Abstract
Objective
To conduct a systematic scoping review of explainable artificial intelligence (XAI) models that use real-world electronic health record data, categorize these techniques according to different biomedical applications, identify gaps of current studies, and suggest future research directions.
Materials and Methods
We searched MEDLINE, IEEE Xplore, and the Association for Computing Machinery (ACM) Digital Library to identify relevant papers published between January 1, 2009 and May 1, 2019. We summarized these studies based on the year of publication, prediction tasks, machine learning algorithm, dataset(s) used to build the models, the scope, category, and evaluation of the XAI methods. We further assessed the reproducibility of the studies in terms of the availability of data and code and discussed open issues and challenges.
Results
Forty-two articles were included in this review. We reported the research trend and most-studied diseases. We grouped XAI methods into 5 categories: knowledge distillation and rule extraction (N = 13), intrinsically interpretable models (N = 9), data dimensionality reduction (N = 8), attention mechanism (N = 7), and feature interaction and importance (N = 5).
Discussion
XAI evaluation is an open issue that requires a deeper focus in the case of medical applications. We also discuss the importance of reproducibility of research work in this field, as well as the challenges and opportunities of XAI from 2 medical professionals’ point of view.
Conclusion
Based on our review, we found that XAI evaluation in medicine has not been adequately and formally practiced. Reproducibility remains a critical concern. Ample opportunities exist to advance XAI research in medicine.
Keywords: Explainable artificial intelligence (XAI), interpretable machine learning, real-world data, electronic health records, deep learning
INTRODUCTION
The emergence of modern data-rich technologies will require physicians to interpret high-dimensional heterogeneous medical data while also making efficient and accurate decisions for diagnosis and treatment.1 Artificial intelligence (AI) techniques are critical tools that can assist physicians with such analyses and decision-making.2 Referring to Norvig and Russel’s classic AI textbook,3 in this review article we define AI as acting humanly through machine learning (ML) and, more specifically, ML-based predictive analytics.
Szolovits4 defines AI in medicine (AIM) as “AI specialized to medical application.” In recent years, AIM has contributed to healthcare in the light of digitized health data.5 The wide adoption of electronic health record (EHR) systems by healthcare organizations and subsequent availability of large collections of EHR data have made the application of AIM more feasible.6,7 EHR data contain rich, longitudinal, and patient-specific information including both structured data (eg, patient demographics, diagnoses, procedures) as well as unstructured data, such as physician notes, among other clinical narratives.8
Despite their promising performance, the production of AIM systems for actual clinical use is challenging.9 A survey of medical professionals in 2018 showed a lack of trust in AIM.10 Limited access to large data, lack of integration to clinical workflows, and, especially, the ambiguity of requirements for regulatory compliance are among the development and deployment challenges of AIM systems.11 In 2017, the Defense Advanced Research Projects Agency (DARPA) released a public update report of their research program on explainable AI (XAI).12 They reported that the new generation of AI systems have limited effectiveness due to the inability of humans to understand why an AI system makes particular decisions.
General Data Protection Regulation (GDPR) in Europe is an example of the increasing needs for XAI from a regulatory perspective. This regulation is a data protection and privacy law for all citizens of the European Union, which regulates any organization that uses personal data including EHRs of European Union residents for automated decision-making. Among other regulations, it requires organizations to provide meaningful explanations about how the algorithm reaches its final decisions.13 However, since there is no concrete formulation and quantification of what an adequate explanation should be, regulatory enforcement seems challenging in this context.
Some researchers argue that if physicians could rely on drugs like aspirin despite the fact that their underlying mechanism was unknown, should they expect AI to give explanations if its performance is promising?14 On the other hand, drugs have to go through rigorously designed and conducted randomized clinical trials for regulatory approval before production. Post-marketing surveillance allows regulatory agencies, such as the Food and Drug Administration in the US, to withdraw them from the market in cases of serious adverse events. AI, built inside labs using potentially biased and limited data with challenges like generalization to new samples, does not have comparable mechanisms to ensure efficacy and safety in the real world. XAI helps to understand whether AIM decisions are valid and come to a consensus with medical professionals and as a result, promote their trust in AIM.15 Thus, XAI for medicine is of vital importance to support the implementation of AI in clinical decision support systems.16,17
The increasing capabilities of AIM married to the necessity of XAI demand a review of the state-of-the-art research in the field. Our review summarizes a decade of research on the enhancement of interpretability in EHR-based AIM. We aim to provide insights into the current research trend by categorizing ML methods, XAI approaches, and targeted ML prediction tasks to identify potential gaps and suggest future research direction in the field. We also assess the reproducibility of the included studies. Finally, we review and evaluate the studies from the medical professional’s perspective on their interpretability enhancement deliverables.
METHOD
Literature selection strategy
We conducted a systematic scoping review of XAI on EHR data using MEDLINE, Web of Science, IEEE Xplore, and Association for Computing Machinery (ACM) databases based on the Preferred Reporting Items for Systematic Review and Meta-Analyses (PRISMA) framework18 to search studies published between January 1, 2009 and May 1, 2019. In this article, we refer to explainable ML methods that are used for predictive analytics as XAI. We used Covidence—a systematic review management system—to conduct this systematic scoping review. We considered different combinations of relevant search keywords in Table 1.
Table 1.
The search queries
| Database | Query | # initial results |
|---|---|---|
| MEDLINE and Web of Science (via Covidence software) | (explainable OR explainability OR interpretable OR interpretability OR understandable OR understandability OR comprehensible OR comprehensibility OR intelligible) AND (machine learning OR artificial intelligence OR prediction model OR predictive model OR deep learning OR AI OR neural network) | 1487 |
| IEEE Xplore | 2208 | |
| ACM digital library | 2734 |
Abbreviations: AI, artificial intelligence.
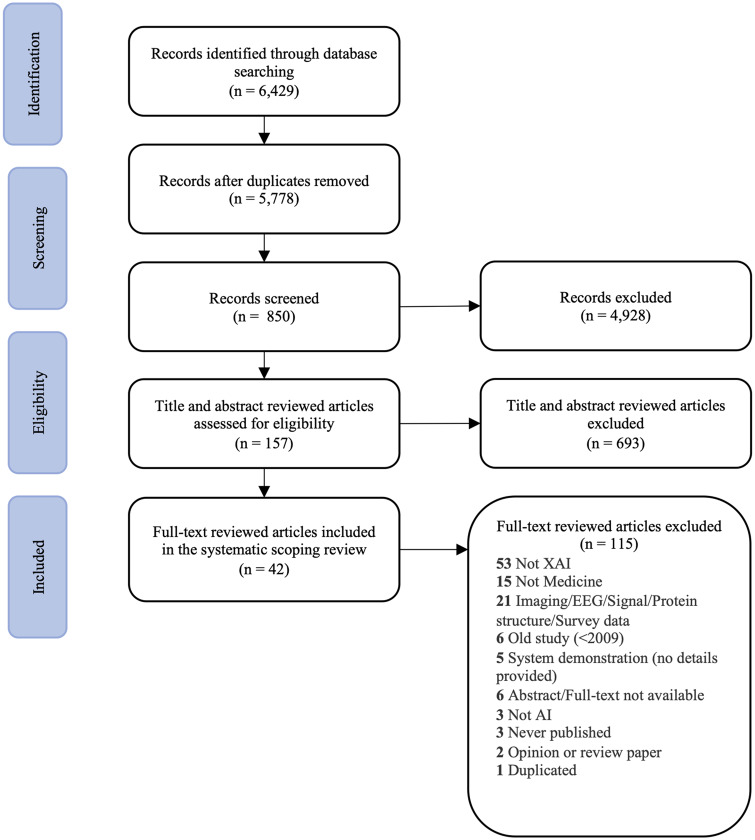
We found 6429 articles from MEDLINE, Web of Science, IEEE Xplore digital library, and ACM digital library. After removing 651 duplicates, 2 authors (SNP and ZC) screened the titles and abstracts of the remaining 5778 studies based on a set of inclusion and exclusion criteria (see Supplementary Appendix Table 1). They retained 157 articles for full-text review and deemed 42 articles relevant to include in the final full-text extraction. The PRISMA flow chart is depicted in Figure 1.
