Abstract
Several MRI measures have been proposed as in vivo biomarkers of myelin, each with applications ranging from plasticity to pathology. Despite the availability of these myelin-sensitive modalities, specificity and sensitivity have been a matter of discussion. Debate about which MRI measure is the most suitable for quantifying myelin is still ongoing. In this study, we performed a systematic review of published quantitative validation studies to clarify how different these measures are when compared to the underlying histology. We analyzed the results from 43 studies applying meta-analysis tools, controlling for study sample size and using interactive visualization (https://neurolibre.github.io/myelin-meta-analysis). We report the overall estimates and the prediction intervals for the coefficient of determination and find that MT and relaxometry-based measures exhibit the highest correlations with myelin content. We also show which measures are, and which measures are not statistically different regarding their relationship with histology.
Research organism: Human, Mouse, Rat
Introduction
Myelin is a key component of the central nervous system. The myelin sheaths insulate axons with a triple effect: allowing fast electrical conduction, protecting the axon, and providing trophic support (Nave and Werner, 2014). The conduction velocity regulation has become an important research topic, with evidence of activity-dependent myelination as an additional mechanism of plasticity (Fields, 2015; Sampaio-Baptista and Johansen-Berg, 2017). Myelin is also relevant from a clinical perspective, given that demyelination is often observed in several neurological diseases such as multiple sclerosis (Höftberger and Lassmann, 2018).
Given this important role in pathology and plasticity, measuring myelin in vivo has been an ambitious goal for magnetic resonance imaging (MRI) for more than two decades (MacKay et al., 1994; Rooney et al., 2007; Stanisz et al., 1999). Even though the thickness of the myelin sheath is in the order of micrometres, well beyond the MRI spatial resolution, its presence influences several physical properties that can be probed with MRI, from longitudinal and transversal relaxation phenomena to water molecule diffusion processes.
However, being sensitive to myelin is not enough: to study how and why myelin content changes, it is necessary to define a specific biomarker. Interestingly, the quest for measuring myelin has evolved in parallel with an important paradigm shift in MRI research, where MRI data are no longer treated as just ‘pictures’, but as actual 3D distributions of quantitative measures. This perspective has breathed new life into an important field of research, quantitative MRI (qMRI), that encompasses the study of how to measure the relevant electromagnetic properties that influence magnetic resonance phenomena in biological tissues (Cercignani et al., 2018; Cohen-Adad and Wheeler-Kingshott, 2014). From the very definition of qMRI, it is clear that its framework applies to any approach for non-invasive myelin quantification.
Similarly to other qMRI biomarkers, MRI-based myelin measurements are indirect, and might be affected by other microstructural features, making the relationship between these indices and myelination noisy. Assessing the accuracy of such measurements, and their sensitivity to change, is essential for their translation into clinical applications. Validation is therefore a fundamental aspect of their development (Cohen-Adad, 2018). The most common approach is based on acquiring MR data from in vivo or ex vivo tissue and then comparing those data with the related samples analyzed using histological techniques. Despite being the most realistic approach, this comparison involves several methodological choices, from the specific technique used as a reference to the quantitative measure used to describe the relationship between MRI and histology. So far, a long list of studies have looked at MRI-histology comparisons (Cohen-Adad, 2018; Laule and Moore, 2018; MacKay and Laule, 2016; Petiet et al., 2019), each of them focusing on a specific pathology and a few MRI measures.
Despite these numerous studies, there is still an ongoing debate on what MRI measure should be used to quantify myelin and as a consequence there is a constant methodological effort to propose new measures. This debate would benefit from a quantitative analysis of all the findings published so far, specifically addressing inter-study variations and prospects for future studies, something that is currently missing from the literature.
In this study, we systematically reviewed quantitative MRI-histology comparisons and we used meta-analysis tools to address the following question: how different are the modalities for myelin quantification in terms of their relationship with the underlying histology?
