Abstract
Objective
In multi-label text classification, each textual document is assigned 1 or more labels. As an important task that has broad applications in biomedicine, a number of different computational methods have been proposed. Many of these methods, however, have only modest accuracy or efficiency and limited success in practical use. We propose ML-Net, a novel end-to-end deep learning framework, for multi-label classification of biomedical texts.
Materials and Methods
ML-Net combines a label prediction network with an automated label count prediction mechanism to provide an optimal set of labels. This is accomplished by leveraging both the predicted confidence score of each label and the deep contextual information (modeled by ELMo) in the target document. We evaluate ML-Net on 3 independent corpora in 2 text genres: biomedical literature and clinical notes. For evaluation, we use example-based measures, such as precision, recall, and the F measure. We also compare ML-Net with several competitive machine learning and deep learning baseline models.
Results
Our benchmarking results show that ML-Net compares favorably to state-of-the-art methods in multi-label classification of biomedical text. ML-Net is also shown to be robust when evaluated on different text genres in biomedicine.
Conclusion
ML-Net is able to accuractely represent biomedical document context and dynamically estimate the label count in a more systematic and accurate manner. Unlike traditional machine learning methods, ML-Net does not require human effort for feature engineering and is a highly efficient and scalable approach to tasks with a large set of labels, so there is no need to build individual classifiers for each separate label.
Keywords: multi-label text classification, biomedical text, deep neural network, biomedical literacutre, clinical notes
INTRODUCTION
Text classification is a common task in natural language processing (NLP) and a building block for many complex NLP tasks. Text classification is the task of classifying an entire text by assigning it 1 or more predefined labels1 and has broad applications in the biomedical domain, including biomedical literature indexing,2,3 automatic diagnosis code assignment,4,5 tweet classification for public health topics,6–8 and patient safety reports classification,9 among others.
Text classification can be further grouped into 2 types: multinomial or multi-class and multi-label. For multinomial or multi-class text classification, each textual document is associated with only 1 label (ie, labels are mutually exclusive). For instance, when only 2 classes are available, binary classification is 1 of the most common multinomial classification tasks. For multi-label text classification, a textual document can be assigned 1 or more labels. For example, in Medical Subject Headings (MeSH) indexing, typically a dozen relevant MeSH terms are assigned to new publications in PubMed.10 Because each textual document can be assigned an indeterminate number of labels, multi-label text classification is often considered more challenging than multinomial classification.11
A traditional approach to solving the multi-label text classification problem is binary relevance, which decomposes the problem into multiple independent binary classification tasks (1 for each label). This method, however, assumes the independence of each label.10,12,13 Label powerset, which creates binary classifiers for each label combination, is able to model potential correlations between labels.14 Both of these approaches, however, could have low throughput when the number of different labels becomes extremely large. There are also other algorithms for multi-label text classification, including learning to rank10 and classifier chains,15 among others. A review of multi-label learning algorithms can be found in Min-Ling & Zhi-Hua.16
In recent years, deep neural networks have been proposed for multi-label text classification tasks. Most of these efforts13,17–21 used a similar framework, which often consists of 2 modules: a neural network and a label predictor. The neural network produces scores for each label, using the multi-layer perceptron (MLP) neural networks,13,17 the convolution neural networks (CNNs),11,18,19 the recurrent neural networks (RNNs),22 or other hybrid neural networks.20 A label predictor splits the label ranking list into the relevant and irrelevant labels by thresholding methods. Under this framework, however, a search for the optimal threshold is often required, and the label decision ignores document context.
Li et al recently incorporated a label-decision module into deep neural networks and achieved state-of-the-art performance in multi-label image classification tasks.12 Motivated by their framework, we propose ML-Net, a novel end-to-end deep learning framework, for multi-label text classification tasks. ML-Net adopts the general label–decision module in Li et al,12 but it changes the image processing framework to text classification (ie, uses an attention-based bidirectional RNN architecture together with deep contextualized word representations). ML-Net combines label prediction and label decision in the same network and is able to determine the output labels based on both label confidence scores and document context. ML-Net aims to minimize pairwise ranking errors of labels and is able to train and predict the label set in an end-to-end manner, without the need for an extra step to determine the output labels. To demonstrate the effectiveness and generalizability of ML-Net, we evaluated the framework on 3 multi-label biomedical text classification tasks in both the biomedical literature domain (2 tasks) and the clinical notes domain (1 task). We compared the proposed framework with both traditional machine learning baseline models and other deep learning models.
