Abstract
Objective
Predictive disease modeling using electronic health record data is a growing field. Although clinical data in their raw form can be used directly for predictive modeling, it is a common practice to map data to standard terminologies to facilitate data aggregation and reuse. There is, however, a lack of systematic investigation of how different representations could affect the performance of predictive models, especially in the context of machine learning and deep learning.
Materials and Methods
We projected the input diagnoses data in the Cerner HealthFacts database to Unified Medical Language System (UMLS) and 5 other terminologies, including CCS, CCSR, ICD-9, ICD-10, and PheWAS, and evaluated the prediction performances of these terminologies on 2 different tasks: the risk prediction of heart failure in diabetes patients and the risk prediction of pancreatic cancer. Two popular models were evaluated: logistic regression and a recurrent neural network.
Results
For logistic regression, using UMLS delivered the optimal area under the receiver operating characteristics (AUROC) results in both dengue hemorrhagic fever (81.15%) and pancreatic cancer (80.53%) tasks. For recurrent neural network, UMLS worked best for pancreatic cancer prediction (AUROC 82.24%), second only (AUROC 85.55%) to PheWAS (AUROC 85.87%) for dengue hemorrhagic fever prediction.
Discussion/Conclusion
In our experiments, terminologies with larger vocabularies and finer-grained representations were associated with better prediction performances. In particular, UMLS is consistently 1 of the best-performing ones. We believe that our work may help to inform better designs of predictive models, although further investigation is warranted.
Keywords: UMLS, terminology representation, predictive modeling, electronic health records
INTRODUCTION
In the current big data era of biomedical informatics, abundant electronic health record (EHR) data are becoming available, leading to the development of predictive modeling algorithms. In the past 5 years, thousands of predictive modeling-related studies have utilized a variety of methods, such as logistic regression (LR) or deep learning, to predict the patient’s risk of developing such diseases as heart failure1–5 and pancreatic cancer (PC).6,7 An important, but unaddressed, research question in regard to predictive modeling is how to efficiently feed the EHR data to models.8–10
Structured diagnosis data in EHR datasets are usually heterogeneous, leading to challenges in data analysis, including interpretability and generalizability issues. For example, different hospitals and departments use different terminologies; thus, to develop a generalizable model, researchers either train the model on all of the different terminologies in use or introduce a standardizer that can normalize the data into a single terminology.
Terminology standards are evolving constantly, and newer versions will introduce additional levels of data redundancy. For example, patient diagnosis information was commonly stored in the International Classification of Diseases-ninth revision (ICD-9) format before 2015; but then, for billing purposes, hospitals had to upgrade it to the tenth revision (ICD-10), which introduced a higher level of details. Currently, an even newer revision, ICD-11, is being released. Further, in many cases, the coding system in EHRs is a mix of multiple ICD terminologies. As a result, it is difficult to organize information represented in heterogeneous formats and for models trained on older terminologies (eg, ICD-9 codes) to generalize to new terminologies without proper normalizations.
EHR vendors also are introducing internal codes that can be mapped to different standard terminologies in a one-to-one manner to facilitate various system functionalities. Using such codes for predictive model training may restrict the generalizability of such models to vendor-specific solutions or even to a single hospital if the mappings are different between sites. In addition, many of the existing terminology mappings are in many-to-many styles, which might hinder the accuracy and the interpretability of the model.
Terminology normalization involves assigning a unique standard medical term to a health condition.11 Most terminology mapping and normalization-related studies concern the development of mappings between different terminologies,12–14 the tools developed for automated mapping suggestions, or the development of concept embeddings based on different terminologies.15–19 There are, however, several practical questions on terminology normalization that have not been addressed. The first is how to find the optimal level of granularity required for predictive modeling, assuming that the data source is homogeneous. For example, it is not known whether we should use the diagnosis information as originally recorded in the dataset or group similar or relevant codes to reduce the input dimension.
The second is how important terminology normalization is when the data source is heterogeneous. Rajkomar et al10 described the advantage of using the Fast Healthcare Interoperability Resources format for interchangeable information representation but acknowledged that the limited semantic consistency from unharmonized data may have a negative impact on the model performance. In our previous work,4 we compared the use of Clinical Classifications Software (CCS) codes with the raw data from Cerner HealthFacts and found that grouping diagnosis codes was not helpful, which conflicts with the findings of other studies2,9 that found the CCS grouping was helpful. Notably, our findings also were supported by those of other studies.18,20 Unified Medical Language System (UMLS) provides a multipurpose knowledge source and attracts more research attention, as it includes mappings to almost all clinical terminologies at different hierarchical levels.21 It also has been broadly used in concept normalizations in the natural language processing domain;17,22,23 thus, we selected it as our most expressive terminology.
