Abstract
Objective
Linking emergency medical services (EMS) electronic patient care reports (ePCRs) to emergency department (ED) records can provide clinicians access to vital information that can alter management. It can also create rich databases for research and quality improvement. Unfortunately, previous attempts at ePCR and ED record linkage have had limited success. In this study, we use supervised machine learning to derive and validate an automated record linkage algorithm between EMS ePCRs and ED records.
Materials and Methods
All consecutive ePCRs from a single EMS provider between June 2013 and June 2015 were included. A primary reviewer matched ePCRs to a list of ED patients to create a gold standard. Age, gender, last name, first name, social security number, and date of birth were extracted. Data were randomly split into 80% training and 20% test datasets. We derived missing indicators, identical indicators, edit distances, and percent differences. A multivariate logistic regression model was trained using 5-fold cross-validation, using label k-fold, L2 regularization, and class reweighting.
Results
A total of 14 032 ePCRs were included in the study. Interrater reliability between the primary and secondary reviewer had a kappa of 0.9. The algorithm had a sensitivity of 99.4%, a positive predictive value of 99.9%, and an area under the receiver-operating characteristic curve of 0.99 in both the training and test datasets. Date-of-birth match had the highest odds ratio of 16.9, followed by last name match (10.6). Social security number match had an odds ratio of 3.8.
Conclusions
We were able to successfully derive and validate a record linkage algorithm from a single EMS ePCR provider to our hospital EMR.
Keywords: electronic patient care records, prehospital care, machine learning, record linkage, emergency medical services, clinical informatics, patient matching
INTRODUCTION
Background
Electronic patient care reports (ePCRs) by emergency medical services (EMS) providers yield critical information that improves patient care in the emergency department (ED) and throughout the entire hospital stay. Through these ePCRs, emergency medicine staff gain insight into the circumstances leading up to the emergency, including information from bystanders on scene, the clinical trajectory of the patient, nature of prehospital treatments administered, and other information that would otherwise be lost.1 The utility of prehospital documentation extends beyond emergency physicians to the interdisciplinary care team. Social workers, case managers, physical therapists, and nurses can develop better care plans if they have information about the patient’s home situation, such as of the number of stairs they must climb, the safety and cleanliness of the surroundings, and contributing factors such as substance abuse and domestic violence. The inpatient team can refer back to prehospital documentation to better correlate new facts that come to light and the patient’s response to treatment (or lack thereof).
Importance
The ongoing transition from traditional paper records to ePCRs has been a critical transformation to allow inclusion of prehospital information into the hospital’s electronic medical record (EMR). However, this integration has been hampered due to the lack of a reliable method to automatically link a patient’s data in the EMS computer to the correct patient record in the EMR. This type of record linkage, matching the same patient’s records across different data sources, is very challenging in the U.S. healthcare system due to the lack of a national patient identifier. Combining data from different data sources increases the breadth and depth of information that can be analyzed.2 Record linkage has been attempted in prior studies for EMS records with limited success.3 Having accurate record linkage can serve many important functions in the ED for both prospective clinical use and retrospective data analysis. In the real-time clinical setting, linkage of the ePCR to the ED EMR provides the clinician access to vital information that can alter management in the ED. Studies have shown that physicians find PCRs to be important for ED care and medical decision making, and the lack of scene data is associated with increased risk of mortality in trauma patients.4 Retrospectively, the ability to match records accurately can improve datasets for research by augmenting existing clinical registries such as the National Trauma Data Bank and Cardiac Arrest Registry to Enhance Survival.5
In the past, the inability to match prehospital data with hospital outcomes has limited the utility of such datasets and potentially hindered advancements in trauma care, cardiac arrest, stroke, and prehospital research in general. Besides the utility in research, record linkage can promote further quality assurance and education to EMS and ED providers.
Receiving feedback on patient outcomes is essential to the honing and calibration of clinical skills. Unfortunately, EMS providers often lack this critical feedback loop. Record linkage of ePCRs to ED records could provide this critical feedback loop of patient outcomes and discharge diagnoses.
Goals of this investigation
The goal of our study is to derive and validate a record linkage algorithm to accurately link EMS ePCRs to hospital EMR systems.