MATERIALS AND METHODS
Deep neural network
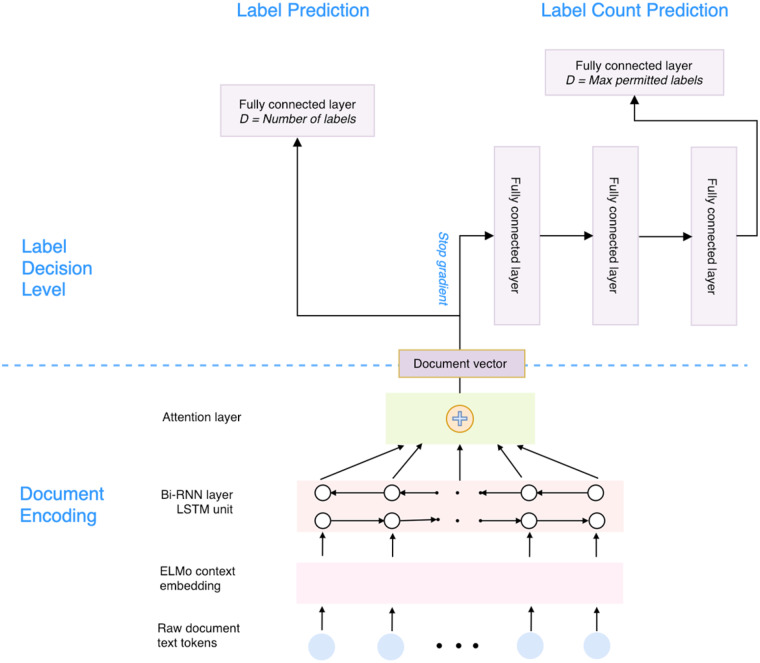
The overall architecture of ML-Net can be seen in Figure 1. The architecture consists of 3 major modules: (1) a document encoding network that takes raw text as the input and outputs the high-dimensional vectors that represent the entire textual document in 2 consecutive steps: (a) the Embeddings from Language Models (ELMo) network23 takes raw text as the input and generates contextualized embeddings for each word, and (b) an attentive RNN network takes the contextualized word embeddings as the input and generates the corresponding document representation; (2) a label prediction network—a fully connected layer with an output layer for which the number of nodes corresponds to the number of unique labels—that takes document vectors as input and outputs a prediction confidence score for each label; (3) a label count prediction network that consists of several fully connected layers with an output layer for which the number of nodes equals the number of maximal permitted labels and that takes the same document vectors as input and outputs the estimation of label counts for each document. The source code of ML-Net is freely available at: https://github.com/ncbi-nlp/ML_Net.
Figure 1.
The framework of ML-Net.
Document encoding network
We propose a document encoding network to encode the textual document to high-dimensional vectors. Traditional word embedding methods assign a static high-dimensional vector to a word, regardless of its context. A word, however, could have multiple context-dependent meanings. Deep contextualized word-embeddings, such as ELMo, can look at the entire context before assigning each word its embedding vector.23 Our document-encoding network adopts the pre-trained ELMo to map each token in the document to high-dimensional vectors and then feeds the vectors to a bi-directional RNN, which is able to capture both forward and backward sequential context. We further add the attention mechanism to augment sequence models by capturing the salient portions and context.24,25
Label prediction network
The label prediction network has a fully connected layer that takes a document vector as the input and outputs a predicted confidence score for each label. We apply the rectified linear unit 26 as the activation function for the output. The intuitive objective for multi-label learning is to minimize the number of misorderings between the pairs of relevant and irrelevant labels.13 Different loss functions have been proposed to model the dependency of individual labels by minimizing the pairwise-ranking errors. We choose log-sum-exp pairwise (LSEP) as our loss function, which has achieved state-of-the-art performance on large-scale multi-label image classification tasks.12 The equation of LSEP can be seen here:
where is the label prediction function that maps the document vector into a K-dimensional label space, which represents the confidence scores of each label (K equals the number of unique labels); and are the and -th element of confidence scores for the -th instance in the data set, respectively; and is the corresponding label set for the -th instance in the data set.