OBJECTIVE
In this study, our objectives are 2-fold. The first objective is to compare simply feeding the models with the raw data, as they were originally collected, versus preprocessing the data when mapping it to a single terminology. The second is to evaluate the performance of predictive models using UMLS and 5 other terminologies commonly used in the healthcare analytics domain. We used 2 clinical prediction tasks: predicting the risk of developing heart failure (DHF) among a cohort of type-II diabetes mellitus (DMII) patients and the risk of developing (PC). Our study cohorts were extracted from the Cerner HealthFacts database, a deidentified EHR database extracted from over 600 hospitals with which Cerner has a data use agreement. The original diagnosis data are coded with a unique diagnosis identifier (Cerner-Diagnosis ID) that can be mapped to ICD-9, ICD-10-CM, or ICD-10-CA codes in a one-to-one manner. For comparison, we further mapped the diagnoses codes to 6 terminologies, including UMLS concept unique identifier (CUI),24 ICD-9,25 ICD-10,25 PheWAS,26 and CCS codes in both the single-level27,28 and its refined version (CCSR).29 We compared the performances using L2 penalized LR (L2LR) and a bidirectional recurrent neural network (RNN)-based predictive model.
MATERIALS AND METHODS
Prediction tasks and cohort description
We evaluated the use of patients’ diagnosis information in different terminology representations on 2 different prediction tasks. The first task is to predict DHF in patients with DMII after at least 1 month of their first DMII diagnosis. The second task is to predict whether the patient will be diagnosed with pancreatic cancer (PC) in the next visit. The second task is more like a diagnosis aid, as we did not specify a prediction window.
We extracted our cohorts from the Cerner HealthFacts dataset version 2017,30 which includes deidentified patient information from more than 600 hospitals for more than a 15-year period. The full cohort for the DHF prediction in DMII patients consists of 70 782 cases and 1 095 412 controls denoted as the “DHF full cohort,” out of which we randomly selected a sample of 60 000 cases and 60 000 controls for terminology evaluations further denoted as the “DHF cohort.” Table 1 shows the descriptive analysis of the selected sample versus the full cohort. For PC prediction, we found 11 486 eligible cases in the population who were 45 years or older and did not report any other cancer diseases before their first PC diagnosis. From a pool of more than 25 million matched controls, we randomly selected 17 919 controls to build our PC experimental cohort, which was denoted as the “PC cohort.” We further randomly split each sample cohort into training, validation, and test sets using the ratio of 7:1:2.
Table 1.
Description of cohortsa
| Characteristic | DHF full cohort |
DHF cohort (study sample) |
PC cohort (study sample) |
|||
|---|---|---|---|---|---|---|
| Case | Controls | Case | Controls | Case | Controls | |
| Cohort size (n) | 70 782 | 1 095 412 | 60 000 | 60 000 | 11 486 | 17 919 |
| Male % | 49% | 47% | 49% | 46% | 47% | 43% |
| Age [mean (std. dev.)] | 70 (12) | 60 (14) | 70 (12) | 60 (14) | 69 (19) | 63 (13) |
| Race | ||||||
| White (%) | 76% | 70% | 77% | 71% | 80% | 75% |
| African American (%) | 17% | 16% | 16% | 16% | 14% | 12% |
| Average number of visits | 13 | 16 | 14 | 15 | 7 | 7 |
| Average number of codes | 28 | 32 | 30 | 31 | 23 | 21 |
Abbreviations: DHF, development of heart failure; PC, pancreatic cancer.
We used the patients’ diagnosis information only before the index visit, which is commonly the last eligible visit before prediction, to train the predictive models. Details of the cohorts’ composition are presented in Supplementary Appendix A.
We used the patients’ diagnosis information only before the index visit, which is commonly the last eligible visit before prediction, to train the predictive models. Details of the cohorts’ composition are presented in Supplementary Appendix A.
