be multiple concepts and assertion/time mentions in 1 sentence. Thus, assigning correct assertions or time attributes to the target concept is critical and challenging. In this work, we used spaCy31 (version 2.0.18) to generate the POS tags and parser dependency tree. The syntactic features were then used to divide the whole sentence into sense groups and calculate the effective scopes of the modification attributes. For example, in the sentence “SOCIAL HISTORY: No tobacco, rare alcohol, and occasional cocaine use.”, the scope of negation mention “No” doesn’t affect the target concept “cocaine use.” Whereas in another sentence, “SOCIAL HISTORY: No tobacco, alcohol, or cocaine use.”, the negation scope contains “cocaine use,” leading to negative evidence of drug abuse. Moreover, we found that a significant amount of assertion patterns and triggers were highly criteria-specific. For instance, “occasional cocaine use” is definitely indicating patients’ drug abuse status and should be regarded as positive evidence for “DRUG-ABUSE” criterion. However, “occasionally drinking alcohol” cannot serve as the positive evidence for “ALCOHOL-ABUSE” according to the criterion definition. Therefore, we designed our rule-based framework to be highly compatible with both universal and criteria-specific rules. Furthermore, some context related to lab test results employed a different format in which test items and values were aligned vertically instead of horizontally. Thus, we developed a rule-based algorithm to automatically detect the vertical alignment format and extract the corresponding lab results.
Meta-information level
Although the lexical and syntactic rules use local information, meta-information rules aim to use section-level, note-level, document-level, and patient-level information such as patient’s gender, date of note, the latest timestamp of the whole record, sections of each note, and so on. For example, many criteria contain time constraints like “Myocardial infarction in the past 6 months,” “taking 2 or more medications to treat CAD,” etc. Thus, a normalization process is required in the Time module to align each time mention with the date of the most recent record which is regarded as “current.” In addition, in the process of error analysis, we found that section information was very useful to rule out some false positives. For instance, a mention of certain medications (eg, aspirin) could indicate either the patient was taking it to treat disease or the patient was allergic to it. However, this information was usually not available locally. Thus, the ability to identify which section the mention belonged to (like “Medication” or “Allergies”) was very useful for disambiguation of the mention. Another example is distinguishing dietary supplements (eg, calcium) from laboratory exam items. In addition, the normal range of certain laboratory test (eg, creatinine) highly depends on gender and age. Thus patient-level rules such as indicating the normal/abnormal ranges of lab tests according to the patients’ genders were generated to locate abnormal lab test results.
General cNLP system
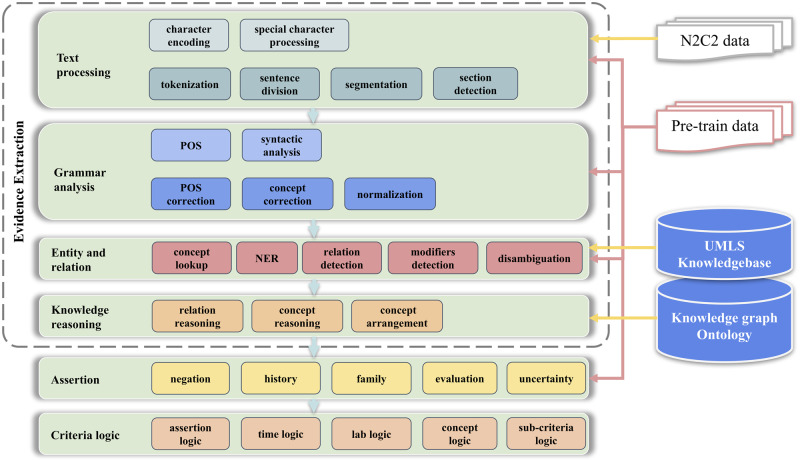
The general cNLP system uses a “top-down” design and is built with the UMLS and UIMA framework. It contains the flowing modules: Text processing, Grammar Analysis, Entity and Relation, and Knowledge Reasoning (Figure 2). In the Text Processing module, all the sub-modules like tokenization, sentence division, section detection use rule/ML hybrid methods and are pretrained with a much larger data set. More specifically, the sentence boundary detection is based on maximum entropy classifier32,33 and manually added rules such as rules considering abbreviations (eg, Mr or Ph.D.), numerical values (eg, 0.92), date and time (eg, 2019.01.01), special format (eg, Alcohol use: None), etc. The word tokenization is mainly based on the Stanford Tokenizer34 modified with rules regarding medical abbreviations (eg, a.c., a.h., b.i.d s/p, w/o). Section detection combines regular expression and the maximum entropy classifier. The data set used for training and generating rules includes open access medical data such as MIMIC III data,35 and data from previous i2b2 challenges.36
Figure 2.
