Abstract
Objective
Recent studies on electronic health records (EHRs) started to learn deep generative models and synthesize a huge amount of realistic records, in order to address significant privacy issues surrounding the EHR. However, most of them only focus on structured records about patients’ independent visits, rather than on chronological clinical records. In this article, we aim to learn and synthesize realistic sequences of EHRs based on the generative autoencoder.
Materials and Methods
We propose a dual adversarial autoencoder (DAAE), which learns set-valued sequences of medical entities, by combining a recurrent autoencoder with 2 generative adversarial networks (GANs). DAAE improves the mode coverage and quality of generated sequences by adversarially learning both the continuous latent distribution and the discrete data distribution. Using the MIMIC-III (Medical Information Mart for Intensive Care-III) and UT Physicians clinical databases, we evaluated the performances of DAAE in terms of predictive modeling, plausibility, and privacy preservation.
Results
Our generated sequences of EHRs showed the comparable performances to real data for a predictive modeling task, and achieved the best score in plausibility evaluation conducted by medical experts among all baseline models. In addition, differentially private optimization of our model enables to generate synthetic sequences without increasing the privacy leakage of patients’ data.
Conclusions
DAAE can effectively synthesize sequential EHRs by addressing its main challenges: the synthetic records should be realistic enough not to be distinguished from the real records, and they should cover all the training patients to reproduce the performance of specific downstream tasks.
Keywords: electornic health records (EHRs), sequential data generation, generative adversarial networks (GANs), generative autoencoder, differential privacy
INTRODUCTION
To deploy electronic health records (EHRs) while protecting sensitive or regulated medical information about patients, healthcare organizations have generated anonymized data by using de-identification techniques.1 Nevertheless, the de-identification only can reduce the privacy risks, but there is no guarantee that attackers cannot find the membership of the patient (ie, reidentification) or link them with external data through inferences (ie, linkage attack or inference attack). A variety of mathematical methods2–5 have been studied to assess and minimize reidentification risk that individuals could be identified by linkage to publicly available data. In particular, the trajectory patterns of individual patients are still very distinctive if there are sufficient observations, and this uniqueness of individuals is known to increase reidentification risk, as the U.S. Department of Health and Human Services referred.6 For this reason, a better way to ensure privacy is to synthesize realistic data by learning real EHRs. Because no synthetic record has a one-to-one relationship to the original patient’s records, privacy attacks can be effectively mitigated, and the adoption of differential privacy (DP) can guarantee to further limit the disclosure of private information.
With the success of generative adversarial networks (GANs), several generative models have been proposed to synthesize the EHR for a wide range of clinical usage, including privacy-preserving cross-institutional data sharing,7,8 exploratory data analysis, and data preparation for hosting a competition. Existing models mainly focused on medical images,9,10 clinical texts,11 knowledge bases,12 and structured records about patients’ visits7,13; however, the most important data sources of the EHR have not been studied, which are the collections of set-valued sequences describing chronological medical conditions of patients. In this work, we focus on generating the realistic sequences of high-dimensional discrete records that represents the set of medical entities assigned to the patients (eg, diagnoses, procedures, medications).
On the one hand, most existing studies on deep generative models basically adopt the architecture of GAN14,15 or variational autoencoder (VAE),16 and they have been successfully applied to high-dimensional continuous data such as images.17–19 On the other hand, training such models for high-dimensional and discrete sequences is known to be much more challenging, for example, the generation of sentences (ie, sequence of word tokens) or EHRs (ie, sequence of patients’ structured records). One approach is to model the sequence generation as a sequential decision-making process and train GAN with policy gradient methods20–22; particularly, these models are mainly applied to the text generation because it can effectively deal with the sampling of a word from an output distribution, which is a nondifferentiable operation.
Owing to the difficulties of modeling this type of nondifferentiable objectives, the autoencoder-based models recently have gained much attention and shown promising results.23–27 However, they are not suitable for being applied to the EHR for the following reasons. First, most of them are designed for the text-generation task, which aims to model the sequence of word tokens. Their sequence decoders map the hidden state at each time step into the probability distribution over all words in the vocabulary by the softmax layer, so they are not able to capture the interactions or co-occurrences among the entities within a single set-valued record. Second, they still have the limited performance in terms of the data generation; for example, some of them using a simple fixed prior (eg, the Gaussian distribution) suffer from the mode collapse problem (ie, produce limited varieties of samples), and some of them output the sequences not realistic enough because of their decoders that are only optimized to capture local sequential contexts (ie, only learns sequence models).
To address these limitations, we propose the dual adversarial autoencoder (DAAE), a deep generative model for the sequences of set-valued medical records. The main difference of our model is that DAAE adversarially learns both the continuous latent (or code) distribution and the discrete data distribution, while the existing models do one of them. Our parametric generator is capable of generating varied latent codes that cover the code space induced by the encoder, and our decoder can produce the realistic discrete sequences by taking the latent codes as its inputs. Unlike the conventional autoencoders that only minimize the reconstruction loss between input and output sequences, our sequence decoder is additionally guided to include global realistic features by a critic.
MATERIALS AND METHODS
Data description
We use 2 EHR datasets with different characteristics: MIMIC-III (Medical Information Mart for Intensive Care-III)28,29 and UT Physicians clinical database (UTP). Both of them contain longitudinal patients’ records including a set of medical entities for each visit. MIMIC-III is a public EHR dataset about intensive care unit patients over 11 years; thus, a single visit includes intense information, such as tens of assigned diagnosis codes, but the length of a patient’s visit sequence is short. On the contrary, UTP is about outpatients of UT Physicians from 2012 to 2015, so the patients visited the hospital more frequently, but the amount of information in each visit is much less compared with MIMIC-III. Among various types of medical entities, we only use diagnosis codes (specifically based on International Classification of Diseases–Ninth Revision [ICD-9]), and it can be easily extended to other types of discrete variables. For holdout tests, we split the set of all patients by 7:1:2 ratio into a training set, a validation set, and a test set. Table 1 summarizes the statistics of the datasets.