MATERIALS AND METHODS
Study design and setting
We conducted a retrospective derivation and internal validation study of a record linkage algorithm between EMS ePCRs and ED EMRs using state-of-the-art machine learning techniques. The study was performed at a 55 000 visits/y Level I trauma center and tertiary academic teaching hospital. The study was performed as part of a quality improvement project and submitted to our institutional review board, and a determination was made that this did not constitute human subjects research and no further review was required.
Selection of participants
All consecutive adult ED patients who arrived in the ED between June 2013 and June 2015 from a single EMS agency were included. No patients were excluded. The hospital uses a locally developed ED information system known as the “ED Dashboard,” which serves as the EMR for ED patients at the institution.
Data collection and processing
Prehospital care occurs in a high pressure environment where providers must record information with limited time, incomplete data, and competing priorities. While errors may be present, we leverage modern machine learning techniques to maximize the value of what was captured, without requiring further manual review.
We electronically extracted 6 commonly found data elements from both the ePCR and EMR: age, gender, last name, first name, social security number (SSN), and date of birth (DOB). The features used in the dataset are listed in Table 1.
Table 1.
Model features
| Feature | Description |
|---|---|
| Identical data features | |
| dob_match | date of birth is identical in both records |
| dob_levenshtien | Levenshtein edit distance between date of birth in each record |
| gender_match | gender is identical in both records |
| lastname_match | last name is identical in both recordsa |
| firstname_match | first name is identical in both recordsa |
| ssn_match | SSN is identical in both records |
| Data comparison features | |
| age_diff | difference in age between 2 records |
| SSN_levenshtein | Levenshtein edit distance between social security numbers in each record |
| last_name_jw | Jaro-Winkler edit distance between the 2 last namesa |
| first_name_jw | Jaro-Winkler edit distance between the 2 first namesa |
| Missing data indicators | |
| missing_emr_age | missing variable indicator if age missing from ED data |
| missing_emr_dob | missing variable indicator if date of birth missing from ED data |
| missing_emr_firstname | missing variable indicator if first name missing from ED data |
| missing_emr_lastname | missing variable indicator if last name missing from ED data |
| missing_emr_gender | missing variable indicator if gender missing from ED data |
| missing_emr_ssn | missing variable indicator if social security number missing from ED data |
| missing_ems_gender | missing variable indicator if gender missing from ePCR |
| missing_ems_firstname | missing variable indicator if first name is missing from ePCR |
| missing_ems_age | missing variable indicator if age missing from ePCR |
| missing_ems_dob | missing variable indicator if date of birth is missing from ePCR |
| missing_ems_ssn | missing variable indicator if social security number missing from ePCR |
| missing_ems_lastname | missing variable indicator if last name missing from ePCR records |
ED: emergency department; ePCR: electronic patient care report; SSN: social security number.
aFirst name/last name transposition are allowed without penalty.
Features were constructed to allow for common transposition errors. For example, the firstname_match and lastname_match features permit the first and last names to be swapped without penalty.
To account for typographical errors in text, we used either a Levenshtein edit distance metric or Jaro-Winkler (J-W) distance metric to quantify how dissimilar 2 pieces of text are to one another.
Typographic errors between the ePCR and EMR SSNs were accounted for using the Levenshtein edit distance metric, which was then scaled to lie between 0 and 1.6 In the following example, the Levenshtein edit distance would be 2 because it would take 2 edits to transform the string “CAR” to “ARK.”
Edit 1. CAR → AR [1 deletion]
Edit 2. AR → ARK [1 addition]
Typographical errors in first and last names were accounted for using the J-W distance2 metric, which is a measure that lies between 0 and 1, where 1 represents a perfect match. For example, we calculate the feature firstname_jw by finding the J-W distance between the EMR first name and the ePCR first name. We also calculate the J-W distance between the EMR first name and the ePCR last name in case the first and last names were accidentally swapped. We use the best (larger) of the 2 J-W distances.
The age_diff was calculated as a percentage error and then divided by 100 to scale the value to lie between 0 and 1 using the following formula:
A percentage error in age was used rather than the absolute error because a difference of 2 years is not that important in a 90-year-old but is very important in a 1 year old. A missing indicator feature variable was created for age, gender, last name, first name, SSN, and DOB for each of the EMR and ePCR datasets to denote missing data.
