Abstract
Objective
As clinical trials evolve in complexity, clinical trial data models that can capture relevant trial data in meaningful, structured annotations and computable forms are needed to support accrual.
Material and Methods
We have developed a clinical trial information model, curation information system, and a standard operating procedure for consistent and accurate annotation of cancer clinical trials. Clinical trial documents are pulled into the curation system from publicly available sources. Using a web-based interface, a curator creates structured assertions related to disease-biomarker eligibility criteria, therapeutic context, and treatment cohorts by leveraging our data model features. These structured assertions are published on the My Cancer Genome (MCG) website.
Results
To date, over 5000 oncology trials have been manually curated. All trial assertion data are available for public view on the MCG website. Querying our structured knowledge base, we performed a landscape analysis to assess the top diseases, biomarker alterations, and drugs featured across all cancer trials.
Discussion
Beyond curating commonly captured elements, such as disease and biomarker eligibility criteria, we have expanded our model to support the curation of trial interventions and therapeutic context (ie, neoadjuvant, metastatic, etc.), and the respective biomarker-disease treatment cohorts. To the best of our knowledge, this is the first effort to capture these fields in a structured format.
Conclusion
This paper makes a significant contribution to the field of biomedical informatics and knowledge dissemination for precision oncology via the MCG website.
Key words
knowledge representation, My Cancer Genome, precision oncology, knowledge curation, cancer informatics, clinical trial data model
INTRODUCTION
Successful clinical trial completion is paramount for drug discovery, and yet about 40% of trials close prematurely due to lack of patient enrollment.1 The rise of biomarker-directed therapies in oncology has resulted in increased complexity of clinical trial eligibility criteria and multi-arm study designs. Clinical trial data models that can store this complex information in meaningful, structured annotations and computable forms can be used in downstream applications to improve trial enrollment and elucidate trends in the oncology treatment space.
The number of cancer clinical trials featuring biomarker-based eligibility criteria has more than quadrupled in less than a decade from 3% in 2006 to 16% in 2013.2 Eligibility criteria have evolved in complexity from single gene alterations in a single disease (eg, BRAF V600E melanoma3) to multi-arm studies evaluating multiple potential single or co-occurring alterations in multiple disease cohorts (eg, NCI-MATCH, ASCO-TAPUR4,5) It is not uncommon for trials to include/exclude groups of gene variants found along specific cell signaling pathways, biomarkers important in drug metabolism, and ones that confer drug resistance. The biomarkers in trials span from well-studied genomic, protein, serological, and cytogenetic markers to newer biomarkers including viral proteins, epigenetic signaling, and tumor mutation burden. As the eligibility criteria for trials become more complex, it has become challenging to accurately match these trials to relevant patient populations. This in turn translates into long and cumbersome recruitment workflows, a high burden of manual review on clinical trial staff, and ultimately low accrual rates.
There have been attempts with varying success to use key word-search and machine-learning approaches to decipher the eligibility criteria of clinical trial documents.6–10 However, nonstandardized and nonstructured clinical trial documents, extensive use of gene and protein aliases, ambiguous sentence structure, and multiple amendments to clinical trial documents make this a challenging task. To address this issue, several groups have created information models that can support clinical trial eligibility criteria curation or annotation. Some of these knowledge bases are Matchminer by Dana-Farber Cancer Institute,11 Trial Prospector by Case Western Reserve University,12 and JAX-CKB by Jackson Labs,13 among others. There are commercial vendors who are creating private versions of these to incorporate in their next generation sequencing workflow (eg, Tempus, Foundation Medicine). However, there are several limitations to existing efforts—both in terms of model expressivity and model content. Expressivity is the ability to express the full range of concepts observed in actual clinical trial documents beyond genomic criteria (ie, cytogenetics, protein expression, serological, epigenetic criteria, etc). Content is the scope of trial curation beyond institutional trials, trials for a specific disease group, etc.
Our prior approaches to clinical trial annotation explored both key word search14 and natural language processing (NLP)-based approaches10 for automated extraction of biomarker eligibility criteria from clinical trial documents. However, we and others with similar efforts concluded that the level of precision and recall achieved by these methods is relatively low.8,9,15–20 We then adopted a combined methodology leveraging both artificial and human intelligence to develop a standardized, structured, and extendable nomenclature model for eligibility criteria curation of oncology trials in collaboration with our software development partner GenomOncology.21
In this article, we provide a detailed description of our clinical trial information model, clinical trial curation information system, standard operating procedure (SOP) for trial curation, evaluation of the model expressivity in curating eligibility criteria for over 5000 cancer clinical trials, and visualization of the model content for both individual trials and aggregate summary statistics. Following an iterative cycle of refinement and development, we have designed an information model that represents various data elements pertaining to a clinical trial. A comprehensive set of annotated instances of this model supports the clinical trial-related content on the My Cancer Genome (MCG) website.22
MATERIALS AND METHODS
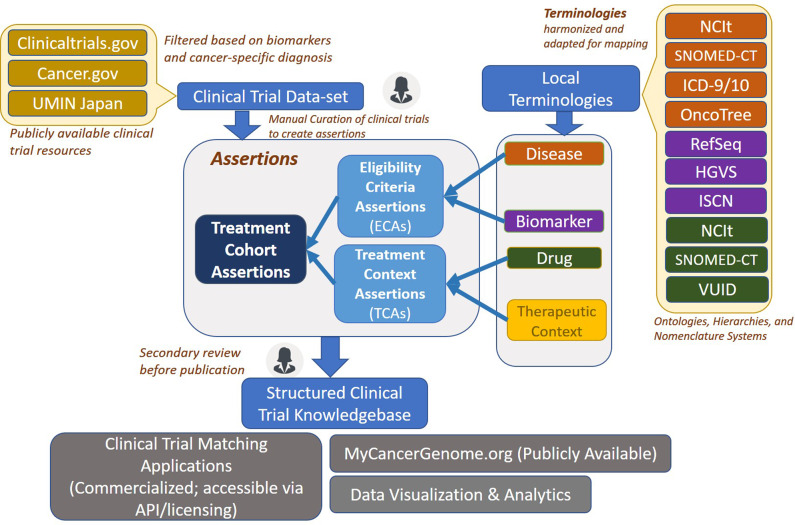
We adopted an assertion-based approach for our model since the computable nature of this approach allows data to be queried and utilized for several downstream applications. Figure 1 shows a high-level overview of the clinical trial curation information system. Clinical trial documents are pulled into the curation system from publicly available sources, such as clinicaltrials.gov,23 cancer.gov,21 and UMIN Japan.24 Using a web-based interface, a curator creates structured assertions related to disease-biomarker eligibility criteria, therapeutic context, and treatment cohorts by leveraging terminologies and concept groups available in the data model. These structured assertions are utilized for clinical trial-related content on the MCG website and other downstream applications.