Figure 1.
The PRISMA diagram depicts the number of records identified, included and excluded, and the reasons for exclusions.
The rest of the study team supervised the screening process, resolved conflicts, and provided clarifications based on their expertise. Since this review focuses on EHR data, studies based on any data other than EHR are not included. Building predictive models using medical images and electroencephalogram data, for instance, do not share similar characteristics with those based on patient EHRs. We refer interested readers to other existing survey literature such as those about AI in medical imaging19,20 and electroencephalogram signal processing.21
Data extraction
We considered the following aspects when evaluating the full text of the 42 articles (referred to as “the articles” throughout the paper) included in this paper: 1) year of publication; 2) ML prediction tasks (eg, incident of a disease, mortality, re-admission, risk assessment); 3) ML algorithm; 4) XAI method; 5) the dataset used to build the model; 6) scope of XAI method (ie, intrinsic/posthoc, local/global, model-specific/model-agnostic); 7) category of XAI method (ie, feature interaction and importance, attention mechanism, data dimensionality reduction, knowledge distillation and rule extraction, and intrinsically interpretable models); and 8) evaluation of XAI method. We also assessed the articles in terms of reproducibility based on 2 objective criteria: 1) if the datasets are accessible to the public (ie, proprietary or not), and 2) if the availability of the source codes/implementations is explicitly mentioned in the manuscript or in the supplementary material.
We reviewed XAI methods in the articles and 2 medical professionals (PRM and JC) evaluated the clinical utility of these methods. This can help identify the potential perception gaps between model builders and the end users of the models. We also identified open issues and challenges in XAI that can serve as suggestions for future work in the field. To the best of our knowledge, this paper is the first attempt to review XAI in AIM with real-world EHR data.
RESULTS
Research trends
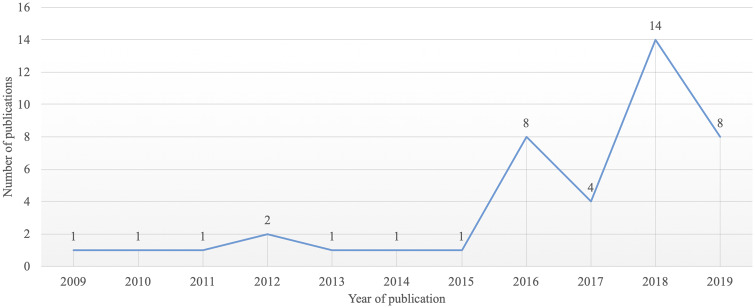
We have seen a surge of XAI studies in AIM applications using EHR data since 2015, with only a small number of studies from 2009 to 2011, as shown in Figure 2. The limited number of publications indicates there is a demand for more research focused on XAI in biomedical applications using EHR data.
Figure 2.
Publication trend of XAI studies using EHR data between January 1st, 2009 and May 1st, 2019.
Prediction tasks, methods, and datasets
As of 2017, cardiovascular diseases, cancer, diabetes, and Alzheimer’s disease are the leading causes of death in the United States.22 The majority of the articles (∼60%) in our review focused on 1 or more of these diseases. Table 2 lists all the articles, the ML methods, the ML prediction tasks, and the datasets. Researchers used different ML methods in the articles including: 1) logistic regression (LR), 2) support vector machines (SVM), 3) decision trees (DT), 4) ensemble, 5) Bayesian networks, 6) fuzzy logic, 7) deep learning (DL), and 8) other approaches.
Table 2.
All the articles grouped based on the ML method along with the associated medical prediction task(s)
| ML Method | Prediction task(s) | Dataset(s)a | Article(s) |
|---|---|---|---|
| Logistic regression | Incidence of Medium-chain acyl-coA dehydrogenase deficiency | A systematic newborn screening by the PCMA screening center (Belgium) | Van den Bulcke et al23 |
| In-hospital mortality (all-cause)/ hospital-acquired infections/ICU admissions/development of pressure ulcers during the patient’s stay | Premier healthcare EHR data | Fejza et al24 | |
| Support vector machines | Incidence of diabetes mellitus | A diabetes dataset in Oman25 | Barakat et al26 |
| Incidence of leukemia/prostate cancer/colon cancer | Unnamed datasets27–29 | Hajiloo et al30 | |
| Gut and skin microbiota/ inflammatory bowel diseases | Unnamed dataset31 | Eck et al32 | |
| Incidence of type 2 diabetes | Federazione Italiana Medici di Medicina Generale | Bernardini et al33 | |
| Hospitalization due to heart diseases or diabetes | Boston Medical Center | Brisimi et al34 | |
| Decision trees | Protein solubility and gene expressions | 40 datasets in the University of California Irvine (UCI) repository, Solubility database of all E. coli proteins, and 9 Gene Expression Machine Learning Repository datasets | Stiglic et al35 |
| Ensemble | Risk of developing Type 2 diabetes | Practice Fusion Diabetes Classification Dataset | Luo36 |
| Stage of acute myeloid leukemia/breast invasive carcinoma | The Cancer Genome Atlas | Jalali and Pfeifer37 | |
| Incidence of central precocious puberty in girls | Pediatric Day Ward of the Endocrinology Department at Guangzhou Women and Children’s Medical Center | Pan et al38 | |
| Incidence of multiple diseases | 13 data sets of life sciences in the UCI Repository | Valdes et al39 | |
| Adverse drug events | Stockholm electronic patient record corpus (HealthBank) | Crielaard and Papapetrou40 | |
| Drug side effect | Side Effect Resource 4 | Zhang et al41 | |
| Bayesian networks | Incidence of heart disease, fetal pathologies | 3 heart disease datasets in the UCI repository | Bouktif et al42 |
| Fuzzy logic | In-hospital mortality (all-cause) | MIMIC-III, Diabetes, Heart Disease and Liver datasets in the UCI repository | Davoodi and Moradi43 |
| Incidence of type 2 diabetes | Pima Indian Dataset in UCI repository | Settouti et al44 | |
| Deep learning | Splice site detection | Unnamed datasets45–47 | Zuallaert et al48 |
| Hospital readmission due to heart failure | Congestive Heart Failure | Xiao et al49 | |
| Illness severity/ in-hospital mortality | University of Florida Health, MIMIC-III | Shickel et al50 | |
| Heart failure/cataract | Health Insurance Review and Assessment National Patient Samples (Republic of Korea) | Kwon et al51 | |
| Cell-type specific enhancer | National Institutes of Health Epigenome Roadmap data; National Human Genome Research Institute ENCODE database; the Encyclopedia of DNA Elements | Kim et al52 | |
| Mortality/ventilator-free days due to acute lung injury. | Pediatric ICU dataset from the Children's Hospital Los Angeles | Che et al53 | |
| Mortality (all-cause) | Asan Medical Center | Ge et al54 | |
| Incidence of autism | N/A | Ghafouri-Fard et al55 | |
| Long-term survival from glioblastoma multiforme | The Cancer Genome Atlas | Hao et al56 | |
| Stage of several cancer | Cancer microarray data sets obtained from Gene Expression Model Selector | Hartono57 | |
| HIV genome integration site | Retrovirus Integration Database | Hu et al58 | |
| Daily sepsis/myocardial infarction/ vancomycin antibiotic administration | MIMIC-III | Kaji et al59 | |
| Heart failure | Sutter Health | Choi et al60 | |
| Incidence of vascular diseases | Seoul National University Bundang Hospital | Park et al61 | |
| Future hospitalization | De-identified EHR data from the University of Virginia Health System | Zhang et al62 | |
| Multifactor affiliation analysis | Dementia stage of Alzheimer’s disease | Open Access Series of Imaging Studies | Aditya and Pande63 |
| Measuring similarity to exemplars of clusters | Stratify patients with stage 1 lung cancer | Xena | Zhao and Bolouri64 |
| Stratify the risk of 30-days mortality in patients with cardiovascular disease | Portuguese real Acute Coronary Syndrome patients’ dataset | Paredes et al65 | |
| Different predictive models on specific clusters of patient population. | Survival from cardiac transplantation | United Network of Organ Sharing | Yoon et al66 |
| Logic optimization for binary input to continuous output | Drug response of cancer cell lines | Genomics of Drug Sensitivity in Cancer, Cancer Therapeutics Response Portal | Knijnenburg et al67 |
| Rule-based | Diabetes and breast cancer stage classification | Wisconsin Breast Cancer Dataset and Pima Indian Diabetes Dataset in the UCI repository | Ming et al.68 |
| Incidence of asthma/diabetes/depression/lung cancer/leukemia/myelofibrosis | Medical diagnosis records of about 150K patients collected by a web-based EHR company and 2 other nonmedical datasets | Lakkaraju et al69 | |
| Diagonal quadratic discriminant analysis | Incidence of leukemia | Unnamed datasets26,70 | Huang71 |
| Artificial hydrocarbon networks | Incidence of breast cancer | Wisconsin Breast Cancer Dataset in the UCI repository | Ponce and Martinez-Villaseñor72 |
| Sparse high-order interaction model with rejection option | Incidence of Alzheimer’s disease | Alzheimer's Disease Neuroimaging Initiative | Das et al73 |
Abbreviations: EHR, electronic health record; ICU, intensive care unit; ML, machine learning; UCI, University of California, Irvine.
