Abstract
Objective
We introduce fold-stratified cross-validation, a validation methodology that is compatible with privacy-preserving federated learning and that prevents data leakage caused by duplicates of electronic health records (EHRs).
Materials and Methods
Fold-stratified cross-validation complements cross-validation with an initial stratification of EHRs in folds containing patients with similar characteristics, thus ensuring that duplicates of a record are jointly present either in training or in validation folds. Monte Carlo simulations are performed to investigate the properties of fold-stratified cross-validation in the case of a model data analysis using both synthetic data and MIMIC-III (Medical Information Mart for Intensive Care-III) medical records.
Results
In situations in which duplicated EHRs could induce overoptimistic estimations of accuracy, applying fold-stratified cross-validation prevented this bias, while not requiring full deduplication. However, a pessimistic bias might appear if the covariate used for the stratification was strongly associated with the outcome.
Discussion
Although fold-stratified cross-validation presents low computational overhead, to be efficient it requires the preliminary identification of a covariate that is both shared by duplicated records and weakly associated with the outcome. When available, the hash of a personal identifier or a patient’s date of birth provides such a covariate. On the contrary, pseudonymization interferes with fold-stratified cross-validation, as it may break the equality of the stratifying covariate among duplicates.
Conclusion
Fold-stratified cross-validation is an easy-to-implement methodology that prevents data leakage when a model is trained on distributed EHRs that contain duplicates, while preserving privacy.
Keywords: federated learning, privacy, validation, duplicated electronic health records, data leakage
INTRODUCTION
The large-scale collection of data and its analysis by artificial intelligence (AI) algorithms have led to new scientific discoveries and huge expectations for the near future.1–5 Although AI algorithms (eg, random forest, gradient boosting, neural networks)6 provide powerful tools, they are difficult to develop, as they generally require large training datasets to reach reasonable performances,7 and often detailed, high-dimensional records about individuals. Beyond the technical challenges that such a data collection represents, storing and analyzing a large amount of personally identifying information (PII) may moreover imply serious risks regarding privacy, and recent research projects have been hindered by public opinion concerns.8,9 These concerns are not unfounded, as reidentification attacks have regularly broken the anonymity of large datasets.10–13 To address these risks, new regulations that impose higher security standards have been introduced.14 Technically, it moreover appears necessary to complement classical anonymization techniques as they are intrinsically limited in the case of high-dimensional data.15,16 Handling and analyzing securely such data therefore requires moving from the anonymize, release, and forget approach to configurations in which a data curator secures and controls the use of data that remain to some extent identifying.17 In the latter configuration, the question arises as to which organization should play the role of the trusted data curator. In the case of medical records, patients and information technology managers have been reluctant to devote this role to centralized private or public organizations,8,9,18 limiting large-scale research on electronic health records (EHRs). To solve this issue, a technique called federated learning has recently been proposed. This technique enables the training of AI models while keeping records in decentralized trusted data warehouses curated for instance by hospitals.19–25 Federated learning appears as a promising privacy-enhancing technique that avoids single points of failure, and it is currently being developed and tested in various projects worldwide.26–30
In addition to privacy concerns, recent controversies indicate that many AI models may have been validated improperly, shedding doubt on the performances that have been advertised.31–35 One of the most frequent sources of bias in performance estimation is the data leakage that occurs when data used for validation and training are correlated.36–39 Avoiding data leakage requires building training and validation datasets in such a way that all the data related to a given individual are exclusively in the former or the latter. This dataset building procedure may be compromised by the presence of different records related to the same individual: this risk is far from being negligible, as it has been shown that up to 15% of records in medical information systems are duplicates,40 and that many patients have records in multiple hospitals.41 Data leakage induced by duplicates may be especially important in the case of AI models, as they are often trained on large real-world datasets such as EHRs that have not been curated for research.4 To address data leakage caused by duplicates, deduplication algorithms, often called record linkage algorithms, have been developed that rely on various deterministic or probabilistic methods.42
Although federated learning and deduplication algorithms address privacy and validation issues, respectively, they cannot be easily combined. Indeed, deduplication relies on the comparison of PII through the computation of a similarity index established between 2 potentially duplicated records, whereas federated learning avoids PII exchange and therefore prevents their comparison when records are located in different data warehouses. Consequently, detecting duplicates of a given record that are present in 2 different hospitals while preserving their privacy appears challenging. Some solutions to this problem have already been proposed.43 A first solution consists of sharing hashes of identifiers among centers to detect duplicates.44 This technique limits the risk of reidentification but can only be used when a shared personal identifier is available. EHRs are often not tagged by such an identifier, limiting the datasets that can be deduplicated by this technique. Moreover, comparison of hashes is vulnerable to some reidentification attacks.43 Another set of solutions consists of cryptographic schemes referred to as secure multiparty computation (SMC). These schemes enable computing in each center the list of its records that are present in at least 1 other center while transferring only encrypted data. SMC has been proven to be secure and scalable but is also limited to the detection of exact matches between 2 identifiers.45,46 It is not straightforward to extend SMC to approximate matching between records that are note tagged by a shared identifier, and additional researches in that direction are necessary.46 In this article, we propose an approach that makes it possible to avoid data leakage in a federated learning setting while not relying on the availability of a shared identifier. Our method is moreover easy to implement and does not rely on cryptographic schemes. We consider the classical cross-validation technique for performance estimation, and complement it by an initial stratification of datasets. Whereas some stratification techniques have already been combined with cross-validation in order to limit disparities between randomly chosen folds,47 we extend the use of stratification to avoid data leakage in the case of undetected data duplicates.