Figure 1.
Clinical trial model and workflow schematic. This high-level schematic describes the curation model components and workflow. Clinical trial documents are pulled into the clinical trial dataset from publicly available sources. Using the web-based interface, a curator creates structured assertions for trials. This is done using the terminologies and concept groups available in the data model. A single clinical trial can be broken down into multiple individual treatment cohort assertions (TCAs), each corresponding to a separate treatment arm. Once the assertions are created, they undergo a secondary manual review before being published into the clinical trial knowledge base. This knowledge base is utilized for clinical trial matching, display on My Cancer Genome website, as well as for multiple downstream applications.
Clinical trial data model
The clinical trial data model is supported by (i) core model assertions, (ii) terminologies, and (iii) concept groups (Table 1). Core model assertions define the relational structure between concepts that make up the set of logical statements pertaining to clinical trial properties. Terminologies from multiple external sources, as well as locally maintained concepts, are used to populate values for individual trial assertions. Concept grouping enables modeling of computable and reusable concepts using logical operators (any, all, none) to enable more consistent and efficient curation. A complete schema of the clinical trial data model (in PDF and JSON format) can be found in the Supplementary Material.
Table 1.
Components of clinical trial eligibility criteria assertion model
| Concepts | Definitions | Examples | |
|---|---|---|---|
| Core model concepts | Eligibility criteria assertion (ECA) | Structured diagnosis with or without associated biomarker criteria as described in the trial document | Breast Cancer: [ER Negative AND PR Negative AND HER2 Negative] AND [NONE: TP53 Germline Mutations] |
| Treatment context assertion (TCA) | Combination of the intention of treatment or sequencing of treatment and the treatment agents themselves as described in the trial document | [Niraparib AND Radiation Therapy] AND [Adjuvant Setting] | |
| Treatment arm assertion (TAA) | Linking the patient population (diagnosis and biomarker criteria) and treatment arms outlined in clinical trial document. Achieved via linking the ECA & TCA | “Breast Cancer: [ER Negative AND PR Negative AND HER2 Negative] AND [NONE TP53 Somatic or Germline Mutations]” & “ [Niraparib AND Radiation Therapy] AND [Adjuvant Setting]” | |
| Terminologies | Diagnosis | Solid tumor, hematologic or lymphoid malignancy. The mapping structure means that multiple disease synonyms will match to the same disease, ie, hepatic cancer, hepatic carcinoma, cancer of the liver, liver cancer, and hepatocellular carcinoma would all map to the concept of hepatocellular carcinoma | breast carcinoma, head and neck squamous cell carcinoma, acute myeloid leukemia |
| Biomarker | Includes biomarkers related to gene variants (mutation, deletion, fusion, amplification, loss); protein variants (expression, over-expression, deficient expression); cytogenetic/chromosomal abnormalities (duplication, deletion, monosomy, trisomy, karyotype, translocations, inversions); viral markers (EBV, HPV, KSHV, MCPyV, etc.), serological (HLA, HLB markers), epigenetic markers (methylation status), specialty markers (microsatellite instability, tumor mutation, MMR status) | MYC amplification, MSH2 loss, ERBB2 overexpression, monosomy 7, complex karyotype, t(9; 11)(p21; q23), KMT2A Fusion, dup(1)(q10qter), EBV positive, HLA-A*02:05, MGMT promoter methylation positive, MSI-High, TMB-Low, dMMR | |
| Therapies | Any therapeutic approach used in clinical trial document and defined on NCIt including, but not limited to, targeted therapy, immunotherapy, hormonal therapy, cytotoxic agents, monoclonal antibodies, antibody-drug conjugates, vaccine therapy, and hematopoietic and bone marrow transplantation (surgical interventions and radiation therapy subtypes are excluded) | osimertinib, larotrectinib, tamoxifen, pembrolizumab, oxaliplatin, trastuzumab, brentuximab vedotin, Lu-177-DOTA-TATE, CAR T-cell therapy | |
| Therapeutic context | Describes the clinical context for treatment as would be relevant to a patient's disease state | Neoadjuvant, adjuvant, metastatic, treatment-naïve, relapse, refractory, etc. | |
| Concept groups | Disease groups | A meaningful grouping of diagnoses usually based on organ systems, similarity of disease biology or other commonalities | Urinogenital Cancer group: Extragonadal Embryonal Carcinoma, Renal Pelvis and Ureter Carcinoma, Urothelial Carcinoma, Uterine Corpus Neuroendocrine Neoplasm, Bladder Small Cell Neuroendocrine Carcinoma, Cervix Carcinoma, Malignant Bladder Neoplasm, Malignant Ovarian Germ Cell Tumor, Malignant Renal Pelvis Neoplasm, Malignant Reproductive System Neoplasm, Malignant Ureter Neoplasm, Malignant Urethral Neoplasm, Ovarian Embryonal Carcinoma, Testicular Embryonal Carcinoma, Transitional Cell Carcinoma, Ureter Small Cell Carcinoma |