Table 1.
Statistics of electronic health record datasets
| Dataset | MIMIC-III | UTP |
|---|---|---|
| Unique ICD-9 codes | 4893 | 3144 |
| Patients | 7537 | 13 025 |
| Patients’ visits | 19 993 | 85 845 |
| Average visits per patient | 2.65 | 6.59 |
| Maximum visits per patient | 42 | 52 |
| Average ICD-9 codes per visit | 13.02 | 2.58 |
| Maximum ICD-9 codes per visit | 39 | 30 |
ICD-9: International Classification of Diseases–Ninth Revision; MIMIC-III: Medical Information Mart for Intensive Care-III; UTP: UT-Physicians.
Dual adversarial autoencoder
Building blocks of the generative autoencoder
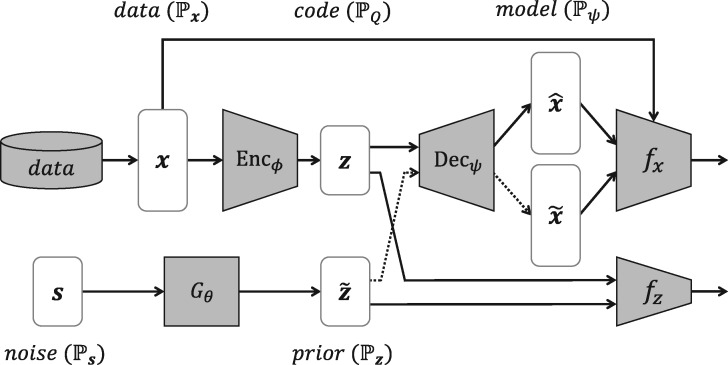
Generative autoencoders aim to learn the underlying distribution of training data by using their encoder and decoder . In general, an encoder is optimized so that the distribution of its output (ie, code distribution ) fits into a simple prior distribution (ie, latent distribution ), and a decoder is trained to make its output distribution (ie, model distribution ) approximate to the original input distribution (ie, data distribution ). The key technique here is effectively matching the distributions, which are and , while minimizing the reconstruction errors for all data inputs. For example, VAE16,23 directly minimizes the Kullback-Leibler divergence between the 2 distributions, whereas adversarial autoencoders24–26 implicitly match them by using a single GAN, referred as to the inner GAN in our work. However, all the existing generative autoencoders fail to generate synthetic data realistic enough not to be distinguished from real ones because their decoders are only trained by the reconstruction errors. To overcome this limitation, our proposed DAAE adopts an additional GAN, referred to as the outer GAN, which adversarially optimizes the decoder to make further close to . That is, DAAE introduces the following deterministic functions: a generator , an inner critic , and an outer critic . The subscript letters denote their trainable parameters; in cases of and , they are parameterized by and , respectively.
As illustrated in Figure 1, DAAE consists of 3 building blocks. First, the sequence-to-sequence (seq2seq) autoencoder learns the hidden code space where semantic features of target sequences are encoded. We employ recurrent neural networks (RNNs) for the encoder and decoder in order to capture the temporal contexts within input sequences. The encoder maps an input sequence from the discrete data distribution into the continuous code distribution , and the decoder generates the model distribution that approximates the original data distribution by serving as a sequence model conditioned on . Second, the inner GAN, including the inner critic and the generator, aims to match the latent distribution with the code distribution . The inner critic distinguishes samples of from those of , and simultaneously, the generator transforms a random noise into a latent vector that can fool the inner critic. Third, similarly, the outer GAN adversarially optimizes the decoder to produce the model distribution , which cannot be separated from the real data distribution by the outer critic. Unlike the conventional GANs whose generator maps each sample from the latent space to the model space, in our outer GAN, the decoder plays the role instead of any additional generators.
Figure 1.
The dual adversarial autoencoder architecture that is composed of the sequence-to-sequence autoencoder, the inner generative adversarial network, and the outer generative adversarial network.
Generative adversarial training
For the optimization of our model, we define a reconstruction loss for the autoencoder and 2 adversarial losses for the inner and outer GANs; each loss alternately optimizes the target building block in our model. The reconstruction loss is defined based on the binary cross entropy between the input sequence and its reconstructed sequence , which is , and the encoder and decoder are trained to minimize this loss. The inner adversarial loss optimizes the inner critic so that it can tell the code samples obtained by the encoder from the latent sample obtained by the generator. At the same time, it tunes the generator to fool the inner critic. Finally, the outer adversarial loss aims to make the outer critic learn the realistic and unrealistic features to discriminate between the real samples and the synthetic samples generated by the decoder, and simultaneously it trains the decoder to generate realistic outputs that can fool the outer critic. The objective of DAAE can be summarized as
| (1) |
and the losses are formulated as
| (2) |
is the set of 1-Lipschitz function set, and the constraint makes our losses correspond to the Wasserstein-1 distance between 2 distributions. We enforce a soft version of the Lipschitz constraint by directly constraining the gradient norm of the critic’s output with respect to its input, known as gradient penalty.30 The detailed algorithm is presented in algorithm 1.
