Abstract
Objective
Machine learning (ML) diagnostic tools have significant potential to improve health care. However, methodological pitfalls may affect diagnostic test accuracy studies used to appraise such tools. We aimed to evaluate the prevalence and reporting of design characteristics within the literature. Further, we sought to empirically assess whether design features may be associated with different estimates of diagnostic accuracy.
Materials and Methods
We systematically retrieved 2 × 2 tables (n = 281) describing the performance of ML diagnostic tools, derived from 114 publications in 38 meta-analyses, from PubMed. Data extracted included test performance, sample sizes, and design features. A mixed-effects metaregression was run to quantify the association between design features and diagnostic accuracy.
Results
Participant ethnicity and blinding in test interpretation was unreported in 90% and 60% of studies, respectively. Reporting was occasionally lacking for rudimentary characteristics such as study design (28% unreported). Internal validation without appropriate safeguards was used in 44% of studies. Several design features were associated with larger estimates of accuracy, including having unreported (relative diagnostic odds ratio [RDOR], 2.11; 95% confidence interval [CI], 1.43-3.1) or case-control study designs (RDOR, 1.27; 95% CI, 0.97-1.66), and recruiting participants for the index test (RDOR, 1.67; 95% CI, 1.08-2.59).
Discussion
Significant underreporting of experimental details was present. Study design features may affect estimates of diagnostic performance in the ML diagnostic test accuracy literature.
Conclusions
The present study identifies pitfalls that threaten the validity, generalizability, and clinical value of ML diagnostic tools and provides recommendations for improvement.
Keywords: machine learning, bias, sensitivity and specificity, research design, diagnostic techniques and procedures
INTRODUCTION
Machine learning (ML) diagnostic tools may improve the delivery of health care. Some ML tools demonstrate promising accuracy versus physicians in diagnosing various diseases.1,2 A variety of ML tools have even successfully received Food and Drug Administration approval for commercialization.3 Some ML models are attractive due to their ability to evaluate massive amounts of data,4–7 low cost of operation,6,7 and potential to be readily updated if new information or training data are available.4,6 The performance of ML tools is expected to improve over time, given the emergence of improved algorithm architectures, computing capabilities, and availability of training data.6
Despite these promises, the evidence on the performance of most ML diagnostic tools is thin. Clinical studies of effectiveness and improved outcomes are sparse. Most ML tests are at best evaluated in diagnostic test accuracy (DTA) studies. Yet, the methodological quality of DTA studies is on average relatively poor.8–11
Specifically, it has been demonstrated in metaepidemiological reviews that various design characteristics, including case-control designs with healthy controls and severe cases, differential verification, and inadequate blinding are associated with higher estimates of accuracy in conventional diagnostic tests.11–13 Moreover, underreporting of key methodological details is common in DTA studies, greatly decreasing their reproducibility and generalizability.14 The STARD (Standards for Reporting of Diagnostic Accuracy Studies) 2015 guidelines were proposed to provide reporting standards for DTA studies,11,15 including aspects of the study design, participant flow, recruitment criteria, and details of index and reference test execution.15 However, the magnitude of potential biases has not been quantitatively determined for ML DTA studies. ML tools could warrant special consideration as they bring new methods and research communities into the diagnostic testing field. These communities may not be as familiar with biases identified in the DTA literature to date, and their work may thus be susceptible to these biases.
ML tools could also be associated with forms of bias that may not be as important or prevalent in traditional diagnostic tests. For example, because ML tools rely heavily on training data, it is crucial that training datasets are large and diverse enough (eg, in severity of disease, demographics) for models to be generalizable in actual clinical practice.16–19 Safeguards should be in place to keep training and test datasets distinct to avoid overinflating accuracy estimates.18–21 Algorithms must also be capable of dealing with missing data in training datasets without introducing bias, especially given that missing data is common in real-world clinical datasets.7 Finally, caution must be exercised when assessing models developed on data that is missing in a nonrandom fashion.22
Some prior work has already started to describe areas for improvement. ML algorithms designed for health care have poor reproducibility when compared with other ML fields.23 Previous analyses of deep learning diagnostic tools have also indicated that few such models are externally validated and that validation is often underreported or prone to bias.24,25 A systematic review of articles published in 2018 examining ML tools for the diagnostic analysis of medical images reported that most studies failed to use external validation, cohort designs, and prospective data collection.26 In this work, we assessed studies without publication year restrictions and examined a greater variety of ML diagnostic tools, beyond deep learning or medical imaging. Additionally, we aimed to systematically address a wide variety of design features, as has been previously done for DTA studies in conventional diagnostic tools. Furthermore, we utilized a metaregression approach to empirically assess the association between design characteristics and estimates of diagnostic accuracy.
MATERIALS AND METHODS
We sought to assess the prevalence of design features and association between these features and estimates of accuracy in a systematically curated sample of ML DTA studies. This sample was compiled by identifying systematic reviews containing meta-analyses of ML DTA studies on PubMed. This approach was selected to identify similar ML DTA studies grouped within meta-analyses in order to facilitate modelling of the association between estimates of accuracy and presence of design features among similar studies.
Inclusion and exclusion criteria for systematic reviews
Systematic reviews containing meta-analyses of ML DTA studies were identified from PubMed using the search strategy detailed in Supplementary Appendix 1, Box S1-1 (search date, January 13, 2019). Studies were excluded if they did not include meta-analyses, did not analyze DTA studies of ML diagnostic tools, had inaccessible primary articles, or contained primary articles not written in English. ML was broadly defined to include artificial neural networks, support vector machines, naive Bayes, decision trees and random forest models, k-nearest neighbors, linear discriminant analysis, Bayesian networks, classification and regression trees, linear classifiers, and logistic regression models. Studies of any disease and any publication date were eligible. Article eligibility was assessed independently by 2 authors (Y.J.T. and R.J.C.). J.P.A.I. adjudicated any unresolved discrepancies.