Results
Literature survey
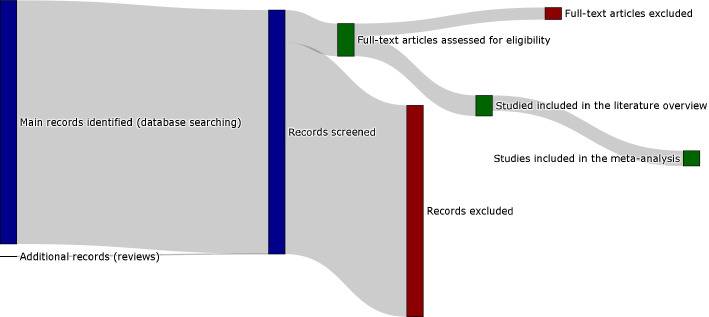
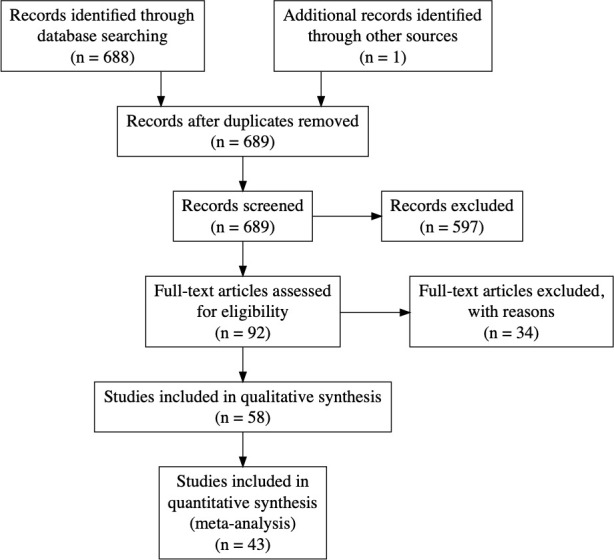
The screening process is summarized in the flowcharts in Figure 1 and Appendix 1—figure 1. The keywords as reported in the appendix returned 688 results on PubMed (last search on 03/06/2020). These results included 50 review articles. From the 50 review articles, six were selected as relevant for both the topics of myelin and related MRI-histology comparisons (Cohen-Adad, 2018; Laule and Moore, 2018; Laule et al., 2007; MacKay and Laule, 2016; Petiet et al., 2019; Turner, 2019). After the assessment, 58 original research studies were considered eligible, as shown in Appendix 1—table 1 (in the appendix) and Figure S2. All the data collected are available in the supplementary materials (Source data 1).
Figure 1. Sankey diagram representing the screening procedure (PRISMA flow chart provided in the appendix).
To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/01/selection.html#figure-1.
In terms of specific modalities, the survey shows that the most common MRI approach compared with histology was diffusion-weighted imaging (used in 28 studies), followed by magnetization transfer (MT, 27 studies), T2 relaxometry (19 studies) and T1 relaxometry (10 studies). Only 20 studies considered more than one approach: among the others, 20 focused exclusively on diffusion, 12 on MT, and six on T2 relaxometry.
From these 58 studies, we then focused only on brain studies and we further excluded studies not reporting either the number of subjects or the number of ROIs per subject. We also excluded one single-subject study that relied on voxels as distinct samples, whereas the other studies in this review are based on ROIs (i.e. including more than one voxel). In the end, 43 suitable studies were identified for the subsequent analyses.