We have either mentioned the name of the dataset/reference, the referenced paper that is indicated by the authors as the source of dataset, or the institute that the dataset is generated by. If there was no information regarding the dataset in a study, we indicated it as N/A in the table.
DL is the most popular ML method as approximately 38% of the articles used DL with different architectures including feed-forward neural networks, convolutional neural networks, recurrent neural networks with long short-term memory or gated recurrent unit. SVM, ensemble techniques, and logistic regression (LR) are the second (∼14%) and third (∼12%, ∼12%) most popular methods, respectively. Other popular techniques in AIM are fuzzy logic and DT.
XAI methods
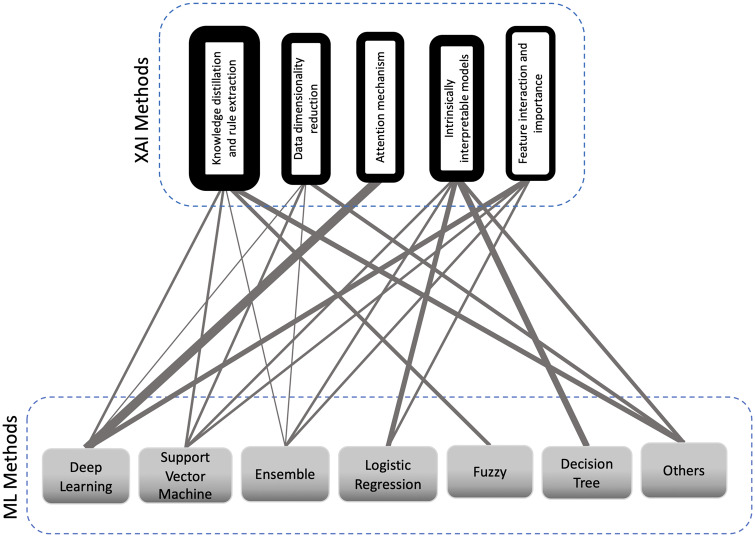
We grouped the XAI methods that were employed in the articles into 5 categories: 1) feature interaction and importance, 2) attention mechanism, 3) data dimensionality reduction, 4) knowledge distillation and rule extraction, and 5) intrinsically interpretable models. We synthesized these categories from extant XAI review papers.74,75Figure 3 depicts the type of XAI methods employed along with different ML methods and Table 3 summarizes different approaches in each category of XAI methods for each of the articles.
Figure 3.
Explainable artificial intelligence (XAI) methods vs machine learning (ML) methods used for interpretability enhancement. The links between ML and XAI methods illustrate that ML method was used with that specific XAI method in a paper. The thicker the links are, the more frequent that combination of ML and XAI method has been practiced. A thicker box around the XAI method shows that it has been applied by more of the articles.
Table 3.
Different explainable artificial intelligence methods, their category, and scope
| XAI Category | Articles | Approach | Intrinsic/Posthoc | Local/Global | Model-specific/-agnostic |
|---|---|---|---|---|---|
| Feature interaction and importance | Ge et al54 | Feature weights in the model | Posthoc | Global | Model-agnostic |
| Zuallaert et al48 | Contribution score of each neuron activation (DeepLIFTundefined18) | ||||
| Eck et al32 | Approximately marginalizing features out | Global, Local | |||
| Pan et al38 | LIME77 | Local | |||
| Ghafouri-Fard et al55 | |||||
| Attention mechanism | Kwon et al51 | The model’s capability to find a set of positions in a sequence with the most relevant information to the prediction task78 | Intrinsic | Global, Local | Model-specific |
| Kaji et al59 | |||||
| Shickel et al50 | |||||
| Hu et al58 | Local | ||||
| Choi et al60 | |||||
| Zhang et al62 | |||||
| Park et al61 | |||||
| Data dimensionality reduction | Zhao and Bolouri64 | Identifying most informative exemplars through cluster analysis and LASSO | Intrinsic | Global | Model-agnostic |
| Kim et al52 | Building a model based on the most important features | ||||
| Hao et al56 | Finding the gene pathways and their interactions using sparse DL | ||||
| Bernardini et al33 | Sparse-balanced SVM | ||||
| Zhang et al41 | Selecting optimal feature subset from the most critical dimensions of features | ||||
| Huang71 | Diagonal quadratic discriminant analysis on chi2 selected features | Local | Model-specific | ||
| Aditya and Pande63 | Affiliation analysis based on capturing inter-feature relationships (knowledge base) | ||||
| Hartono57 | Providing clearer mathematical description | ||||
| Knowledge distillation and rule extraction | Xiao et al49 | Distilling complex relationships between hospital readmission and potential risk factors | Posthoc | Global | Model-specific |
| Settouti et al44 | Fuzzy rules | Intrinsic | Global, Local | ||
| Davoodi and Moradi43 | Global | ||||
| Hajiloo et al30 | |||||
| Che et al53 | Mimic learning | Posthoc | Global, Local | ||
| Barakat et al26 | Intelligible representation of the SVM’s classification decision | Intrinsic | Global | ||
| Das et al73 | sparse high-order interaction model with rejection option | Global, Local | |||
| Crielaard and Papapetrou40 | Rule induction from transparent oracle-coached predictive models | Global | |||
| Paredes et al65 | Rule extraction from regions of belonging associate with each class | ||||
| Lakkaraju et al69 | Decision-sets (nonhierarchical if-then rules) | Local | |||
| Ming et al68 | Visualizing rules | Posthoc | Global | ||
| Ponce and Martinez-Villanseñor72 | Rule extraction based on predictive features (polynomial weights) | Intrinsic | |||
| Luo36 | Automatically pruning and manually refining association rules | Posthoc | Global, Local | Model-agnostic | |
| Intrinsically interpretable models | Fejza et al24 | Distributed logistic regression framework | Intrinsic | Global | Model-specific |
| Van den Bulcke et al23 | Parameter and threshold optimization of decision tree, LR, and ridge LR | ||||
| Bouktif et al42 | Ant colony optimization of combining Bayesian classifiers | ||||
| Brisimi et al34 | Alternating clustering and classification | ||||
| Yoon et al66 | A tree of clusters with base learners associated with each cluster | ||||
| Valdes et al39 | Accurate decision trees based on boosting | Global, Local | |||
| Knijnenburg et al67 | Logic optimization for binary input to continuous output | Global | |||
| Jalali and Pfeifer37 | Ensemble of regularized linear SVM | Global, Local | |||
| Stiglic et al35 | Automated pruning for decision tree. | Global |
Abbreviations: DL, deep learning; LIME, local interpretable model-agnostic explanation; LR, logistic regression; SVM, support vector machine; XAI, explainable artificial intelligence.