Inclusion and exclusion criteria for individual DTA studies
Only DTA studies with binary classification results (ie, 2 × 2 tables) analyzed in meta-analyses within the identified systematic reviews were eligible for inclusion. The grouping of specific binary classification tables within specific published meta-analyses was retained. Binary classification results of studies were excluded if the index test was not based on a ML algorithm or if binary classification tables were not available or possible to reliably reconstruct. Binary classification tables that were present in more than 1 meta-analysis (n = 3) were retained in our analysis to preserve the structure of the meta-analyses. Independent assessment was conducted by 2 authors.
Data extraction
A variety of different study characteristics were extracted from primary research articles and systematic reviews by 2 authors independently.
Sensitivity, specificity, and numbers of true positives, true negatives, false positives, and false negatives were extracted from primary research publications or systematic reviews. Occasionally, in DTA studies, many estimates of diagnostic performance are provided with subtle differences and different reviewers may extract data from different binary classification result tables. In cases in which the data within systematic reviews differed from the data that we extracted ourselves from the primary publication, the values from the systematic review were used for analysis in order to align with the overall intention that each systematic review had in summarizing the data across different studies.
Information on index test modality and the ML algorithm was extracted. Modality refers to the method by which index tests diagnose diseases (eg, by processing images or by evaluating patient characteristics such as biomarkers), while algorithm refers to specific ML approaches (eg, artificial neural networks, support vector machines). We determined if authors of the DTA study had also designed the algorithm they were evaluating. Additional information on reference test modality was also extracted.
We also extracted information relevant to 5 design features shown in prior metaepidemiological studies to be associated with higher estimates of accuracy in conventional diagnostic tests12: study design, recruitment criteria, blinded interpretation of tests, verification procedure, and reporting of population details. Scoring of these characteristics was performed in the same manner as in the corresponding previous metaepidemiological review showing statistically significant evidence for the association of the characteristic with bias.11,13 Additionally, methodological characteristics not previously evaluated and considered to be of importance to ML were also assessed, including appropriateness of validation method and reporting of both the number of model features and the number of observations in the training dataset. Specific coding of each feature is shown in Supplementary Appendix 1 (Supplementary Table S1-1; Supplementary Figure S1-1).
Metaregression
A metaepidemiologic regression approach utilizing a mixed model was fit to quantitatively assess the association between study design features and estimates of diagnostic accuracy. This model is an extension of the summary receiver-operating characteristic model previously used to assess the degree of design-related bias in conventional diagnostic tests.27 The diagnostic odds ratio (DOR) was utilized as the measurement of diagnostic tool performance. The DOR incorporates both sensitivity and specificity as:
As previously applied to conventional diagnostic tests,11,13 the log (DOR) of a particular DTA was modeled as a normally distributed variable and as a function of the pooled summary DOR for that meta-analysis, the threshold for positivity used in the particular DTA study, the effect of design features, and residual error. With this regression approach, the effect of individual methodological factors was adjusted for the potentially confounding effect of other design features. For the a priori defined primary analysis, we specified random effects for only the intercept and the positivity threshold and fixed effects for all design feature covariates.13 Design feature covariates were also defined a priori and included the previously described 5 design features shown in prior studies to be associated with higher estimates of accuracy, along with the 2 novel methodological characteristics considered to be of importance to ML. No feature selection was conducted, and interactions between design feature covariates were not explored. After antilogarithm transform, the coefficients of the terms representing individual design features can be interpreted as a relative DOR (RDOR). The RDOR indicates the diagnostic accuracy of a test in studies lacking a specific design feature, relative to studies with the corresponding feature. For RDORs >1, studies that fail to implement the design feature are associated with larger estimates of accuracy as compared with studies with that design feature. Several secondary analyses were also conducted (modeling design feature covariates as random effects; univariable analysis; and complete-cases analysis). Supplementary Appendix 1 contains additional details on the meta-regression approach (Supplementary Box S1-2) and secondary analyses (Supplementary Box S1-3).
Data availability
Datasets and analysis scripts used in this study have been deposited on the Open Science Framework and are available for access online (https://osf.io/2csz4/?view_only=45566b4ff7524234947fb4c62b9f90d8).