Diagnosis terminology
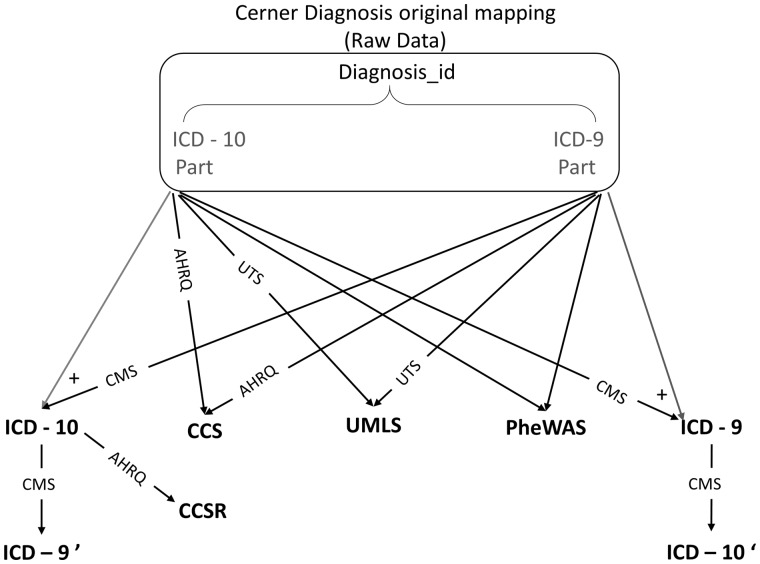
Cerner HealthFacts v. 2017 includes 17 629 ICD-9 codes, 94 044 ICD-10-CM codes, and 16 044 ICD-10-CA codes, each of which is mapped to a unique Cerner-Diagnosis ID that is used to unify the representation of diagnosis among all hospitals’ diagnoses data within the Cerner HealthFacts database. The patient’s diagnosis information is stored mainly through the use of Cerner-Diagnosis ID. The main advantage of using this raw data is that they include the information of the original code types used for documentation and can be directly used without any further processing. This dilutes the actual value of the patient diagnosis, however, as the same diagnosis may be represented by multiple codes. We also included the raw data using the Cerner-Diagnosis ID as a baseline terminology. Figure 1 shows our diagnosis terminology mapping roadmap.
Figure 1.
Terminology roadmap.
We used official resources for code mappings. For example, we used the Center of Medicare and Medicaid Services’ (CMS) most recent general equivalence mapping GEM version 201825 to map between ICD-9 and ICD-10 codes. For UMLS mapping, we used the UMLS knowledge resources available on the UMLS Terminology Services website,24 and we used the latest version of ICD-9 and ICD-10 to CCS single-level mapping available on the Agency of Healthcare Research and Quality website27,28 as well as the CCS refined version.29 For PheWAS mapping, we used the Phecode maps available in the PheWAS catalog.26 We had to review the mappings to the raw data due to some typos in the Cerner diagnosis dictionary table that led to ICD-9/10 codes not exactly matching the corresponding records in different mapping files (mainly missing the last digit, which can be either 0 or 9).
All of the Cerner-Diagnosis IDs in our cohort were successfully mapped to CCS and ICD-9 codes, regardless of those that were mapped to “noDx” for no mapping as existing in the original mapping files. There were approximately 300 ICD-10 codes in our cohort that were not mapped that were associated with approximately 100 UMLS codes. We decided to ignore those codes, as they appeared only a maximum of 10 times in our cohort. Details of the different terminologies used and the mapping are provided in Supplementary Appendix B.
To understand whether the difference in the predictive model accuracy is due to the terminology representation itself or to the information loss induced by the mapping process, we focused on the ICD-9/ICD-10 conversion as an example. We converted the previously converted ICD-9 codes to ICD-10 and named them ICD-10 revert prime (ICD-10′). We did the same for the previously converted ICD-10 codes and converted them back to ICD-9 and named them ICD-9 revert prime (ICD-9′). For the revert prime mappings, we used only the original mapping files provided by CMS without any further review or improvement. For reproducibility, we share our codebase and mappings on https://github.com/ZhiGroup/terminology_representation.
Tasks and models
We evaluated the usefulness of the terminologies described above for 2 tasks. The first task is the prediction of the DMII patients’ risk of DHF after 30 days from their first diabetes diagnosis. The second task is the calculation of the risk score of the patient to be diagnosed with PC based on the patient’s history until the most recent visit.
For both tasks, we evaluated 2 models: L2LR and RNN. LR is a popular model for its accuracy and interpretability. The majority of currently implemented predictive models are based on LR. We used one-hot encoding for the presence/absence of any diagnosis code as input for LR. We used the default LR implementation available in the Scikit-Learn31 package which includes L2 penalty for regularization. We also experimented with hyperparameter grid search for the L2 penalty. In addition, we evaluated a bidirectional RNN. RNNs are appropriate for modeling the sequential nature of patient medical records and have been shown to provide high predictive accuracy in the healthcare domain.1,2,4,32 Following Choi et al,2–4,33 we represented a patient record as a sequence of visits (encounters) and each encounter as a set of diagnosis codes. We used an embedding layer to transform one-hot input diagnosis vectors into dense vectors and then used a bidirectional gated recurrent unit for propagating information across visits and a fully connected layer for the output label. Hyperparameters were chosen by Bayesian optimization. This architecture was shown to be very competitive in our previous benchmark.34 We used our previously published code on https://github.com/ZhiGroup/pytorch_ehr. A detailed description of our model implementation is available in Supplementary Appendix C.