The article is organized as follows. In the Materials and Methods, we describe the fold-stratified cross-validation methodology we propose and the simulation methodology we follow to assess its validity on both synthetic and real-world MIMIC-III (Medical Information Mart for Intensive Care-III) records. In the Results, we simulate a data analysis in a federated learning setting following different validation strategies, the performances and limitations of which are detailed in Discussion section.
MATERIALS AND METHODS
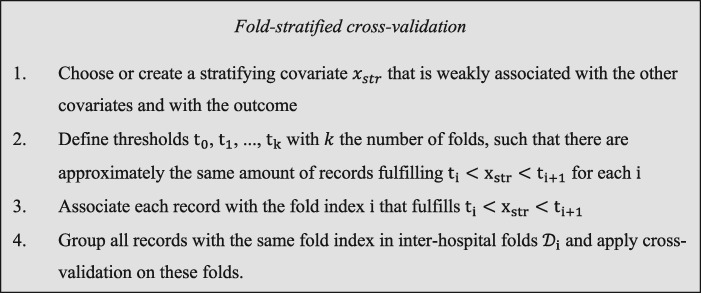
Fold-stratified cross-validation
We consider a model that computes a predicted of an outcome using covariates : . We consider a performance metric that we want to maximize and that is computed as the expectation of a function . In the case of accuracy , where the function equals 1 if is true and 0 otherwise. A record is a point in a mathematical space that gathers for an individual her covariates and her realized outcome , and a dataset is a collection of records. A dataset contains duplicates when there are 2 records and with such that . For the sake of simplicity, we limit ourselves to exact duplicates but as discussed later our conclusions apply also to inaccurate duplicates, caused for instance by flawed or incomplete recording of data related to a given individual. We consider hereafter that there are never 2 individuals with perfectly identical records, implying that equal records always relate to the same individual that has been registered more than once. We moreover consider that individuals whose records are duplicated are distributed uniformly in the population.
Cross-validation is a statistical technique commonly used to estimate the performances of a model.6 In cross-validation a dataset is partitioned in folds (disjoint subsets) , with . For each fold the statistical model is trained on all the folds apart from fold (training), and its performances are estimated on fold (validation). The average of per-fold estimates is used as the estimate of the performances of the same model that would have been trained on all the records. Classical cross-validation often relies on a random partitioning of the dataset in folds. In that case, duplicated records may be simultaneously present in training and validation folds thus inducing data leakage and yielding overoptimistic estimates of performance compared with cross-validation without duplicates, that we consider hereafter as the unbiased estimate.
We consider training and validation of a model in a federated learning setting in which records are distributed in different hospitals. In that case, many datasets related to different hospitals are jointly analyzed by an external data scientist without exchanging PII (see Figure 1). Cross-validation in a federated learning setting involves partitioning the dataset of each hospital in folds with . Folds are then merged over hospitals to obtain global folds: . To better characterize the presence of duplicates in a federated learning setting we introduce the following definitions:
Figure 1.
Privacy-preserving federated learning: analysis by a data scientist of medical records (blue and red individuals) distributed in 2 hospitals without extracting personally identifying information. One individual’s record is duplicated in the 2 hospitals (red) (ID4), due for instance to multiple admissions. The performances of a model are estimated through cross-validation, partitioning the datasets in training and validation folds either randomly (left) or through stratification (ie, grouping similar patients in folds) (right). Whereas duplicated records (red) may be simultaneously in training and validation folds when random partitioning is applied, thus causing data leakage, this risk is circumvented by stratification.
Definition 1: Intrahospital deduplication: For all records in a hospital with we have .
Definition 2: Interhospital deduplication: For all records , in different hospitals we have .
Datasets that jointly fulfill definitions 1 and 2 are completely deduplicated (ie, there are no 2 identical records with in the whole dataset). As explained previously, although many deduplication techniques have been developed to fulfill definition 1, fulfilling definition 2 remains challenging without losing the privacy-enhancing advantage of federated learning. We therefore consider a weaker definition of deduplication that is sufficient to avoid data leakage between folds:
Definition 3: Interfold deduplication: For all records , related to different fold indexes we have .