| Biomarker groups | A grouping of biomarker concepts usually to accommodate biomarkers that frequently appear together in clinical trials, or are related via a single pathway, or are usually altered in a particular disease. Can also be used to accommodate NCCN-approved risk biomarker groups for prognostic risk or diagnostic classification | 11q23 abnormalities: del(11)(q10), KMT2A-AFF1 Fusion, KMT2A Fusion, KMT2A-MLLT3 Fusion, inv(11)(p15q23), KMT2A-ELL Fusion, KMT2A-MLLT10 Fusion, KMT2A-MLLT1 Fusion, KMT2A-MLLT4 Fusion, t(10; 11)(p12; q23), t(11; 19)(q23; p13.1), t(11; 19)(q23; p13.3), t(4; 11)(q21; q23), t(6; 11)(q27; q23), t(9; 11)(p21; q23), Trisomy 11 | |
| Drug groups | A grouping of drugs based on drug categories/classes that have similar mechanism of action or usually appear together in trial documents. Can accommodate drug groups that cannot be directly derived from the drug ontology in NCIt | FDA approved aromatase inhibitors: exemestane, anastrozole, letrozole. |
Core model assertions
Eligibility criteria assertion
We have adopted the approach to curate the diagnosis and associated biomarker criteria for oncology trials. Curating the full set of eligibility criteria was out of scope for the current curation effort, but the eligibility criteria assertion (ECA) model is extensible to include other types of eligibility criteria. An ECA specifies the relationships between the cancer diagnosis of interest (including diagnoses that were excluded) and the associated biomarker eligibility criteria for that diagnosis. Gene, protein, cytogenetic, serological, viral, as well as epigenetic biomarkers are recognized and modeled within the system.
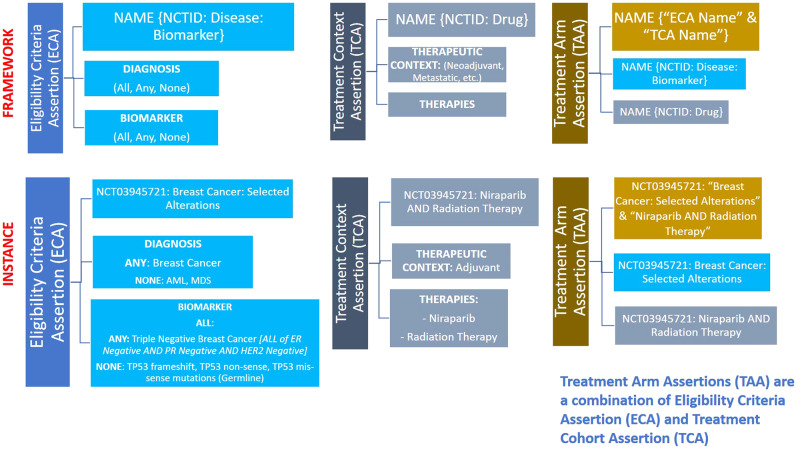
An ECA can support hierarchical nesting of biomarkers to replicate the complex eligibility criteria of oncology trials and can be used to define several cohorts, or trial arms, presented in a trial. This can be done by using the top-level or higher-level operator and assigning it 1 of the logical operators (any, all, none). For consistency, we have adopted the following nomenclature format for ECAs: “Clinical Trial Identifier,” “Disease,” and “Biomarker” Positive/Negative (+/-) (eg, NCT03945721: Breast Cancer: Selected Alterations) (Figure 2).
Figure 2.
Core model concepts: framework and instance. The figure shows the components of the assertions defined as core model concepts. The top section indicates the framework for eligibility criteria assertion, therapeutic context assertion, and treatment arm assertion; and the bottom section presents a real-world trial example.
Treatment context assertions
Treatment context assertions (TCAs) define the relationship between the therapeutic context and associated therapies/interventions in a clinical trial. The therapeutic context combines the intention of treatment (eg, curative, palliative, and supportive care) with concepts representing the sequencing of treatments (eg, Induction, Neoadjuvant, Adjuvant, and First Line Metastatic). A full list of the locally developed therapeutic context concepts supporting solid tumor, hematologic, and lymphoid malignancies can be found in the SOP (see Supplementary Material). A TCA can model multiple interventional arms, multimodality treatments (eg, surgery and radiation therapy), and multidrug treatments. We have adopted the following nomenclature format for TCAs: “Clinical Trial Identifier” AND “Therapies” (eg, NCT03945721: Niraparib AND Radiation Therapy) (Figure 2).
Treatment arm assertions
Treatment arm assertions (TAAs) establish the relationship between the eligibility criteria assertions (ECAs) and the treatment context assertions (TCAs) in a many-to-many model. Within a single trial, multiple trial arms or subcohorts can be modeled, each with these properties; that is, a single clinical trial can be broken down into multiple individual TCAs, each corresponding to a separate treatment arm. However, each TAA is a unique combination of individual ECA and TCA, respectively. For consistency, we have adopted the following nomenclature format for TAAs: “ECA Name” AND “TCA Name” (eg, “NCT03945721: Breast Cancer: Selected Alterations” and “Niraparib AND Radiation Therapy”) (Figure 2).
Terminologies
For consistent curation and downstream mapping to routinely used clinical and genomics terminologies, several external and internal terminologies have been integrated and harmonized for use in the curation information system.
Disease terminology
The multiple-parent disease terminology is derived from the National Cancer Institute Thesaurus (NCIt)25 and the WHO diagnostic classifications for hematologic disorders.26 Each disease is mapped to multiple synonyms and concept identifiers in several commonly used disease hierarchies (eg, OncoTree27), ontologies (eg, NCIt25), and nomenclatures (eg, SNOMEDCT,26 UMLS,28 ICD-9/10,29 etc.).