RESULTS
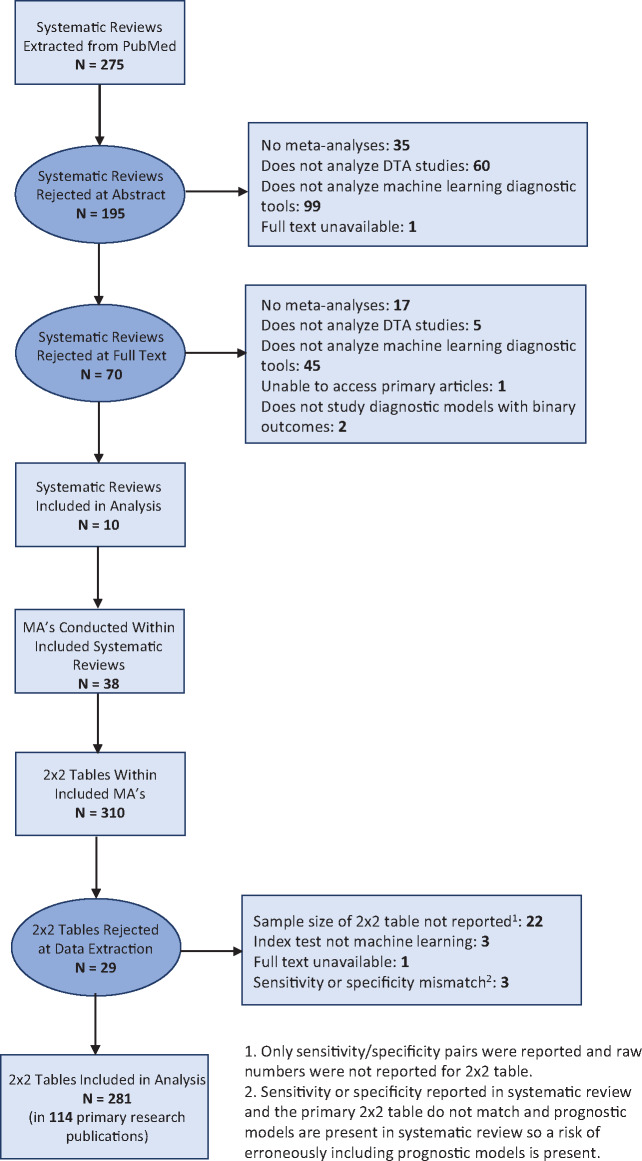
A total of 275 publications were initially identified from PubMed. Following exclusion of ineligible publications, 10 systematic reviews, each containing 1 or more meta-analyses of DTA studies on ML diagnostic tools, remained (Figure 1). Publication dates of the systematic reviews ranged from 2009 to 2018. The topics of the systematic reviews were breast cancer (n = 2), skin cancer (n = 2), intracranial malignancies (n = 2), ovarian cancer (n = 1), Down syndrome (n = 1), ectopic pregnancies (n = 1), and respiratory disorders (n = 1). The 38 meta-analyses present within the systematic reviews contain 310 sets of binary classification results. Each systematic review publication had a median of 2.5 (interquartile range [IQR], 1-5.75) meta-analyses, and each meta-analysis consisted of a median of 4 (IQR, 2-14.25) sets of binary classification results. Supplementary Appendix 2 contains the references of the included systematic reviews.
Figure 1.
Study flowchart showing publications excluded or included in the analysis. DTA: diagnostic test accuracy; MA: meta-analysis.
After further screening for data eligibility and availability, 281 sets of binary classification results were retained. These results belonged to a total of 114 primary research publications, with publication dates ranging from 1992 to 2018. Tables 1-4 list additional characteristics of the included primary research articles and sets of binary classification results. Supplementary Appendix 3 lists the references of these primary research publications.
Table 1.
Key characteristics of primary research articles included in the analysis (n = 114 unique primary research articles)
| Primary article characteristics | n | % |
|---|---|---|
| Index test type | ||
| Patient characteristics | 34 | 30 |
| Image processing | 75 | 66 |
| Combination of above | 3 | 3 |
| Other | 2 | 2 |
| Type of algorithm of index test | ||
| Regression | 31 | 27 |
| ANN | 27 | 24 |
| SVM | 14 | 12 |
| KNN | 3 | 3 |
| Decision trees | 3 | 3 |
| Bayesian methods | 1 | 1 |
| Other methods | 14 | 12 |
| Multiple types within 1 primary article | 11 | 10 |
| Unclear | 10 | 9 |
| Reference standard type | ||
| Histology | 66 | 58 |
| Genetic marker | 11 | 10 |
| Physician interpretation | 2 | 2 |
| Combination of above | 16 | 14 |
| Other | 5 | 4 |
| Unclear | 14 | 12 |
| Recruitment of study population | ||
| Recruited by authors of primary paper | 73 | 64 |
| Sourced from existing dataset | 21 | 18 |
| Unclear | 19 | 17 |
| Multiple types within 1 primary article | 1 | 0 |
| Designing of index test algorithm | ||
| Designed by authors of primary paper | 87 | 76 |
| Not designed by authors of primary paper | 27 | 23 |
ANN: artificial neural network; KNN: k-nearest neighbors; SVM: support vector machine.
Table 2.
Breakdown of study design characteristics (n = 114 unique primary research articles)
| Primary article study design characteristics | n | % |
|---|---|---|
| Study design | ||
| Cohort | 55 | 48 |
| Case control | 27 | 24 |
| Unclear | 32 | 28 |
| Recruitment criteria | ||
| Symptoms and signs | 36 | 32 |
| Other test results | 33 | 29 |
| Referral for index test | 7 | 6 |
| Unclear | 37 | 33 |
| Multiple types within 1 primary article | 1 | 0 |
| Reporting of population characteristics | ||
| Age | 58 | 51 |
| Gender | 64 | 56 |
| Distribution of symptoms | 76 | 67 |
| Ethnicity | 12 | 11 |
| Blinded interpretation of test results | ||
| Blinded as reported | 23 | 20 |
| Not blinded as reported | 20 | 18 |
| Unclear | 70 | 61 |
| Multiple types within 1 primary article | 1 | 0 |
| Reporting of number of features and training data sample size | ||
| Reported | 69 | 57 |
| Not reported | 45 | 42 |
| Verification procedure | ||
| Same reference standard | 69 | 61 |
| Differential verification | 31 | 27 |
| Unclear | 14 | 12 |
| Validation | ||
| External validation | 27 | 24 |
| Appropriate internal validation | 37 | 33 |
| Inappropriate internal validation | 50 | 44 |
Table 4.