Note that our decoder is optimized by 2 different losses. On the one hand, the reconstruction loss makes the decoder copy the input to the output conditioned on the latent code, so the decoder learns the local context features for sequence modeling. On the other hand, the outer adversarial loss computes the gradient to capture additional features that make the output realistic by the help of the outer critic.
Differentially private training
DP31 has demonstrated itself as a strong standard to provide rigorous privacy guarantees for aggregate dataset analysis algorithms.
Definition 1.
(-Differential privacy) Let and be 2 neighboring datasets that differ in at most 1 entry. A randomized algorithm is -differentially private if for all :
where represents the output of with an input of .
To guarantee the synthetic samples of our generative model do not leak private information about the training data, we adopt the differentially private stochastic gradient descent32 with its variant (ie, DP-Adam) as the optimizer, which injects the calibrated noise by the Gaussian mechanism to the clipped gradient during the model training process. The privacy cost for each epoch is bounded by the Gaussian mechanism, while the total privacy cost during the model training is accumulated by using a moments accountant.
Theorem 1.
(Gaussian mechanism) For and , the Gaussian mechanism with the parameter which adds noise scaled by to each component of the output of algorithm , is -differentially private.
Because DAAE generates its synthetic samples by using the decoder and the generator of the inner GAN, we adopt DP-Adam only for these 2 functions when updating their parameters during the model training. Therefore, the overall privacy cost for training our model is the sum of the privacy cost induced by the decoder and generator. Owing to the postprocessing property of the DP, after the generative model is trained with differential privacy guarantee, it can generate as many synthetic samples as we need without increasing the privacy leakage of the private training data.
Model architecture
Notations
We assume that there exist M distinct medical entities identified from a dataset (ie, ICD-9 diagnosis codes), and we consider 2 additional entities to represent the start and the end of each sequence (denoted by and , respectively). Then, each set-valued record can be represented as a multihot vector of the size ; ie, a binary vector whose -th entry indicates the corresponding entity is included or not in the record. Formally, the sequential record of the -th patient is represented as where is the maximum length of the original sequences, and its -th record is denoted by . The lengths of sequences are all different, but we use just for notational convenience. Note that 2 set-valued records and are respectively inserted at the beginning and end of each sequence.
Sequence-to-sequence autoencoder
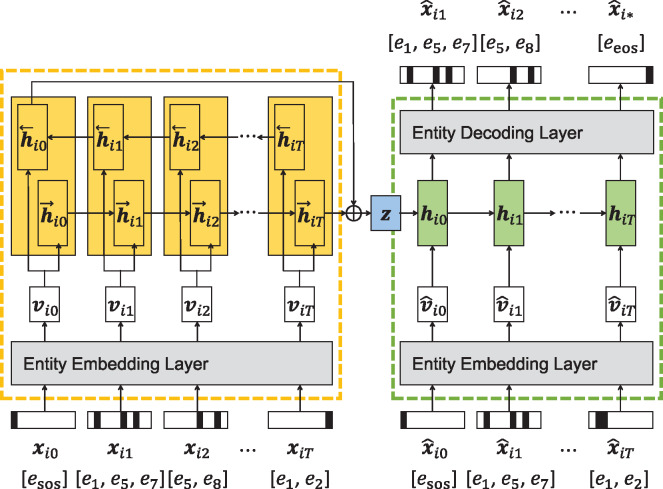
We employ gated recurrent unit (GRU) for our sequence encoder and decoder. The encoder adopts a bidirectional structure to encode both the forward and backward contexts of the sequences into low-dimensional code vectors. At first, the entity embedding layer which learns the semantics of all the entities maps a high-dimensional set-valued record to the low-dimensional embedding vector of the record . Then, the GRU layer takes the sequence of embedding vectors as its inputs and produces a final code vector by concatenating the last hidden states of the forward and backward GRUs. We finally apply the tanh activation to this encoder output.
Similarly, the GRU layer of the decoder sequentially takes the embedding vector of the set-valued record, which is generated at the previous time step. Its entity decoding layer outputs final high-dimensional vectors whose values are the assignment scores of all the entities, which represent how likely each entity would be assigned in the next record. Using the sigmoid activation, the entity decoding layer computes the scores that range in (0, 1), and we filter out the entities with the scores less than the predefined threshold. This decoding is repeated until the generated record contains the entity eeos. Figure 2 illustrates how our seq2seq autoencoder encodes a sequence into a code vector and decodes (or reconstructs) the sequence from the code vector.
Figure 2.
The detailed architecture of the sequence-to-sequence autoencoder in dual adversarial autoencoder architecture. The embedding layer encodes the semantics of all the medical entities, and the gated recurrent unit layer learns the temporal contexts within patients’ sequential records.
Inner GAN and outer GAN
Our generator and inner critic in the inner GAN use a multilayer perceptron equipped with the batch normalization and the ReLU activation for each layer. The last layer of the generator adopts the tanh activation to match its outputs with the encoder’s. In case of the outer critic, we choose a convolutional neural network (CNN) to effectively extract discriminative features for the real and fake sequences. A simple CNN model designed for sentence classification33 showed a high accuracy while employing a simple architecture, so its architecture has been widely used for modeling various sequential data.34–36
More specifically, the CNN critic takes the score vector (rather than the multihot vector or ) from the decoder as its inputs, so that the gradients from the outer GAN loss can be delivered to the GRU decoder. It utilizes the entity embedding layer, which maps the input score vectors into the low-dimensional vectors, and this layer shares the parameters with the embedding layer in the seq2seq autoencoder.