Meta-analysis
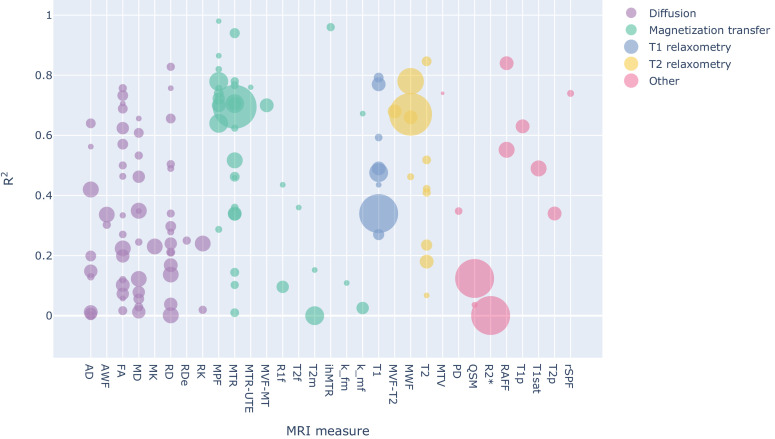
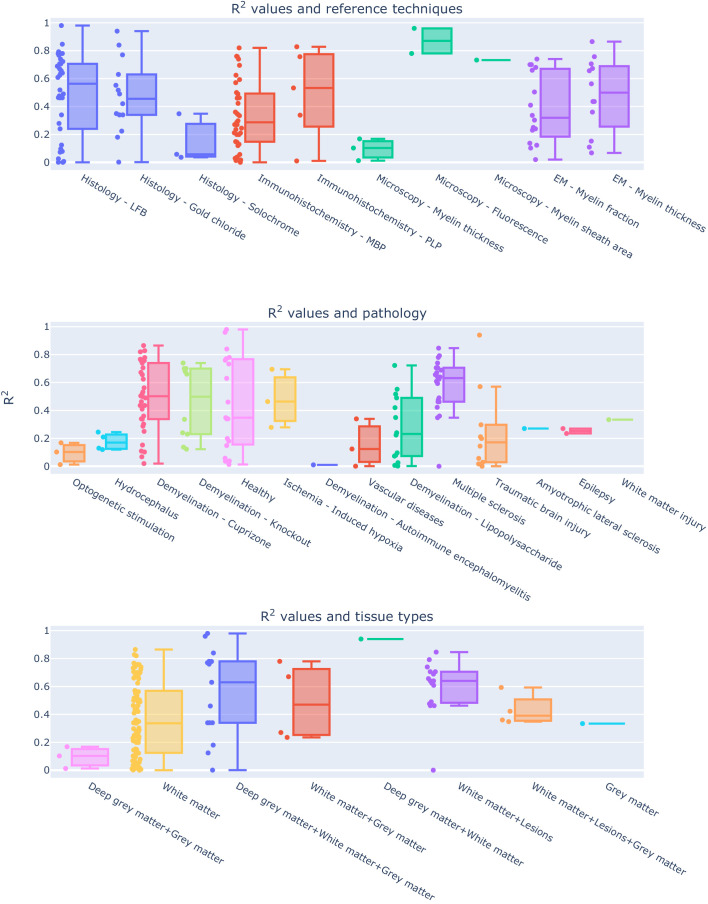
To compare the studies of interest, we first organized them according to the MRI measure used. Figure 2 and Figure 3 (and also Figure S3-S4) show the R2 values for the selected studies across measures: the highest values (R2 >0.8) are obtained mostly from MT measures, but they are associated with small sample sizes (with an average of 32 sample points). The studies with largest sample sizes are associated with R2 values between 0.6 and 0.8 for MT and T2 relaxometry, but with lower values for T1 relaxometry and other approaches.
Figure 2. Bubble chart of R2 values between a given MRI measure and histology for each study across MRI measures, with the area proportional to the number of samples.
To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/02/closer_look.html#figure-3.
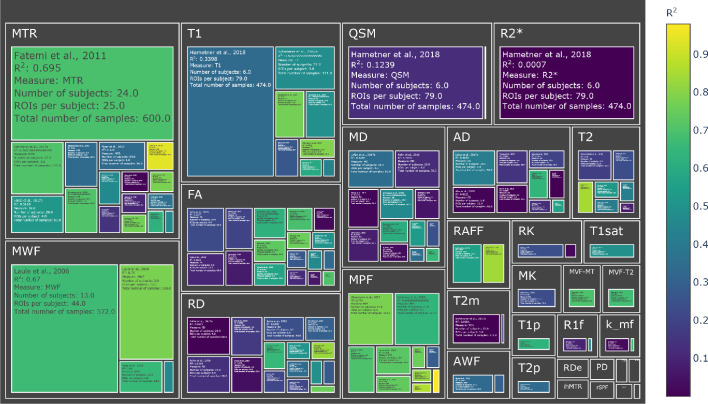
Figure 3. Treemap chart of the studies considered for the meta-analysis, organized by MRI measure.
The color of each box represents the reported R2 value while the size box is proportional to the sample size. To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/02/closer_look.html#figure-4.