Breakdown of covariates used in the metaregression (n = 281 sets of binary classification results)
| Metaregression covariates | n | % |
|---|---|---|
| Study design | ||
| Cohort | 185 | 66 |
| Case-control | 52 | 19 |
| Unclear | 44 | 16 |
| Recruitment criteria | ||
| Symptoms and signs | 167 | 59 |
| Referral for index test | 10 | 4 |
| Other test results | 50 | 18 |
| Unclear | 54 | 19 |
| Reporting of population characteristics | ||
| Age | 167 | 59 |
| Gender | 213 | 76 |
| Distribution of symptoms | 186 | 66 |
| Blinded interpretation of test results | ||
| Blinded as reported | 28 | 10 |
| Not blinded as reported or unclear | 253 | 90 |
| Reporting number features and number of observations in training data | ||
| Reported | 109 | 39 |
| Not reported | 172 | 61 |
| Verification procedure | ||
| Same reference standard | 202 | 72 |
| Differential verification or unclear | 79 | 28 |
| Validation | ||
| External validation | 151 | 54 |
| Appropriate internal validation | 48 | 17 |
| Inappropriate internal validation | 82 | 29 |
Characteristics of primary research publications
Among the 114 primary research articles included, the majority of publications (n = 75, 66%) investigated ML tools that aimed to diagnose diseases by analyzing images such as scans. The top 3 algorithm types used were regressions (n = 31, 27%), artificial neural networks (n = 27, 24%), and support vector machines (n = 14, 12%). The majority of the ML diagnostic tools were compared against histological reference standards (n = 66, 58%). In 87 (76%) articles, authors investigated the performance of a diagnostic tool that they had designed within the same publication.
In most studies (n = 73, 64%), authors directly recruited the study population. A total of 21 studies (18%) used an existing, published dataset instead. The source of the study population was not reported in 19 (17%) studies. The majority of these study populations were recruited through a cohort study design (n = 55, 48%). A total of 27 (24%) studies used a case-control design, and the study design was not clearly reported in the remainder (n = 32, 28%). Subjects were most commonly recruited solely based on clinical symptoms and signs (n = 36, 32%) or based on other test results (n = 33, 29%). A total of 7 (6%) studies recruited patients referred specifically for the index test. In 37 (33%) studies, recruitment criteria were unclear.
A total of 23 (20%) studies used blinding in the interpretation of test results. A total of 20 (18%) studies reported no blinding, 1 study used different blinding practices for different sets of binary classification results, and the remainder did not report blinding (n = 70, 61%). Most studies used the same reference test for all individuals (n = 69, 61%), and 31 (27%) studies used different reference tests for subsets of participants. The remainder failed to report sufficient experimental details to determine if a single reference standard was used (n = 14, 12%). A total of 37 (33%) studies trained and tested ML algorithms using internal validation with appropriate safeguards. A total of 50 (44%) studies used internal validation without appropriate safeguards. The remaining studies used external validation (n = 27, 24%). These characteristics, delineated on the level of sets of binary classification results, are described in Supplementary Appendix 4 (Supplementary Boxes S4-1, S4-2). To determine if research practices changed over time, we also compared the characteristics of studies that were published before and after the median publication date, as shown in Supplementary Appendix 4 (Supplementary Tables S4-1 to S4-4).
Sample sizes
The sample size distributions of the assessed studies are right-skewed, with most studies having sample sizes toward the lower end as described in Supplementary Figure S4-1 and Table 3. The median number of participants in the training dataset was 300 (IQR, 102-681), and the median sample size for the reported test dataset was 120 (IQR, 54-300). In terms of the number of events, defined as the minimum of the number of diseased or healthy individuals, medians were 27 (IQR 12-47) for the test dataset and 47.5 (IQR 26.8-98) for the training dataset.
Table 3.
Key characteristics of diagnostic tests included in this analysis (n = 281 sets of binary classification results)
| Diagnostic test characteristics | Median | IQR |
|---|---|---|
| Diagnostic odds ratio | 20.5 | 10.7-45.7 |
| Sample size of reported binary classification result tables | 120 | 54-300 |
| Sample size of training dataa | 300 | 102-681 |
| Number of featuresb | 4 | 3-7 |
| Index test type | n | % |
| Patient characteristics | 179 | 64 |
| Image processing | 94 | 34 |
| Combination of above | 5 | 2 |
| Other | 3 | 1 |
| Type of algorithm of index test | n | % |
| Regression | 148 | 53 |
| ANN | 67 | 24 |
| SVM | 21 | 8 |
| KNN | 6 | 2 |
| Bayesian method | 6 | 2 |
| Decision trees | 5 | 2 |
| Other methods | 17 | 6 |
| Unclear | 11 | 4 |
| Reference standard type | n | % |
| Histology | 200 | 71 |
| Genetic marker | 16 | 6 |
| Physician interpretation | 3 | 1 |
| Combination of above | 26 | 9 |
| Other | 15 | 5 |
| Unclear | 21 | 8 |
| Recruitment of study population | n | % |
| Recruited by authors of primary paper | 205 | 73 |
| Sourced from existing dataset | 49 | 17 |
| Unclear | 27 | 10 |
| Designing of index test algorithm | n | % |
| Designed by authors of primary paper | 130 | 46 |
| Not designed by authors of primary paper | 151 | 54 |
ANN: artificial neural network; KNN: k-nearest neighbors; SVM: support vector machine.
Training dataset sample size was not reported for 157 sets of binary classification results.
Number of features was not reported for 29 sets of binary classification results.