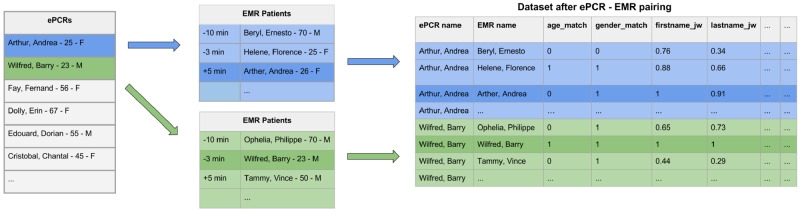
The dataset was comprised of every pair of ePCR and EMR records that was registered within ±2 hours of the EMS arrival time indicated in the ePCR that is described in detail in (Figure 1).ePCR-EMR pairs were randomly allocated to a training (80%) or test (20%) dataset, in which the unit of randomization was at the level of the ePCR records to ensure that all sets of ePCR-EMR pairs were randomized to the same dataset.
Figure 1.
Dataset generation. EMR: emergency medical record; ePCR: electronic patient care report.
Outcome measure
A human reviewer manually matched records to determine a gold standard match between ePCRs and ED visits. For each ePCR, the primary reviewer (CR) was presented a time ordered list of ED visits that began within a 2-hour window around the reported time of EMS arrival. The reviewer matched each ePCR to an ED visit patient record to create a gold standard linkage. This linkage was based on the demographic fields from each source as discussed previously (name, DOB, age, gender, and SSN) as well as additional data elements (arrival time, chief complaint, patient home address, EMS historical narrative, and the ED nursing triage summary). A positive example was defined if the reviewer linked an ePCR to that EMR record. A negative example was defined if the reviewer did not link the ePCR. As there can only be 1 positive example per ePCR and many negative examples, there exists a class imbalance between positive and negative classes that is described in the Model Derivation section.
To ensure that our labeling method was accurate, a second reviewer (LAN) randomly oversampled the primary reviewer’s linkages (n = 1400, 10%) and a Cohen's kappa was calculated.
Primary data analysis
Statistical analysis was performed using the python scikit-learn package.7 Means with 95% confidence intervals were reported for normally distributed variables and medians with interquartile ranges were reported for non-normal variables.
Model derivation and analysis
We used the python scikit-learn package7 to train a multivariate logistic regression model using 5-fold cross-validation on the training dataset. We used the “label k-fold” feature of sci-kit learn’s cross-validation package to ensure that all ePCR-EMR pairs were assigned to the same dataset. Given the large number of features, we used L2 regularization to prevent overfitting to fully utilize all the features, rather than an automated feature selection method. The optimal regularization parameter C was chosen using cross-validation. We also assigned sample weights to negative samples to account for the large class imbalance between positive and negative samples using the following formula:
The default threshold of 0.5 was used as a decision boundary. We report the sensitivity, positive predictive value, area under the receiver-operating characteristic curve (AUC) on both the 80% training dataset and the 20% held-out test dataset.
We also performed an error analysis on the test dataset, analyzing all false positives and false negatives. We also perform a sensitivity analysis of false positives and false negatives using different uncertainty thresholds.
RESULTS
Characteristics of study subjects
A total of 14 032 patients were enrolled during the study period. The mean age was 53.0 years of age and 47.9% were men. For urgency to scene, 14% patients had priority 1, which represents a time sensitive or life-threatening event; 13.4% patients had priority 2, which represents a non–life-threatening event; 7.8% had priority 3, which represents nonacute injury; 0.2% patients had priority 4, which represents hold until verified need by another responding agency such as police or fire, and 64.4% had an undocumented priority. For urgency from scene, 5.7% patients had priority 1, 26.8% patients had priority 2, 25.8% had priority 3, and 41.7% had an undocumented priority. The EMS skillset was advanced life support in 10.9% patients and basic life support in 88.7%. 84.7% of the patient population spoke English, with a total of 39 languages. More detailed patient demographics are shown in Table 2.ePCRs were matched against a total of 90 937 unique ED visits. As the ePCRs are matched against EHR records within 2 hours of the EMS arrival time, each ePCR was matched against a mean of 15.4 (95% confidence interval, 15.2-15.5) ED visits. Each ED visit was matched against a mean of 2.3 (95% confidence interval, 2.3-2.3) ePCRs.