We analyzed the XAI methods’ scope and categorized them into 1) intrinsic/posthoc (ie, interpretation as a result of inherited characteristics of the ML method/interpretation as an additional step on top of the ML model), 2) global/local (ie, interpretation of the whole logic of the model/interpretation of a specific decision for an instance), and 3) model-specific/-agnostic (ie, interpretation method limited to a specific model/interpretation method not tied to a specific model). We referred to the definition of these categories in previously published XAI review papers.74,76 Visualization techniques are often used as a complementary tool to facilitate the interpretation of the results in most of the articles. Thus, we did not consider visualization as a separate interpretability enhancement method.
According to the articles, the majority of researchers chose “if-then” rules (∼28%) to enhance the interpretability of complex ML methods. Another major trend is to preserve the interpretability of less complex ML methods while boosting their performance and applying optimization (∼21%). These 2 major trends are followed by dimensionality reduction techniques (∼19%).
Feature interaction and importance
Researchers have used feature importance and pairwise feature interaction strengths to provide interpretability to ML models.75 The level of contribution of the input features to the output prediction has been extensively used for XAI in AIM.76 Ge et al54 used feature weights to report top-10 contributing features for intensive care unit (ICU) mortality prediction. Researchers have also used sensitivity analysis for deriving the feature importance.80 Based on sensitivity analysis, the most important features are those to which the output is most sensitive. Eck et al32 determined the most important features for microbiota-based diagnosis by approximately marginalizing features out and evaluating the effect on the model’s output.
Ribeiro et al77 introduced the local interpretable model-agnostic explanation (LIME) method. LIME produces explanations for any classifier by approximating the reference model with a “locally faithful” interpretable representation. To produce explanations, LIME perturbs an instance, generates neighborhood data, and learns linear models in that neighborhood. Pan et al38 used LIME to investigate the contribution level of features of new instances for predicting central precocious puberty in girls. Ghafouri-Fard et al55 took the same approach for diagnosing autism spectrum disorder.
Shrikumar et al79 introduced DeepLIFT, which is a backpropagation-based interpretability approach. Backpropagation approaches calculate the gradient of an output with respect to the input via the backpropagation algorithm to report the feature importance. Zuallaert et al48 used DeepLIFT to build interpretable deep models for splice site prediction by calculating the contribution score of each nucleotide.
Attention mechanism
The main idea behind the attention mechanism78 is the model’s capability to find a set of positions in a sequence with the most relevant information to the prediction task. This idea is proved to apply to interpretability enhancement as well.81
Attention mechanism has been used to 1) highlight the specific times when the input features have mostly influenced the predictions of clinical events in ICU patients,59 2) present an interpretable acuity score framework based on DL (DeepSOFA) that can evaluate a patient’s severity of illness during an ICU stay50; 3) provide “mechanistic explanations” on accurate prediction of HIV genome integration sites (DeepHINT)58; 4) feed gradient-weighted class activation mapping (Grad-CAM) with feature representations that include embedded time intervals information to recurrent neural networks to predict vascular diseases61; and 5) to learn a representation of EHR data which captures the relationships between clinical events for each patient (Patient2Vec).62
Choi et al60 introduced a reverse time attention model (RETAIN), which uses 2 sets of attention weights, 1 for visit-level (to capture each visit’s influence) and the other for variable-level. RETAIN is a reverse attention mechanism to preserve interpretability, mimic medical professional’s behavior, and incorporate sequential information. Kwon et al51 developed a visually interpretable DL model for heart failure and cataract risk prediction based on RETAIN (RetainVis). The commonality of these articles is in their aim to enhance the interpretability of DL models by highlighting specific position(s) within a sequence (eg, time, visits, DNA) in which certain input features influence the prediction outcome.
Data dimensionality reduction
Researchers used data dimensionality reduction to build models only by including the most important features. Bernardini et al,33 for instance, used the least absolute shrinkage and selection operator (LASSO)82 to induce sparsity for SVMs for type 2 diabetes early diagnosis. Hao et al56 developed pathway-associated sparse DL to find the gene pathways and their interactions in patients with glioblastoma multiforme. Kim et al52 selected the most important input features from the datasets based on domain knowledge and built a DL model. Then, they ranked the features based on their weights in the model and visualized the result for predicting cell-type-specific enhancers.
In another work, Zhao and Bolouri64 stratified patients with stage-1 lung cancer by identifying the most informative exemplars through supervised learning. They proposed a hybrid approach for dimensionality reduction by integrating pattern discovery and regression analytics to identify a group of “exemplars” and create a “dense data matrix.” Then, they included those exemplars that are the most predictive of the outcome in the final model.
Zhang et al41 built a model for drug side effects prediction based on the optimal dimensions of input features by combining multi-label k-nearest neighbor and genetic algorithm techniques. To provide more transparency to the model, Hartono57 visualized cancer classification using a clearer mathematical description. This was achieved by introducing Softmax restricted radial basis function networks. Huang71 developed an integrated method for cancer classification. He reduced the feature dimension by selecting important genes using the Chi2 algorithm. Then, he applied diagonal quadratic discriminant analysis for classification. Finally, he used general rule induction to extract association rules.
Knowledge distillation and rule extraction
In their influential work, Hinton et al83 proposed a knowledge distillation technique for neural networks. This technique transfers knowledge from a complex and accurate model to a smaller and less complex one which is faster but still accurate. Che et al53 used knowledge distillation to build an interpretable prediction model for ICU outcome (ie, mortality, ventilator-free days) by feeding learned features from the base model into the helper classifier (mimic model) and reporting feature importance of the mimic model to deliver interpretability to the basic complex model. A similar approach was taken by Ming et al68 to extract rules by approximating a complex model using model induction on several tasks, such as breast cancer diagnosis and diabetes classification.
Xiao et al49 developed a DL model (CONTENT) that distills complex relationships between hospital readmission and potential risk factors for patients by transforming patients' EHR events into clinical concept embeddings. As a result, they produced a context vector that characterizes the overall condition of the patient. Also, classification rules were derived as a means to provide human interpretable representations of the black-box predictive models. Other researchers used rule extraction techniques to 1) provide decision-sets (nonhierarchical if-then rules) with use case on several diseases diagnosis69; 2) automatically and manually prune association rules for explanations of type 2 diabetes risk prediction36; 3) classify micro-arrays,30 predict mortality in ICUs,43 and classify diabetes44 by fuzzy rule extraction; 4) diagnose Alzheimer’s disease73 by adding a rejection option (on hard-to-classify samples); 5) diagnose diabetes mellitus26; and 6) stratify patients with cardiovascular disease risk.65
Intrinsically interpretable models
Besides common interpretability enhancement techniques described earlier, many researchers have taken a different strategy to provide interpretability to their predictive models. These approaches mainly rely on preserving the interpretability of less complex ML methods while enhancing their performance by boosting and optimization techniques. Researchers implemented 1) distributed logistic regression framework to enhance the accuracy of logistic regression dealing with large data with application to daily in-hospital mortality prediction during the patient stay,24 2) automated pruning of decision trees for multiple disease classification,35 3) ensembles of regularized linear SVMs for gene expressions,37 4) logic optimization for binary input to continuous output to infer logic models for drug response in cell lines,67 5) accurate decision trees based on boosting for stratification of patients into subpopulations,39 6) clusters of base learners (LR, linear perceptron, Cox regression) to create a tree of classifiers with application to mortality prediction after cardiac transplant,66 7) alternating clustering and classification optimization using sparse linear SVM framework for hospitalization prediction due to heart disease and diabetes,34 8) ant colony optimization of combining Bayesian classifiers with application to heart diseases and cardiotocography-based fatal pathologies prediction,42 and 9) parameter and threshold optimization of DT, LR, and ridge LR for medium-chain acyl-CoA dehydrogenase deficiency classification.23
Reproducibility assessment
Many research fields, including AI, have been struggling with a reproducibility crisis over the past decade.84 A survey of 400 algorithms presented in the 2 top AI conferences shows that only 6% of the presenters have shared their implementation code, around 30% shared data, and only 50% shared “pseudocode” with the public.85 Another recent study emphasizes the importance of reproducibility in AIM research to ensure safety and effectiveness.86
We believe the reproducibility in this field deserves more attention. In general, ∼43% (18/42) of the articles did not explicitly (ie, mentioned in the manuscript or in the supplementary material) make their datasets accessible to the public; in ∼57% (24/42) of the studies, source codes were not made publicly available; and ∼31% (13/42) did not meet both criteria (see Supplementary Appendix Table 2). Interested audiences should refer to the original work for more details about their reproducibility. Nevertheless, more in-depth analyses are required to rigorously evaluate the reproducibility of the articles, which is out of the scope of this review paper.