Reporting of experimental details
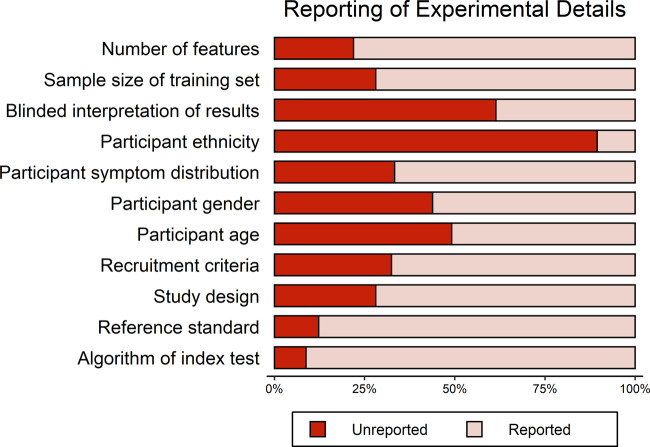
The level of unreported experimental details in primary research articles varied across the different reporting items assessed (Figure 2, Supplementary Table S5-1). To assess reporting, we looked only at the main text and supplementary material of the primary publication and not in any cited references. The 3 categories with highest unreported details were ethnicities of participants (90%, n = 102), blinding in interpretation of test results (62%, n = 70), and age of participants (49%, n = 56). Underreporting was also relatively high for rudimentary methodological characteristics including study design type (28%, n = 32), number of features in the algorithm (22%, n = 25), recruitment criteria (33%, n = 37), and the sample size of the training set (28%, n = 32). The proportion of unreported details on the level of binary classification result tables is described in Supplementary Appendix 5.
Figure 2.
Plot describing the proportion of unreported experimental details across primary research articles included in the analysis (n = 114 primary research articles).
Among the 73 studies that directly recruited their study population, 7 (10%) studies did not report the type of study design and 12 (16%) did not report the recruitment criteria. In contrast, among the remaining 41 studies that either used a previously published dataset or did not clearly define the source of their dataset, the corresponding number of studies was 25 (61%) for both of these categories.
In the 87 studies in which the authors assessed the performance of a diagnostic test they had designed within the same publication, 7 (8%) failed to report the sample size of the training dataset. Conversely, among the remaining 27 studies that assessed the performance of a previously published diagnostic test, the corresponding number of studies was 25 (93%).
Association of study design characteristics with estimates of diagnostic accuracy
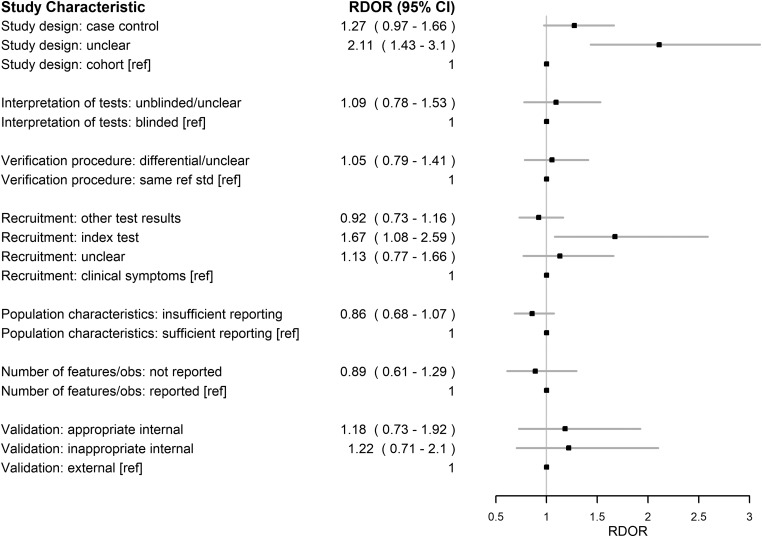
The relative effects of all assessed characteristics are depicted in Figure 3 for the primary analysis using the multivariable mixed model with design covariates modeled as fixed effects.
Figure 3.
Forest plot describing the effect of study design characteristics on estimates of diagnostic accuracy. Plotted are relative diagnostic odds ratios (RDORs) and their 95% confidence intervals (95% CIs) estimated in a multivariable mixed-effects metaregression with the design covariates modeled as fixed effects. The reference categories are indicated with [ref].
Most of the assessed characteristics were associated with a RDOR point estimate above 1, indicating a trend toward potential association with higher estimates of accuracy (Figure 3). The 3 design characteristics with the greatest RDOR magnitudes were having an unreported study design (RDOR, 2.11; 95% CI, 1.43-3.1), recruiting participants specifically for the index test (RDOR, 1.67; 95% CI, 1.08-2.59), and employing a case control study design (RDOR, 1.27; 95% CI, 0.97-1.66).
Results for the secondary analyses are detailed in Supplementary Appendix 6.
DISCUSSION
Our systematic review of ML DTA studies covered a diverse set of different diagnostic tools that utilized a broad range of different algorithm types and test modalities, and were designed to diagnose a variety of diseases. The majority of assessed diagnostic tools aimed to detect various malignancies, although other diseases such as Down syndrome and ectopic pregnancies were also represented. We found that there were no substantial differences in research practices between studies published before or after the median publication date, indicating that our observations are likely broadly generalizable over time.
Reporting of experimental details
Overall, we observed significant underreporting of study design characteristics. The ethnicities of participants and the presence of blinding in the interpretation of test results were not reported for the overwhelming majority of publications. Many articles do not even report enough information to effectively describe the basic structure of the ML algorithm or the DTA study. For instance, details regarding the type of study design, number of features included in the algorithm, recruitment criteria, and sample size of the training set were frequently unreported. A recent systematic review of ML prognostic tools found similarly poor reporting of experimental details,25 and poor reporting may be a systemic problem within the ML field currently. This lack of methodological transparency is a considerable impediment to physicians seeking to evaluate ML diagnostic tools in terms of their validity, generalizability, and clinical value.13 Furthermore, this underreporting hinders future replication efforts.
In terms of population characteristics of the participants enrolled, underreporting was highest for ethnicity, compared with age, gender, and distribution of symptoms. The somewhat higher, though still lacking, awareness in reporting of the latter 3 characteristics might stem from long-standing emphasis on the importance of such reporting in meta-epidemiological studies conducted on conventional diagnostic tests.12,13 However, ethnicities of participants remains an important characteristic to report. Different racial groups have differential susceptibility to a variety of diseases.28,29 Additionally, ML tools in various applications sometimes use training datasets skewed toward particular ethnic groups.30 As such, reporting of participant ethnicity remains a key factor in determining generalizability of a diagnostic tool.