Definition 3 is weaker than definitions 1 and 2 as it can be fulfilled without removing all duplicates if one ensures instead that duplicates of a given record are present in the same fold. We propose hereafter a technique to create folds that fulfill definition 3. We consider a partition of the record space in subspaces : and if . Such a partition can be realized stratifying relatively to 1 covariate, and we refer to such a partition as a stratification. Once a stratification has been defined, each hospital dataset can be partitioned in folds as follows:
| (1) |
Partitioning each hospital dataset using a given stratification leads for to: , and definition 3 is therefore fulfilled. Combining stratification technique equation 1 with classical cross-validation constitutes the validation methodology that we call fold-stratified cross-validation.
Although fold-stratified cross-validation prevents overoptimistic estimates of performance induced by duplicates, it does not systematically provide an unbiased estimator compared with cross-validation in the absence of duplicates. A stratification procedure may indeed induce training and validation folds featuring different covariate distributions and covariate-outcome associations. A model trained and validated on such folds tends to overfit the training population and to be unfit for a generalization to the validation population, and fold-stratified cross-validation appears closer to the external validation on a new population. Validating externally a model on a population coming from a different hospital is commonly recognized as a proof of quality, as it measures the generalizability of a model to new care contexts, but fold-stratified cross-validation unfortunately does not measure this relevant interhospital generalizability, as folds cannot be identified with hospitals. The stratification procedure should therefore be defined as to minimize the irrelevant pessimistic bias associated with the heterogeneity of folds populations. An ideal stratifying covariate would therefore be a covariate shared by duplicates but independent of the other covariates and of the outcome, as it would provide folds that would be statistically equivalent (Figure 2). Such a stratifying covariate is often not available since covariates extracted from EHRs are associated with a patient’s medical condition to some extent. A challenge of fold-stratified cross-validation consists in finding a surrogate stratifying covariate that is weakly correlated to the other covariates and to the outcome. In the following section, we run simulations to investigate and discuss the impact of various stratification strategies.
Figure 2.
Description of fold-stratified cross-validation.
Simulation
In order to study the properties of fold-stratified cross-validation, we simulate data analysis in a federated learning setting in presence of duplicates. We generate synthetic datasets in which a binary outcome depends on 10 covariates . Covariates are generated randomly following a multivariate gaussian distribution:
| (2) |
with the covariates means and the covariance matrix. To generate , we choose 10 eigenvalues , and sample a random orthogonal matrix of size . An intermediate covariance matrix is obtained through:
| (3) |
The final covariance matrix is then generated concatenating with in a block-diagonal matrix:
| (4) |
Generating covariates according to equations 2-4 provides a set of 9 correlated covariates and an independent covariate . Once covariates have been generated, we randomly generate their associated outcomes through a logistic model. We consider a situation where the logarithm of the odds is a strongly nonlinear function of the covariates:
| (5) |
with a set of constants. Covariates , , …, are associated with the outcome contrary to that remains independent of all other variables. The strongly nonlinear case given by equation 5 corresponds to a generic situation with complex interactions that cannot be accounted for by simple generalized linear models.
Each simulation consists in generating randomly records and then in adding randomly duplicates (17% of the total number of records). Each of the original records is drawn from equations 2 and 5, and is then attributed randomly to 1 of the hospitals with uniform probability. To generate duplicates, we randomly draw 1 of the original records and 1 of the hospitals. We then add a duplicate of the drawn record to the drawn hospital unless a duplicate of the original record already exists in the hospital that has been drawn, thus ensuring that definition 1 is fulfilled. We repeat this procedure until duplicated records have been added.
Unless stated otherwise, we consider centered covariates generated through equation 2 using eigenvalues . Outcomes are generated using equation 5 with parameters , leading to of records associated to a positive outcome . Cross-validation is realized with folds. The code used for the simulations is available in the Supplementary Appendix.
We complement the model analysis of synthetic datasets by an analysis of real-world medical records included in the MIMIC-III dataset.48 Each record corresponds to the admission of a patient in an intensive care unit (ICU). We focus on admissions either in the medical ICU or in the surgical ICU. When multiple records are related to a given patient, to avoid data leakage we keep at random only 1 of these records, leading to 15 446 and 7232 records in the medical and surgical ICUs, respectively. We duplicate at random 4644 records, thus mimicking data duplication that may be caused by imperfectly traced transfers of patients between 2 ICUs. For each record, we compute the 15 explanatory variables used in the SAPS-II (Simplified Acute Physiology Score-II),49 and use them as the training covariates. As stratifying covariate we use (1) the age at admission, (2) the weight at admission, (3) the lowest creatinine value from the first 24 hours after admission, (4) the lowest blood urea nitrogen value from the first 24 hours after admission, or (5) the highest hemoglobin value from the first 24 hours after admission.