Statistical analysis for model comparison
We used the area under the receiver operating characteristic curve (AUROC) as the evaluation metric for the model prediction accuracy. For deterministic methods, such as L2LR, we apply the Delong test35 to calculate the significance of the difference between different models’ AUROC. For probabilistic methods, such as RNN (due to random initialization of model parameters), we repeated the analyses for RNN models of each terminology 10 times, and multi-group 1-way ANOVA tests (unpaired t-tests for 2 groups) were used for comparing the means of each terminology. All-pairwise Tukey-Kramer analysis was used to identify significant group-wise differences.
RESULTS
As noted, the description of both cohorts is presented in Table 1. Also as noted, we lost some patient information for the incomplete terminology mapping, mainly for the primary ICD-10 to 9 code mappings and the reversed prime conversions. Nevertheless, that rarely leads to loss of a complete patient sample for the initial rules of the minimum number of visits, and original diagnosis codes were redefined before the random sample selection in the DHF cohort. Thus, our test set of 24 000 patients remains consistent along with the evaluations of all of our models. For the PC cohort, only a couple of patients from our test set of 5881 patients were not included in the ICD-9 mappings; those patients were excluded from the reported results.
As shown in Table 2, for the DHF prediction, the test AUROC ranges between 78% and 81% for L2LR and between 83% and 85% for RNN. For the PC prediction, the test AUROC ranges between 77% and 80.5% for L2LR and between 79% and 82.5% for RNN. The difference between the RNN and L2LR AUROCs remains nearly the same among all diagnosis terminologies, approximately 4.9% on average for DHF and 1.5% for PC. The best L2LR models’ AUROC is associated with the use of UMLS-CUI on both tasks, whereas single-level CCS shows the worst AUROC in all tasks and models. The findings remain consistent even with the DHF full cohort (Supplementary Appendix D), for which UMLS showed the highest AUROC (82%). Also, our results remained consistent using LR with different regularization hyperparameters for both L1 and L2 regularizations (Supplementary Appendix E).Using the Delong test to understand the difference in the AUROC significance and with a P value of .0024 after Bonferroni correction (Figure 2A), we find that the UMLS results are significantly better than those for the other terminologies except for the raw data in PC prediction and PheWAS in DHF prediction. For RNN models, UMLS showed the highest AUROC for PC prediction, whereas PheWAS was associated with the best AUROC for DHF prediction. These pairwise comparison results are statistically significant based on the Tukey-Kramer procedure, as shown in Figure 2B and Supplementary Appendix F. We train and test the RNN models only once on the DHF full cohort (Supplementary Appendix D). UMLS was the second-best performer, with an AUROC of 85.52%, which is 0.34% less than that of the raw data, which showed the highest AUROC at 85.86%. The AUROC of PheWAS was lower at 85.07%.
Table 2.
Prediction performance of different diagnosis terminologies for the DHF and PC tasksa
| Diagnosis terminology | Diabetes heart failure cohort (DHF) |
Pancreatic cancer cohort (PC) |
||||
|---|---|---|---|---|---|---|
| Number of unique codes | L2LR | RNN | Number of unique codes | L2LR | RNN | |
| Raw data (Cerner-Diagnosis ID) | 26 427 | 80.61 | 85.48 (0.10) | 13 071 | 80.30 | 81.43 (0.37) |
| CCS-single level | 284 | 78.07 | 82.96 (0.15) | 253 | 77.23 | 79.03 (0.36) |
| CCSR | 538 | 78.87 | 84.17 (0.21) | 538 | 77.92 | 79.63 (0.34) |
| ICD-9 | 11 187 | 80.12 | 85.20 (0.13) | 7055 | 79.15 | 80.78 (0.32) |
| ICD-10 | 22 893 | 79.78 | 84.35 (0.20) | 13 620 | 78.95 | 79.27 (0.44) |
| PheWAS | 1820 | 80.71 | 85.87 (0.10) | 1715 | 78.82 | 81.15 (0.31) |
| UMLS CUI | 29 491 | 81.15 | 85.55 (0.06) | 14 551 | 80.53 | 82.24 (0.29) |
Abbreviations: AUROC, area under the receiver operating characteristics; DHF, development of heart failure; L2LR, L2 penalized LR; PC, pancreatic cancer; RNN, recurrent neural network.