For type of study design and recruitment criteria, lack of reporting is especially severe when authors used a previously published dataset instead of recruiting study participants on their own. Similarly, the proportion of studies that did not report training dataset sample size was much higher for publications assessing the performance of a previously published algorithm. This may result from difficulty in elucidating these characteristics from published resources; a perception that simply citing the reference of the resource is a sufficient substitute, especially for more established resources; a lack of appreciation of the value in reporting these characteristics; or worse, an outright failure of authors to consider how these characteristics relate to bias when using published resources for convenience. Authors who use such resources should do so with due consideration of whether they are valid and appropriate for the questions they want to address.
Effect of study design characteristics on estimates of diagnostic accuracy
Overall, we observed that various study design characteristics were associated with different estimates of diagnostic accuracy. The results largely trended in the same direction as previous metaepidemiological studies of traditional diagnostic tests.11–13 These design features likely play a similar role in both ML and conventional diagnostic tools. For example, features such as study design, blinded interpretation of tests, differential verification, and recruitment criteria are likely to be important considerations regardless of what test is being evaluated, even though substandard studies may not always be biased to the same extent in their results.
In addition to the covariates previously assessed, we also investigated the effect of validation methods. We found that external validation was less common than internal validation. A recent systematic review of artificial intelligence algorithms for image analysis also found similar results.26 Compared with external validation, studies employing either appropriate or inappropriate internal validation were possibly more likely to be associated with higher estimates of accuracy, though this was not statistically significant at the 5% level of significance. This observation is in line with a recent systematic review assessing deep learning algorithms as diagnosis tools.24 Inappropriate internal validation was also more commonly used than appropriate internal validation. Qualitatively, we noted that authors frequently tuned their models (in terms of inputs, hyperparameters, or algorithm types) on the same dataset that was used to derive the reported measure of diagnostic test accuracy.31–34 Additionally, we observed that some authors used different datasets for training and tuning the algorithm (through cross-validation or split sample) but selectively reported only the algorithm that performed optimally on the tuning dataset as their measure of diagnostic test accuracy.35–38 These observations are corroborated by a recent systematic review on ML clinical prediction models demonstrating widespread use of validation procedures that could be prone to bias, such as conducting tuning or variable selection on data eventually used to test models.25 Encouragingly, however, we observed that newer studies appear to be adopting better validation practices, with higher proportions of studies utilizing external validation or appropriate internal validation as compared with older studies. Overall, we believe that validation methods merit great consideration as well as continued investigation to track changes in practices over time.
Recruitment of participants specifically for the index test was demonstrated to be associated with a larger estimate of accuracy in our study but was previously associated with lower estimates of accuracy.11 The authors of the prior metaepidemiological study proposed that patients referred for the index test are likely to have uncertain disease status, thus leading to a decrease in proportion of true positives and true negatives.11 Perhaps, specifically for ML diagnostic tools, investigators who refer patients for the index test might be selecting patients for whom the algorithm is most likely to perform well. For instance, investigators might select patients with characteristics most similar to the training dataset.
Similarly, in our study, insufficient reporting of population characteristics was associated with lower estimates of accuracy (though not significant at the 5% level), while in a prior study the same covariate was associated with a higher estimate of accuracy.13 The prior study’s authors highlight that it is unclear how this covariate influences measures of accuracy, as reporting practices are not directly related to methodological design.13 Indeed, unreported experimental details are a soft indicator of quality that can be difficult to interpret.
Recommendations
Throughout this study, we identified areas of possible improvement that we believe should be emphasized.
First, for authors of ML DTA studies, we recommend a greater emphasis on methodology reporting and awareness of potential biases. The STARD 2015 guidelines for general DTA studies is a natural starting point.15 Items of extra importance for ML DTA studies may include details of validation methods, description of the feature selection methods, and details of missing data and missing data handling methods.
Second, for metaresearchers in the field, transparency can be improved. The binary classification result tables reported in systematic reviews do not always match those reported within the primary research publication.34,39–43 Often, documentation of reasons behind this discrepancy was absent or poor. Careful definitions and prespecification thereof may avoid having multiplicity and ambiguity in diagnostic performance estimates.
Third, there are no consensus identifiers for publicly available datasets or published algorithms, unlike in other fields of research with formalized databases (eg, PubChem). This makes it extremely difficult for researchers to track the performance of specific algorithms across different patient populations. Improved tracking through consensus identifiers would greatly improve transparency and ease evidence synthesis.
Finally, DTA studies provide important metrics when assessing the diagnostic accuracy of ML diagnostic tools, but accuracy may not directly correspond to improved clinical outcomes. Hence, we recommend that, following DTA studies, researchers should also conduct clinical studies of effectiveness and improved outcomes to assess clinical efficacy.
Limitations
We wish to highlight several limitations of the present study. First, we used unclear reporting as inherently imperfect, soft indicators of poor quality for several covariates. When studies failing to report experimental details are associated with an RDOR >1, the implicit assumption is that these studies implemented suboptimal practices that were not reported. However, having an RDOR around 1 for such studies does not necessarily indicate that studies reporting details and studies not reporting details both utilized optimal practices. It is also possible that both categories of studies implemented suboptimal practices.