Baseline models
To evaluate the performance of DAAE in terms of data generation, we choose baseline models from (1) variants of EHR-GANs: such as medical GAN (medGAN)7; and (2) generative autoencoders: VAE,16 VAE-GAN,37 Wasserstein autoencoder (WAE),25 and adversarially regularized autoencoder (ARAE).26 As medGAN learns independent set-valued records, not a sequence, we randomly select 2 noises and sequentially synthesize the samples by using interpolated noises. This setting can be interpreted that a patient’s medical condition changes from a random point to the another random point during time steps. Each length is sampled from the length distribution of the original data. Note that medGAN is the only baseline model designed for EHRs because generative models for the set-valued sequences from the EHR do not exist. Although several variants of medGAN have been proposed to improve their performances, all of them are not able to synthesize the sequential clinical records, which is necessary for representing the chronological medical history of each patient; for this reason, we only choose medGAN as our baseline.13,38 Thus, for fair comparisons, we employ the same architecture of the seq2seq autoencoder for all the other baseline models (ie, VAE, VAE-GAN, WAE, and ARAE) with that of DAAE.
Evaluation strategy
In our experiments, we demonstrated the superiority of our model to other baseline models by quantitatively and qualitatively comparing them in terms of the following aspects: (1) the quality of synthetic sequences, (2) the obtained code distributions, and (3) the DP guarantee.
Evaluation of synthetic sequences
We first compared the accuracy of prediction models trained using synthetic sequences obtained from our model and other baseline models. To be specific, we trained single-layer long-short-term memory (LSTM) (to be specific, we employ the model architecture of DoctorAI, which is designed for the same task based on the EHR)39 to predict top- diagnosis codes in the next set-valued record within a sequence (ie, many-to-many LSTM). The same number of synthetic sequences as the number of real sequences were used to train each model, and we measured top- recall and precision values as performance metrics. We repeated the experiments 5 times using different sets of generated sequences, and reported the average results.
Then, we measured the classification accuracy of 2 different discriminators trained to classify the real and fake samples. A CNN with a single convolutional layer33 and a single-layer bidirectional LSTM (ie, many-to-one LSTM) were selected as the classifiers in order to consider context features as well as realistic and unrealistic features. They were trained using the synthetic samples mixed with the same number of real samples. In general, the more similar the data and model distributions are, the worse performance a discriminator would achieve. In the ideal case that the model distribution is exactly identical to the data distribution, their classification accuracy should be 0.5, which means that a classifier cannot distinguish the 2 distributions at all.
Finally, we compared the plausibility scores of synthetic sequences, qualitatively assessed by domain experts. We synthesized sequences (40 for each model) and asked 3 medical experts, who do not know the model each sequence is generated by, to score how realistic each sequence is using the scale [0, 10]. The raters conducted comprehensive evaluation on the realistic-ness of the sequences in terms of (1) the plausibility of set-valued records (eg, co-occurrences of diagnosis codes), (2) the sequential contexts within a sequence, and (3) the characteristics of the target dataset (eg, intensive care unit patients or outpatients).
Qualitative analysis on code distribution
Analyzing the fidelity and coverage of the generated samples on the data space is challenging because of the discreteness and high dimensionality of the EHR, so we instead investigated the continuous code space obtained by each generative model. We observed that the sequences in the EHR do not have distinctly different classes (or modes), unlike the other benchmark datasets that have few representative classes (eg, MNIST [modified National Institute of Standards and Technology). Thus, we first ran the DBSCAN (density-based spatial clustering of applications with noise) algorithm40 on each code space to identify several significant modes. We visualized the code distribution (ie, a marginalized posterior) induced by the encoder by using t-distributed stochastic neighbor embedding (t-SNE).41
Analysis on DP guarantee
We also investigated the performance of DAAE under different privacy cost induced by the DP-Adam optimizer. The privacy cost is represented by the value of , as we fixed to 0.001 and varied the noise scale (defined in theorem 1) from 0.5 to 10. Note that a smaller value indicates the stronger privacy protection guaranteed by the model. We reported the test recall values of the subsequent code prediction task (ie, sequence modeling) on real sequences, after training the LSTM model using the synthetic sequences.
RESULTS
Evaluation of synthetic sequences
Table 2 shows how effective synthetic sequences are to train the sequence model for subsequent code prediction, compared with real sequences. DAAE achieves the best accuracy among all the baselines, and notably, training on our synthetic data shows the comparable accuracy to the case of training directly on real data. DAAE significantly improves the performance against ARAE, and it implies that our outer GAN is helpful to improve the sample quality.
Table 2.
Recall@ and Precision@ for the subsequent code prediction task (%). The sequence models are trained on the synthetic samples generated from each model, and evaluated on real data
| MIMIC-III |
UTP |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Recall@ |
Precision@ |
Recall@ |
Precision@ |
|||||||||
| Metric | 10 | 20 | 30 | 10 | 20 | 30 | 5 | 10 | 15 | 5 | 10 | 15 |
| medGAN | 13.3 | 19.5 | 23.8 | 17.5 | 13.1 | 10.9 | 18.9 | 27.2 | 33.4 | 10.4 | 7.3 | 5.8 |
| VAE | 21.7 | 29.0 | 33.7 | 26.5 | 18.6 | 14.7 | 17.3 | 21.3 | 23.9 | 9.5 | 5.8 | 4.3 |
| VAE-GAN | 22.4 | 30.5 | 35.5 | 27.9 | 19.7 | 15.6 | 12.1 | 13.4 | 13.8 | 6.8 | 3.8 | 2.6 |
| WAE | 26.1 | 35.9 | 42.0 | 32.1 | 23.1 | 18.4 | 31.4 | 37.4 | 41.1 | 16.5 | 9.9 | 7.3 |
| ARAE | 26.3 | 36.1 | 42.6 | 31.8 | 23.1 | 18.5 | 31.0 | 40.3 | 45.2 | 16.7 | 10.5 | 7.9 |
| DAAE | 26.7 a | 36.9 a | 43.3 a | 32.8 a | 23.7 a | 19.0 a | 32.4 a | 40.8 a | 46.2 a | 17.0 a | 10.7 a | 8.1 a |
| Real | 26.8 | 37.2 | 43.9 | 32.9 | 23.8 | 19.1 | 33.6 | 42.4 | 47.8 | 17.5 | 11.1 | 8.3 |
ARAE: adversarially regularized autoencoder; DAAE: dual adversarial autoencoder; medGAN: medical generative adversarial network; MIMIC-III: Medical Information Mart for Intensive Care-III; UTP: UT-Physicians; VAE: variational autoencoder; VAE-GAN: variational autoencoder with generative adversarial network; WAE: Wasserstein autoencoder.