Interpretability evaluation
We observed that more than ∼26% (11/42) of the articles did not explicitly report any evaluation of the XAI method they used, ∼28% (12/42) have either referred to the common medical knowledge and medical literature or compared the results with hypotheses, and only ∼7% (3/42) reported human expert confirmation of the results. The rest of the papers took different strategies for the XAI method evaluation, especially regarding effectiveness measurement. Barakat et al,26 for instance, measured the fidelity of the rules they derived from the model against the original model as a way of measuring the effectiveness of the proposed interpretability method. They also measured “comprehensibility” which they define as the number of rules. Ponce and Martinez-Villaseñor72 compared different ML methods for breast cancer classification, reporting accuracy percentage and interpretability level (low, medium, high). However, the logic behind this categorization is not clear.
Lakkaraju et al69 defined several metrics for evaluation including 1) fraction overlap (the extent of overlap between every pair of rules of a decision set); 2) fraction uncovered (the fraction of data points not covered by any rule); 3) average rule length (average number of predicates a human needs to parse in a decision set); 4) number of rules in a decision set; and 5) fraction of classes (the fraction of class labels predicted by at least 1 rule). To qualitatively analyze their XAI method, Kwon et al51 verified whether the medical codes that were highly predictive of heart failure in their model are supported by general medical knowledge. Both of these studies51,69 performed a user study to evaluate different aspects of interpretability enhancement in their proposed approach. Kwon et al51 concluded that AIM applications should incorporate more human interactions with the system.
DISCUSSION AND CONCLUSIONS
Medical professionals’ perspectives of XAI for medicine
We studied the articles included in this systematic scoping review from the lenses of 2 medical professionals (co-authors PRM and JC), aiming to highlight the 1) general opportunities and challenges in XAI for medicine and 2) examples of specific pros and cons regarding each XAI category from the end-users' perspective rather than from the XAI researchers’ perspective. We summarized our findings in Table 4. Nevertheless, more extensive studies are required to systematically collect feedback from medical professionals (eg, Diprose et al88) and analyze potential pros and cons, opportunities, and challenges from their perspective. Such a study can assist to identify the gaps between XAI researchers’ and end-users' needs in real-world scenarios.
Table 4.
Potential pros and cons of explainable artificial intelligence (XAI) categories from the medical professional’s point of view
| XAI Category | Pros | Cons |
|---|---|---|
| Feature interaction and importance | Illustrates not only important features, but also their relative importance toward clinical interpretation | Numerical weights are often not easily interpretable, or might be misinterpreted |
| Attention mechanism | Does not directly inform the clinical end user of the answer but does highlight the areas of most interest to support easier decision-making. Thus, user might be more tolerant of imperfect accuracy | Simply providing this information to a clinical end user might not be useful. Major issues are information overload, alert fatigue, etc. Providing areas of attention without clarity on what to do with the results can potentially be even more confusing if the end user is unsure of what to make of a highlighted section (and also likely to miss nonhighlighted areas that are sometimes crucial) |
| Data dimensionality reduction | Simplifying the data down to a small subset can make the model’s underlying behavior comprehensible. It also can be generally advantageous with potentially more robust regularized models that are less likely to overfit training data | Risk of missing other features that can still be important in individual cases, but the reduced models inadvertently do not include them |
| Knowledge distillation and rule extraction | Potentially more robust models with summarized representations of complex data that allows clinical end users to naturally infer meaning from87 | If clinical end users cannot intuitively interpret the meaning of these representations, then the representations are likely to make it even harder for the end users to interpret and explain |
| Intrinsically interpretable models | Simple models that are more familiar and intuitive to clinical end users. Even if they don't understand how these types of models are constructed, many medical professionals will at least have some familiarity with how to apply them | If ensemble of simple models is used to enhance the accuracy, then a clinical end user is not able to interpret the results |
Challenges: 1) Not all visualizations are interpretable by medical professionals. In other words, visualization does not necessarily provide better interpretability. 2) There is a need to incorporate more longitudinal features in XAI (as opposed to just using aggregated values of a lab feature in a period) to improve the robustness of the models. 3) The absence of a definition for sufficient explainability, and how it can vary substantially in different use cases, is an ongoing issue. 4) Including more features may help improve model accuracy. However, this may also result in overfitting that is not robust to variations and, thus, less usable and trustworthy for medical professionals. 5) Predictive analytics on uncommon diseases might reveal some causations that are not known now and can be used to prevent extensive and expensive workups.
Opportunities identified by these medical professionals are: 1) more transparent predictive models for major diseases (eg, diabetes, cancer) that are the reason for extensive pathologies in the field of preventive medicine, 2) more emphasis on studying uncommon diseases for possible etiologies in predictive analytics to prevent extensive and expensive workups, 3) rerouting healthcare funds to outpatient care and implementing preventive strategies using explainable hospital readmission prediction due to chronic diseases, 4) educating new generations of medical professionals with basic AI knowledge to overcome the gap between AI systems and medical professionals, as an ultimate goal of XAI, 5) using XAI to assist medical professionals overcome their medical knowledge biases and become more objective, 6) enforcing more regulations to ensure AI methods are evaluated rigorously, reproducible, and accompanied by clear circumstances under which the methods are applicable, and 7) more focus on integrating causal inference with AI to provide explanations.
Potential gap in the perspectives of designers and medical professionals in XAI for medicine
We observed that knowledge distillation and rule extraction is the most popular approach, followed by intrinsically interpretable models. XAI can assist designers to debug model development and do sanity checks for spurious associations. Medical professionals, on the other hand, may not require specific explanations of predictions and recommendations if they have been empirically validated through other mechanisms (eg, randomized clinical trials). However, being informed of the features/elements of a prediction model that are important for risk assessment can itself be instructive to medical professionals.
XAI can provide transparency to the prediction models that are built on a cohort that excluded certain types of patients (eg, pregnant patients), thus, help medical professionals understand when it may be unfair to directly apply the XAI methods to individual patients. A closer look at the scope of the approaches proposed in different XAI method categories reveals that the majority of current approaches focus on the global scope. While valuable, more methods for local explanation need to be explored. Medical professionals work with individual patients more often and need specific explanations tailored to each patient’s situation to assess how the XAI results do, or do not, apply to individual patient contexts.
If we look further at each XAI method category, we can see that the attention mechanism (100%) and feature interaction and importance (60%) are the top approaches supporting local explanations. This trend is followed by knowledge distillation and rule extraction (∼38%), intrinsically interpretable models (∼22% local), and data dimensionality reduction (12.5%) focus on the local explanations, while they appear to be quite popular for XAI in medicine. This can represent a potential gap between theory and practice.