Table 2.
Patient demographics
| Age, y | 53.0 (52.6-53.3) |
| Male | 6724 (47.9) |
| Urgency to scene | |
| Priority 1 (time sensitive or life-threatening event) | 1976 (14) |
| Priority 2 (non–life-threatening event) | 1884 (13.4) |
| Priority 3 (nonacute injury) | 1097 (7.8) |
| Priority 4 (hold until verified need by another responding agency such as police or fire) | 30 (0.2) |
| Priority not documented | 9040 (64.4) |
| Urgency from scene | |
| Priority 1 | 805 (5.7) |
| Priority 2 | 3770 (26.8) |
| Priority 3 | 3589 (25.8) |
| Priority not documented | 5863 (41.7) |
| Skillset | |
| Advanced life support | 1535 (10.9) |
| Basic life support | 12 447 (88.7) |
| Unknown | 5 (0) |
| Languages | |
| English | 11 306 (84.7) |
| Spanish | 782 (5.8) |
| Russian | 377 (2.8) |
| Cape Verdean | 337 (2.5) |
| Haitian Creole | 72 (0.5) |
| Cantonese | 62 (0.4) |
| Portuguese | 32 (0.2) |
| Mandarin | 32 (0.2) |
| American Sign Language | 20 (0.1) |
| Polish | 14 (0.1) |
| Greek | 14 (0.1) |
| Vietnamese | 13 (0.1) |
| Italian | 12 (0.1) |
| Hebrew | 11 (0.1) |
| Persian | 10 (0.1) |
| Other | 73 (0.5) |
Values are mean (95% confidence interval) or n (%).
Main results
The matching algorithm, when evaluated on the training set (10 727 ePCRs; 165 138 ePCR-EMR pairs; 80%), had a sensitivity of 99.4%, a positive predictive value of 99.9%, and an AUC of 0.99. When applied to the test set (2682 ePCRs; 40 774 ePCR-EMR pairs; 20%), it had an identical sensitivity of 99.4%, a positive predictive value of 99.9%, and an AUC of 0.99. Model features and their weights are shown in Table 3.
Table 3.
Feature weights
| Feature | n (%) | Coefficient | Odds ratio |
|---|---|---|---|
| dob_match | 12 750 (95.1) | 2.8 | 16.9 |
| lastname_match | 12 267 (91.5) | 2.4 | 10.6 |
| firstname_match | 10 728 (80) | 1.9 | 6.9 |
| first_name_jw | n/a | 1.8 | 6.3 |
| dob_levenshtien | 12 750 (95.1) | 1.8 | 6.3 |
| last_name_jw | n/a | 1.8 | 6.2 |
| ssn_match | 8208 (61.2) | 1.3 | 3.8 |
| gender_match | 13 240 (98.8) | 1.2 | 3.4 |
| missing_ems_age | 106 (0.8) | 1.2 | 3.2 |
| missing_ems_dob | 104 (0.8) | 1.2 | 3.2 |
| missing_ems_firstname | 49 (0.3) | 1.0 | 2.6 |
| missing_ems_ssn | 4210 (31.4) | 0.6 | 1.9 |
| SSN_levenshtein | n/a | 0.6 | 1.8 |
| missing_ems_lastname | 2 (0) | 0.0 | 1.0 |
| age_diff | n/a | 0.0 | 1.0 |
| missing_emr_age | 0 (0) | 0.0 | 1.0 |
| missing_emr_dob | 0 (0) | 0.0 | 1.0 |
| missing_emr_firstname | 0 (0) | 0.0 | 1.0 |
| missing_emr_lastname | 0 (0) | 0.0 | 1.0 |
| missing_emr_gender | 0 (0) | 0.0 | 1.0 |
| missing_emr_ssn | 0 (0) | 0.0 | 1.0 |
| missing_ems_gender | 0 (0) | 0.0 | 1.0 |
n/a: not applicable.
The feature dob_match had the highest odds ratio (OR) of 16.9 of predicting if an ePCR matched an EMR record followed by lastname_match (OR, 10.6), firstname_match (OR, 6.9), and firstname_jw (OR, 6.3).