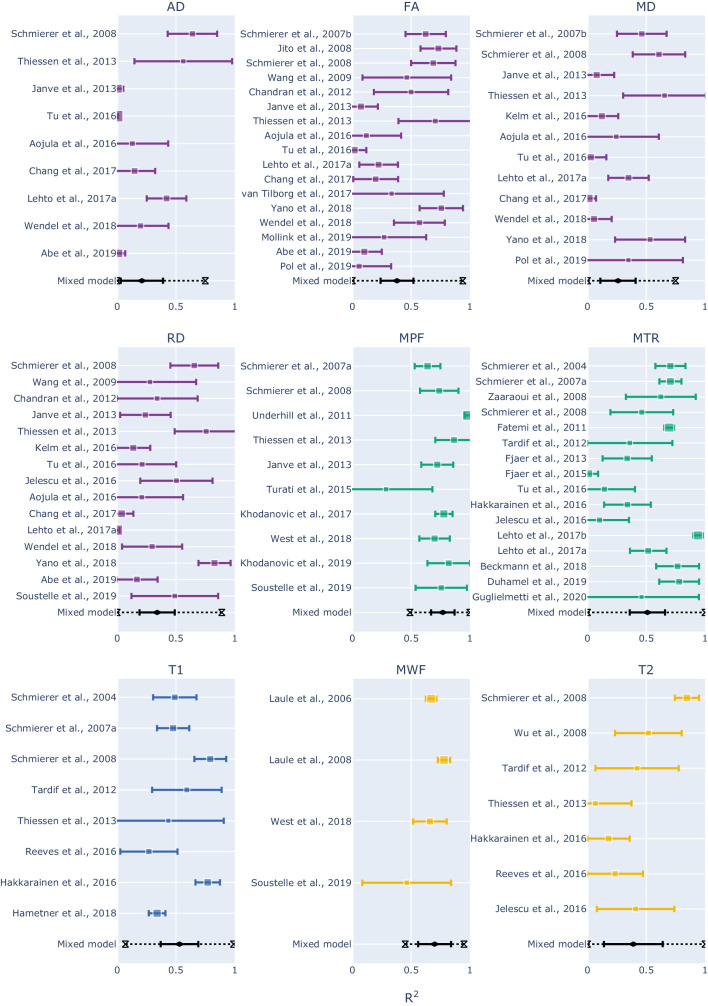
To combine the results for each measure, we then used a mixed-effect model: in this way we were able to express the overall effect size in terms of a range of R2 values within a confidence interval, but also to assess prediction intervals and inter-study differences. The results are shown as forest plots in Figure 4 (and also Figure S5).
Figure 4. Forest plots showing the R2 values reported by the studies and estimated from the mixed-effect model for each measure.
The hourglasses and the dotted lines in the mixed-effect model outcomes represent the prediction intervals. To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/03/meta_analysis.html#figure-5.
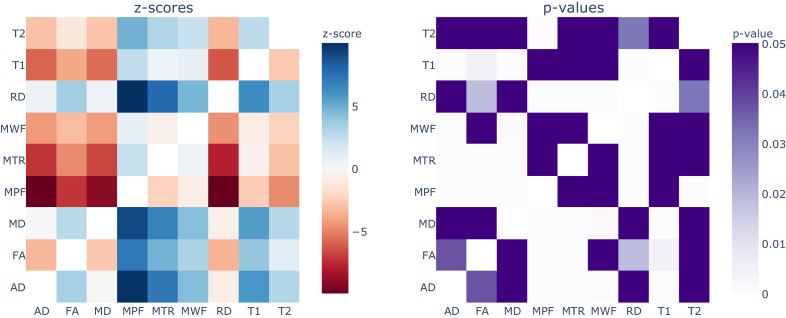
Apart from MPF and MWF, all the measures showed R2 overall estimates in the range 0.21–0.53. To investigate the significance of the differences between measures, we conducted a repeated measures meta-regression on every R2 estimate recorded (98 in total over 43 studies). As shown in Figure 5 (and also Figure S6), the measures can be roughly subdivided in two groups: MT- and relaxometry-based measures gave significantly higher R2 estimates compared to diffusion-based measures. Within the diffusion-based measures, FA shows slightly higher estimates than the others, with marginal significance over RD and AD or no significance in case of MD.