Potential limitations of XAI methods
Despite the valuable effort in providing interpretability for black-box models such as neural networks, researchers call for more caution and evaluation while applying these methods. Here we provide some examples of these concerns. Ghorbani et al89 were able to compute small adversarial perturbations (in a similar way to computing adversarial examples to neural networks90) that cause substantial changes to feature importance maps of several interpretation methods. However, such adversarial perturbations in the case of real-world datasets such as EHR and their impact on XAI methods for EHR-based models need to be studied further.
Sokol and Flach91 emphasize the importance of deriving more meaningful concepts when using an XAI method by personalizing the explanations through user’s input. Miller et al92 also note that people maintain mental models of each other to tailor explanations to individuals. This has not been the focus of much XAI research. From another point of view,93 using attention mechanisms does not necessarily provide more transparency to the black-box model. Based on their experiments, the relationship between the attention weights and model output is unclear. Another similar study94 argued that whether or not attention is explanation depends on the definition of explanation. However, both studies confirmed that researchers should be cautious when using attention distributions for explanations.
Rudin95 refers to post hoc XAI methods as “problematic” in the case of high-risk decision-making. From her point of view, the way forward is to create inherently interpretable models rather than creating methods that explain black-box models. She also calls the accuracy and interpretability trade-off a myth. However, from our perspective, we argue that such a statement might not always be valid in medical predictive modeling using EHR data. Further, an inherently interpretable model does not give an explainable decision. A large decision tree may still require explanation to a nontechnical user; and concepts such as contrastive and counterfactual explanations are independent of the interpretability of the model. Nevertheless, XAI and, more specifically, XAI for medicine, is a relatively new topic that is still in its initial stage of formation. Thus, diverse points of view and approaches to addressing existing limitations and shortcomings of different XAI methods should be welcomed.
XAI evaluation issue
Few researchers have considered XAI evaluation in their work. There is no consensus on the definition of interpretability yet; thus, there is no agreed-upon approach to evaluate the results of the XAI methods. We argue that making effective XAI methods for medicine requires more interdisciplinary collaboration between different professionals such as AI researchers and medical professionals. Other researchers also have emphasized the importance of including human expertise in the explanation process.92 The most recent report on XAI program of DARPA96 explicitly mentioned that “an XAI system’s explanation effectiveness must be assessed according to how its explanations aid human users. This requires human-in-the-loop psychologic experiments to measure the user’s satisfaction, mental model, task performance, and appropriate trust.”
Reproducibility issue
Based on the results of this systematic review, there is not enough emphasis on the reproducibility of the research work published in this field. Considering the issue of XAI evaluation, in addition to the critical nature of AI applications for medical practices, research reproducibility is crucial.86 To evaluate their new ideas, researchers in the field need to compare their work to the previous work done by other researchers. It would be easier and faster to examine and compare ideas if researchers use publicly available datasets, describe how they select specific features, clearly mention the dimensionality of the dataset as well as the infrastructure they use, and provide the source code. Thus, we suggest that publication venues should require authors to meet certain reproducibility criteria before publishing their research work.
FUNDING
This project was partially supported by the National Institute on Aging of the National Institutes of Health (NIH) under award number R21AG061431, the National Cancer Institute under award number R01CA246418, and the University of Florida Clinical and Translational Science Institute, which is supported in part by the NIH National Center for Advancing Translational Sciences under award number UL1TR001427. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
AUTHOR CONTRIBUTIONS
SNP and ZH conceived the main conceptual idea. SNP, ZH, ZC, TM, and JB contributed to research design. SNP and ZC performed the literature search, record screening, and data analysis. PRM and JHC acted as medical professionals and provided medical insights. XL, TM, and JB acted as experts on artificial intelligence, machine learning, and deep learning throughout. SNP wrote the initial draft of the manuscript. All the authors reviewed and edited the paper iteratively for important intellectual content. All the authors approved the version to be published. ZH supervised this research and takes primary responsibility for the research reported here.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Johnson KW, Torres Soto J, Glicksberg BS, et al. Artificial intelligence in cardiology. J Am Coll Cardiol 2018; 71 (23): 2668–79. [DOI] [PubMed] [Google Scholar]
- 2. Krittanawong C, Zhang H, Wang Z, Aydar M, Kitai T.. Artificial intelligence in precision cardiovascular medicine. J Am Coll Cardiol 2017; 69 (21): 2657–64. [DOI] [PubMed] [Google Scholar]
- 3. Stuart R, Peter N. . Artificial Intelligence:A Modern Approach. 3rd ed.Berkeley: Pearson; 2009. [Google Scholar]
- 4. Szolovits P. Artificial Intelligence in Medicine. Abingdon, UK: Routledge; 2019. [Google Scholar]
- 5. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1 (1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choi E, Bahadori MT, Schuetz A, Stewart WF, Sun J. Doctor AI: predicting clinical events via recurrent neural networks. In Machine Learning for Healthcare Conference; December 10, 2016: 301–18. [PMC free article] [PubMed] [Google Scholar]
- 7. Mesko B. The role of artificial intelligence in precision medicine. Exp Rev Precis Med Drug Dev 2017; 2 (5): 239–41. [Google Scholar]
- 8. Jensen PB, Jensen LJ, Brunak S.. Mining electronic health records: towards better research applications and clinical care. Nat Rev Genet 2012; 13 (6): 395–405. [DOI] [PubMed] [Google Scholar]
- 9. Dreyer KJ, Geis JR.. When machines think: radiology’s next frontier. Radiology 2017; 285 (3): 713–8. [DOI] [PubMed] [Google Scholar]
- 10.U.S. Healthcare leaders expect widespread adoption of artificial intelligence by 2023. Intel Newsroom.https://newsroom.intel.com/news-releases/u-s-healthcare-leaders-expect-widespread-adoption-artificial-intelligence-2023/ Accessed October 24, 2019.
- 11. He J, Baxter SL, Xu J, Xu J, Zhou X, Zhang K.. The practical implementation of artificial intelligence technologies in medicine. Nat Med 2019; 25 (1): 30–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gunning D. Explainable artificial intelligence (XAI). Defense Advanced Research Projects Agency (DARPA); 2017: 36. [Google Scholar]
- 13. Kim TW, Routledge BR. Informational privacy, a right to explanation, and interpretable AI. In: proceedings of 2018 IEEE Symposium on Privacy-Aware Computing (PAC); September 26–28, 2018; Washington, DC.
- 14. Wang F, Kaushal R, Khullar D.. Should health care demand interpretable artificial intelligence or accept ‘black box’ medicine. Ann Intern Med 2020; 172 (1): 59. [DOI] [PubMed] [Google Scholar]
- 15. Nundy S, Montgomery T, Wachter RM.. Promoting trust between patients and physicians in the era of artificial intelligence. JAMA 2019; 322 (6): 497–8. [DOI] [PubMed] [Google Scholar]
- 16. Holzinger A, Langs G, Denk H, Zatloukal K, Müller H.. Causability and explainability of artificial intelligence in medicine. Wiley Interdiscip Rev: Data Min Knowl Discov 2019; 9 (4): e1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vellido A. The importance of interpretability and visualization in machine learning for applications in medicine and health care. Neural Comput Appl 2019. doi: 10.1007/s00521-019-04051-w. [Google Scholar]
- 18. Moher D, Liberati A, Tetzlaff J, Altman DG The PRISMA Group. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med 2009; 6 (7): e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu J, Pan Y, Li M, et al. Applications of deep learning to MRI images: a survey. Big Data Min Anal 2018; 1 (1): 1–18. [Google Scholar]
- 20. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL.. Artificial intelligence in radiology. Nat Rev Cancer 2018; 18 (8): 500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iftikhar M, Khan SA, Hassan A. A survey of deep learning and traditional approaches for EEG signal processing and classification. In: 2018 IEEE 9th Annual Information Technology, Electronics and Mobile Communication Conference (IEMCON); November 1–3, 2018; Vancouver, BC, Canada.