Biomarker terminology
The biomarker terminology supports multiple biomarker classes (Table 1). All genomic biomarker criteria are annotated using the RefSeq gene database (GRCh37, annotation release 10530) with variant names derived from the Human Genome Variation Society (HGVS)31 and gene names from the Human Genome Organization (HUGO)32 and HUGO Gene Nomenclature Committee (HGNC).33 Cytogenetic biomarkers are represented using the International System for Human Cytogenetic Nomenclature (ISCN)31 grammar and, when appropriate, mapped to specific gene fusion events. Variants are mapped to codons as well as exons, while translocations are mapped to fusions for seamless and accurate translation of information. Each genomic biomarker concept is programmatically mapped via proprietary technology to all known gene synonyms and can be manually mapped to colloquial terms, common clinically-used terms, and/or relevant protein concepts (eg, the ERBB2 overexpression concept maps to HER2 Positive, HER2+, Her2-overexpressing, etc.) to facilitate document curation decision support algorithms. Protein biomarkers map to protein expression, overexpression, equivocal, deficient, or no expression. We added certain biomarker concepts as a parser-override term since these cannot be modeled in our system yet (ie, viral, serological, and epigenetic markers).
Drug terminology
Systemic therapy interventions are modeled at the level of drug name. Specifications of drug dose, route, or frequency are not modeled or curated at this time. The multiparent drug ontology from NCIt25 is used as the primary drug terminology given the breadth and depth of investigational drug concepts used in humans compared to other terminologies which focus on drugs approved by regulatory agencies (eg, RxNorm34). The import includes drug definitions, drug parents, synonyms, mapping codes, gene targets (where applicable) and spans individual drugs, drug classes, and drug regimens. The NCI-metathesaurus35 enables mapping the NCIt drug concepts to other drug terminologies (SNOMED CT,27 Drug Bank36) and nomenclature systems such as Veterans Health Administration National Drug File (VANDF).37
The NCIt drug concepts include extensive synonyms essential for identifying drugs as they transition names through the drug development process towards regulatory approval. While the “preferred term” is used as the display name, the extensive synonyms facilitate consistent curation of drug interventions throughout the drug development life cycle.
Concept groups
A key feature of the clinical trial curation model are the concept groups that allow for grouping of multiple concepts using logical operators of “Any,” “All,” and “None” to create computable and reusable sets of terms. Concept groups are used extensively to create groups of co-occurring biomarkers that commonly appear in clinical trial documents (eg, EGFR sensitizing mutations, 11q23 abnormalities, BRCA1/2 frameshift/nonsense mutations). Both these terms encompass a substantial number of mutations. It would be cumbersome to add these qualifying mutations individually every time they appear in trials. The concept grouping feature allows the user to create reusable groups that are saved in the system and can be reused multiple times, improving annotation consistency and efficiency. It can also be used to create disease groups (eg, urinogenital cancers) and drug groups (eg, FDA-approved aromatase inhibitors).
Curation methodology
As described in Figure 1, clinical trial documents are pulled into the curation information system from various publicly available sources. At the highest level, the manual curation workflow consists of 1) the creation of manually generated assertions, 2) a quality assurance process, and 3) publication.
Data loading process
In a nightly refresh, clinical trial documents are loaded into the information system from multiple public sources.21,23,24 Clinical trials with any of the following key words: cancer, tumor, neuroblastoma, melanoma, leukemia, sarcoma, lymphoma, carcinoma, or malignancies are loaded during this refresh. There are no restrictions based on trial phase or recruiting status. During this loading process, documents are automatically parsed for biomarker concepts and diseases that would qualify as cancer by leveraging the terminology synonyms. Trial metadata (trial title, recruiting status, phase, locations, trial sponsor, last change date, etc.) are parsed automatically from structured fields in the clinical trial documents for inclusion in the trial-level data model. During each import, the trial document loader completes a word-by-word check of the trial title, trial arm, and eligibility criteria information currently stored in the information system against the current document on ClinicalTrials.gov and imports the most recent document if any of these fields have changed. New documents are stored in a series of versioned documents, and the amended trials are automatically flagged for re-review within the user interface.
Curation workflow
Trials are manually annotated by curators using a cloud-based curation interface. To standardize best curation practices, we have developed a detailed SOP (see Supplementary Material). Primarily, the curator creates assertions for each trial which are manually reviewed by a secondary curator before they are published and used for downstream applications. To support the secondary review of curation and ongoing auditing, the system requires the curator to highlight relevant portions of text in the clinical trial document and attach these to the respective manual annotation.
When a trial document uses ambiguous language in the biomarker eligibility criteria, our curation philosophy is to model the eligibility criteria to be more inclusive and less restrictive for a potential patient population. Trial documents often contain vague language about permissible genomic criteria—this is sometimes to maintain the intellectual property of the protocol and other times to provide room for exceptions at the discretion of the trial investigator. By erring towards inclusivity, we ensure these trials will be returned in downstream search and trial-matching use cases.
Evaluation of model expressivity
All biomarker eligibility criteria were curated. Genomic alteration, protein expression, and cytogenetic terms have structured components and standard terminologies underlying the concepts. Other biomarkers were modeled as local terminologies with free text string concepts. This achieved the goal of comprehensive biomarker eligibility criteria curation and demonstrates remaining opportunity to further extend the expressivity of the structured biomarker terminology in the future.