We observe that the sequence model trained on the samples synthesized by medGAN cannot predict subsequent codes as accurately as the others do. Existing generative models designed for EHRs, including medGAN, learn medical records independently while excluding their temporal contexts, and consequently they fail to generate sequential records that are similar to real ones in the training set. In other words, even though medGAN turned out to be effective in generating realistic EHRs compared with naïve baselines,2 such as random noise and independent sampling, it still has a limitation to reproduce complex relationships (or co-occurrences of diagnosis codes) among different records within a single patient trajectory.
Table 3 presents the indistinguishability of synthetic and real samples based on binary classification using parametric classifiers. As shown in the results, both the classifiers achieve the worst performance on our synthetic samples. This result indirectly shows that DAAE can generate more realistic sequences that are not easy to be distinguished from real sequences, compared with the other baselines.
Table 3.
Binary classification accuracies of simple discriminators trained to distinguish real samples from fake samples generated by each model
| Dataset | Discriminators | medGAN (%) | VAE (%) | VAE-GAN (%) | WAE (%) | ARAE (%) | DAAE (%) |
|---|---|---|---|---|---|---|---|
| MIMIC-III | CNN | 98.7 | 97.5 | 95.5 | 78.2 | 74.5 | 71.3 a |
| Bi-LSTM | 97.6 | 94.3 | 95.4 | 76.3 | 74.1 | 70.7 a | |
| UTP | CNN | 99.4 | 99.4 | 99.3 | 99.5 | 86.2 | 83.8 a |
| Bi-LSTM | 99.5 | 99.6 | 99.5 | 99.8 | 86.7 | 84.3 |
ARAE: adversarially regularized autoencoder; Bi-LSTM: bidirectional long-short-term memory; CNN: convolutional neural network; DAAE: dual adversarial autoencoder; GAN: generative adversarial network; medGAN: medical generative adversarial network; MIMIC-III: Medical Information Mart for Intensive Care-III; UTP: UT-Physicians; VAE: variational autoencoder; WAE: Wasserstein autoencoder.
Before comparing the plausibility scores of synthetic sequences evaluated by medical experts, we first tested the Kendall coefficient to investigate the interrater reliability on ordinal rating scores, and the results of 0.66 (for MIMIC-III) and 0.83 (for UTP) indicate the 3 raters consistently evaluate the samples. Figure 3 summarizes the plausibility scores as a boxplot, and the sequences of DAAE achieve higher scores than do those of ARAE, the state-of-the-art generative model. Interestingly, in case of MIMIC-III, the sequences of VAE are ranked as the best, but most of their records turn out to contain only 1 or 2 diagnosis codes (ie, not informative enough); this is similar to the situation where a mode-collapsed model synthesize only few types of realistic images easy to learn. In addition to the evaluation by the parametric classifiers, this medical review emphasizes the qualitative aspect of DAAE.
Figure 3.
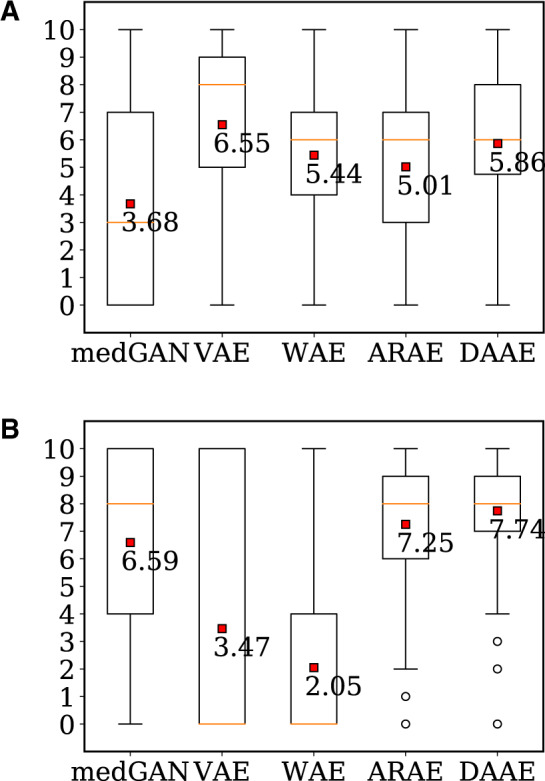
Plausibility scores evaluated by medical experts. (a) Dataset: MIMIC-III (b) Dataset: UTP. ARAE: adversarially regularized autoencoder; DAAE: dual adversarial autoencoder; medGAN: medical generative adversarial network; VAE: variational autoencoder; WAE: Wasserstein autoencoder.