Model definition and dataset partitioning strategies
High-dimensional nonlinear problems on tabular data are commonly modeled using gradient boosting, random forests, or support vector machines.6 We focus hereafter on gradient boosting and we use its implementation in the XGBoost library.50 We consider trees of depth 3 that are added successively during 200 boosting iterations. The learning parameter is set to 0.6 and we use the binary logistic loss function. Although it is currently not possible to apply directly XGBoost in a federated learning setting, protocols are being developed to circumvent this difficulty.51,52 These computational considerations are not related to the data leakage issue under consideration, and for the sake of simplicity, in our simulations XGBoost is applied on physically centralized datasets, simulating gradient boosting in a federated learning setting.
For each analysis, we start with a deduplicated dataset on which we fit and validate a gradient boosting model using the classical cross-validation methodology. We measure thus an unbiased estimate of the performance reached by a model with a dataset that has been perfectly deduplicated. Having computed this baseline, we consider the case of imperfect deduplication. We add duplicates to the dataset and consider various cross-validation strategies. Random partitioning consists in partitioning randomly each hospital dataset in folds of the same size. Stratified partitioning consists in choosing first a stratifying covariate and a set of thresholds . Each record is then attributed to the fold that fulfills . We choose thresholds in such a way that the number of records in each global fold is the same. Once fold partitioning is realized, gradient boosting models are fitted and validated on these folds.
RESULT
Model training and validation
We consider first the analysis of synthetic datasets. For each , the model is trained on all the folds apart from fold , and its training performances are measured on the same folds. Figure 3 shows the training learning curves obtained during boosting for unbiased (squares), random (circles), and stratified along (triangles) strategies. Training accuracies increase monotonously as the model learns from the training dataset, and the learning speed does not depend on the fold-partitioning strategy that is adopted. Indeed, training accuracies do not depend on strategy-dependent data leakage between training and validation folds.
Figure 3.

Accuracies computed through cross-validation as a function of the number of boosting iterations in the case of synthetic datasets. Symbols and curves correspond respectively to training accuracies and validation accuracies. Green, red, and blue colors correspond to unbiased, random, and stratified, respectively, along fold-partitioning strategies. Unbiased validation accuracy lies between the overoptimistic random and the pessimistic -stratified estimates. The horizontal black dashed line indicates the theoretical optimal accuracy .
For each the model trained on folds with is validated on fold . Figure 3 shows the variation of validation accuracies during training (solid and dashed lines). When a deduplicated dataset is used (green dashed curve), the unbiased validation accuracy increases during the first 50 boosting iterations and then saturates at an that is lower than the optimal that a predictive model could reach (straight dashed black line, see Supplementary Appendix). As expected the validation accuracy remains lower than the optimal accuracy. When duplicates are added and random fold-partitioning strategy is used, the estimated validation accuracy (solid red curve) is biased by data leakage, and it increases monotonously until falsely reaching a high accuracy. When stratified along strategy is adopted, duplicates of a given record are grouped in the same fold and definition 3 is fulfilled: there is consequently no overoptimistic bias, and the estimated accuracy remains close to the unbiased one, but a small pessimistic bias is observed that is due to interfold heterogeneity. Applying a random fold-partitioning strategy on a dataset with duplicates therefore misses the saturation of the performances observed in unbiased and stratified cases.
Bias and feature importance
We ran additional simulations to better understand the implications of the choice of a stratifying covariate. Figure 4 shows validation accuracies obtained after 200 boosting iterations for 30 simulations using the same set of generating parameters and but applying various fold-partitioning strategies. Whereas random fold partitioning always leads to overoptimistic estimates of accuracy (red) compared with the unbiased estimates obtained without duplicates (green), other stratification strategies lead to accuracy estimates that feature pessimistic biases of variable importance (blue). Whereas stratification leads to a pessimistic bias of roughly 5%, stratifying along , , , , , or leads to estimates that are close to the unbiased ones. The arbitrary choice of a stratifying covariate may consequently lead to important pessimistic biases. The closeness with the unbiased estimates of the estimates obtained having stratified along was predictable, as is an independent variable that does not induce interfold heterogeneity.
Figure 4.