Architecture of the general cNLP system. This system is constructed with the UIMA framework and UMLS knowledge base. It also utilizes multiple machine learning/rule-based models pretrained with a much larger medical data set.
Abbreviations: cNLP, clinical natural language processing; UIMA, Unstructured Information Management Architecture; UMLS, Unified Medical Language System.
In the Entity and Relation module, the medical entities such as diseases, symptoms, lab items, medications, and treatments are identified through modified Lucene37 lookup in the form of CUIs in the UMLS database. Time mentions and lab results are identified through deep neural network-based NER (more specifically, bidirectional long short-term memory-conditional random field [LSTM-CRF] models38,39) We also use knowledge graphs derived from UMLS (eg, CUI-CUI relations,40 such as “treats” and “prevents”) and pretrained deep learning models based on bidirectional LSTM41 to assign the relationship between entities, such as treatment relations between drugs and diseases. The knowledge reasoning module then normalizes these entities and relations to facilitate accurate data analysis. In the Assertion module, we use rule/deep learning hybrid methods combing LSTM models and rules based on sentence pattern and POS which were generated during error analysis. All the machine learning based models are pretrained with a much larger medical data set as previously mentioned.
RESULTS AND DISCUSSION
The systems were evaluated as part of the 2018 n2c2 challenge. The evaluation was conducted using a script released by n2c2 organizers, which reports Precision, Recall, and F1score for the “met”/“not met” classes of each criterion. In addition, overall microaverage F1 score is also generated and considered as the main evaluation in the challenge. Table 3 shows the results (microaverage F1 score) of our systems submitted to the 2018 n2c2 challenge, where we won fourth place. The best, mean, and median results of all the 109 submissions from 45 teams are also included. As shown in Table 3, our challenge-oriented rule-based system achieved a high score of 0.9028, which had less than 1% difference compared to the best submission of the challenge. Compared with the performance on the training data set, we observed a 3.4% drop in the rule-based system. This performance drop is expected especially considering the relatively small data set size (202 records for training and 86 records for testing), which indicates that the rule-based system actually worked very well to generalize the cohort selection task.
Table 3.
Overall systems’ performance
| System on Test Data | Score |
|---|---|
| Best n2c2 submission | 0.91 |
| Our rule-based system | 0.9028 |
| Median n2c2 submission | 0.8227 |
| Our hybrid system | 0.8145 |
| Mean n2c2 submission | 0.7988 |
| System on Train Data | Score |
| Our rule-based system | 0.9388 |
| Our hybrid system | 0.8495 |
The system performances on each criterion are established in Table 4. As shown in Table 4, the criteria “ABDOMINAL,” “ENGLISH,” and “HBA1C” are in the high-performance tier, where the overall F1 scores are higher than 90%. However, “ALCOHOL-ABUSE,” “DRUG-ABUSE,” “KETO-1YR,” and “MAKES-DECISIONS” are in the low-performance tier, where the overall F1 scores are lower than 70%. This result may be due to the imbalanced population of data in “met” or “not met” classes. For example, there is only 1 “met” sample in training data for “KETO-1YR,” and there is not a single “met” sample in the testing data. Those low-instance criteria are very sensitive to prediction variance but have a relatively small influence on the final micro-F1 evaluation.
Table 4.