L2LR and RNN show the average and the standard deviation for AUROC on the test set. Bold indicates the values with the highest AUROC per task/model.
Figure 2.
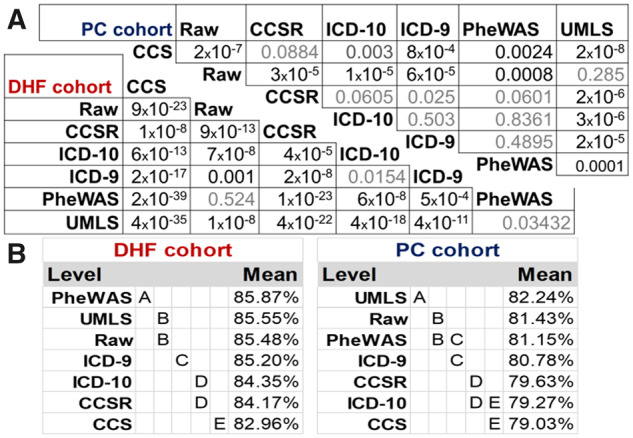
Significance of AUROC difference. (A) Logistic regression pairwise AUROC difference significance calculated using Delong test; P values less than .0024 are significantly different. (B) For the Tukey-Kramer honest significant difference test value, levels not connected by the same letter are significantly different.
The mapping to ICD-9 is always better than mapping to ICD-10, although those differences were not significant for L2LR models, based on the Delong test, but were significant for RNN models. We hypothesize that the result is due to the majority of the original data having been recorded in ICD-9, and, thus, mapping to ICD-10 will incur a loss of information during the terminological translation. We further investigated this loss-in-translation effect and report the results in the next section.
Effect of information loss due to terminology mapping
Mapping back from earlier converted ICD-10 codes to ICD-9 was associated with clear information loss that can be seen in the difference in the number of codes in our cohort; for example, our cohort originally had a 26 427-diagnosis code that mapped to a combination of ICD-9 and ICD-10 codes (Table 2). Approximately 70% of our patient diagnosis data already were coded in ICD-9 codes; so, after mapping the ICD-10 codes to ICD-9 and combining the codes with those data originally mapped to ICD-9 codes, we had 11 187 ICD-9 codes in our cohort. Thus, we can explain the decrease in the number of codes as a result of the grouping effect of the lower dimension ICD-9 codes, but, on mapping back to ICD-10 codes, the number of codes increases only to 14 644 codes, which is approximately 50% of the number of original diagnosis codes, or a little higher percentage of the primarily converted ICD-10 codes (22 893 codes). Such information loss may explain the significant decrease in AUROC, using the ICD-9′ and ICD-10′ sets (Table 3).
Table 3.
Difference in AUROC between primary mapping to ICD-9/10 codes and reversed mapping to ICD-9′/10′ codes
| Number of Codes | L2LR AUROC | Delong P value | RNN AUROC | Unpaired t-test P-value | |
|---|---|---|---|---|---|
| ICD-9 | 11 187 | 80.12 | P < .0001 | 85.20 (0.13) | P < .0001 |
| ICD-9′ | 9063 | 79.28 | 84.18 (0.09) | ||
| ICD-10 | 22 893 | 79.78 | P < .0001 | 84.35 (0.20) | P < .0001 |
| ICD-10′ | 14 644 | 79.23 | 83.12 (0.21) |
Abbreviations: AUROC, area under the receiver operating characteristics; L2LR, L2 penalized LR; RNN, recurrent neural network.
DISCUSSION
For L2LR models, the results were consistent between the 2 prediction tasks. UMLS showed the best performance, whereas CCS single-level mapping was associated with the lowest AUROC on both prediction tasks and on both models, which is consistent with our previous experiments.4 There were no significant differences between ICD-9 and ICD-10 code mapping, although ICD-9 mappings are always higher in our experiments. The findings remain consistent even when we evaluated the differences using the DHF full cohort (Supplementary Appendix D). There were no significant differences between CCS and CCSR codes in PC prediction, but the difference was significant for DHF prediction, which can be explained by the larger test set in the DHF cohort. In general, although LR models are not longitudinal, they are simple to use and have been the most commonly used models in EHR predictive modeling. Our results indicated that UMLS is often the top choice for predictive modeling when using LR models in our datasets.