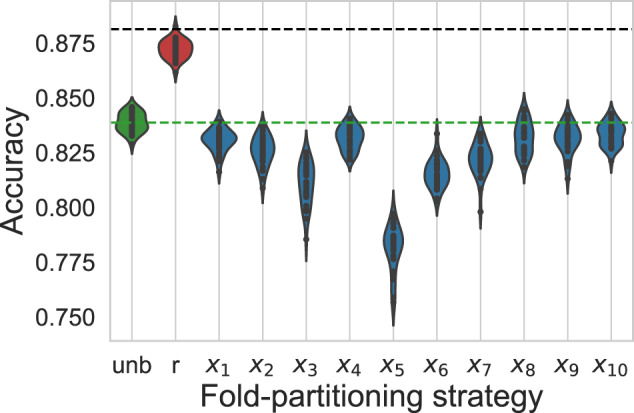
Violin plots for cross-validation estimates of accuracy adopting either an unbiased (green), random (red), or –stratified (blue) fold-partitioning strategy and running 30 simulations in the case of synthetic datasets. Horizontal black and green dashed lines correspond to the optimal accuracy that a model could reach and to the mean unbiased estimate of the accuracy reached by the model under consideration, respectively. Whereas random fold partitioning leads to overoptimistic estimates of accuracy, –stratified estimates feature pessimistic biases of various sizes.
To better characterize pessimistic biases, we run additional simulations varying also the covariate covariance matrix and the outcome generating parameters . We draw 100 different sets of parameters (see Supplementary Material for details), and for each set of generating parameters, we generate a dataset. For each dataset, we fit and validate a gradient boosting model using classical cross-validation and measuring its unbiased validation accuracy . We then add duplicates and fit successively 10 gradient boosting models corresponding each to fold-stratified cross-validation having stratified along 1 of the 10 covariates. Figure 5 shows for each one of the 1000 models the validation accuracy measured having stratified along divided by the unbiased estimate obtained on the same dataset, plotted with respect to normalized feature importance of measured during the training of the unbiased model—see Hastie et al6 for feature importance computation. We observe a negative Pearson correlation coefficient 5 between the pessimistic bias on accuracy and the normalized importance of the stratifying covariate.
Figure 5.
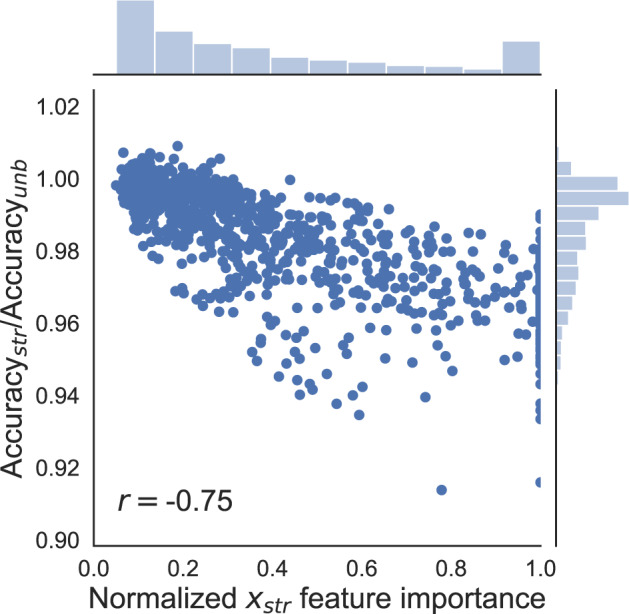
Ratio of -stratified estimate of accuracy over the unbiased estimate of accuracy plotted with respect to the normalized importance of the stratifying covariate (see text) in the case of synthetic datasets. A total of 100 datasets are generated corresponding to different , and for each dataset, each covariate is taken successively as stratifying covariate. The Pearson correlation coefficient is .
Simulations using real data
We consider now the case of MIMIC-III database.48 We reproduce the training and validation of a gradient boosting model,53 that predicts hospital mortality using as performance metric the area under the curve (AUC) of the receiver-operating characteristic curve (Figure 6). We run 30 simulations corresponding each to duplicated records being chosen at random. Applying random cross-validation on the deduplicated dataset, we reproduce the original performance (AUC = 0.826).53 Considering the dataset with added duplicates, random cross-validation leads to a performance (AUC = 0.826) that is overoptimistic compared with the unbiased case, and fold-stratified cross-validation leads to pessimistic performances AUC = 0.806, 0.823, 0.807, 0.798, and 0.820 using age, weight, creatinine, blood urea nitrogen, and hemoglobin, respectively, as stratifying covariates. However, this bias is limited using weight or hemoglobin.
Figure 6.

Violin plots for cross-validation estimates of accuracy adopting either an unbiased (green), a random (red) or a stratified (blue) fold-partitioning strategy and running 30 simulations in the case of MIMIC-III (Medical Information Mart for Intensive Care-III)–based datasets. The horizontal green dashed line corresponds to the mean unbiased estimate of the accuracy reached by the model under consideration. Whereas random fold partitioning leads to overoptimistic estimates of accuracy, stratified estimates feature pessimistic biases of various sizes when the age at admission (age), the weight at admission (wei), the lowest creatinine value from the first 24 hours after admission (cre), the lowest blood urea nitrogen value from the first 24 hours after admission (bun), or the highest hemoglobin value from the first 24 hours after admission (hem) are used as stratifying covariates (see text). Inset shows the ratio of stratified estimate of accuracy over the unbiased estimate of accuracy plotted with respect to the normalized importance of the stratifying covariate see text). The Pearson correlation coefficient is . AUC: area under the curve.