The interrater reliability between the primary and secondary reviewers was extremely reliable, with a kappa of 0.9.
Error analysis
When validating the model using the test set, there were 14 (0%) false negatives, which were not matched by the algorithm. Three ePCR records were missing the first name, last name, and DOB fields. The 11 remaining patients had inaccuracies with multiple fields such as name misspellings, incorrect first and last name, or incorrect DOB. There were 0 (0%) false positives that were incorrectly matched by the algorithm. A histogram of predicted probabilities for all ePCR-EMR pairs in the test set is shown in Figure 2A. As shown in this histogram, the large majority of predicted probabilities are around 0 and 1. To better illustrate the distribution of pairs around the decision threshold of 0.5, we also report a censored histogram of probabilities from 0.05 to 0.95 in Figure 2B. Given the small number of misclassifications, we show a histogram of predicted probabilities for both training and test datasets in Figure 3.
Figure 2.
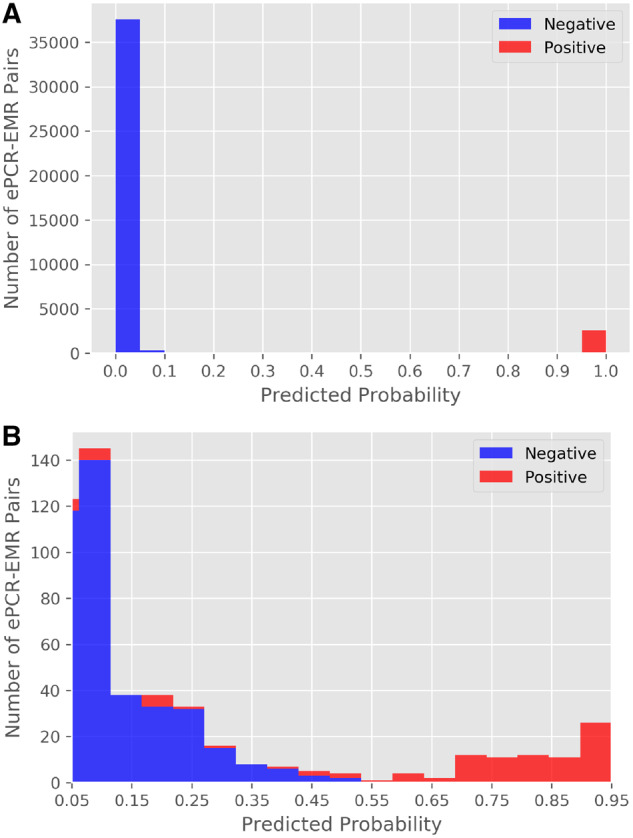
(A) Histogram of predicted probability for all electronic patient care report (ePCR)-emergency medical record (EMR) pairs in the test set. (B) Censored histogram of predicted probability from 0.05 to 0.95 for all ePCR-EMR pairs in the test set.
Figure 3.
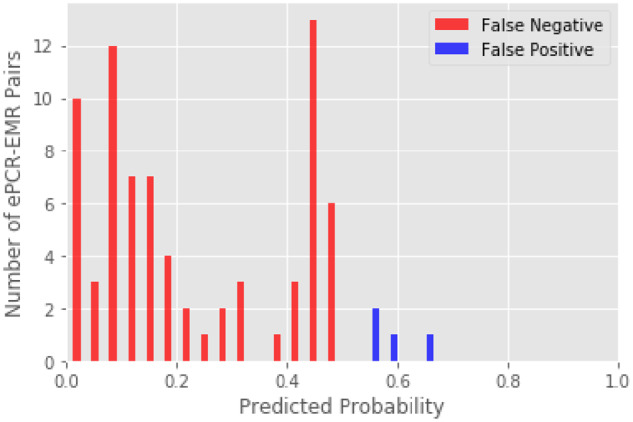
Histogram of predicted probability of misclassifications for the training and test datasets. EMR: emergency medical record; ePCR: electronic patient care report.