Systems’ performance of each criterion on test data set
| Met |
Not met |
Overall | ||||||
|---|---|---|---|---|---|---|---|---|
| System | Criteria | Prec. | Rec. | F1 | Prec. | Rec. | F1 | F1 |
| Rule-based system | ABDOMINAL | 0.9600 | 0.8000 | 0.8727 | 0.9016 | 0.9821 | 0.9402 | 0.9064 |
| ADVANCED-CAD | 0.6780 | 0.8889 | 0.7692 | 0.8148 | 0.5366 | 0.6471 | 0.7081 | |
| ALCOHOL-ABUSE | 0.0000 | 0.0000 | 0.0000 | 0.9647 | 0.9880 | 0.9762 | 0.4881 | |
| ASP-FOR-MI | 0.8500 | 1.0000 | 0.9189 | 1.0000 | 0.3333 | 0.5000 | 0.7095 | |
| CREATININE | 0.8696 | 0.8333 | 0.8511 | 0.9365 | 0.9516 | 0.9440 | 0.8975 | |
| DIETSUPP-2MOS | 0.8333 | 0.9091 | 0.8696 | 0.8947 | 0.8095 | 0.8500 | 0.8598 | |
| DRUG-ABUSE | 0.2500 | 0.6667 | 0.3636 | 0.9872 | 0.9277 | 0.9565 | 0.6601 | |
| ENGLISH | 0.9865 | 1.0000 | 0.9932 | 1.0000 | 0.9231 | 0.9600 | 0.9766 | |
| HBA1C | 1.0000 | 0.8571 | 0.9231 | 0.9107 | 1.0000 | 0.9533 | 0.9382 | |
| KETO-1YR | 0.0000 | 0.0000 | 0.0000 | 1.0000 | 1.0000 | 1.0000 | 0.5000 | |
| MAJOR-DIABETES | 0.8043 | 0.8605 | 0.8315 | 0.8500 | 0.7907 | 0.8193 | 0.8254 | |
| MAKES-DECISIONS | 0.9630 | 0.9398 | 0.9512 | 0.0000 | 0.0000 | 0.0000 | 0.4756 | |
| MI-6MOS | 0.7000 | 0.8750 | 0.7778 | 0.9868 | 0.9615 | 0.9740 | 0.8759 | |
| Overall (micro) | 0.8639 | 0.9129 | 0.8877 | 0.9368 | 0.8998 | 0.9180 | 0.9028 | |
| Overall (macro) | 0.6842 | 0.7408 | 0.7017 | 0.8652 | 0.7849 | 0.8093 | 0.7555 | |
| Hybrid system | ABDOMINAL | 0.4906 | 0.8667 | 0.6265 | 0.8788 | 0.5179 | 0.6517 | 0.6391 |
| ADVANCED-CAD | 0.7458 | 0.9778 | 0.8462 | 0.9630 | 0.6341 | 0.7647 | 0.8054 | |
| MAJOR-DIABETES | 1.0000 | 0.3023 | 0.4643 | 0.5890 | 1.0000 | 0.7414 | 0.6028 | |
Abbreviations: Prec, precision; Rec, recall.
The general cNLP system was initially expected to be a competitor of our rule-based system, which might benefit from the larger knowledge base and pretrained models, and was applied to criteria “ABDOMINAL,”, “ADVANCED-CAD,” and “MAJOR-DIABETES.” Thus, a hybrid system, which combines the general cNLP system on those 3 criteria and the rule-based system for the rest of the criteria, was also evaluated. The performances of this hybrid system are included in Tables 3 and 4. As shown in Table 3, it seems that the hybrid system overall didn’t work as well as the rule-based system, though it shows more robustness to data variances. However, it’s interesting to see that the hybrid system actually worked better on “ADVANCED-CAD” as established in Table 4. On criterion “ADVANCED-CAD,” the hybrid system outperformed the rule-based system on almost every aspect including precision, recall, and F1 score.
An error analysis was conducted and indicated that the UMLS-based general cNLP system indeed worked better on finding clinical concepts, especially for those that didn’t present in the training data set (such as new medication, and new expression of certain symptoms or disease. This is the main reason why the hybrid system worked better on “ADVANCED-CAD.” However, accurate concept lookups in UMLS requires the expressions match between the mention and the CUIs. In criterion “MAJOR-DIABETES,” the hybrid system failed to find the evidence indicating “skin conditions” or “kidney damage” because the mention in the context is too general. That explains why we observed a high precision but low recall for that criterion. This seemingly insignificant result illustrates the challenges researchers face when developing knowledge-based general cNLP systems. Moreover, these results also infer that a hybrid system combining a knowledge-based general cNLP approach and a task-specific rule-based system is promising to leverage the performance to the next level, though it may require efforts to translate the selection criteria into the feasible language of the CUI system in UMLS.
CONCLUSION
In this study, we described an integrated clinical NLP system which consisted of various NLP components (concepts extraction, assertion, temporal information extraction, lab results extraction, and meta-information extraction) enabled by a generic rule-based framework with lexical-, syntactic-, and meta-level domain knowledge inputs. We also demonstrated 1 real-world practice of this system with task-specific knowledge inputs, as submitted to the 2018 n2c2 challenge on cohort selection for clinical trials. In the 2018 n2c2 challenge, our system achieved an overall micro-F1 score of 0.9028 which was ranked fourth and had less than 1% difference from the best system, which indicates that our approach is very promising. In addition, the investigation of a UMLS-based general cNLP system on this task suggests that a hybrid system, which combines general cNLP and task-specific rule-based systems, hold the potential to leverage the performance to the next level, though extra efforts are required to fill the gap between task-specific language and the feasible language of the CUI system in UMLS.