Label count prediction network
Deciding the proper label set from the predicted label set is a key challenge in multi-label classification. In common practice, a threshold function is trained to split the ranking of the labels into relevant vs irrelevant labels.13,17 Such a thresholding method, however, ignores the document context in decision-making. Inspired by a framework from multi-label image classification,12 our label count prediction network takes the document vector as the input and casts the label count estimation as an -way classification task, where is a hyper-parameter for the maximum number of permitted labels that can be returned by the neural network. For a document that has a number of labels fewer than or equal to , the model keeps the exact number of labels as the label count for that document; for a document that has a number of labels greater than , the model uses as the label count for that document. We designed a MLP network for the label count prediction. This network consists of several fully connected layers and an output layer with Softmax function for classification.
There are 2 training steps. We first train the label prediction network. During training, the label prediction and the document encoding networks are updated through back propagation. Then, we train the label count prediction network. However, different from the training label prediction network, only the MLP part is updated as the gradient descent stops at the layer of the document vector. For prediction, we first rank all of the individual labels by their corresponding confidence scores generated from the label prediction network, and then the top ( ≤ , decided by the label count prediction network) labels are used as the final output.
Evaluation design
Evaluation tasks
We evaluate ML-Net on 3 different text classification tasks with publicly available data sets in 2 text genres: biomedical literature and clinical notes.
Task 1. Hallmarks of cancer classification. The hallmarks of cancer consist of a small number of underlying principles that describe its complexity.27 Baker et al introduced a corpus of 1580 PubMed abstracts manually annotated according to the scientific evidence of 10 currently known hallmarks of cancer.28 The data set is available at: https://www.cl.cam.ac.uk/∼sb895/HoC.html.
Task 2. Chemical exposure assessments. The vast amount of chemical-specific exposure information available in PubMed is of critical significance; however, the manual collection of such information from the biomedical literature can be labor intensive. Larsson et al proposed an exposure taxonomy that includes 32 classes and introduced a corpus of 3661 abstracts with annotated chemical exposure information.29 The data set is available at: https://figshare.com/articles/Corpus_and_Software/4668229.
We note that the annotation of Tasks 1 and 2 was originally performed at the sentence level. As very few sentences are annotated with multi-labels, we aggregated all of the unique labels for every sentence in an abstract as the labels for that abstract and performed the multi-label classification on the abstract level. This is also consistent with Hanahan & Weinberg27 and Larsson et al,29 for which the authors built binary classifiers for each label and performed the classification on the abstract level.
Task 3. Diagnosis codes assignment. The automatic assignment of diagnosis codes to medical notes is a useful task that could benefit the computational modeling of patient status. Due to the extremely large label collection, the diagnosis codes assignment task can be considered an extreme multi-label text classification problem.30 Perotte et al proposed a hierarchy-based classification to automatically assign Internation Classification of Diseases (ICD)-9 codes to the discharge summaries from the publicly available Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC II) data set,4 using a hierarchical support vector machine (SVM). In this work, we followed the same steps to augment the label set, using the hierarchy of the ICD-9 codes in Perotte et al.4 That is, if an ICD code is in the label set of a document, all of its ancestors are also included in the label set for that document. Their data set and label augmentation script are publicly available at: https://physionet.org/works/ICD9CodingofDischargeSummaries.
The overall statistics of the 3 data sets can be seen in Table 1. Tasks 1 and 2 have similar characteristics in terms of number of tokens and sentences in a document, which is not surprising, as they are both collected from PubMed abstracts. In comparison, Task 2 has a relatively larger number of unique labels and corpus size. The Task 3 corpus has very distinct characteristics from the PubMed abstracts, with a significantly larger number of unique labels (over 7000), and each document is assigned many more labels (37 on average, after label augmentation).
Table 1.
Descriptors and basic statistics for 3 text classification tasks
| Task No. | Number of unique labels | Corpus size | Number of tokens in document |
Number of sentences in document |
Number of labels in document |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Max | Min | SD | Mean | Max | Min | SD | Mean | Max | Min | SD | |||
| 1 | 10 | 1580 | 209.29 | 638 | 44 | 58.32 | 9.44 | 27 | 2 | 2.87 | 1.56 | 5 | 1 | 0.78 |
| 2 | 32 | 3661 | 233.66 | 622 | 49 | 60.41 | 9.88 | 34 | 1 | 2.81 | 2.05 | 8 | 0 | 1.30 |
| 3 | 7042 | 22 815 | 1039.73 | 5882 | 8 | 623.21 | 165.71 | 904 | 4 | 95.86 | 36.68 | 127 | 5 | 16.16 |
Note. Task 1: Hallmarks of cancer classification; Task 2: Chemical exposure assessments; Task 3: Diagnosis code assignment (data after label augmentation).