Data visualization & dissemination
Our model supports visualization and dissemination of the curated clinical trial data both at the individual trial level and in aggregated form. This is done using standard python data science tools, including the panda library for data aggregation and the bokeh library for data visualization. Aggregated data is visualized to show treatment trends in by disease, drug, or biomarker. The individual assertions and select aggregate analysis related to trials have been disseminated for public use on the MCG website.22
RESULTS
Clinical trial curation content
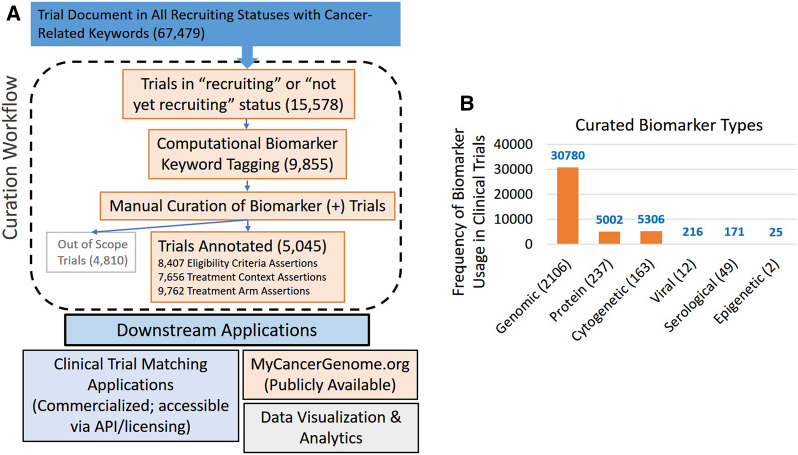
Figure 3A summarizes the outcomes of the curation workflow. Between 11/15/2015 and 10/30/2019, 9855 trials were curated and 8407 eligibility criteria assertions, 7656 treatment context assertions, and 9762 treatment arm assertions were created for these eligible trials. The full clinical trial document and its detailed curation for a set of 5 clinical trials in JSON format can be found in the Supplementary Material. Figure 3B shows the usage of various biomarker classes encountered in clinical trials. Although genomic biomarkers made up the majority of biomarkers curated in clinical trials, biomarkers related to protein expression, cytogenetic alterations, serological markers, viral particles, and epigenetic regulation were frequently encountered and modeled. Table 2 shows the number and frequency of usage of various concept groups as well as germline and parser-override biomarkers.
Figure 3.
Clinical trial curation workflow and results. The figure above shows (A) a broad overview of the curation workflow. There are currently 67 479 cancer-related clinical trials loaded into the system (as of 10/30/2019). Of these, 15 578 were found to have a status of recruiting or not-yet-recruiting. Of these, 9855 trials were automatically flagged for manual review as possibly containing biomarker key words. According to the curation SOP, of the 9855 manually reviewed clinical trials, 5045 met criteria for manual curation of disease-biomarker eligibility criteria and treatment context. A total of 4810 trials were considered out of scope based on the curation SOP. The trials that had a biomarker-driven eligibility criterion were curated. To date, we have manually curated and created structured annotations for 5045 clinical trials. A detailed copy of the SOP is provided in the Supplementary Material. Trials included for manual curation have a recruiting status of “Recruiting” or “Not yet recruiting,” that are (i) interventional (ii) directed toward treating cancer (not for treating side-effects or toxicities caused by cancer treatments), and (iii) contain biomarker-driven eligibility criteria (patient’s tumor is required to have a specific biomarker to enroll on the trial). (B) the different biomarker type supported by the system for clinical trial curation. Genomic biomarker makes up the largest category followed by protein, cytogenetic, viral, serological, and epigenetic-related biomarkers. The numbers in parentheses on the X-axis indicate the actual number of defined concepts in each category, while the instances of cumulative use across clinical trial curations are shown on the y-axis. The curated trial dataset (n = 5045) was used to calculate these numbers.
Table 2.
Number of entities for concept groups and germline biomarkers with usage data
| Number of entities | Cumulative Usage in Unique Trials | |
|---|---|---|
| Biomarker groups | 355 | 2404 |
| Drug groups | 257 | 277 |
| Disease groups | 3 | 8 |
| Germline biomarkers | 104 | 250 |
Dissemination of content on My cancer genome
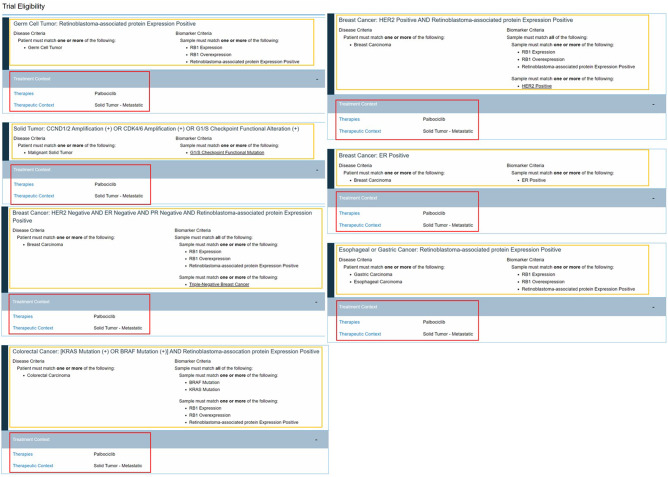
All trial assertion data is available to view publicly on the MCG website.22 The website features an improved trial search feature using multifaceted filtering. Trials can be filtered based on diagnosis, biomarkers, phase, recruiting status, and drug categories using dynamic sorting. Each clinical trial page contains a trial description, recruiting status, and phase information imported from the clinical trial source document along with the respective manually curated assertions. The curated ECAs and TCAs are displayed as logical statements with dynamic content that allows the user to quickly navigate to content on related entities (Figure 4).22
Figure 4.
Screenshot of display of curated clinical trial assertions. This screenshot from the My Cancer Genome website shows the grouping of eligibility criteria assertions (enclosed by yellow boxes) with the treatment context assertions (enclosed by red boxes). Accessed 2/2/2020.