Although we demonstrated our systems on 1 specific task, our system architectures and frameworks were designed to be generic and friendly for other applications, as long as the domain knowledge inputs are provided. This approach can be applied to other applications especially for those interested in health informatics fields, such as Computer-Assisted Coding (CAC)28 and automated Healthcare Effectiveness Data and Information Set (HEDIS) measurement.42
FUNDING
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
AUTHOR CONTRIBUTIONS
LC devised the main idea for the work, designed the study, carried out the data collection, developed the systems, analyzed the results, and wrote the article, with YH, and YG contributing significant edits. YG assisted with study design, module development, and analysis. XJ, CL, ZS, and HL contributed to the NLP system design and implementation. YH and YG supervised this study. All the authors discussed the results and contributed to the final manuscript. LC and YG contributed equally.
ACKNOWLEDGMENTS
The authors would like to thank the 2018 n2c2 challenge organizers for organizing the challenge and providing the data used in training and testing. The authors also thank the participants of the 2018 n2c2 workshop for helpful discussions.
Conflict of interest statement
None declared.
REFERENCES
- 1. Casey JA, Schwartz BS, Stewart WF, et al. Using electronic health records for population health research: a review of methods and applications. Annu Rev Public Health 2016; 371: 61–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geneletti S, Richardson S, Best N.. Adjusting for selection bias in retrospective, case-control studies. Biostatistics 2008; 101: 17–31. [DOI] [PubMed] [Google Scholar]
- 3. Abrahão MTF, Nobre MRC, Gutierrez MA.. A method for cohort selection of cardiovascular disease records from an electronic health record system. Int J Med Inform 2017; 102: 138–49. [DOI] [PubMed] [Google Scholar]
- 4. Friedman C, Alderson PO, Austin JHM, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994; 12: 161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kreimeyer K, Foster M, Pandey A, et al. Natural language processing systems for capturing and standardizing unstructured clinical information: a systematic review. J Biomed Inform 2017; 73: 14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y, Wang L, Rastegar-Mojarad M, et al. Clinical information extraction applications: a literature review. J Biomed Inform 2018; 77: 34–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Glicksberg BS, Miotto R, Johnson KW, et al. Automated disease cohort selection using word embeddings from electronic health records. Pac Symp Biocomput 2018; 23: 145–56. http://www.ncbi.nlm.nih.gov/pubmed/29218877 Accessed January 15, 2019. [PMC free article] [PubMed] [Google Scholar]
- 8. N2C2: National NLP Clinical Challenges. https://n2c2.dbmi.hms.harvard.edu/ Accessed April 9, 2019.
- 9. Unified Medical Language System (UMLS). https://www.nlm.nih.gov/research/umls/ Accessed January 15, 2019.
- 10. Apache UIMA. https://uima.apache.org/ Accessed January 15, 2019.
- 11. Aronson AR, Lang F-M.. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc 2010; 173: 229–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010; 175: 507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goryachev S, Sordo M, Zeng QT.. A suite of natural language processing tools developed for the I2B2 project. Annual Symposium Proceedings AMIA Symposium; 2006: 931 http://www.ncbi.nlm.nih.gov/pubmed/17238550 Accessed January 15, 2019). [PMC free article] [PubMed] [Google Scholar]
- 14. Liu H, Bielinski SJ, Sohn S, et al. An information extraction framework for cohort identification using electronic health records. AMIA Joint Summits on Translational Science; 2013: 149–53. http://www.ncbi.nlm.nih.gov/pubmed/24303255 Accessed January 15, 2019. [PMC free article] [PubMed] [Google Scholar]
- 15. Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001; 345: 301–10. [DOI] [PubMed] [Google Scholar]
- 16. Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009; 425: 839–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mehrabi S, Krishnan A, Sohn S, et al. DEEPEN: a negation detection system for clinical text incorporating dependency relation into NegEx. J Biomed Inform 2015; 54: 213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Uzuner Ö, South BR, Shen S, et al. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc 2011; 185: 552–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Farkas R, Vincze V, Móra G, et al. The CoNLL-2010 shared task: learning to detect hedges and their scope in natural language text. In: Proceedings of the fourteenth conference on Computational Natural Language Learning - Shared Task. Uppsala, Sweden: Association for Comput Linguistics 2010. 1–2.