Abbreviation: SD, standard deviation.
Evaluation metric
The example-based metrics evaluate the multi-label learning system’s performance on each test example (ie, each document) separately by comparing the predicted labels with the gold standard labels for each test example. We focus on 3 major example-based metrics, as defined in Min-Ling & Zhi-Hua16:
where is the number of instances in the test set; refers to the true label set for the -th instance in the test set; and refers to the predicted label set for the -th instance in the test set.
For Tasks 1 and 2, we define the true positives (TP) as the labels that are identical to the gold standard labels, false positives (FP) as labels that are not true positives, and false negatives (FN) as the gold standard labels that were missed in the prediction results. For Task 3, considering the hierarchical structure of ICD-9 codes, we follow the same definition of TP in Perotte et al,4 in which TP are predicted codes that are ancestors of, descendants of, or identical to an assigned code, and FP are predicted labels that are not true positives. FN are the gold standard labels for which the labels or their descendants are not predicted.
Text preprocessing
ML-Net requires little effort for text preprocessing. For Tasks 1 and 2, we removed punctuation marks and tokens that have only 1 character and then concatenated all tokens in the abstract together. For Task 3, we first removed some common pattern strings, such as “admission date,” and “signed electronically by,” as described in the previous effort,4 and then followed the same preprocessing steps in Tasks 1 and 2.
System implementation
For Tasks 1 and 2, we used the same implementation as follows. We split the annotated corpus into training, validation, and test sets with a ratio of 7: 1: 2, respectively. We loaded the pretrained ELMo model from TensorFlow Hub (https://tfhub.dev/google/elmo/2), and ELMo was set as trainable. We chose long short-term memory (LSTM) as the RNN unit. We set the number of hidden units in the RNN layer and the dimension of attention output both at 50. Dropout (rate at 0.5) was added on bi-RNN layer to avoid overfitting. The maximum number of permitted labels was set at 5 and 8 for Tasks 1 and 2, respectively. The number of neurons in the MLP in the label count prediction network were set at 128, 128, and 64, respectively. We first trained the label prediction network with the hierarchical attention network for 50 epochs. We then applied early stopping while training in the label count prediction network (20 epochs at most). We adopted the Adam optimizer31 and set the learning rate at 0.001. The hyper-parameter tuning was performed on the validation set.
For Task 3, due to computation limitations, we took the first 1500 tokens from each clinical note as input. We followed almost the same hyper-parameters in Tasks 1 and 2. Considering the large collection of labels (7024 unique ICD codes), we set the number of neurons in the MLP of the label count prediction network at 7024 (total number of unique labels), 7024, and 128, respectively. The maximum number of permitted labels was set at 70. To make our model comparable with previous efforts, we followed the same data preprocessing steps and used the same data sets for training and testing.4 The major parameters setting for 3 tasks can be seen in Table 2.
Table 2.
Major parameter settings in ML-Net for the 3 tasks
| Parameter | Setting |
||
|---|---|---|---|
| Task 1 | Task 2 | Task 3 | |
| Maximum permitted labels | 5 | 8 | 70 |
| Maximum tokens in documents | All tokens | First 1500 tokens | |
| Neurons in the MLP | 128, 128, 64 | 7024, 7024, 128 | |
| Batch size | 32 | 16 | |
| Training epochs (label prediction network) | 50 | 30 | |
| RNN unit (dimension) | LSTM (50) | ||
| Attention layer dimension | 50 | ||
| Dropout rate | 0.5 | ||
| Optimizer (learning rate) | Adam (0.001) | ||
Abbreviations: LSTM, long short-term memory; MLP, multi-layer perceptron; RNN, recurrent neural networks;
Machine-learning baseline
For traditional machine learning algorithms, we framed the multi-label classification task as a binary relevance task. We used term frequency-inverse document frequency as features and trained a separate binary classifier for each label. We compared multiple machine learning algorithms, including SVM, logistic regression, random forest, and extra trees. We report only the results of SVM with linear kernel here, as it obtained better performance than did other algorithms. For Task 3, we report the best SVM-based results (ie, hierarchy-based SVM) in Perotte et al.4
Deep-learning baseline
To assess the effect of the document encoding network, we replaced it in ML-Net with 2 neural networks: (1) the classic CNNs proposed by Kim32 (which we term ML-CNN), and (2) the hierarchical attention network (HAN) described in Du et al33 (which we term ML-HAN), while keeping the label prediction network intact.