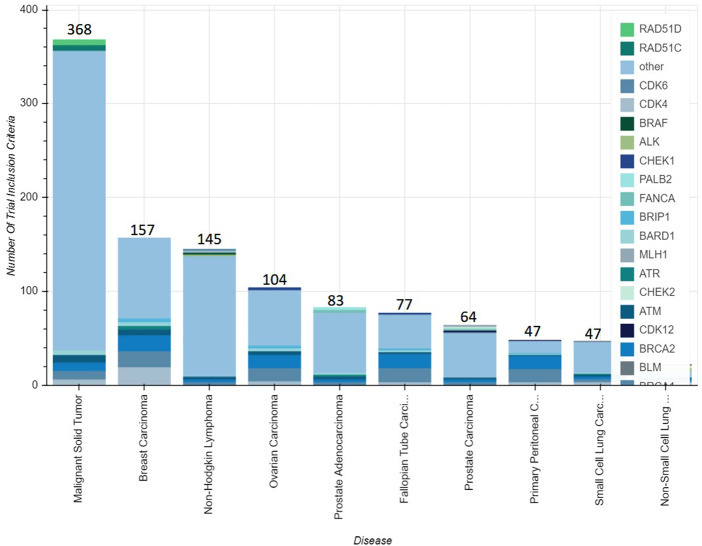
Aggregated analysis of the curated data is also visualized on the website.22 This includes a series of dynamic bar charts counting the number of open and closed clinical trials oriented to disease, biomarker, or investigational drug. This type of analysis allows users to understand drugs being studied across multiple cancer types and a spectrum of biomarker criteria. For example, as of April 2019, there were 60 clinical trials exploring the use of olaparib across 10 tumor types and spanning dozens of biomarkers (Figure 5).
Figure 5.
Screenshot of gene and disease inclusion criteria for open trials investigating olaparib. This screenshot from the My Cancer Genome website shows the grouping of trials by disease and biomarker eligibility criteria that are investigating the use of olaparib. The total number of trials associated with each disease is shown on the data labels. The breakdown of the trials for each disease category can be viewed by hovering over the area of interest at the relevant My Cancer Genome web page (https://www.mycancergenome.org/content/drugs/olaparib/). The curated trial dataset (n = 5045) was used to calculate these numbers. Accessed 06/26/2019.
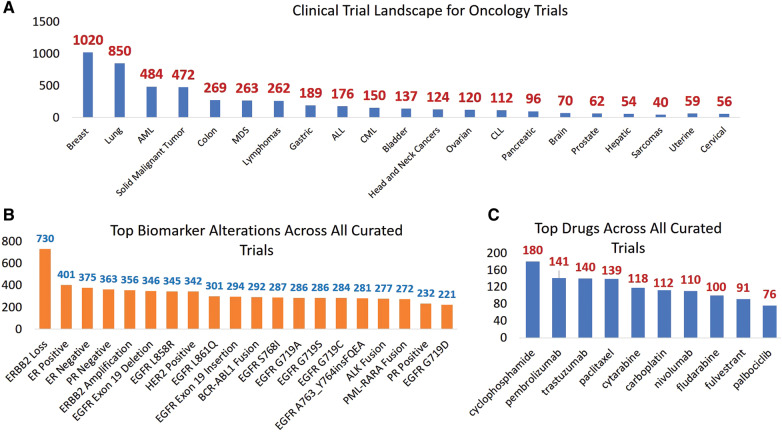
Landscape analysis of clinical trials
Querying our structured knowledge base, we performed a deeper landscape analysis across all curated cancer clinical trials. The top diseases, biomarker alterations, biomarker alteration types, and drugs featured across all trials are shown in Figures 6A, B, and C, respectively (curated between 11/2015 and 04/2019). Breast cancer, lung cancer, and adult acute myeloid leukemia had the highest number of curated clinical trials, owing to the rapid discovery of biomarker-driven therapies in these diseases. Since a vast majority of phase I trials allow patients with any advanced solid malignant tumor, there were a substantial number of trials (n = 472) annotated as solid cancer trials. There were 5 diseases (pancreatic, brain, hepatic, prostate, and sarcomas) with fewer than 100 curated trials. This may represent a lack of known biomarkers associated with the disease, fewer active trials for rare cancers, or availability of standard of care options. Not surprisingly, biomarkers related to breast cancer (ERBB2 loss, ER/PR/HER2 expression) and lung cancer (EGFR alterations) clinical trials constituted the top biomarkers alterations (Figure 6B). Finally, Figure 6C shows the most widely used drugs across all clinical trials: cyclophosphamide, pembrolizumab, and trastuzumab. It is noteworthy that although trials used in the analysis were biomarker-driven trials, targeted therapy was not the top drug category given that they are often combined with or compared to cytotoxic therapies.
Figure 6.
Landscape analysis of all cancer trials shows the (A) top diseases (B) top biomarker alterations, and (C) top drugs in our curated knowledge base. The curated trial dataset (n = 5045) was used to calculate these numbers.
DISCUSSION
Because clinical trial accrual remains a consistent barrier to precision oncology, it is important to develop supportive infrastructure to expedite patient-matching to clinical trials. In this article, we describe an extensible clinical trial data-model, curation information system, and standard operating procedure for manually generating assertions of clinical trial biomarker eligibility criteria, treatment context, and treatment arms. Using this approach, we have created assertions for over 5000 biomarker-driven oncology trials and present an analysis of the treatment trends and biomarker profiles.