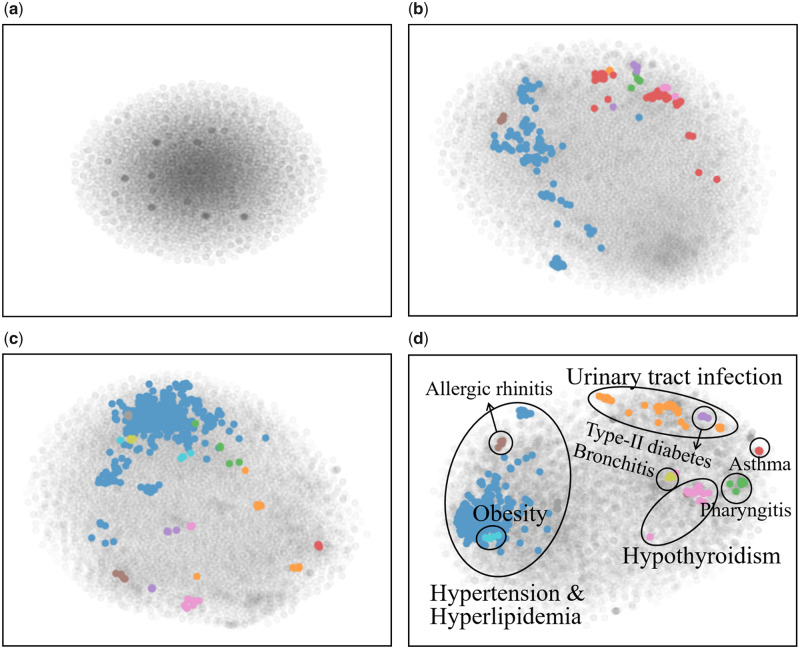
Qualitative analysis on code distribution
Figure 4 illustrates the code distributions induced by the encoder of DAAE and other generative models. A few modes are identified from the code spaces of WAE, ARAE, and DAAE, whereas DBSCAN cannot find any modes from that of VAE and VAE-GAN. This implies that VAE-based models suffer from the posterior collapse problem, whose encoder carries no information and decoder degenerates into an unconditional sequence model. On the contrary, specific disease-related patient groups are identified in case of DAAE, which demonstrates that DAAE is capable of effectively encoding the semantics of sequences into the code space. From these observations, we can conclude that DAAE effectively learns the underlying latent distribution of the data by adversarially optimizing the inner critic and the generator.
Figure 4.
The code distribution (gray points) with identified modes (colored points). (a) VAE, (b) WAE, (c) ARAE, and (d) DAAE. Dataset: UT Physicians. ARAE: adversarially regularized autoencoder; DAAE: dual adversarial autoencoder; medGAN: medical generative adversarial network; VAE: variational autoencoder; WAE: Wasserstein autoencoder.
Analysis on differential privacy guarantee
Figure 5 shows the performance changes of our model with respect to its privacy guarantees. DAAE is successfully optimized to guarantee the differential privacy while slightly compromising the performance. In case of UTP, our synthetic sequences with a very tight privacy guarantee (=1.06, =0.001) achieve the recall value of 44.8, showing that DAAE preserves a good model utility (see Table 2 for the performances without DP). In conclusion, the DP optimizer enables DAAE to publish synthetic patients’ sequences realistic enough for research purposes, with the rigorous privacy guarantee.
Figure 5.
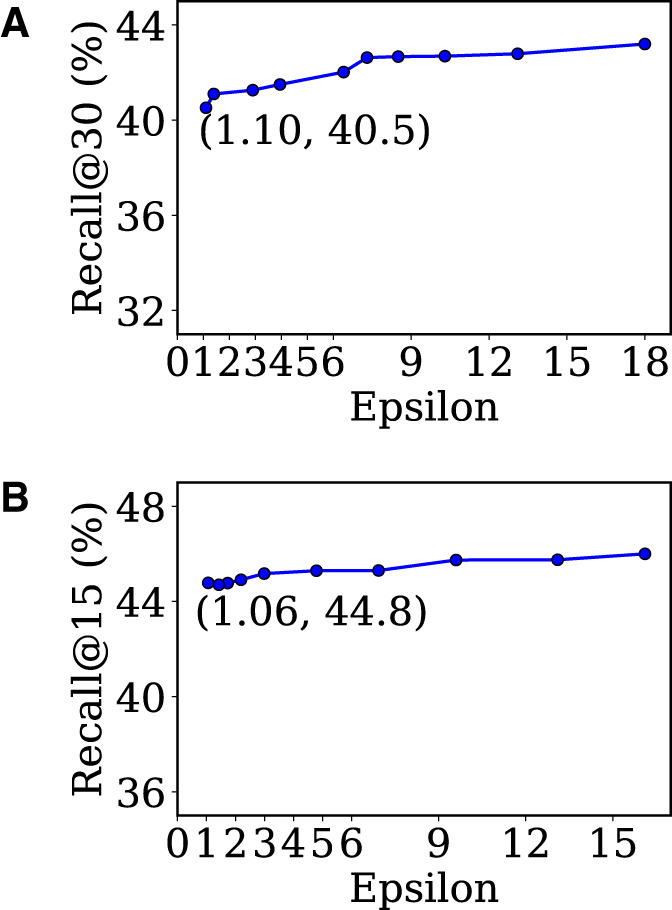
Sequence modeling accuracies achieved by synthetic sequences with different privacy cost. (a) Dataset: MIMIC-III, (b) Dataset: UTP.