- 20. Uzuner O, Solti I, Cadag E.. Extracting medication information from clinical text. J Am Med Inform Assoc 2010; 175: 514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun W, Rumshisky A, Uzuner O.. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc 2013; 205: 806–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stubbs A, Kotfila C, Xu H, et al. Identifying risk factors for heart disease over time: overview of 2014 i2b2/UTHealth shared task Track 2. J Biomed Inform 2015; 58: S67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uzuner O, Goldstein I, Luo Y, et al. Identifying patient smoking status from medical discharge records. J Am Med Inform Assoc 2008; 151: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chiticariu L, Li Y, Reiss FR. Rule-based information extraction is dead! Long live rule-based information extraction systems! Proceedings of the 2013 Conference on Empirical Methods in Natural Language Processing;2013: 827–32. https://aclanthology.info/papers/D13-1079/d13-1079 Accessed January 15, 2019).
- 25. Karystianis G, Dehghan A, Kovacevic A, et al. Using local lexicalized rules to identify heart disease risk factors in clinical notes. J Biomed Inform 2015; 58: S183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lobach DF, Johns EB, Halpenny B, et al. Increasing complexity in rule-based clinical decision support: the symptom assessment and management intervention. JMIR Med Inform 2016; 44: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jiang Y, Qiu B, Xu C, et al. The research of clinical decision support system based on three-layer knowledge base model. J Healthcare Eng 2017; 2017: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crawford M. Truth about computer-assisted coding: a consultant, HIM professional, and vendor weigh in on the real CAC impact. J AHIMA 2013; 84: 24–7. http://library.ahima.org/doc? oid=106663#.XD8mDxNKhTY Accessed 16 January, 2019). [PubMed] [Google Scholar]
- 29. RxList. Supplement Information-Vitamins, Herbs, and Dietary Supplements on RxList. https://www.rxlist.com/supplements/article.htm Accessed January 16, 2019.
- 30. Woodward English. Countries and Nationalities: English Vocabulary. https://www.vocabulary.cl/Basic/Nationalities.htm Accessed January 16, 2019.
- 31. spaCy. Industrial Strength Natural Language Processing: In Python. https://spacy.io/ Accessed January 16, 2019.
- 32. Reynar JC, Ratnaparkhi A.. A maximum entropy approach to identifying sentence boundaries In: Proceedings of the Fifth Conference on Applied Natural Language Processing, Morristown, NJ: Association for Computational Linguistics; 1997: 16–9. [Google Scholar]
- 33. Agarwal N, Ford KH, Shneider M. Sentence boundary detection using a maxent classifier. In: Proceedings of MISC;2005: 1–6. https://nlp.stanford.edu/courses/cs224n/2005/agarwal_herndon_shneider_final.pdf Accessed April 9, 2019.
- 34. Standford Natural Language Processing Group. Stanford Tokenizer. https://nlp.stanford.edu/software/tokenizer.shtml Accessed April 9, 2019.
- 35. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Scientific Data2016; 3:160035. doi: 10.1038/sdata.2016.35 [DOI] [PMC free article] [PubMed]
- 36.i2b2 NLP Research Data Sets. https://www.i2b2.org/NLP/DataSets/Main.php Accessed April 9, 2019.
- 37. Lucene. Welcome to Apache Lucene. http://lucene.apache.org/ Accessed January 16, 2019.
- 38. Lample G, Ballesteros M, Subramanian S, et al. Neural Architectures for Named Entity Recognition In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. Stroudsburg, PA: Association for Computational Linguistics; 2016. 260–70. doi: 10.18653/v1/N16-1030 [Google Scholar]
- 39. Huang Z, Xu W, Yu K. Bidirectional LSTM-CRF Models for Sequence Tagging. 2015. http://arxiv.org/abs/1508.01991 Accessed April 9, 2019.
- 40. US Department of Health and Human Services. UMLS: Current Relations in the Semantic Network. https://www.nlm.nih.gov/research/umls/META3_current_relations.html Accessed April 9, 2019.
- 41. Miwa M, Bansal M. End-to-end relation extraction using LSTMs on sequences and tree structures. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers) Stroudsburg, PA: Association for Computational Linguistics; 2016: 1105–16.
- 42. NCQA. HEDIS and Performance Measurment. https://www.ncqa.org/hedis/ Accessed January 16, 2019.