To assess the effect of the label count prediction network, we replaced it with an alternative thresholding mechanism for determining the final predicted labels. Specifically, for ML-Net, ML-CNN and ML-HAN, we trained the label prediction network first. Then, we searched the optimal global threshold (ie, 1 threshold score for all of the labels in all examples in 1 task) for the confidence scores generated from the label prediction network. The labels, whose confidence scores were higher than the global threshold, were included in the predicted label set. We name these 3 networks ML-Net-threshold, ML-CNN-threshold and ML-HAN-threshold, respectively. We searched the optimal threshold on the validation set for Tasks 1 and 2. For Task 3, due to the lack of a validation set, we searched the optimal threshold on the training set.
For these deep learning-based baselines, we first leveraged the Natural Language Toolkit (NLTK 3.3) to perform the tokenization and then removed the stop words. Next, we used the pretrained word embedding to map tokens in the text to high-dimensional vectors, which are then fed to the following networks: for Tasks 1 and 2, we used the pretrained PubMed word2vec34 (dimension: 200); and for Task 3, we used the word embedding trained from MIMIC III corpus, using the word2vec algorithm35 (dimension: 300).
RESULTS
The performance of the proposed ML-Net and other baseline models is summarized in Table 3. As we can see, ML-Net has the best F score in both Tasks 1 and 2. For the hallmarks of cancer task, all of the deep learning-based approaches outperformed the binary relevance baseline methods. The ML-Net outperformed the baseline model by more than 16%. For chemical exposure assessments, all of the models with proposed label count prediction network (ML-Net, ML-CNN, and ML-HAN) outperformed the binary relevance baseline methods in the F score; for thresholding methods, only ML-Net-threshold outperformed the binary–relevance baseline. For these 2 tasks, consistent with other findings, the label count prediction network can make better decisions as compared to the thresholding methods. In addition, the models with the proposed document encoding network (ML-Net) achieved higher F score than did the convolutional neural network (ML-CNN) and hierarchical attention network (ML-HAN) in both cases.
Table 3.
Comparison of various algorithms for multi-label classification on 3 tasks
| Task 1. Hallmarks of cancer classification |
Task 2. Chemical exposure assessments |
Task 3. Diagnosis code assignment |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Algorithm | Precision | Recall | F score | Precision | Recall | F score | Precision | Recall | F score |
| Binary-relevance (SVM with TFIDF) | 0.742 | 0.688 | 0.714 | 0.778 | 0.677 | 0.724 | 0.577 | 0.300 | 0.395 |
| ML-Net | 0.848 | 0.811 | 0.829 | 0.784 | 0.724 | 0.753 | 0.404 | 0.374 | 0.389 |
| ML-Net-threshold | 0.765 | 0.850 | 0.805 | 0.714 | 0.779 | 0.745 | 0.506 | 0.347 | 0.412 |
| ML-HAN | 0.813 | 0.817 | 0.815 | 0.753 | 0.724 | 0.738 | 0.355 | 0.338 | 0.346 |
| ML-HAN-threshold | 0.752 | 0.837 | 0.793 | 0.700 | 0.735 | 0.717 | 0.492 | 0.360 | 0.416 |
| ML-CNN | 0.843 | 0.778 | 0.809 | 0.778 | 0.689 | 0.731 | 0.311 | 0.442 | 0.365 |
| ML-CNN-threshold | 0.764 | 0.817 | 0.790 | 0.662 | 0.774 | 0.713 | 0.501 | 0.373 | 0.428 |
Note. For the diagnosis code assignment, the binary-relevance scores are the best results reported in Perotte et al.4Abbreviations: CNN, convolutional neural networks; HAN, hierarchical attention network; SVM, support vector machine; TFIDF, term frequency-inverse document frequency;
For Tasks 1 and 2, we analyzed the errors on the label count prediction made by the ML-Net and ML-Net-thresholds. Compared to ML-Net, the best ML-Net-threshold model generated more labels per example in the test set (Task 1: 1.73 vs 1.54; Task 2: 2.00 vs 1.91) and made higher errors per example in the test set (Task 1: 0.87 vs 0.44; Task 2: 0.86 vs 0.72, the absolute difference of predicted counts with gold standard counts).