There have been several other data models for structuring clinical trial data with varying scope and expressivity. In a prior review of the existing models,38 a key differentiator is our model’s depth of expressivity of biomarkers and scope of trial curation. Unlike other models that only leverage genomic and protein biomarkers, our model is capable of representing cytogenetic biomarkers. As a result, 13% of curated clinical trials in our knowledge base include cytogenetic eligibility criteria (Figure 3B). Cytogenetic biomarkers are widely used for diagnostic and prognostic evaluations in hematologic malignancies, highlighting their importance and the need to include these as high-value biomarkers in data-model design. Protein and cytogenetics-related biomarkers were used to curate 25% of the trials in our dataset, highlighting the importance of designing models that can accurately capture these biomarker classes. Designing models with sufficient expressivity to handle these classes of biomarkers are crucial for broader curation of biomarker criteria in trial documents and improving the precision of clinical trial-matching applications. Although our current model does not support structured representation of viral, serologic, or epigenetic markers, we have circumvented this limitation by creating parser-override biomarkers which account for less than 2.5% of all biomarker concepts and are used in curation of less than 1% of trials (Figure 3B). This represents a potential limitation of our model expressivity but allows for complete curation of biomarker eligibility criteria; these curations can be evaluated and used to inform priorities for model extension to support real-world applications.
Further, we have extended our model to support the curation of treatment context assertions and treatment arm assertions in addition to biomarker eligibility criteria assertions. This allows us to record the treatment setting and investigational drugs in a structured form—allowing for deeper and more enriched treatment-level data as visualized on the MCG website.22 Adding structured assertions for drugs and correlating this with biomarker eligibility criteria represents a nontrivial effort, especially in multi-arm or umbrella studies. Our curation for treatment arm assertions gives us a unique understanding of the depth of eligibility criteria and allows us to perform drug-biomarker analyses in clinical trials, which may be otherwise hard to compute. Finally, this supports aggregate analyses to illuminate drug-biomarker-disease associations and to process complex data that would otherwise remain cloaked (eg, imatinib is being investigated in 18 clinical trials spanning 7 diagnoses and 6 biomarkers). One potential limitation to our model is the adopted curation scope for therapies; for example, we curated drugs or biologics at a more granular level than radiation or surgical interventions. This choice of model priority was guided by our overarching goal of advancing precision oncology.
An important contribution of this work is the deep knowledge that has been added to the public domain. All of the data generated through the model—curated clinical trial eligibility criteria assertions, therapeutic context assertions, drug usage data, biomarker landscape—are available on the MCG website22 for public use. The website also has several visualizations that include aggregated analysis on individual biomarkers, drugs, trials, etc. that can be downloaded instantly (no login needed). The figures presented in the paper are derived from the dataset created using the described model. However, not all of these figures are available on the MCG website.22 An application programming interface (API) for searching and filtering through the trial dataset allows us to do these additional analytics on the dataset. APIs are available via GenomOncology39 for deep use of this dataset, making the public/private nature of the model a potential limitation.
This knowledge base could also be used as a training and validation set for automated or semiautomated methods for extracting clinical trial data from clinical trial documents. It is to be noted that full access to the dataset is only possible through a licensed API at present. There is, however, an ongoing need for national and global efforts to standardize and structure publicly available clinical trial content for multiple downstream uses.
CONCLUSIONS
Precision oncology is a rapidly evolving field, and the ability to process large amount of clinical trial data and derive meaningful insights can spur interesting research and open potentially new avenues for patients as well as researchers. Clinical trial data processing and analysis requires consistent, structured, and detailed biomarker eligibility criteria nomenclature in trial documents that allow stakeholders the ability to query and assess trial criteria computationally. This article outlines our model for curation of precision oncology clinical trials and dissemination of this structured trial information via the MCG website.1 This work is part of a larger effort to improve trial accrual and advance the forefront of precision oncology.
FUNDING
This work was supported by the Susan G. Komen grant (SAC160070), the NHGRI–IGNITE I3P grant (NHGRI U01 HG007253), and the GE Healthymagination award.
AUTHOR CONTRIBUTIONS
NJ, KF, and MH worked on creating trial related assertions; NJ worked on the data analysis, MN worked on disease ontology harmonization, IM, C. Miller, MS, MB, JC worked on the software development and updates to the curation tool. C. Micheel, ML provided design input and supervised the entire effort. All authors contributed toward the design of the curation tool and writing the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
NJ, KF, MH, and MN have no competing interests to declare. IM, C. Miller, MS, MB, and JC are employees of GenomOncology LLC, which is a for-profit entity. C. Micheel is a consultant for Roche and is the personal investigator on the GenomOncology professional services agreement with Vanderbilt University Medical Center (VUMC). ML is a consultant for GenomOncology LLC and Roche, receives royalties from GenomOncology LLC, and is on the external scientific advisory board of Personalis.
Supplementary Material
References
- 1. Unger JM, Cook E, Tai E, Bleyer A.. The role of clinical trial participation in cancer research: barriers, evidence, and strategies. Am Soc Clin Oncol Educ Book 2016; 36 (36): 185–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roper N, Stensland KD, Hendricks R, Galsky MD.. The landscape of precision cancer medicine clinical trials in the United States. Cancer Treat Rev 2015; 41 (5): 385–90. [DOI] [PubMed] [Google Scholar]
- 3.LCCC 1128: Open Label Phase II Trial of the BRAF Inhibitor (Dabrafenib) and the MEK Inhibitor (Trametinib) in Unresectable Stage III and Stage IV BRAF Mutant Melanoma; Correlation of Resistance With the Kinome and Functional Mutations—Full Text View—ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT01726738 Accessed October 4, 2019
- 4. McNeil C. NCI-MATCH launch highlights new trial design in precision-medicine era. J Natl Cancer Inst 2015; 107 (7): djv193. [DOI] [PubMed] [Google Scholar]
- 5. Redig AJ, Jänne PA.. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J Clin Oncol 2015; 33 (9): 975–7. [DOI] [PubMed] [Google Scholar]
- 6. Luo Z, Johnson SB, Weng C.. Semi-automatically inducing semantic classes of clinical research eligibility criteria using UMLS and hierarchical clustering. AMIA Ann Symp Proc 2010; 2010: 487–91. [PMC free article] [PubMed] [Google Scholar]
- 7. Luo Z, Miotto R, Weng C.. A human–computer collaborative approach to identifying common data elements in clinical trial eligibility criteria. J Biomed Inform 2013; 46 (1): 33–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zeng J, Wu Y, Bailey A, et al. Adapting a natural language processing tool to facilitate clinical trial curation for personalized cancer therapy. In: AMIA Joint Summits Translational Science Proceedings 2014: 126–31. [PMC free article] [PubMed] [Google Scholar]
- 9. Xu J, Lee H-J, Zeng J, et al. Extracting genetic alteration information for personalized cancer therapy from ClinicalTrials. J Am Med Inform Assoc 2016; 23 (4): 750–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu Y, Levy MA, Micheel CM, et al. Identifying the status of genetic lesions in cancer clinical trial documents using machine learning. BMC Genomics 2012; 13 (Suppl 8): S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lindsay J, Fitz CDV, Zwiesler Z, et al. MatchMiner: An Open Source Computational Platform for Real-Time Matching of Cancer Patients to Precision Medicine Clinical Trials Using Genomic and Clinical Criteria. bioRxiv2017: 199489.