DISCUSSION
The data leakage phenomenon at play in our simulations is specific neither to the XGBoost model nor to the data under scrutiny, and the risk of duplicate-caused data leakage should be addressed whatever the scientific problem at stake. Fold-stratified cross-validation methodology avoids data leakage between folds by fulfilling interfold deduplication (definition 3) without requiring full deduplication (definitions 1 and 2), thus providing a validation methodology that is robust to the presence of undetected duplicates (Figures 3 and 4). Although fold-stratified cross-validation avoids an overoptimistic bias due to data leakage between folds, it may be subject to a pessimistic bias due to interfold heterogeneity (Figures 3, 4, and 6). In order to limit this undesired pessimistic bias, it appears optimal to choose a stratifying covariate that is weakly associated to the other covariates and to the outcome (Figures 5 and 6). Determining a priori which covariate is weakly or strongly associated to the other covariates and to the outcome is challenging, and can rely either on prior knowledge of the problem under scrutiny or on preliminary fitting of the model on a local dataset. In the special case of the date of birth or a personal identifier being available, hashing it provides an ideal stratifying covariate that is shared by duplicates. Another difficulty might arise when records related to a given individual are not perfectly equal, as it is the case when duplicated records correspond to different recording events such as hospital admissions. But perfect equality of duplicated records is not necessary to apply fold-stratified cross-validation: identifying a single stratifying covariate, the value of which is shared among records related to a given individual, is indeed sufficient.
Fold-stratified cross-validation methodology presents generic limitations. First, when duplicated records are so different that no shared covariate can be found to stratify, it appears impossible to ensure through stratification that 2 records related to a given individual are present in the same fold. Second, when data curators are not hospitals but instead are directly individuals, such as for instance in the case of data stored in mobile phones,25 it is impossible to partition datasets in folds as each dataset corresponds to a single record. Third, pseudonymization is often applied to records prior to their analysis (eg, date shifting, quantization, noise addition).54,55 Applying hospital-specific or probabilistic pseudonymization may break the equality of stratifying covariates among duplicates. Last, a subpopulation may be more subject to data duplication, thus inducing its overrepresentation in the validation fold. We moreover underline that the presented simulations are mostly illustrative: the size of biases measured in this article depend strongly on the data, the model, and the metric at stake.
CONCLUSION
When a model is trained and validated in a privacy-preserving federated learning setting, the presence of duplicated records may lead to overoptimistically biased estimates of its performances. We have shown that fold-stratified cross-validation methodology can be used to avoid this overoptimistic bias while not relying on deduplication algorithms, the application of which is often limited to records that are tagged by a shared personal identifier. Fold-stratified cross-validation presents drawbacks, as it may be subject to a pessimistic bias, but this bias can be minimized by carefully choosing the stratifying covariate. We underline that fold-stratified cross-validation, although of special importance in the case of federated learning in which full deduplication is often unfeasible, also applies to the case of a centralized dataset with undetected duplicates and can therefore be used as an easy-to-implement sanity check. Although of possibly broad application, fold-stratified cross-validation is only a partial solution to duplicate problems: tagging records by shared identifiers and applying deduplication algorithms remains optimal to ensure databases integrity.
FUNDING
This project is supported by Bpifrance as part of the HealthChain project, which resulted from the Digital Investments Program for the major challenges of the future RFP. As part of the HealthChain project, a consortium coordinated by Owkin (a private company) has been established, including the Substra association, Apricity (a private company), the Assistance Publique-Hôpitaux de Paris, University Hospital Center of Nantes, Léon Bérard Center, French National Center for Scientific Research, École Polytechnique, Institut Curie, and University of Paris.
AUTHOR CONTRIBUTIONS
RB designed the methodology, conducted the simulations, and wrote the article. RG and MB provided technical advice and article feedback. FG realized the analysis based on the MIMIC-III dataset and provided article feedback. RP oversaw the project and helped with writing the article .