For the task of diagnosis codes assignment, the ML-CNN-threshold achieved the highest F score (0.428) among all models. Note that the thresholding method generally achieved better performance than did the label count prediction network for this task. We suspect that this is due to the inclusion of additional codes based on the hierarchical relations in the ICD-9 codes. By doing so, the count of label set might largely depend on the hierarchical structure of ICD-9 codes, instead of the context of the document. As our proposed label count prediction network takes only the document vectors as the input, the label-count estimation is not less accurate in this case. All of the deep learning models with thresholding methods outperformed the binary–relevance baseline, which again demonstrated the superiority of a deep neural network for the diagnosis code assignment.
DISCUSSION
We reviewed FP and FN in the test sets for Tasks 1 and 2 and found that the most common errors of ML-Net are in the most frequent labels. For example, in Task 1, the label “sustaining proliferative signaling” is the most frequent label in the data set28 and the most frequent FP and FN among all of the labels. For Task 2, the label “effect marker-physiological parameter” is the most frequent label in the data set29 and the second most frequent FP and most frequent FN among all of the labels. As the data distribution is imbalanced in both tasks, it is understandable that ML-Net tends to include the common labels in prediction as FP. For example, in 1 abstract (PMID: 23257893) in Task 1, the gold standard labels are “genomic instability and mutation” and “resisting cell death.” Although ML-Net accurately predicted the count of labels, it wrongly predicted the labels to be “sustaining proliferative signaling” and “resisting cell death.” In addition, the inaccurate prediction of label count can lead to FP and FN. For example, in 1 abstract (PMID: 20184723), the gold standard labels are “tumor promoting inflammation” and “genomic instability and mutation,” whereas ML-Net output includes only “tumor promoting inflammation.”
Compared to the first 2 tasks, the performance of Task 3 is much lower (by all methods), as the task is inherently more challenging. Perotte et al found a slight relationship between the diagnosis code prevalence in the training data and performance.4 The prevalence of diagnosis codes indeed varies in this corpus. Following the label preprocessing and code augmentation steps in Perotte et al, we find that the top 100 most frequent codes account for more than half of the total occurrences (115 268 out of 215 805). We also find noticeable differences in diagnosis code co-occurrences in the training vs test set. For instance, the count of the co-occurring codes of 403.90 and 585.9 ranks 144th in the training set, while 12th in the test set. Taken together, the unbalanced code distribution and the differences of co-occurring codes in the training vs test sets may have a negative impact on the system performance. When we further examined the differences in prediction and gold standard codes in the test set, we found that the system can more easily predict diagnosis codes that are close to each other. For example, the count of co-occurrence of codes 412 and 414.01 is 155 in the prediction and 88 in the gold standard; and co-occurrence of codes 413.9 and 414.01 is 139 in the prediction and 49 in the gold standard. It is understandable that some closely related codes can be both highly related to the document and, thus, included together in the prediction results by the system, whereas, in practice, the nurses or physicians might choose only 1 from these code pairs.
In addition, we evaluated the distance between predicted labels with gold standard labels in Task 3 by calculating the shared path, the depth in the ICD-9 tree of the deepest common ancestor between a gold standard code and a predicted code.4 ML-Net was found to be able to predict further along the correct path to the gold standard codes than the hierarchy-based SVM reported in Perotte et al4: the most common shared paths for ML-Net are levels 3 and 6, while the most common shared paths for hierarchy-based SVM are levels 2 and 3.
Compared with binary relevance methods with traditional machine learning algorithms, the proposed deep learning model alleviates human effort for feature engineering and avoids building individual classifiers for each label, especially when the label collection is large (eg, over 1000 labels). ML-Net advances the state of the art by combining the label prediction network with a label count prediction network, which can not only avoid the manual searching of optimal thresholds for label prediction confidence scores, but also dynamically estimate the label count based on the document context in a more accurate manner.
This study had certain limitations. Due to the limitations of computation resources, we did not perform a thorough hyper-parameter tuning (ie, the current parameters setting may not be optimal). In addition, our proposed label count prediction network takes only the document vector as the input. However, the counts of labels also might depend on other information, for example, the hierarchical structure of the labels. Our current network is not able to model such information. As seen, the label count prediction does not work well for the labels with a hierarchical structure, such as those in Task 3. We also evaluated Task 2 when expanding the labels by their hierarchical structures,29 and a similar result was found: The ML-Net does not demonstrate superiority over the binary-relevance method on the labels with a hierarchical structure. In future research, we will further investigate new architectures that can better model the hierarchical relation among labels.