- 22. National Vital Statistics Reports Deaths: Final Data for 2017, 2017; 68 (9): 77. https://www.cdc.gov/nchs/data/nvsr/nvsr68/nvsr68_09-508.pdf Accessed November 1, 2019 [PubMed] [Google Scholar]
- 23. Van den Bulcke T, Broucke PV, Hoof VV, et al. Data mining methods for classification of Medium-Chain Acyl-CoA dehydrogenase deficiency (MCADD) using non-derivatized tandem MS neonatal screening data. J Biomed Inform 2011; 44 (2): 319–25. [DOI] [PubMed] [Google Scholar]
- 24. Fejza A, Genevès P, Layaïda N, Bosson JL. Scalable and interpretable predictive models for electronic health records. In: proceedings of 2018 IEEE 5th International Conference on Data Science and Advanced Analytics (DSAA); October 1–3, 2018; Turin, Italy.
- 25. Asfour MG, Lambourne A, Soliman A, et al. High prevalence of diabetes mellitus and impaired glucose tolerance in the Sultanate of Oman: results of the 1991 national survey. Diabet Med 1995; 12 (12): 1122–5. [DOI] [PubMed] [Google Scholar]
- 26. Barakat N, Bradley AP, Barakat MNH.. Intelligible support vector machines for diagnosis of diabetes mellitus. IEEE Trans Inform Technol Biomed 2010; 14 (4): 1114–20. [DOI] [PubMed] [Google Scholar]
- 27. Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999; 286 (5439): 531–7. [DOI] [PubMed] [Google Scholar]
- 28. Singh D, Febbo PG, Ross K, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 2002; 1 (2): 203–9. [DOI] [PubMed] [Google Scholar]
- 29. Alon U, Barkai N, Notterman DA, et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc Natl Acad Sci U S A 1999; 96 (12): 6745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hajiloo M, Rabiee HR, Anooshahpour M.. Fuzzy support vector machine: an efficient rule-based classification technique for microarrays. BMC Bioinform 2013; 14 (S13): S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meij TGJ, Budding AE, Groot EFJ, et al. Composition and stability of intestinal microbiota of healthy children within a Dutch population. FASEB J 2016; 30 (4): 1512–22. [DOI] [PubMed] [Google Scholar]
- 32. Eck A, Zintgraf LM, de Groot EFJ, et al. Interpretation of microbiota-based diagnostics by explaining individual classifier decisions. BMC Bioinform 2017; 18 (1): 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bernardini M, Romeo L, Misericordia P, Frontoni E.. Discovering the type 2 diabetes in electronic health records using the sparse balanced support vector machine. IEEE J Biomed Health Inform 2020; 24 (1): 235–1. [DOI] [PubMed] [Google Scholar]
- 34. Brisimi TS, Xu T, Wang T, Dai W, Adams WG, Paschalidis IC.. Predicting chronic disease hospitalizations from electronic health records: an interpretable classification approach. Proc IEEE 2018; 106 (4): 690–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stiglic G, Kocbek S, Pernek I, Kokol P.. Comprehensive decision tree models in bioinformatics. PLoS ONE 2012; 7 (3): e33812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo G. Automatically explaining machine learning prediction results: a demonstration on type 2 diabetes risk prediction. Health Inf Sci Syst 2016; 4 (1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jalali A, Pfeifer N.. Interpretable per case weighted ensemble method for cancer associations. BMC Genomics 2016; 17 (1): 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan L, Liu G, Mao X, et al. Development of prediction models using machine learning algorithms for girls with suspected central precocious puberty: retrospective study. JMIR Med Inform 2019; 7 (1): e11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Valdes G, Luna JM, Eaton E, Simone CB, Ungar LH, Solberg TD.. MediBoost: a patient stratification tool for interpretable decision making in the era of precision medicine. Sci Rep 2016; 6 (1): 37854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Crielaard L, Papapetrou P. Explainable predictions of adverse drug events from electronic health records via oracle coaching. In: 2018 IEEE International Conference on Data Mining Workshops (ICDMW); November 17–20, 2018: 707–14; Singapore.
- 41. Zhang W, Liu F, Luo L, Zhang J.. Predicting drug side effects by multi-label learning and ensemble learning. BMC Bioinform 2015; 16 (1): 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bouktif S, Hanna EM, Zaki N, Khousa EA.. Ant colony optimization algorithm for interpretable Bayesian classifiers combination: application to medical predictions. PLoS ONE 2014; 9 (2): e86456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Davoodi R, Moradi MH.. Mortality prediction in intensive care units (ICUs) using a deep rule-based fuzzy classifier. J Biomed Inform 2018; 79: 48–59. [DOI] [PubMed] [Google Scholar]
- 44. Settouti N, Chikh MA, Saidi M.. Generating fuzzy rules for constructing interpretable classifier of diabetes disease. Australas Phys Eng Sci Med 2012; 35 (3): 257–70. [DOI] [PubMed] [Google Scholar]
- 45. Degroeve S, Saeys Y, De Baets B, Rouzé P, Van de Peer Y.. SpliceMachine: predicting splice sites from high-dimensional local context representations. Bioinformatics 2005; 21 (8): 1332–8. [DOI] [PubMed] [Google Scholar]
- 46. Sonnenburg S, Schweikert G, Philips P, Behr J, Rätsch G.. Accurate splice site prediction using support vector machines. BMC Bioinform 2007; 8 (Suppl 10): S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bari G,, Reaz MR, Jeong B-S. Effective DNA encoding for splice site prediction using SVM; 2014.
- 48. Zuallaert J, Godin F, Kim M, Soete A, Saeys Y, De Neve W.. SpliceRover: interpretable convolutional neural networks for improved splice site prediction. Bioinformatics 2018; 34 (24): 4180–8. [DOI] [PubMed] [Google Scholar]
- 49. Xiao C, Ma T, Dieng AB, Blei DM, Wang F.. Readmission prediction via deep contextual embedding of clinical concepts. Plos ONE 2018; 13 (4): e0195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shickel B, Loftus TJ, Adhikari L, Ozrazgat-Baslanti T, Bihorac A, Rashidi P.. DeepSOFA: a continuous acuity score for critically ill patients using clinically interpretable deep learning. Sci Rep 2019; 9 (1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kwon BC, Choi M-J, Kim JT, et al. RetainVis: visual analytics with interpretable and interactive recurrent neural networks on electronic medical records. IEEE Trans Vis Comput Graph 2019; 25 (1): 299–309. [DOI] [PubMed] [Google Scholar]
- 52. Kim SG, Theera-Ampornpunt N, Fang C-H, Harwani M, Grama A, Chaterji S.. Opening up the blackbox: an interpretable deep neural network-based classifier for cell-type specific enhancer predictions. BMC Syst Biol 2016; 10 (S2): 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Che Z, Purushotham S, Khemani R, Liu Y.. Interpretable deep models for ICU outcome prediction. AMIA Annu Symp Proc 2017; 2016: 371–80. [PMC free article] [PubMed] [Google Scholar]
- 54. Ge W, Huh J-W, Park YR, Lee J-H, Kim Y-H, Turchin A.. An interpretable ICU mortality prediction model based on logistic regression and recurrent neural networks with LSTM units. AMIA Annu Symp Proc 2018; 2018: 460–9. [PMC free article] [PubMed] [Google Scholar]
- 55. Ghafouri-Fard S, Taheri M, Omrani MD, Daaee A, Mohammad-Rahimi H, Kazazi H.. Application of single-nucleotide polymorphisms in the diagnosis of autism spectrum disorders: a preliminary study with artificial neural networks. J Mol Neurosci 2019; 68 (4): 515–21. [DOI] [PubMed] [Google Scholar]
- 56. Hao J, Kim Y, Kim T-K, Kang M.. PASNet: pathway-associated sparse deep neural network for prognosis prediction from high-throughput data. BMC Bioinform 2018; 19 (1): 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hartono P. A transparent cancer classifier. Health Inform J doi:10.1177/1460458218817800. [DOI] [PubMed]
- 58. Hu H, Xiao A, Zhang S, et al. DeepHINT: understanding HIV-1 integration via deep learning with attention. Bioinformatics 2019; 35 (10): 1660–7. [DOI] [PubMed] [Google Scholar]
- 59. Kaji DA, Zech JR, Kim JS, et al. An attention based deep learning model of clinical events in the intensive care unit. PLoS ONE 2019; 14 (2): e0211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Choi E, Bahadori MT, Sun J, Kulas J, Schuetz A, Stewart W.. Retain: an interpretable predictive model for healthcare using reverse time attention mechanism. In: Presented at the Advances in Neural Information Processing Systems; 2016: 3504–12. [Google Scholar]
- 61. Park S, Kim YJ, Kim JW, Park JJ, Ryu B, Ha JW. Interpretable prediction of vascular diseases from electronic health records via deep attention networks. In: 2018 IEEE 18th International Conference on Bioinformatics and Bioengineering (BIBE); October 29–31, 2018; Taichung, Taiwan.