DISCUSSION
Comparison with prior studies
Prior studies have attempted to match prehospital records to inpatient records with varying degrees of success, reporting match rates ranging from 14% to 87%.5,8 Manual matching provides a higher success rate8 but is resource-intensive, inaccurate,9 and slow. Most recently in 2015, Mumma et al3 successfully linked 34% of hospital cardiac arrest patients using an unsupervised probabilistic algorithm.
To our knowledge, we are the first to propose a matching algorithm based on a supervised machine learning approach. To do so, we needed to collect gold standard labels, which was time-consuming, but needed only to be done once. In Mumma’s work, they developed probabilistic algorithms using unsupervised techniques, using visual inspection of population statistics to help classify patients. Their unsupervised technique is limited by its assumption that all features are of equal importance, and does not allow for standard interrogation techniques of supervised models to uncover collinearity or nonlinearity.
Clinical impact
Having the ability to link ePCR with hospital records is crucial for improving patient care. Prior studies have demonstrated a lack of EMS documentation, leading to increased mortality in trauma patients.4 A successful linkage algorithm would increase the amount of data available to hospital providers and potentially reduce trauma mortality. Another advantage to using a linkage algorithm is the increase in speed that the ePCR are available to be used by providers in the ED.
Linking the ePCR and hospital record allows hospitals to leverage prehospital care for both pay for performance metrics as well as publicly available hospital performance reports. CMS rules allow hospitals to record interventions provided in the field, such as pain medication for long bone fracture or electrocardiography acquisition and aspirin administration in nontraumatic chest pain, as having occurred during the hospital visit and occurring at time 0 for process metrics provided the prehospital documentation is part of the hospital record. This has the potential to significantly improve reported metrics, and will have direct implications for reimbursement as payers continue the transition to quality payment programs. In addition to the direct implications on reimbursement, studies have shown publicly reported measures have significant impact on hospital reputation and market share, which further increases the potential financial benefits for hospitals and providers.10,11
One result of this study is the ability to accurately and reliably merge EMS records with hospital records in real time to create an integrated database. This allows us to monitor and research previously difficult prehospital questions such as on scene time in acute coronary syndrome,12 or the impact of EMS interventions in trauma, or airway management in cardiac arrest.13,14
Incorrect matches
These cases were matched by the reviewer based on a comparison of the EMS historical narrative and ED nursing triage note. Given the omitted or erroneous fields, it is our impression that a computer algorithm would not be able to match these patients without including additional textual elements from the narrative section of the ePCR and ED triage note.
Failed or incorrect matches have the potential for significant impact on clinical care provided to the patient.15 For example, a patient with chest pain could incorrectly be matched to a patient that received aspirin by EMS, and inadvertently have delayed administration of aspirin, or not at all. A patient could also have a first-line medication withheld because of another patient’s allergy from an incorrect match. Despite these risks, we still believe that our algorithm could be used in real-time for clinical care as we had both a high positive predictive value and negative predictive value that would result in very few failed or incorrect matches.
Real-time clinical implementation
Given the excellent performance characteristics of the algorithm, we have already implemented this algorithm as a fully automated system without a human in the loop. However, the user interface was constructed so that it is immediately obvious when a false positive match occurs, and effectively still has a human in the loop, but as an opt-out rather than opt-in. As shown in Figure 3, the algorithm is not perfect, and still has false positives and false negatives, which could lead to clinical errors. However, we believe that a missing record linkage is more likely to lead to patient harm. In our ED, we already perform record linkage for a variety of tasks such as call-ins from primary care physicians, radiology images from outside hospitals, and laboratory results from outside hospitals and have already developed cognitive forcing strategies to mitigate this risk.
In other EDs in which there is a higher risk of collisions (false positives or false negatives), a semiautomated system could be used in which less certain predictions would require manual confirmation by a human first. A sensitivity analysis of this tradeoff between the false positives, false negatives, and uncertain matches for such a semiautomated system using uncertainty thresholding is reported in Table 4. Machine learning algorithms are rarely perfect and present a challenge to a clinical informatician who must integrate them into clinical workflows. These implementation decisions should be made on a case-by-case basis depending on the specific use case, performance metrics of the algorithm, prevalence of the outcome, harm of an incorrect prediction and harm of having no algorithm at all, and culture of the facility.
Table 4.