DISCUSSION
This research has been conducted to build synthetic EHRs, specifically the sequences of high-dimensional and discrete clinical records. First of all, by designing a novel seq2seq autoencoder for set-valued records and combining it with GANs, this work makes it possible to synthesize a bunch of clinical trajectories of fake patients. Second, the outer GAN and inner GAN in our proposed model successfully address the important challenges of sequential EHR generation, which are that (1) the synthetic records should be realistic enough not to be distinguished from the real records and (2) they should cover all the training patients to reproduce the performance of specific downstream tasks. Finally, our model is trained with rigorous privacy guarantee by a differentially private optimizer, while preserving the good model utility. These implications mitigate the difficulties in obtaining real EHR data as well as handling their privacy issues, and consequently contribute to the technical progress of machine learning in medicine.
Nevertheless, there still remain several limitations that this study has not taken into consideration. First, we mainly validate our proposed model by using the subsequent code prediction task, which indirectly evaluates the synthetic records. Although the comparable performance of our synthetic sequences to that of real sequences strongly indicates that they accurately capture sequential contexts within the trajectories of training patients, the evaluation depends on the specific sequence model (ie, LSTM) and does not consider other properties of clinical records. Furthermore, we only modeled and learned ICD-9 codes for medical entities in EHR. There exist other types of structured codes, including medication codes and procedure codes, so our model should be extended so that it can model the causal relationships between different types of codes. We leave these challenges as our future work.
CONCLUSION
This article proposes DAAE, a novel deep generative model to learn set-valued sequences about patients’ longitudinal records. DAAE improves both the mode coverage and quality of synthetic samples with the help of 2 adversarial networks. As a result, our synthetic sequences achieve the comparable results to real data for predictive modeling, and they are assessed to be realistic enough by parametric classifiers and domain experts. Finally, the fake sequences generated by DAAE with strict privacy guarantee are expected to be widely utilized for machine learning researches on the medical domain, without increasing the privacy leakage of real patients’ data.
FUNDING
This research was supported by Institute for Information & communications Technology Protmotion (IITP) grant funded by the Korea government (no. 2018-0-00584). XJ is the Cancer Prevention and Research Institute of Texas Scholar in Cancer Research (RR180012), and was supported in part by a Christopher Sarofim Family Professorship, UT Stars award, UTHealth startup, the National Institutes of Health under award numbers R01GM114612, R01GM118574, and U01TR002062.
AUTHOR CONTRIBUTIONS
DL implemented the work, carried out experiments, analyzed results, and drafted the manuscript. XJ and HY substantially contributed to the work by supervising the whole research and advising in drafting the manuscript. DR, MG, and MT, who are medical experts, assessed the plausibility of synthetic records generated by the proposed model and baselines with their clinical judgment. QZ and LX helped to implement differentially private optimization of the generative model.
CONFLICT OF INTEREST STATEMENT
The authors have no competing interests to declare.
REFERENCES
- 1. El Emam K, Rodgers S, Malin B. Anonymising and sharing individual patient data. BMJ 2015; 350: h1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. El Emam K, Jonker E, Arbuckle L, et al. A systematic review of re-identification attacks on health data. PLoS One 2011; 6 (12): e28071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. El Emam K, Dankar FK, Neisa A, et al. Evaluating the risk of patient re-identification from adverse drug event reports. BMC Med Inform Decis Mak 2013; 13 (1): 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dankar FK, El Emam K, Neisa A, et al. Estimating the re-identification risk of clinical data sets. BMC Med Inform Decis Mak 2012; 12 (1): 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Simon GE, Shortreed SM, Coley RY, et al. Assessing and minimizing re-identification risk in research data derived from health care records. EGEMS (Wash DC) 2019; 7 (1): 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Department of Health and Human Services. Standards for privacy of individually identifiable health information. Federal Register 2002. https://www.federalregister.gov/documents/2002/08/14/02-20554/standards-for-privacy-of-individually-identifiable-health-information [PubMed] [Google Scholar]
- 7. Choi E, Biswal S, Malin B, et al. Generating multi-label discrete patient records using generative adversarial networks. Proc Machine Learn Healthcare 2017; 286–305. [Google Scholar]
- 8. Beaulieu-Jones BK, Wu ZS, Williams C, et al. Privacy-preserving generative deep neural networks support clinical data sharing. Circ Cardiovasc Qual Outcomes 2019; 12 (7): e005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nie D, Trullo R, Lian J, et al. Medical image synthesis with context-aware generative adversarial networks. In: proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention; 2017: 417–425. [DOI] [PMC free article] [PubMed]
- 10. Xue Y, Xu T, Zhang H, et al. Adversarial network with multi-scale l1 loss for medical image segmentation. Neuroinformatics 2018; 16 (3–4): 383–92. [DOI] [PubMed] [Google Scholar]
- 11. Spinks G, Moens MF. Generating continuous representations of medical texts. In: proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Demonstrations; 2018: 66–70.
- 12. Zhang C, Li Y, Du N, et al. On the generative discovery of structured medical knowledge. In: proceedings of the ACM SIGKDD International Conference on Knowledge Discovery Data Mining; 2018: 2720–8.
- 13. Baowaly MK, Lin CC, Liu CL, et al. Synthesizing electronic health records using improved generative adversarial networks. J Am Med Inform Assoc 2019; 26 (3): 228–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodfellow I, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. In: proceedings of the 27th International Conference on Neural Information Processing Systems (NIPS’14); 2014: 2672–2680.
- 15. Arjovsky M, Bottou L. Towards principled methods for training generative adversarial networks. arXiv:1701.04862; 2017.