For RNN models, the results vary between the different prediction tasks or between different cohort sizes. Whereas UMLS and PheWAS were the top-performing terminologies, their relative rankings change depending on the tasks. PheWAS was the best-performing model for DHF in the selected sample cohort, whereas UMLS was the best-performing for PC prediction. When evaluated using the DHF full cohort, raw data were associated with the best AUROC.
We note that it is not our main goal to benchmark models for realistic clinical tasks; therefore, the performance documented here does not necessarily translate to applicability in the real world. For example, PC risk prediction is a notoriously difficult task. Our PC performance may be due to biases in the data preparation. Nonetheless, our reported AUROCs are consistent with the range reported in previous studies for both DHF prediction1,2,4 and PC prediction.6,7
We admit that, although the differences among groups are often statistically significant, the actual effect sizes are not necessarily large. For RNN models, the maximum difference in mean AUROC among UMLS, PheWAS, and raw data in the DHF, and the PC cohorts were 0.4% and 1%, respectively. The lower difference seen in the DHF cohort were owing to the larger cohort size, as RNN models can easily overfit on smaller cohorts. Nonetheless, the effect of terminologies appears independent of model architecture, and thus terminology choice has a real impact on predictive modeling.
Although the choice of model architecture (LR vs RNN) has a major impact on prediction performance, the choice of terminologies also has a small but significant impact. Moreover, this impact is on top of the performance difference for model architectures. Therefore, terminology choice is a decision that has real-world impact.
To understand the key factors of terminologies that have an impact on prediction performance, we look at the characteristics of the best- and the worst-performing terminology mappings. There are 2 factors related to terminology mapping’s influence on the accuracy of clinical prediction models from EHRs: quality of the terminology and the quality of mapping. Although mapping to more expressive terminology is a common practice for expressive deep-learning models, it is common for traditional machine-learning methods, such as LR, to reduce dimensionality in search of a parsimonious model. Our results showed that, for both L2LR and RNN, large vocabulary sizes are associated with better performance. UMLS showed both the best performance in LR models and high performance with deep-learning models; it is the vocabulary with the highest number of codes and has the advantage of better semantic consistency and hierarchical relationships. Surprisingly, PheWAS, with a vocabulary size of only 1820, showed good performance compared to other terminologies with higher levels of granularity. This can be attributed to the careful definitions of the mapping, as it was revised based on statistical co-occurrence, code frequency, and human review.12,13,26 While the performance of the CCS- and CCSR-trained models were suboptimal during our experiments—mainly due to their smaller vocabulary sizes (284 and 538, respectively)—they may be still a good choice in practice due to their human readability. Not surprisingly, the use of raw data provides 1 of the best results as compared to other terminology-mapping exercises. Such a conclusion can give us assurance that models can learn from the current data without any further preprocessing. We can explain the good performance of the raw data-based models through 2 factors. First, the original coding type includes a level of important information for our prediction tasks. Second, the preprocessing and mapping exercise, although of high quality and including attention to detail, introduces some noise that may have impact on the model’s learning ability. In our study, the raw data are represented by the Cerner-Diagnosis ID that maps to different terminologies, such ICD-9 and ICD-10.
Mapping structured raw data to UMLS-CUI can lead to better integration with diagnosis information extracted from the unstructured text as well as data recorded in other terminologies, such as SNOMED-CT. Further, it will be easier to embed knowledge about relationships between different clinical entities, including diseases, medications, procedures, laboratory tests, and so forth.
There are several limitations to this study. The first is the lack of measurement of the quality of the codes’ mapping. We had observed a few incorrect ICD-9/ICD-10 codes in the Cerner diagnosis dictionary table, which could be due to data-entry typos. In addition, as we kept the hierarchical mapping especially when using the UMLS codes, the one-to-many mappings of codes may require extra scrutiny. The second is that the prediction labels are derived from the raw data that are coded in either ICD-9 or ICD-10. This is reflected by the superior performance of ICD-9 over ICD-10, as the majority of data were coded in ICD-9. In addition, this may create a bias that favors ICD-9 or ICD-10 over other terminologies. The third is that our findings are shown to be valid for only the tested terminologies, tasks, models, and data sets. The generalizability of our results to other scenarios warrants further study. The fourth is that, for the sake of simplicity, we focused on only a single element of the EHR data: the diagnosis information. Future work that evaluates the terminology representation on other elements, including medication, procedures, and laboratory tests, as well as the interactions between the terminology of those elements is warranted. We also plan to evaluate the same on different tasks to validate the generalizability of our conclusion.