Sensitivity analysis of uncertainty thresholding
| Lower threshold | Upper threshold | False positives | False negatives | Uncertain |
|---|---|---|---|---|
| 0.50 | 0.50 | 0 | 14 | 0 |
| 0.45 | 0.55 | 0 | 9 | 9 |
| 0.40 | 0.60 | 0 | 5 | 19 |
| 0.35 | 0.65 | 0 | 5 | 23 |
| 0.30 | 0.70 | 0 | 5 | 36 |
| 0.25 | 0.75 | 0 | 5 | 68 |
| 0.20 | 0.80 | 0 | 3 | 108 |
| 0.15 | 0.85 | 0 | 1 | 151 |
| 0.10 | 0.90 | 0 | 1 | 226 |
| 0.05 | 0.95 | 0 | 0 | 455 |
| 0.00 | 1.00 | 0 | 0 | 40 936 |
Future directions
In this work, we used a supervised machine learning algorithm using edit distances of 14 032 labeled patient visits. However, there are several hundred thousand additional patient visits that are unlabeled that we could not use for training using supervised machine learning methods. An alternative approach would be to learn from these unlabeled patient visits the types of names and misspellings that occur, and then fine-tune the model using the 14 032 labeled patient visits, in a method called semisupervised learning. For example, we could use methods such as bidirectional character-level embeddings and deep learning.
In this work, we labeled consecutive patient visits. Given the already high performance of our model, exhaustively labeling new patient visits would be inefficient. Instead, we could use active learning to help choose which patients to label next.
Last, an error analysis of the 14 false negatives revealed that the record linkages could only have been made if the free text sections of the ePCR assessment and ED triage note were used. This would be an obvious next step, specifically given the recent advancements in clinical natural language processing using machine learning and word embeddings, in which a word or phrase is represented as a vector, with the capability of capturing context, relationships, and similarity.
Limitations
A significant limitation to our study is the reliance on a single EMS agency. Prior studies that have looked at multiple EMS agencies have demonstrated significant variations in practice among different companies.3 The use of a single agency could lead to a less generalizable algorithm due to less variability in data entry practices and quality.
In this study, we were only able to study a single EMS agency, as we could not access the ePCRs of other EMS agencies electronically. Other EMS agencies either print the ePCR and leave a physical copy or fax the run sheet to a virtual fax number for the ED, after which they are eventually scanned into the system. In future work, we will apply optical character recognition and attempt to match those ePCRs as well. As optical character recognition will likely introduce new types of errors in the data, new algorithms will need to be developed using similar methodology.
Some EDs may also have a workflow where the EMS run sheet number (a unique number corresponding to every run sheet) is recorded either manually or scanned via barcode. Although this is not part of the workflow at this institution, inclusion of this feature where it is recorded could further improve the match rate.
Another limitation is the retrospective nature of our study, which will need to be prospectively validated. While we were able to internally validate our probabilistic matching algorithm, its generalizability will be unknown until it is externally validated with different ePCR systems at different study settings.
CONCLUSION
The inability to link prehospital ePCRs has previously been identified as a key barrier to the transfer of information for clinical care, as well as for the creation of integrated research databases, registries, quality improvement, and quality assurance.
Prior published work to link EMS to ED records approached only a 34% success rate,3 which improved to 87% with manual review.8 Our automated method that requires no manual review is a substantial improvement over prior published work, with a 99.4% sensitivity, 99.9% positive predictive value, and an AUC of 0.99.
These excellent performance characteristics would enable the previously reported use cases that would otherwise have previously been infeasible.
AUTHOR CONTRIBUTIONS
SH and LAN conceived and designed the study. CR and AT collected the data. AT, YH, DAS, and SH performed the analysis. CR, AT, and SH drafted the manuscript, and all authors contributed substantially to its revision. SH takes responsibility for the article as a whole.
ACKNOWLEDGMENTS
We presented a preliminary version of this algorithm at the May 2016 Society of Academic Emergency Medicine Annual Meeting. The algorithm presented in this article is significantly different.
CONFLICT OF INTEREST STATEMENT
SH receives grant funding by Philips Healthcare in the areas of heart failure risk stratification, imaging analysis, and big data. The submitted manuscript has no relationship to the grants. LAN has stock in Forerun Systems, an emergency department information system. The submitted manuscript did not use this system.