Second, in our metaregression, sets of binary classification results are grouped in the same meta-analyses that they appeared in within the published literature. In previous metaepidemiological studies using the same approach, grouped studies for the metaregression utilized the exact same diagnostic test applied to different populations, settings, or experimental designs. This was not possible to replicate, as not every published meta-analysis comprised studies evaluating the exact same ML test. Frequently, these were distinct tests deemed similar enough to group within a meta-analysis by respective authors, based on algorithm type, test modality, or other characteristics. We preserved this grouping, as any attempts to regroup binary classification results would have been arbitrary.
Third, our PubMed search strategy is unable to capture unpublished ML models and ML models not included in prior meta-analyses. The prevalence of design features and their true effects on accuracy could well be different if the population of models not captured is markedly different from the assessed studies. To quantify the generalizability of our findings, we compared the fields of study captured using our search strategy with those captured by recent systematic reviews of ML tools that were not limited to meta-analyses.25,26 Our search strategy captured some of the main fields that use ML tools for diagnosis including radiology, oncology, and dermatology.25,26 However, it missed some fields such as cardiovascular medicine, critical care, ophthalmology, and endocrinology. Studies from each of these fields represented about 10% to 14% of studies captured by the other systematic reviews.25,26
Last, DOR is an imperfect measurement of accuracy. Individual study design characteristics may have opposing effects on sensitivity and specificity.12,44,45 These opposing effects on sensitivity and specificity may result in a relatively unchanged DOR and are not readily captured by analyses using the DOR as a metric of accuracy.
CONCLUSION
Our review of ML diagnostic tools reveals key areas for improvement, spanning multiple groups of stakeholders. For authors of DTA studies, reporting of key methodological details is poor, especially among studies using publicly available datasets or algorithms. In addition, classically emphasized methodological pitfalls first described in conventional diagnostic tests likely hold true for ML tools as well.11,13 We also urge designers of algorithms to critically evaluate the suitability of their validation procedures. Finally, in addressing the field as a whole, we argue that there is great value in improving the tracking of databases and algorithms. Our work builds on the existing body of evidence examining pitfalls in both conventional diagnostic tools and other applications of ML11,13,22,24–26 and underscores the importance of addressing these shortcomings as ML grows in prominence within medicine.
FUNDING
YJT’s work is supported by a Stanford Graduate Fellowship. RJC’s work is funded by a Stanford Major Grant.
AUTHOR CONTRIBUTIONS
RJC and YJT conceived the project, extracted data, conducted analyses, interpreted results, and wrote the article. JPAI contributed to study design, analysis, and interpretation. All authors revised and approved the article.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542 (7639): 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hannun AY, Rajpurkar P, Haghpanahi M, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 2019; 25 (1): 65–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019; 25 (1): 44–56. [DOI] [PubMed] [Google Scholar]
- 4. Waljee AK, Higgins P.. Machine learning in medicine: a primer for physicians. Am J Gastroenterol 2010; 105 (6): 1224–6. [DOI] [PubMed] [Google Scholar]
- 5. Deo RC. Machine learning in medicine. Circulation 2015; 132 (20): 1920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yu K-H, Beam AL, Kohane IS.. Artificial intelligence in healthcare. Nat Biomed Eng 2018; 2 (10): 719–31. [DOI] [PubMed] [Google Scholar]
- 7. Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med 2001; 23 (1): 89–109. [DOI] [PubMed] [Google Scholar]
- 8. Reid MC, Lachs MS, Feinstein AR.. Use of methodological standards in diagnostic test research: getting better but still not good. JAMA 1995; 274 (8): 645–51. [PubMed] [Google Scholar]
- 9. Harper R, Reeves B.. Compliance with methodological standards when evaluating ophthalmic diagnostic tests. Invest Ophthalmol Vis Sci 1999; 40 (8): 1650–7. [PubMed] [Google Scholar]
- 10. Morris RK, Selman TJ, Zamora J, et al. Methodological quality of test accuracy studies included in systematic reviews in obstetrics and gynaecology: sources of bias. BMC Womens Health 2011; 11: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rutjes AWS, Reitsma JB, Di NM, et al. Evidence of bias and variation in diagnostic accuracy studies. CMAJ 2006; 174 (4): 469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Whiting PF, Rutjes AWS, Westwood ME, et al. A systematic review classifies sources of bias and variation in diagnostic test accuracy studies. J Clin Epidemiol 2013; 66 (10): 1093–104. [DOI] [PubMed] [Google Scholar]
- 13. Lijmer JG, Mol BW, Heisterkamp S, et al. Empirical evidence of design-related bias in studies of diagnostic tests. JAMA 1999; 282 (11): 1061–6. [DOI] [PubMed] [Google Scholar]
- 14. Estrada CA, Bloch RM, Antonacci D, et al. Reporting and concordance of methodologic criteria between abstracts and articles in diagnostic test studies. J Gen Intern Med 2000; 15 (3): 183–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016; 6 (11): e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dietterich T. Overfitting and undercomputing in machine learning. ACM Comput Surv 1995; 27 (3): 326–7. [Google Scholar]
- 17. Thrall JH, Li X, Li Q, et al. Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol 2018; 15 (3): 504–8. [DOI] [PubMed] [Google Scholar]
- 18. Kassraian-Fard P, Matthis C, Balsters JH, et al. Promises, pitfalls, and basic guidelines for applying machine learning classifiers to psychiatric imaging data, with autism as an example. Front Psychiatry 2016; 7: 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kubota KJ, Chen JA, Little MA.. Machine learning for large-scale wearable sensor data in Parkinson’s disease: concepts, promises, pitfalls, and futures. Mov Disord 2016; 31 (9): 1314–26. [DOI] [PubMed] [Google Scholar]
- 20. Bone D, Goodwin MS, Black MP, et al. Applying machine learning to facilitate autism diagnostics: pitfalls and promises. J Autism Dev Disord 2015; 45 (5): 1121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cawley GC, Talbot N.. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res 2010; 11: 2079–107. [Google Scholar]
- 22. Gianfrancesco MA, Tamang S, Yazdany J, et al. Potential biases in machine learning algorithms using electronic health record data. JAMA Intern Med 2018; 178 (11): 1544–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDermott MBA, Wang S, Marinsek N, et al. Reproducibility in machine learning for health. arXiv: 1907.01463 http://arxiv.org/abs/1907.01463 Accessed January 12, 2020.