- 12. Sahoo SS, Tao S, Parchman A, et al. Trial prospector: matching patients with cancer research studies using an automated and scalable approach. Cancer Inform 2014; 13: CIN.S19454–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patterson SE, Liu R, Statz CM, Durkin D, Lakshminarayana A, Mockus SM.. The clinical trial landscape in oncology and connectivity of somatic mutational profiles to targeted therapies. Hum Genomics 2016; 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levy MA, Lovly CM, Pao W.. Translating genomic information into clinical medicine: lung cancer as a paradigm. Genome Res 2012; 22 (11): 2101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borlawsky T, Payne PRO.. Evaluating an NLP-based approach to modeling computable clinical trial eligibility criteria. AMIA Ann Symp Proc 2007; 2007: 878. [PubMed] [Google Scholar]
- 16. Doods J, Dugas M, Fritz F.. Analysis of eligibility criteria from ClinicalTrials.Gov. Stud Health Technol Inform 2014; 205: 853–7. [PubMed] [Google Scholar]
- 17. Kang T, Elhadad N, Weng C.. Initial readability assessment of clinical trial eligibility criteria. AMIA Ann Symp Proc 2015; 2015: 687–96. [PMC free article] [PubMed] [Google Scholar]
- 18. Ni Y, Wright J, Perentesis J, et al. Increasing the efficiency of trial-patient matching: automated clinical trial eligibility pre-screening for pediatric oncology patients. BMC Med Inform Decis Mak 2015; 15: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pfiffner PB, Oh J, Miller TA, Mandl KD.. ClinicalTrials.Gov as a data source for semi-automated point-of-care trial eligibility screening. PLoS One 2014; 9 (10): e111055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ross J, Tu S, Carini S, Sim I.. Analysis of eligibility criteria complexity in clinical trials. Summit Transl Bioinform 2010; 2010: 46–50. [PMC free article] [PubMed] [Google Scholar]
- 21.Find NCI-Supported Clinical Trials. https://www.cancer.gov/about-cancer/treatment/clinical-trials/search Accessed October 10, 2019
- 22.Home—My Cancer Genome. https://www.mycancergenome.org/ Accessed October 10, 2019
- 23.Home—ClinicalTrials.gov. https://clinicaltrials.gov/ct2/home Accessed October 10, 2019
- 24.UMIN Clinical Trials Registry. https://upload.umin.ac.jp/cgi-open-bin/icdr_e/ctr_view.cgi? recptno=R000030315 Accessed October 10, 2019
- 25.NCI Thesaurus. https://ncit.nci.nih.gov/ncitbrowser/ Accessed October 25, 2019
- 26. Williams ME. Revised WHO classification of hematologic malignancies. NEJM J. Watch 2016; 2016. [Google Scholar]
- 27.SNOMED. Home page/. http://www.snomed.org/snomed-ct/sct-worldwide Accessed October 25, 2019.
- 28.Unified Medical Language System (UMLS). https://www.nlm.nih.gov/research/umls/index.html Accessed October 25, 2019
- 29.ICD-ICD-10-CM—International Classification of Diseases, (ICD-10-CM/PCS Transition https://www.cdc.gov/nchs/icd/icd10cm_pcs_background.htm Accessed October 25, 2019
- 30.Homo sapiens (ID 51)—Genome—NCBI. https://www.ncbi.nlm.nih.gov/genome/? term=txid9606[orgn] Accessed October 25, 2019
- 31.Sequence Variant Nomenclature. https://varnomen.hgvs.org/ Accessed October 25, 2019
- 32.Human Genome Organisation (HUGO) International Ltd. http://www.hugo-international.org/ Accessed October 25, 2019
- 33.Home | HUGO Gene Nomenclature Committee. https://www.genenames.org/ Accessed October 25, 2019
- 34.RxNorm. https://www.nlm.nih.gov/research/umls/rxnorm/index.html Accessed October 25, 2019
- 35.NCI Metathesaurus. https://ncim.nci.nih.gov/ncimbrowser/ Accessed Oct 25, 2019
- 36. Wishart DS, Feunang YD, Guo AC, et al. DrugBank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res 2018; 46 (D1): D1074–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veterans Health Administration National Drug File (VANDF). Source information https://www.nlm.nih.gov/research/umls/rxnorm/sourcereleasedocs/vandf.html Accessed Oct 25, 2019
- 38. Jain NM, Culley A, Knoop T, Micheel C, Osterman T, Levy M.. Conceptual framework to support clinical trial optimization and end-to-end enrollment workflow. JCO Clin Cancer Inform 2019; (3): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Precision Oncology Solutions | GenomOncology. https://www.genomoncology.com/ Accessed November 1, 2019
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.