The document encoding network that maps the text to a high-dimensional representation can be further improved, and a different architecture could be exploited and evaluated. For example, Transformer36-based language representation models, including Generative Pre-Training37 and Bidirectional Encoder Representations,38 have significantly advanced major NLP tasks recently. An intuitive change is to replace the document encoding network of ML-Net with these advanced language representation models. As these models require quite large computation resources, we leave this to future work. In addition, we plan to apply our proposed network on other larger scale multi-label biomedical text classification tasks, including automatic MeSH indexing, which aims to assign a small set of relevant terms (∼12 on average) to a given document from more than 27 000 unique concepts.10
CONCLUSION
ML-Net is a novel end-to-end deep learning framework for multi-label classification of biomedical texts. Unlike traditional machine learning methods, ML-Net does not require human effort for feature engineering nor the need to build individual classifiers for each separate label. ML-Net is a highly efficient and scalable approach to tasks with a large set of labels and tasks with different biomedical genres.
AUTHOR CONTRIBUTIONS
Study concept and design: JD, ZL. Drafting of the manuscript: JD, ZL. Acquisition, analysis, or interpretation of data: All authors. Critical revision of the manuscript for important intellectual content: All authors. Study supervision: ZL.
ACKNOWLEDGMENTS
This research was supported by the National Center for Biotechnology Information (NCBI) Scientific Visitors Program; Intramural Research Program (IRP) of the National Institutes of Health (NIH), National Library of Medicine (NLM); NLM of the NIH award nos. 2R01LM010681–05 and R01 LM011829, and the Cancer Prevention Research Institute of Texas (CPRIT) Training Grant #RP160015.
Dicslaimer: The content is the sole responsibility of the authors and does not necessarily represent the official views of NCBI, NLM, NIH, or CPRIT.
Conflict of interest statement
None declared.
REFERENCES
- 1. Jurafsky D, Martin JH.. Speech and Language Processing. London: Pearson; 2014. [CrossRe.f] [Google Scholar]
- 2. Huang M, Névéol A, Lu Z.. Recommending MeSH terms for annotating biomedical articles. J Am Med Inform Assoc 2011; 185: 660–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peng S, You R, Wang H, et al. DeepMeSH: Deep semantic representation for improving large-scale MeSH indexing. Bioinformatics 2016; 3212: i70–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perotte A, Pivovarov R, Natarajan K, et al. Diagnosis code assignment: models and evaluation metrics. J Am Med Inform Assoc 2014; 212: 231–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baumel T, Nassour-Kassis J, Cohen R, Elhadad M, et al. Multi-label classification of patient notes: case study on ICD code assignment. InWorkshops at the Thirty-Second AAAI Conference on Artificial Intelligence 2018 Jun 20.
- 6. Du J, Tang L, Xiang Y, et al. Public perception analysis of tweets during the 2015 measles outbreak: comparative study using convolutional neural network models. J Med Internet Res 2018; 207:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Du J, Zhang Y, Luo J, et al. Extracting psychiatric stressors for suicide from social media using deep learning. BMC Med Inform Decis Mak 2018; 18 (Suppl 2): 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bian J, Zhao Y, Salloum RG, et al. Using social media data to understand the impact of promotional information on laypeople’s discussions: a case study of Lynch syndrome. J Med Internet Res 2017; 1912: e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liang C, Gong Y.. Automated classification of multi-labeled patient safety reports: a shift from quantity to quality measure. Stud Health Technol Inform 2017; 245: 1070–4. [PubMed] [Google Scholar]
- 10. Mao Y, Lu Z.. MeSH now: automatic MeSH indexing at PubMed scale via learning to rank. J Biomed Semantics 2017; 8: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gargiulo F, Silvestri S, Ciampi M. Deep convolution neural network for extreme multi-label text classification. In: Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies . 2018: 641–50.
- 12. Li Y, Song Y, Luo J. Improving pairwise ranking for multi-label image classification. In: Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition; July 2017; Hawaii. doi: 10.1109/CVPR.2017.199.
- 13. Nam J, Kim J, Mencia EL, et al. Large-scale multi-label text classification - revisiting neural networks. In: Proceedings of Joint European Conference on Machine Learning and Knowledge Discovery in Databases 2014 Sep 14 (pp. 437-452). Springer, Berlin, Heidelberg.