- 62. Zhang J, Kowsari K, Harrison JH, Lobo JM, Barnes LE.. Patient2Vec: a personalized interpretable deep representation of the longitudinal electronic health record. IEEE Access 2018; 6: 65333–46. [Google Scholar]
- 63. Aditya CR, Pande MBS.. Devising an interpretable calibrated scale to quantitatively assess the dementia stage of subjects with Alzheimer’s disease: a machine learning approach. Inform Med Unlocked 2017; 6: 28–35. [Google Scholar]
- 64. Zhao LP, Bolouri H.. Object-oriented regression for building predictive models with high dimensional omics data from translational studies. J Biomed Inform 2016; 60: 431–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Paredes S, Henriques J, Rochar T, et al. A clinical interpretable approach applied to cardiovascular risk assessment. In: 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC); July 18–21, 2018; Honolulu, HI. [DOI] [PubMed]
- 66. Yoon J, Zame WR, Banerjee A, Cadeiras M, Alaa AM, van der Schaar M.. Personalized survival predictions via trees of predictors: an application to cardiac transplantation. PLoS ONE 2018; 13 (3): e0194985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Knijnenburg TA, Klau GW, Iorio F, et al. Logic models to predict continuous outputs based on binary inputs with an application to personalized cancer therapy. Sci Rep 2016; 6 (1): 36812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ming Y, Qu H, Bertini E.. RuleMatrix: visualizing and understanding classifiers with rules. IEEE Trans Vis Comput Graph 2019; 25 (1): 342–52. [DOI] [PubMed] [Google Scholar]
- 69. Lakkaraju H,, Bach SH, Leskovec J. Interpretable decision sets: a joint framework for description and prediction. In: proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining-KDD ‘16, San Francisco, California, USA; August 13–17, 2016: 1675–84. [DOI] [PMC free article] [PubMed]
- 70. Armstrong SA, Staunton JE, Silverman LB, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 2002; 30 (1): 41–7. [DOI] [PubMed] [Google Scholar]
- 71. Huang L-T. An integrated method for cancer classification and rule extraction from microarray data. J Biomed Sci 2009; 16 (1): 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ponce H, de Lourdes Martinez-Villaseñor M. Interpretability of artificial hydrocarbon networks for breast cancer classification. in 2017 International Joint Conference on Neural Networks (IJCNN).
- 73. Das D, Ito J, Kadowaki T, Tsuda K.. An interpretable machine learning model for diagnosis of Alzheimer’s disease. PeerJ 2019; 7: e6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Du M, Liu N, Hu X.. Techniques for interpretable machine learning. Commun ACM 2019; 63 (1): 68–77. 10.1145/3359786 [DOI] [Google Scholar]
- 75. Carvalho DV, Pereira EM, Cardoso JS.. Machine learning interpretability: a survey on methods and metrics. Electronics 2019; 8 (8): 832. [Google Scholar]
- 76. Adadi A, Berrada M.. Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 2018; 6: 52138–60. [Google Scholar]
- 77. Ribeiro MT, Singh S, Guestrin C. Model-agnostic interpretability of machine learning. arXiv:1606.05386 [cs, stat]; 2016. http://arxiv.org/abs/1606.05386 Accessed December 30, 2019
- 78. Bahdanau D, Cho K, Bengio Y. Neural machine translation by jointly learning to align and translate. arXiv: 1409.0473 [cs, stat]; 2014. http://arxiv.org/abs/1409.0473 Accessed October 03, 2019
- 79. Shrikumar A, Greenside P, Kundaje A. Learning important features through propagating activation differences. In: proceedings of the 34th International Conference on Machine Learning—Volume 70; August 6–11, 2017; Sydney, NSW, Australia. http://dl.acm.org/citation.cfm? id=3305890.3306006 Accessed October 06, 2019 [Google Scholar]
- 80. Montavon G, Samek W, Müller K-R.. Methods for interpreting and understanding deep neural networks. Digit Signal Process 2018; 73: 1–15. [Google Scholar]
- 81. Mascharka D,, Tran P,, Soklaski R, Majumdar A. Transparency by design: closing the gap between performance and interpretability in visual reasoning. In: presented at the Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. http://openaccess.thecvf.com/content_cvpr_2018/html/Mascharka_Transparency_by_Design_CVPR_2018_paper.html Accessed January 01, 2020
- 82. Tibshirani R. Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B 1996; 58 (1): 267–88. [Google Scholar]
- 83. Hinton G,, Vinyals O, Dean J. Distilling the knowledge in a neural network. arXiv:1503.02531 [cs, stat]; 2015. http://arxiv.org/abs/1503.02531 Accessed October 07, 2019
- 84. Hutson M. Artificial intelligence faces reproducibility crisis. Science 2018; 359 (6377): 725–6. [DOI] [PubMed] [Google Scholar]
- 85. Gundersen OE, Gil Y, Aha DW.. On reproducible AI: towards reproducible research, open science, and digital scholarship in AI publications. AI Mag 2018; 39 (3): 56–68. [Google Scholar]
- 86. Beam AL, Manrai AK, Ghassemi M.. Challenges to the reproducibility of machine learning models in health care. JAMA 2020; 323 (4): 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen JH, Goldstein MK, Asch SM, Mackey L, Altman RB.. Predicting inpatient clinical order patterns with probabilistic topic models vs conventional order sets. J Am Med Inform Assoc 2017; 24 (3): 472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Diprose WK, Buist N, Hua N, Thurier Q, Shand G, Robinson R.. Physician understanding, explainability, and trust in a hypothetical machine learning risk calculator. J Am Med Inform Assoc 2020; 27 (4): 592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ghorbani A, Abid A, Zou J.. Interpretation of neural networks is fragile. AAAI 2019; 33 (01): 3681–8. [Google Scholar]
- 90. Szegedy C, Zaremba W, Sutskever I, et al. Intriguing properties of neural networks, arXiv:1312.6199 [cs], FeCb; 2014. http://arxiv.org/abs/1312.6199 Accessed March 27, 2020
- 91. Sokol K, Flach P.. One explanation does not fit all. Künstl Intell 2020. doi:10.1007/s13218-020-00637-y. [Google Scholar]
- 92. Miller T,, Howe P, Sonenberg L. Explainable AI: beware of inmates running the asylum or: how I learnt to stop worrying and love the social and behavioural sciences. arXiv:1712.00547 [cs]; 2017. http://arxiv.org/abs/1712.00547 Accessed December 09, 2019
- 93. Jain S, Wallace BC. Attention is not explanation. arXiv:1902.10186 [cs]; 2019. http://arxiv.org/abs/1902.10186 Accessed March 19, 2020
- 94. Wiegreffe S, Pinter Y. Attention is not not explanation. In: proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP); November 3–7, 2019; Hong Kong. https://www.aclweb.org/anthology/D19-1002/
- 95. Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell 2019; 1 (5): 206–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Gunning D, Aha D.. DARPA’s Explainable Artificial Intelligence (XAI) Program. AIMag 2019; 40 (2): 44–58. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.