Figure 5. Results from the repeated measures meta-regression, displayed in terms of z-scores (left) and p-values (right) for each pairwise comparison across all the MRI measures.
In the z-score heatmap, each element refers to the comparison between the measure on the x axis with the one on the y axis. For example, MPF and FA (z-score = 7.14; p-value<0.0001) are statistically different, while MPF and T1 (z-score = 2.51; p-value=0.43) are not statistically different. To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/03/meta_analysis.html#figure-6.
Within MT- and relaxometry-based measures, the trends follow those in the forest plots (Figure 4), but most differences are not significant (Figure 5). However, the results in terms of z-score give a measure of distance between the R2 distributions. From this perspective, MPF has higher R2 estimates compared to all the other measures, but it is only marginally higher than MWF (z-score = 0.77; p-value=1) so we cannot claim that one is superior to the other. Following the same reasoning, MTR and T1 are not statistically different (z-score = 0.47; p-value=1).
When considering the prediction intervals calculated using τ2 (the variance of the effect size parameters across the population of studies), for most measures the interval spanned from 0.1 to 0.9 (Figure 4 and Figure S5). This implies that future studies relying on such measures can expect, on the basis of these studies, to obtain any R2 value in this broad interval. The only exceptions were MPF (0.49–1) and MWF (0.45–0.95), whose intervals were narrower than the alternatives. Finally, I2 (a measure of how much of the variability in a typical study is due to heterogeneity in the experimental design) was generally quite high (Table 1). MWF showed the lowest I2 across measures (I2 = 73.19%), but this may be misleading considering that it was based on only four studies, while the other measures included around 10 studies. Excluding MWF, MPF also showed a relatively low I2 (I2 = 83.18%). Qualitative comparisons across experimental conditions and methodological choices highlighted differences across pathology models, targeted tissue types and reference techniques (Figure 6 and Figure S7). Other factors such as magnetic field, co-registration, specific tissue and the related conditions (Figure S8) showed comparable distributions.
Table 1. Results from the mixed-effect models: for each measure we reported the number of studies, the estimate and standard error of the overall R2 distribution, the τ2 and the I2.
| Measure | Number of studies | Estimate | Standard error | Tau2 | I2 |
|---|---|---|---|---|---|
| MTR | 16 | 0.508 | 0.0691 | 0.07 | 96.03% |
| MPF | 10 | 0.7657 | 0.0455 | 0.0128 | 83.18% |
| FA | 17 | 0.3766 | 0.0663 | 0.0652 | 87.49% |
| RD | 15 | 0.3364 | 0.0679 | 0.0615 | 92.30% |
| MD | 12 | 0.2639 | 0.0679 | 0.044 | 87.35% |
| T1 | 8 | 0.5321 | 0.0692 | 0.0328 | 86.51% |
| AD | 9 | 0.2095 | 0.0802 | 0.048 | 97.69% |
| T2 | 7 | 0.3938 | 0.1023 | 0.0651 | 84.49% |
| MWF | 4 | 0.6997 | 0.0432 | 0.0041 | 73.19% |
Figure 6. Experimental conditions and methodological choices influencing the R2 values (top: reference techniques; middle: pathology model; bottom: tissue types).
To see the interactive figure: https://neurolibre.github.io/myelin-meta-analysis/04/other_factors.html#figure-7.
Discussion
Indirect measures are the most popular (for better or worse)
The literature survey offers an interesting perspective on popular research trends (Figure S2). The first consideration one can make is that every myelin imaging technique achieves myelin sensitivity through different means. A clear example is offered by the two most common approaches in this meta-analysis, DWI and MT: the MT effect is driven by saturation pulses interacting with myelin macromolecules that transfer their magnetization to water, whereas in diffusion experiments myelin is just not part of the picture. Diffusion acquisitions are blind to direct myelin measurement because the TEs used are too long (~100 ms) to be influenced by the actual macromolecules – with T2 of ~ 10 us (Stanisz et al., 1999) – or even the water molecules trapped in the myelin sheath – with T2 of ~ 30 ms (MacKay et al., 1994). To infer myelin content, one needs to rely on the interaction between intracellular and extracellular water compartments. The majority of diffusion studies included in this analysis used tensor-based measures (with fractional anisotropy being the most common), but some also used kurtosis-based analysis. The main issue with this approach is that other factors affect those measures (Beaulieu, 2002; Beaulieu, 2009), making it difficult to specifically relate changes in water compartments to changes in myelin.