REFERENCES
- 1. Landman AB, Lee CH, Sasson C, Van Gelder CM, Curry LA.. Prehospital electronic patient care report systems: early experiences from emergency medical services agency leaders. PLoS One 2012; 7 (3): e32692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jaro MA. Probabilistic linkage of large public health data files. Stat Med 1995; 14 (5–7): 491–8. [DOI] [PubMed] [Google Scholar]
- 3. Mumma BE, Diercks DB, Danielsen B, Holmes JF.. Probabilistic linkage of prehospital and outcomes data in out-of-hospital cardiac arrest. Prehosp Emerg Care 2015; 19 (3): 358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laudermilch DJ, Schiff MA, Nathens AB, et al. Lack of emergency medical services documentation is associated with poor patient outcomes: a validation of audit filters for prehospital trauma care. J Am Coll Surg 2010; 210 (2): 220–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newgard C, Malveau S, Staudenmayer K, et al. Evaluating the use of existing data sources, probabilistic linkage, and multiple imputation to build population-based injury databases across phases of trauma care. Acad Emerg Med 2012; 19 (4): 469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Levenshtein V. Binary codes capable of correcting deletions, insertions, and reversals. Soviet Phys Doklady 1966; 10 (8): 707–10. [Google Scholar]
- 7. Pedregosa F. e A. Scikit-learn: machine learning in python. J Mach Learn Res 2011; 12: 2825–30. [Google Scholar]
- 8. Hettinger AZ, Cushman JT, Shah MN, Noyes K.. Emergency medical dispatch codes association with emergency department outcomes. Prehosp Emerg Care 2013; 17 (1): 29–37. doi: 10.3109/10903127.2012.710716 [DOI] [PubMed] [Google Scholar]
- 9. Waien SA. Linking large administrative databases: a method for conducting emergency medical services cohort studies using existing data. Acad Emerg Med 1997; 4 (11): 1087–95. [DOI] [PubMed] [Google Scholar]
- 10. Schwartz L M, Woloshin S, Birkmeyer JD.. How do elderly patients decide where to go for major surgery? Telephone interview survey. BMJ 2005; 331 (7520): 821.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hibbard Judith H, Stockard J, Tusler M.. Hospital performance reports: impact on quality, market share, and reputation. Health Aff (Millwood) 2005; 24 (4): 1150–60. [DOI] [PubMed] [Google Scholar]
- 12. Fosbøl EL, Granger CB, Peterson ED, et al. Prehospital system delay in ST-segment elevation myocardial infarction care: a novel linkage of emergency medicine services and in hospital registry data. Am Heart J 2013; 165 (3): 363–70. [DOI] [PubMed] [Google Scholar]
- 13. McMullan J, Gerecht R, Bonomo J, et al. Airway management and out-of-hospital cardiac arrest outcome in the CARES registry. Resuscitation 2014; 85 (5): 617–22. [DOI] [PubMed] [Google Scholar]
- 14. Hasegawa K, Hiraide A, Chang Y, Brown DF.. Association of prehospital advanced airway management with neurologic outcome and survival in patients with out-of-hospital cardiac arrest. JAMA 2013; 309 (3): 257–66. [DOI] [PubMed] [Google Scholar]
- 15. Edward J, Dunn Paul J. Moga. Patient misidentification in laboratory medicine: a qualitative analysis of 227 root cause analysis reports in the Veterans Health Administration. Arch Pathol Lab Med 2010; 134 (2): 244–55. [DOI] [PubMed] [Google Scholar]
- 16.U.S. Department of Health and Human Services. Emergency Department (ED): Median Time From ED Arrival to Time of Initial Oral, Intranasal or Parenteral Pain Medication Administration for ED Patients With a Principal Diagnosis of Long Bone Fracture. Rockville, MD: Agency for Healthcare Research and Quality; 2016. [Google Scholar]
- 17.U.S. Department of Health and Human Services. Acute Myocardial Infarction (AMI)/Chest Pain: Percentage of ED Patients With AMI or Chest Pain Who Received Aspirin Within 24 Hours Before ED Arrival or Prior to Transfer. Rockville, MD: Agency Agency for Healthcare Research and Quality; 2016. [Google Scholar]