- 16. Kingma DP, Welling M. Auto-encoding variational Bayes. In: Proceedings of the International Conference on Learning Representations; 2014. https://arxiv.org/abs/1312.6114
- 17. Chen X, Duan Y, Houthooft R, Schulman J, Sutskever I, Abbeel P. InfoGAN: Interpretable representation learning by information maximizing generative adversarial nets. In: proceedings of the 30th International Conference on Neural Information Processing Systems (NIPS’16); 2016; 2172–80.
- 18. Isola P, Zhu JY, Zhou T, et al. Image-to-image translation with conditional adversarial networks. In: proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2017: 1125–1134.
- 19. Nguyen T, Le T, Vu H, Phung D. Dual discriminator generative adversarial nets. In: proceedings of the 31th International Conference on Neural Information Processing Systems (NIPS’17); 2017: 2670–80.
- 20. Yu L, Zhang W, Wang J, Yu Y. SeqGAN: Sequence generative adversarial nets with policy gradient. In: proceedings of the 31st AAAI Conference on Artificial Intelligence (AAAI-17); 2017: 2852–8.
- 21. Che T, Li Y, Zhang R, et al. Maximum-likelihood augmented discrete generative adversarial networks. arXiv:1702.07983; 2017. https://arxiv.org/abs/1702.07983
- 22. Li J, Lan Y, Guo J, Xu J, Cheng X. Long text generation via adversarial training with leaked information. In: proceedings of the 33rd AAAI Conference on Artificial Intelligence (AAAI-19); 2019: 6682–9.
- 23. Bowman SR, Vilnis L, Vinyals O, Dai A, Jozefowicz R, Bengio S. Generating sentences from a continuous space. In: proceedings of the 20th SIGNLL Conference on Computational Natural Language Learning 2016: 10–21.
- 24. Makhzani A, Shlens J, Jaitly N, Goodfellow I, Frey B. Adversarial autoencoders. In: Proceedings of the International Conference on Learning Representations (Workshop); 2016. https://arxiv.org/abs/1511.05644
- 25. Tolstikhin I, Bousquet O, Gelly S, Schoelkopf B. Wasserstein auto-encoders. In: Proceedings of the International Conference on Learning Representations; 2018. https://arxiv.org/abs/1711.01558
- 26. Zhao J, Kim Y, Zhang K, Rush A, LeCun Y. Adversarially regularized autoencoders. Proc Mach Learn Res 2018; 80: 5902–11. [Google Scholar]
- 27. Subramanian S, Mudumba SR, Sordoni A, Trischler A, Courville A, Pal C. Towards text generation with adversarially learned neural outlines. In: proceedings of the 32nd Conference on Neural Information Processing Systems (NIPS 2018); 2018: 7562–74.
- 28. Goldberger AL, Amaral LAN, Glass L, et al. Physiobank, physiotoolkit, and physionet: components of a new research resource for complex physiologic signals. Circulation 2000; 101 (23): e215–20. [DOI] [PubMed] [Google Scholar]
- 29. Johnson AEW, Pollard TJ, Shen L, et al. Mimic-iii, a freely accessible critical care database. Sci Data 2016; 3 (1): 160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gulrajani I, Ahmed F, Arjovsky M, Dumoulin V, Courville A. Improved training of wasserstein GANS. In: proceedings of the 31st Conference on Neural Information Processing Systems (NIPS’17); 2017; 5769–79.
- 31. Dwork C, Roth A. The algorithmic foundations of differential privacy. Foundations Theor Comput Sci 2013; 9 (3–4): 211–407. [Google Scholar]
- 32. Abadi M, Chu A, Goodfellow I, et al. Deep learning with differential privacy. In: proceedings of the 2016 ACM SIGSAC Conference on Computer and Communications Security (CCS ’16); 2016: 308–18.
- 33. Kim Y. Convolutional neural networks for sentence classification. In: proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP); 2014: 1746–51. [DOI] [PMC free article] [PubMed]
- 34. Abdel-Hamid O, Mohamed AR, Jiang H, et al. Convolutional neural networks for speech recognition. IEEE/ACM Trans Audio Speech Lang Process 2014; 22 (10): 1533–45. [Google Scholar]
- 35. Kim D, Park C, Oh J, et al. Convolutional matrix factorization for document context-aware recommendation. In: proceedings of the 10th ACM Conference on Recommender Systems (RecSys ’16); 2016: 233–240.
- 36. Chan PPK, Hu X, Zhao L, Yeung DS, Liu D, Xiao L. Convolutional neural networks based click-through rate prediction with multiple feature sequences. In: proceedings of the 27th International Joint Conference on Artificial Intelligence (ICJAI ’18); 2018; 2007–13.
- 37. Larsen ABL, Sønderby SK, Larochelle H, Winther O. Autoencoding beyond pixels using a learned similarity metric. Proc Mach Learn Res 2016; 48: 1558–66. [Google Scholar]
- 38. Zhang Z, Yan C, Mesa DA, et al. Ensuring electronic medical record simulation through better training, modeling, and evaluation. J Am Med Inform Assoc 2020; 27 (1): 99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi E, Bahadori MT, Schuetz A, Stewart WF, Sun J. Doctor AI: predicting clinical events via recurrent neural networks. Proc Mach Learn Res 2016; 56: 301–18. [PMC free article] [PubMed] [Google Scholar]
- 40. Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. In: proceedings of the 2nd International Conference on Knowledge Discovery & Data Mining (KDD’96); 1996: 226–31.
- 41. Maaten LVD, Hinton G. Visualizing data using t-SNE . J Machine Learn Res 2008; 9: 2579–605. [Google Scholar]