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We thank the partners of HealthChain consortium for fruitful discussions and Elise Diard for her help with figure design.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542 (7639): 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hosny A, Parmar C, Quackenbush J, et al. Artificial intelligence in radiology. Nat Rev Cancer 2018; 18 (8): 500–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Komorowski M, Celi LA, Badawi O, et al. The Artificial Intelligence Clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med 2018; 24 (11): 1716–20. [DOI] [PubMed] [Google Scholar]
- 4. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rahimian F, Salimi-Khorshidi G, Payberah AH, et al. Predicting the risk of emergency admission with machine learning: Development and validation using linked electronic health records. PLoS Med 2018; 15 (11): e1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hastie T, Tibshirani R, Friedman J.. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. 2nd ed New York, NY: Springer-Verlag; 2009. [Google Scholar]
- 7. van der Ploeg T, Austin PC, Steyerberg EW.. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol 2014; 14 (1): 137.doi : 10.1186/1471-2288-14-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Powles J, Hodson H.. Google DeepMind and healthcare in an age of algorithms. Health Technol 2017; 7 (4): 351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Caldicott F. Review of data security, consent and opt-outs. National Data Guardian. 2016. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/535024/data-security-review.PDF Accessed April 29, 2020.
- 10. Homer N, Szelinger S, Redman M, et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet 2008; 4 (8): e1000167. CrossRef][10.1371/journal.pgen.1000167] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bohannon J. Genealogy databases enable naming of anonymous DNA donors. Science 2013; 339 (6117): 262. [DOI] [PubMed] [Google Scholar]
- 12. Gymrek M, McGuire AL, Golan D, et al. Identifying personal genomes by surname inference. Science 2013; 339 (6117): 321–4. [DOI] [PubMed] [Google Scholar]
- 13. Rocher L, Hendrickx JM, de Montjoye Y-A.. Estimating the success of re-identifications in incomplete datasets using generative models. Nat Commun 2019; 10 (1): 3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Price WN, Cohen IG.. Privacy in the age of medical big data. Nat Med 2019; 25 (1): 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aggarwal CC. On K-anonymity and the Curse of Dimensionality. In: proceedings of the 31st International Conference on Very Large Data Bases. VLDB Endowment; 2005: 901–9.
- 16. Brickell J, Shmatikov V. The cost of privacy: destruction of data-mining utility in anonymized data publishing. In: proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2008: 70–8.
- 17. de Montjoye Y-A, Gambs S, Blondel V, et al. On the privacy-conscientious use of mobile phone data. Sci Data 2018; 5 (1): 180286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vest JR, Simon K.. Hospitals’ adoption of intra-system information exchange is negatively associated with inter-system information exchange. J Am Med Inf Assoc 2018; 25 (9): 1189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu Y, Jiang X, Kim J, et al. Grid Binary LOgistic REgression (GLORE): building shared models without sharing data. J Am Med Inf Assoc 2012; 19 (5): 758–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lu C-L, Wang S, Ji Z, et al. WebDISCO: a web service for distributed cox model learning without patient-level data sharing. J Am Med Inf Assoc 2015; 22: 1212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shokri R, Shmatikov V. Privacy-preserving deep learning. In: proceedings of the 22nd ACM SIGSAC Conference on Computer and Communications Security; 2015: 1310–21.
- 22. McMahan HB, Moore E, Ramage D, Hampson S, Agüera y Arcas B. Communication-efficient learning of deep networks from decentralized data. arXiv:160205629v3. http://arxiv.org/abs/1602.05629 Accessed December 16, 2019.
- 23. Bonawitz K, Ivanov V, Kreuter B, et al. Practical secure aggregation for privacy-preserving machine learning. In: proceedings of the 2017 ACM SIGSAC Conference on Computer and Communications Security; 2017: 1175–91.
- 24. Kairouz P, McMahan HB, Avent B, et al. Advances and open problems in federated learning. arXiv:1912.04977v1. http://arxiv.org/abs/1912.04977 Accessed December 13, 2019.
- 25. Bonawitz K, Eichner H, Grieskamp W, et al. Towards federated learning at scale: system design. arXiv:1902.01046v2. http://arxiv.org/abs/1902.01046 Accessed November 26, 2019.
- 26. Raisaro JL, Tramèr F, Ji Z, et al. Addressing Beacon re-identification attacks: quantification and mitigation of privacy risks. J Am Med Inf Assoc 2017; 24 (4): 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raisaro JL, Troncoso-Pastoriza JR, Misbach M, et al. MedCo: enabling secure and privacy-preserving exploration of distributed clinical and genomic data. IEEE/ACM Trans Comput Biol Bioinf 2019; 16 (4): 1328–41. [DOI] [PubMed] [Google Scholar]
- 28. Ryffel T, Trask A, Dahl M, et al. A generic framework for privacy preserving deep learning. arXiv:1811.04017v2. http://arxiv.org/abs/1811.04017 Accessed November 9, 2019.
- 29. Galtier MN, Marini C. Substra: a framework for privacy-preserving, traceable and collaborative machine learning. arXiv:1910.11567v1. http://arxiv.org/abs/1910.11567 Accessed November 6, 2019.