- 14. Boutell MR, Luo J, Shen X, et al. Learning multi-label scene classification. Pattern Recognit 2004; 379: 1757–71. [Google Scholar]
- 15. Read J, Pfahringer B, Holmes G, et al. Classifier chains for multi-label classification. Mach Learn 2011; 853: 333. [Google Scholar]
- 16. Min-Ling Z, Zhi-Hua Z.. A review on multi-label learning algorithms. Knowl Data Eng IEEE Trans 2014; 26: 1819–37. [Google Scholar]
- 17. Zhang M, Zhou Z-H, Member S.. Multilabel neural networks with applications to functional genomics and text categorization. IEEE Trans Knowl Data Eng 2006; 18: 1338–51. [Google Scholar]
- 18. Li M, Fei Z, Zeng M, et al. Automated ICD-9 coding via a deep learning approach. IEEE/ACM Trans Comput Biol Bioinforma pp. 1–1, 2018. [DOI] [PubMed] [Google Scholar]
- 19. Baker S, Korhonen A.. Initializing neural networks for hierarchical multi-label text classification. BioNLP 2017: 307–15. [Google Scholar]
- 20. Lenc L, Král P. Ensemble of neural networks for multi-label document classification. In: proceeding of 17th Information Technologies - Applications and Theory: Conference on Theory and Practice of Information Technologies. Martinske Hole, Slovakia.
- 21. Moro PL, Broder K, Zheteyeva Y, et al. Adverse events in pregnant women following administration of trivalent inactivated influenza vaccine and live attenuated influenza vaccine in the Vaccine Adverse Event Reporting System, 1990-2009. Am J Obstet Gynecol 2011; 204: 146–e1–7.. [DOI] [PubMed] [Google Scholar]
- 22. Nigam P. Applying deep learning to ICD-9 multi-label classification from medical records. Technical report, Stanford University.
- 23. Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations. arXiv preprint arXiv:1802.05365. 2018 Feb 15.
- 24. Yang Z, Yang D, Dyer C, et al. Hierarchical attention networks for document classification. In: Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies; June 2016, San Diego, California: 1480–9.
- 25. Wang X, Peng Y, Lu L, et al. Tienet: text-image embedding network for common thorax disease classification and reporting in chest x-rays. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, June 2018, Salt Lake City, UT: 9049–58.
- 26. Nair V, Hinton GE. Rectified linear units improve restricted boltzmann machines. In: Proceedings of the 27th international conference on machine learning (ICML-10), June, 2010, Haifa, Israel: 807–14.
- 27. Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell 2011; 1445: 646–74. [DOI] [PubMed] [Google Scholar]
- 28. Baker S, Silins I, Guo Y, et al. Automatic semantic classification of scientific literature according to the hallmarks of cancer. Bioinformatics 2016; 323: 432–40. [DOI] [PubMed] [Google Scholar]
- 29. Larsson K, Baker S, Silins I, et al. Text mining for improved exposure assessment. PLoS One 2017; 123: e0173132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J, Chang W-C, Wu Y, et al. Deep learning for extreme multi-label text classification. In: Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval, August, 2017, Tokyo, Japan: 115–24.
- 31. Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014 Dec 22.
- 32. Rios A, Kavuluru R. Convolutional neural networks for biomedical text classification. In: Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics. September, 2017. Atlanta, Georgia; 2015: 258–67. doi: 10.1145/2808719.2808746. [DOI] [PMC free article] [PubMed]
- 33. Du J, Chen Q, Peng Y, et al. ML-Net: multi-label classification of biomedical texts with deep neural networks. arXiv preprint arXiv:1811.05475. 2018 Nov 13. [DOI] [PMC free article] [PubMed]
- 34. Moen S, Ananiadou TSS. Distributional semantics resources for biomedical text processing. In: Proceedings of the 5th International Symposium on Languages in Biology and Medicine, Tokyo, Japan, Dec., 2013, Tokyo Japan: 39–43.
- 35. Wu Y, Xu J, Jiang M, et al. A study of neural word embeddings for named entity recognition in clinical text. In: AMIA Annual Symposium Proceedings, Nov., 2015. Chicago, Illinois: 1326. [PMC free article] [PubMed]
- 36. Vaswani A, Shazeer N, Parmar N, et al. Attention is all you need. In: Advances in Neural Information Processing Systems Proceedings; Dec, 2017, Long Beach, CA: 5998–6008.
- 37. Radford A, Narasimhan K, Salimans T, et al. Improving language understanding by generative pre-training. 2018. https://s3-us-west-2.amazonaws.com/openai-assets/research-covers/language-unsupervised/language_understanding_paper.pdf .
- 38. Devlin J, Chang MW, Lee K, et al. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805. 2018 Oct 11.