CONCLUSION
Through benchmarking, we found that the normalization of EHR diagnosis data to the UMLS standard was the best (or second best) performing among tested terminologies for both prediction tasks and both prediction models. For research purposes or local model development, raw data, when the sample size is large enough, are often sufficient to achieve decent accuracy. If there is a need for diagnosis code grouping for dimension reduction, however, PheWAS, with fewer than 2000 codes, is the best option. The quality of mapping had an impact on our study findings. In our data set, ICD-9 had better results did than ICD-10 mainly because a larger proportion of the raw data was coded in ICD-9.
For a real-world project, when generalizability is a priority and the quality of terminology mapping is assured, we recommend normalization of terminologies to an expressive common terminology, such as UMLS. Due to information loss in translation in existing mapping tools, however, evaluation of mapping quality may be needed before determining the optimal target terminology for predictive modeling.
FUNDING
This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT) Grant No. RP170668; UTHealth Innovation for Cancer Prevention Research Training Program Pre-Doctoral Fellowship (CPRIT Grant No. RP160015); the National Cancer Institute Grant No. 1U24CA194215; and the National Institutes of Health Grant No. R01AI130460.
AUTHOR CONTRIBUTIONS
LR carried out the experiments and led the writing of the manuscript. FT developed the Cerner UMLS mappings. LR and YZ extracted the EHR data. YX and CT participated in the writing. HX and DZ conceived the original idea, contributed to the writing, and supervised the project. LR and DZ finalized the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
ACKNOWLEDGMENTS
We would like to acknowledge the use of the Cerner HealthFacts dataset and the assistance provided by the University of Texas Health Science Center in Houston School of Biomedical Informatics Data Service team.
CONFLICT OF INTEREST STATEMENT
None declared.
REFERENCES
- 1. Maragatham G, Devi S. LSTM model for prediction of heart failure in big data. J Med Syst 2019; 43 (5): 111. [DOI] [PubMed] [Google Scholar]
- 2. Choi E, Bahadori MT, Kulas JA, et al. RETAIN: an interpretable predictive model for healthcare using reverse time attention mechanism. Adv Neural Inf Process Syst 2016: 3504–12. http://papers.nips.cc/paper/6321-retain-an-interpretable-predictive-model-for-healthcare-using-reverse-time-attention-mechanism Accessed December 29, 2017 [Google Scholar]
- 3. Choi E, Schuetz A, Stewart WF, et al. Using recurrent neural network models for early detection of heart failure onset. J Am Med Inform Assoc 2017; 24 (2): 361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rasmy L, Zheng WJ, Xu H, et al. A study of generalizability of recurrent neural network-based predictive models for heart failure onset risk using a large and heterogeneous EHR data set. J Biomed Inform 2018; 84: doi: 10.1016/j.jbi.2018.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jin B, Che C, Liu Z, et al. Predicting the risk of heart failure with EHR sequential data modeling. IEEE Access 2018; 6: 9256–61. [Google Scholar]
- 6. Muhammad W, Hart GR, Nartowt B, et al. Pancreatic cancer prediction through an artificial neural network. Front Artif Intell 2019; 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hsieh MH, Sun L-M, Lin C-L, et al. Development of a prediction model for pancreatic cancer in patients with type 2 diabetes using logistic regression and artificial neural network models. Cancer Manag Res 2018; 10: 6317–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ayala Solares JR, Diletta Raimondi FE, Zhu Y, et al. Deep learning for electronic health records: A comparative review of multiple deep neural architectures. J. Biomed. Inform 2020; 101: 103337. [DOI] [PubMed] [Google Scholar]
- 9. Min X, Yu B, Wang F. Predictive modeling of the hospital readmission risk from patients’ claims data using machine learning: a case study on COPD. Sci Rep 2019; 9 (1): doi: 10.1038/s41598-019-39071-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rajkomar A, Oren E, Chen K, et al. Scalable and accurate deep learning with electronic health records. NPJ Digit Med 2018; 1: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subramanyam KK, S S. Deep contextualized medical concept normalization in social media text. Proc Comput Sci 2020; 171: 1353–62. [Google Scholar]
- 12. Wei W-Q, Bastarache LA, Carroll RJ, et al. Evaluating phecodes, clinical classification software, and ICD-9-CM codes for phenome-wide association studies in the electronic health record. PLoS One 2017; 12 (7): e0175508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu P, Gifford A, Meng X, et al. Developing and evaluating mappings of ICD-10 and ICD-10-CM codes to Phecodes. bioRxiv 2018: 462077.doi: 10.1101/462077 [Google Scholar]
- 14. Thompson WK, Rasmussen LV, Pacheco JA, et al. An evaluation of the NQF quality data model for representing electronic health record driven phenotyping algorithms. AMIA Ann Symp Proc 2012; 2012: 911–20. [PMC free article] [PubMed] [Google Scholar]
- 15. Choi E, Xiao C, Stewart W, et al. MiME: multilevel medical embedding of electronic health records for predictive healthcare. 2018: 4547–57. http://papers.nips.cc/paper/7706-mime-multilevel-medical-embedding-of-electronic-health-records-for-predictive-healthcare Accessed November 21, 2019
- 16. Beam AL, Kompa B, Fried I, et al. Clinical concept embeddings learned from massive sources of multimodal medical data. 2018. http://arxiv.org/abs/1804.01486 Accessed March 13, 2019 [PMC free article] [PubMed]
- 17. Alawad M Hasan SMS, Blair Christian J,. et al. Retrofitting word embeddings with the UMLS metathesaurus for clinical information extraction. In: proceedings of the 2018 IEEE International Conference on Big Data (Big Data); December 10–13, 2018; Seattle, WA.