- 30. Duan R, Boland MR, Liu Z, et al. Learning from electronic health records across multiple sites: a communication-efficient and privacy-preserving distributed algorithm. J Am Med Inf Assoc 2020; 27 (3): 376–85. doi: 10.1093/jamia/ocz199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lazer D, Kennedy R, King G, et al. Big data. The parable of Google Flu: traps in big data analysis. Science 2014; 343 (6176): 1203–5. [DOI] [PubMed] [Google Scholar]
- 32. Dressel J, Farid H.. The accuracy, fairness, and limits of predicting recidivism. Sci Adv 2018; 4 (1): eaao5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Király FJ, Mateen B, Sonabend R. NIPS-not even wrong? A systematic review of empirically complete demonstrations of algorithmic effectiveness in the machine learning and artificial intelligence literature. arXiv:1812.07519v1 . http://arxiv.org/abs/1812.07519 Accessed November 6, 2019.
- 34. Park SH, Han K.. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology 2018; 286 (3): 800–9. [DOI] [PubMed] [Google Scholar]
- 35. Vollmer S, Mateen BA, Bohner G, et al. Machine learning and AI research for patient benefit: 20 critical questions on transparency, replicability, ethics and effectiveness. arXiv:1812.10404v1. http://arxiv.org/abs/1812.10404 Accessed November 6, 2019. [DOI] [PMC free article] [PubMed]
- 36. Kaufman S, Rosset S, Perlich C, et al. Leakage in data mining: formulation, detection, and avoidance. ACM Trans Knowl Discov Data 2012; 6 (4): 1–21. [Google Scholar]
- 37. Harron K, Wade A, Gilbert R, et al. Evaluating bias due to data linkage error in electronic healthcare records. BMC Med Res Methodol 2014; 14 (1): 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo W, Phung D, Tran T, et al. Guidelines for developing and reporting machine learning predictive models in biomedical research: a multidisciplinary view. J Med Internet Res 2016; 18 (12): e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Saeb S, Lonini L, Jayaraman A, et al. The need to approximate the use-case in clinical machine learning. Gigascience 2017; 6 (5): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCoy AB, Wright A, Kahn MG, et al. Matching identifiers in electronic health records: implications for duplicate records and patient safety. BMJ Qual Saf 2013; 22 (3): 219–24. [DOI] [PubMed] [Google Scholar]
- 41. Everson J, Adler-Milstein J.. Gaps in health information exchange between hospitals that treat many shared patients. J Am Med Inf Assoc 2018; 25 (9): 1114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harron K, Goldstein H, Dibben C.. Methodological Developments in Data Linkage. New York, NY: Wiley; 2015. [Google Scholar]
- 43. Vatsalan D, Christen P, Verykios VS.. A taxonomy of privacy-preserving record linkage techniques. Inf Syst 2013; 38 (6): 946–69. [Google Scholar]
- 44. Weber GM. Federated queries of clinical data repositories: the sum of the parts does not equal the whole. J Am Med Inform Assoc 2013; 20 (e1): e155–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yigzaw KY, Michalas A, Bellika JG.. Secure and scalable deduplication of horizontally partitioned health data for privacy-preserving distributed statistical computation. BMC Med Inf Decis Mak 2017; 17(1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Laud P, Pankova A.. Privacy-preserving record linkage in large databases using secure multiparty computation. BMC Med Genomics 2018; 11 (S4): 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diamantidis NA, Karlis D, Giakoumakis EA.. Unsupervised stratification of cross-validation for accuracy estimation. Art Int 2000; 116 (1–2): 1–16. [Google Scholar]
- 48. Johnson AEW, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Le Gall JR, Lemeshow S, Saulnier F.. A new Simplified Acute Physiology Score (SAPS II) based on a European/North American multicenter study. JAMA 1993; 270 (24): 2957–63. [DOI] [PubMed] [Google Scholar]
- 50. Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. In: proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining-KDD 16; 2016: 785–94.
- 51. Liu Y, Ma Z, Liu X, et al. Boosting privately: privacy-preserving federated extreme boosting for mobile crowdsensing. arXiv:1907.10218v2. http://arxiv.org/abs/1907.10218 Accessed December 4, 2019.
- 52. Cheng K, Fan T, Jin Y, et al. SecureBoost: a lossless federated learning framework. arXiv:1901.08755v1. http://arxiv.org/abs/1901.08755 Accessed December 4, 2019.
- 53. Pirracchio R, Petersen ML, Carone M, et al. Mortality prediction in intensive care units with the Super ICU Learner Algorithm (SICULA): a population-based study. Lancet Respir Med 2015; 3 (1): 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Emam KE, Arbuckle L.. Anonymizing Health Data: Case Studies and Methods to Get You Started. Newton, MA: O’Reilly Media; 2013. [Google Scholar]
- 55. Dwork C, Roth A.. The algorithmic foundations of differential privacy. FNT Theor Comput Sci 2013; 9 (3–4): 211–407. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.