- 24. Liu X, Faes L, Kale AU, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 2019; 1 (6): e271–97. [DOI] [PubMed] [Google Scholar]
- 25. Christodoulou E, Ma J, Collins GS, et al. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol 2019; 110: 12–22. [DOI] [PubMed] [Google Scholar]
- 26. Kim DW, Jang HY, Kim KW, et al. Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol 2019; 20 (3): 405–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moses LE, Shapiro D, Littenberg B.. Combining independent studies of a diagnostic test into a summary roc curve: Data-analytic approaches and some additional considerations. Stat Med 1993; 12 (14): 1293–316. [DOI] [PubMed] [Google Scholar]
- 28. Glicksberg BS, Li L, Badgeley MA, et al. Comparative analyses of population-scale phenomic data in electronic medical records reveal race-specific disease networks. Bioinformatics 2016; 32 (12): i101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cooper R, Cutler J, Desvigne-Nickens P, et al. Trends and disparities in coronary heart disease, stroke, and other cardiovascular diseases in the United States: findings of the national conference on cardiovascular disease prevention. Circulation 2000; 102 (25): 3137–47. [DOI] [PubMed] [Google Scholar]
- 30. Merler M, Ratha N, Feris RS, et al. Diversity in faces. arXiv: 1901.10436 http://arxiv.org/abs/1901.10436 Accessed December 5, 2019.
- 31. Biagiotti R, Cariati E, Brizzi L, et al. Maternal serum screening for Down’s syndrome in the first trimester of pregnancy. Br J Obstet Gynaecol 1995; 102 (8): 660–2. [DOI] [PubMed] [Google Scholar]
- 32. Forest JC, Massé J, Moutquin JM.. Screening for Down syndrome during first trimester: a prospective study using free beta-human chorionic gonadotropin and pregnancy-associated plasma protein A. Clin Biochem 1997; 30 (4): 333–8. [DOI] [PubMed] [Google Scholar]
- 33. Juntu J, Sijbers J, De Backer S, et al. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. J Magn Reson Imaging 2010; 31 (3): 680–9. [DOI] [PubMed] [Google Scholar]
- 34. Mayerhoefer ME, Breitenseher M, Amann G, et al. Are signal intensity and homogeneity useful parameters for distinguishing between benign and malignant soft tissue masses on MR images? Objective evaluation by means of texture analysis. Magn Reson Imaging 2008; 26 (9): 1316–22. [DOI] [PubMed] [Google Scholar]
- 35. Abdolmaleki P, Buadu LD, Naderimansh H.. Feature extraction and classification of breast cancer on dynamic magnetic resonance imaging using artificial neural network. Cancer Lett 2001; 171 (2): 183–91. [DOI] [PubMed] [Google Scholar]
- 36. Abdolmaleki P, Buadu LD, Murayama S, et al. Neural network analysis of breast cancer from MRI findings. Radiat Med 1997; 15 (5): 283–93. [PubMed] [Google Scholar]
- 37. Vergnaghi D, Monti A, Setti E, et al. A use of a neural network to evaluate contrast enhancement curves in breast magnetic resonance images. J Digit Imaging 2001; 14 (S1): 58–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee SH, Kim JH, Cho N, et al. Multilevel analysis of spatiotemporal association features for differentiation of tumor enhancement patterns in breast DCE-MRI. Med Phys 2010; 37 (8): 3940–56. [DOI] [PubMed] [Google Scholar]
- 39. Alldred SK, Takwoingi Y, Guo B, et al. First trimester serum tests for Down’s syndrome screening. Cochrane Database Syst Rev 2015; 11: CD011975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kagan KO, Cicero S, Staboulidou I, et al. Fetal nasal bone in screening for trisomies 21, 18 and 13 and Turner syndrome at 11-13 weeks of gestation. Ultrasound Obstet Gynecol 2009; 33 (3): 259–64. [DOI] [PubMed] [Google Scholar]
- 41. Fusco R, Sansone M, Filice S, et al. Pattern recognition approaches for breast cancer DCE-MRI classification: a systematic review. J Med Biol Eng 2016; 36 (4): 449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee SH, Kim JH, Park JS, et al. Characterizing time-intensity curves for spectral morphometric analysis of intratumoral enhancement patterns in breast DCE-MRI: comparison between differentiation performance of temporal model parameters based on DFT and SVD. In: 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro. 2009:65–8.
- 43. Sinha S, Lucas-Quesada FA, DeBruhl ND, et al. Multifeature analysis of Gd-enhanced MR images of breast lesions. J Magn Reson Imaging 1997; 7 (6): 1016–26. [DOI] [PubMed] [Google Scholar]
- 44. Philbrick JT, Heim S.. The D-dimer test for deep venous thrombosis: gold standards and bias in negative predictive value. Clin Chem 2003; 49 (4): 570–4. [DOI] [PubMed] [Google Scholar]
- 45. Punglia RS, D'Amico AV, Catalona WJ, et al. Effect of verification bias on screening for prostate cancer by measurement of prostate-specific antigen. N Engl J Med 2003; 349 (4): 335–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets and analysis scripts used in this study have been deposited on the Open Science Framework and are available for access online (https://osf.io/2csz4/?view_only=45566b4ff7524234947fb4c62b9f90d8).