Despite this issue, the use of diffusion as a proxy for myelin is quite widespread, specifically outside the field of quantitative MRI. This is probably a consequence of how popular DWI has become and how widely available are the related acquisition sequences. MT, the second most popular technique for quantifying myelin, estimates myelin by acquiring data with and without saturating the macromolecular proton pool. The simplest MT measure, MT ratio (MTR), incorporates non-myelin contributions in the final measurement. Recent acquisition variations include computing MTR from acquisitions with ultra-short echo times (Du et al., 2009; Guglielmetti et al., 2020; Wei et al., 2018) or relying on inhomogeneous MT (Duhamel et al., 2019; Varma et al., 2015). More complex experiments, for example quantitative MT, are based on fitting two compartments to the data, the free water and the macromolecular compartments, or pools. In this way, one is able to assess myelin through MPF with higher specificity, although still potentially including contributions from other macromolecules. Additional measures have also been considered (including the T2 of each pool, the exchange rate between the pools). The drawback of qMT is the requirement for a longer and more complex acquisition. Recently, there have been alternative techniques to estimate only MPF, resulting in faster acquisitions with similar results (Khodanovich et al., 2019; Khodanovich et al., 2017; Yarnykh, 2012). Despite being focused on macromolecular contributions, these approaches are not strictly specific to myelin (Sled, 2018): in this sense, an important limitation is that MT effects are sensitive to the pH of the targeted tissue and therefore changes in the pH (caused for example by inflammation processes) will affect MT-based measures of myelin (Stanisz et al., 2004).
Following diffusion and MT, the most popular approach is T2 relaxometry. Unlike diffusion and MT, in T2 relaxometry experiments one can directly observe the contribution from the water trapped between the myelin bilayers, and can therefore estimate the myelin water fraction. A simpler but less specific approach consists in estimating the transverse relaxation time considering the decay to be mono-exponential. A historical and practical drawback of these approaches is that they require longer acquisitions, although faster alternatives have been developed (Does and Gore, 2000; Prasloski et al., 2012). A more subtle but nevertheless important limitation lies in the multi-compartment model used in multi-exponential T2 relaxometry (Does, 2018): this model generally assumes slow water exchange between compartments, but it has been showed that water exchange actually contributes to T2 spectra variations (Dula et al., 2010; Harkins et al., 2012).
Finally, other studies used a diverse collection of other measures, including T1 relaxometry, apparent transversal relaxation rate (R2*), proton density (PD), macromolecular tissue volume (MTV), relaxation along a fictitious field (RAFF), and quantitative susceptibility mapping (QSM).
After this general overview, it is clear that each modality could be a suitable candidate for a quantitative myelin biomarker. To then make a choice informed by the studies here reported, it becomes necessary to consider not only effect sizes in terms of correlation, but also sample sizes and acquisition times.
There is no myelin MRI measure true to histology
When looking at the R2 values across the different measures, the first detail that catches one’s eye is how most measures present a broad range of values (Figure 2 and Figure 3). When taking into account the sample size, the largest studies show higher correlations for MT and T2 relaxometry studies than any other approach (Figure S3 and Figure S4). In quantitative terms, the meta-analysis corroborates this idea, showing that MPF and MWF tend to be more specific to myelin compared to the other measures (respectively with R2 = 0.7657 and R2 = 0.6997), in line with the underlying theory. Notably, diffusion-based measures show the lowest overall estimates (with values between R2 = 0.3766 for FA and R2 = 0.2095 for AD): this could be due to the fact, as already mentioned, that DWI does not specifically measure myelin properties, and despite FA and RD being influenced by the myelin content, they are also influenced by other factors that make them unsuitable as measures of myelin. The repeated measure meta-regression confirms this overall picture, clearly distinguishing MT- and relaxometry-based measures from diffusion-based ones (Figure 5).