- 18. Xiang Y, Xu J, Si Y, et al. Time-sensitive clinical concept embeddings learned from large electronic health records. BMC Med Inform Decis Mak 2019; 19 (S2): 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng Y, Min X, Chen N, et al. Patient outcome prediction via convolutional neural networks based on multi-granularity medical concept embedding. ieeexplore.ieee.orghttps://ieeexplore.ieee.org/abstract/document/8217753/ Accessed September 30, 2019
- 20. Jung K, Sudat SEK, Kwon N, et al. Predicting need for advanced illness or palliative care in a primary care population using electronic health record data. J Biomed Inform 2019; 92: 103115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32 (Database issue): D267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi Y, Chiu CY-I, Sontag D. Learning low-dimensional representations of medical concepts. In: AMIA Joint Summits Translational Science Proceedings; 2016: 41–50. [PMC free article] [PubMed] [Google Scholar]
- 23. Maldonado R, Yetisgen M, Harabagiu SM. Adversarial learning of knowledge embeddings for the unified medical language system. AMIA Jt Summits Transl Sci Proc 2019; 2019: 543–52. [PMC free article] [PubMed] [Google Scholar]
- 24. UMLS Knowledge Sources: File Downloads. https://www.nlm.nih.gov/research/umls/licensedcontent/umlsknowledgesources.html Accessed March 13, 2019
- 25. 2018-ICD-10-CM-and-GEMs; 2017. https://www.cms.gov/medicare/coding/icd10/2018-icd-10-cm-and-gems.html Accessed March 13, 2019
- 26. PheWAS-Phenome Wide Association Studies. https://phewascatalog.org/phecodes Accessed March 13, 2019
- 27. Beta Clinical Classifications Software (CCS) for ICD-10-CM/PCS. https://www.hcup-us.ahrq.gov/toolssoftware/ccs10/ccs10.jsp Accessed March 13, 2019
- 28. HCUP CCS. Healthcare Cost and Utilization Project (HCUP). Rockville, MD: Agency for Healthcare Research and Quality; 2017. www.hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp Accessed March 13, 2019.
- 29. Clinical Classifications Software Refined (CCSR) for ICD-10-CM Diagnoses. Accessed March 1330. 201920. [Google Scholar]
- 30.Cerner—Cerner Health Facts ®—Data Sets—SBMI Data Service—The University of Texas Health Science Center at Houston (UTHealth) School of Biomedical Informatics. https://sbmi.uth.edu/sbmi-data-service/data-set/cerner/ Accessed November 25, 2018
- 31. sklearn.linear_model.LogisticRegression—scikit-learn 0.20.3 documentation. https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html Accessed March 13, 2019
- 32. Ma F, Chitta R, Zhou J, et al. Dipole: diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks 2017. doi: 10.1145/3097983.3098088
- 33. Ma F, Chitta R, Zhou J, et al. Dipole: diagnosis prediction in healthcare via attention-based bidirectional recurrent neural networks 2017. doi: 10.1145/3097983.3098088
- 34. Rasmy L, Zhu J, Li Z, et al Medinfo 2019 (podium abstract submitted Nov 2018). Simple Recurrent Neural Networks is all we need for clinical events predictions using EHR data. Lyon, France: MedInfo; 2019.
- 35. DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988; 44 (3): 837–45. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.