Despite these considerations on the advantages of MPF and MWF, one should refrain from concluding that they are the ‘true’ MRI measures of myelin. The reason for this caution is given not by the overall effect sizes observed here, but by the collateral outcomes of the meta-analysis. The first one is given by the prediction intervals: most measures exhibit large intervals (Figure 4), not supporting the idea of them being robust biomarkers. MPF and MWF seem to be again the most suitable choices for future studies, but a range between 0.5 and 1 is still quite large.
The second important aspect to consider is given by the differences across studies: the meta-analysis showed how such differences strongly limit inter-study comparisons for a given measure (Figure 6). This result should be expected, given that the studies here examined are inevitably influenced by the specific experimental constraints and methodological choices. Given the limited number of studies, it is not possible to quantitatively study interactions between MRI measures and the other factors (e.g. modality used as a reference, tissue types, magnetic field strength). For further qualitative insights, we invite the reader to explore the interactive figures S7-S8. A first important factor to consider is the validation modality used as a reference, which will be dictated by the equipment availability and cost. However, such a choice has an impact on the actual comparison: histology and immunochemistry, despite being specific to myelin, do not offer a volumetric measure of myelin, but rather a proxy based on the transmittance of the histological sections. So far, the only modality able to give a volumetric measure would be electron microscopy, which is an expensive and resource-consuming approach. Also, electron microscopy has several limitations, including tissue shrinkage, degradation of the myelin sheath structure due to imperfect fixation, imperfect penetration of the osmium stain, polishing, keeping focus over large imaging regions. All these effects contribute to the lack of precision and accuracy when quantifying myelin content with EM-based histology (Cohen-Adad, 2018). Another important observation is that none of the studies here reviewed considered histology reproducibility, which is hard to quantify as a whole given that a sample can be processed only once: collateral factors affecting tissue processing (e.g. sectioning distortions, mounting and staining issues) constitute an actual limitation for histology-based validation. A further example of influential factor often dictated by equipment availability is the magnetic field strength of the MRI scanner: figure S8 shows that most studies were conducted at 7T and 9.4T, with some pioneering studies at 1.5T and even fewer ones at other field strengths.
In addition to differences in experimental and methodological designs, there are also several considerations that arise out of the lack of shared practices in MRI validation studies. The first evident one is the use of correlations: despite being a simple measure that serves well the purpose of roughly characterizing a relationship, Pearson correlation is not the right tool for quantitative biomarkers, as it does not characterize the actual relationship between histology and MRI. Linear regression is a step forward but has the disadvantage of assuming a linear relationship. Despite Pearson correlation and linear regression being the most common measures used in the studies here reviewed, it is still not clear if the relationship is actually linear. Only one study among the considered ones computed both Pearson and Spearman correlation values (Tardif et al., 2012), and reported higher Spearman correlations, pointing out that non-linear relationships should actually be considered. One last consideration regarding the use of correlation measures for validating quantitative biomarkers is about the intercept in the MRI-histology relationship. Notably, only MWF is expected to assume a value equal to zero when myelin is absent (West et al., 2018). For the other measures, it would be necessary to estimate the intercept, which leads to the calibration problem in the estimate of myelin volume fraction. Notably, calculating Pearson correlation does not provide any information for such calibration. Another arbitrary practice that would benefit from some harmonization is the choice of ROIs. The studies reported here examined a diverse list of ROIs, in most cases hand-drawn on each modality, encompassing different types of tissue, and the most common approach is to report a single, pooled correlation. This is problematic, as different types of tissue (e.g. grey matter and white matter) will show different values for MRI-based measures but also for histology-based ones, making linearity assumptions about the two modalities. However, with this approach gross differences between tissues drive the observed correlation, without actually showing if the MRI-based measure under analysis is sensitive to subtle differences and therefore a suitable quantitative biomarker for myelin. The effect of considering different types of tissues is showed in Figure 6 and Figure S7, where correlation ranges change when considering different types of tissue. However, the large correlation range in white matter, the most common tissue studied, suggests that other factors also affect the correlation.
It should be clear at this point that any debate about a universal MRI-based measure of myelin is pointless, at least at the moment, as the overall picture provided by previous studies does not point to any such ideal measure. Nevertheless, is debating about a universal measure helpful for future studies?
Better biomarkers require more reproducibility studies
We hope this meta-analysis convinces the reader that a holy grail of myelin imaging does not exist, at least as long as we consider histology to be the ground truth. Given that we all have to pick our poison, the upside is that measures based on MT and relaxometry are not statistically different, and therefore, future studies have an actual choice among candidate measures. For further progress, rather than debating about a perfect measure, we would argue that what is missing at the moment is a clear picture of what can be achieved with each specific MRI modality. The studies examined here focus on a large set of different measures, and more than half of them considered at most two measures, highlighting how the field is mostly focused on formulating new measures. While it is understood that novel measures can provide new perspectives, it is also fundamentally important to understand the concrete capabilities and limitations of current measures. From this meta-analysis, what the literature clearly lacks is reproducibility studies, specifically answering two main questions: (1) what is the specificity of each measure? We should have a practical validation of our theoretical understanding of the relevant confounds; (2) what is the ‘parameter sensitivity’ of each measure? Here, we refer to parameter sensitivity in a broad sense, that includes also experimental conditions and methodological choices. The results here presented show how certain conditions (e.g. pathology) seem to affect the coefficient of determination more than others but given the limited number of studies for each modality, we refrained from additional analyses to avoid speculation. A warning message that is evident from these results is the inherent limitation of DWI for estimating myelin content: this is not by any means a novel result (Beaulieu, 2002; Beaulieu, 2009), but it is nevertheless worth reiterating given the outcomes of our analysis. If estimating myelin content is relevant in a diffusion study, it is important to consider complementing the diffusion measure with one of the modalities here reviewed; in this way, it would be possible to decouple the influence of myelin content from the many other factors that come into play when considering diffusion phenomena.
Finally, an important factor to take into account when choosing a biomarker of myelin is the actual application. For animal research, long acquisitions are not a major issue. However, when considering biomarkers for potential clinical use, the acquisition time can become a relevant issue. An example is the well-established multi-echo spin-echo implementation of MWF, that can only be used for a specific slice in a hypothetical clinical scenario. Faster techniques have been proposed for estimating it with gradient- and spin-echo (GRASE) sequences (Does and Gore, 2000; Feinberg and Oshio, 1991; Prasloski et al., 2012). Even in this case, the acquisition time still reaches 15 min for acquiring roughly the whole brain with an isotropic resolution of 2 mm. Complex MT acquisitions such as qMT suffer from the same problem, although it is possible to use optimized and faster protocols to focus specifically on MPF (Khodanovich et al., 2019; Khodanovich et al., 2017; Yarnykh, 2012).
Conclusions
Several MRI measures are sensitive to myelin content and the current literature suggests that most of them are not statistically different in terms of their relationship with the underlying histology. Measures highly correlated with histology are also the ones with a higher expected specificity. This suggests that future studies should try to better address how specific each measure is, for the sake of clarifying suitable applications.
Materials and methods
Review methodology
The Medline database (https://pubmed.ncbi.nlm.nih.gov) was used to retrieve the articles. The keywords used are specified in the appendix. We followed the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) guidelines for record screening and study selection. The results were first screened to remove unrelated work. Specifically we discarded: work relying only on MRI; work relying only on histology or equivalent approaches; work reporting only qualitative comparisons. After this first screening, the remaining papers were assessed. At this stage, we discarded: studies using MRI-based measures in arbitrary units (e.g. T1-weighted or T2-weighted data); studies using measures of variation in myelin content (defined either as the difference between normal and abnormal myelin content) either for MRI or for histology; studies using arbitrary assessment scales; studies comparing MRI-based absolute measures of myelin with histology-based relative measures (e.g. g-ratio); studies reporting other quantitative measures than correlation or R2 values; studies comparing histology from one dataset and MRI from a different one. As an additional source for potential candidate studies, we screened the review articles in the initial results, and we selected the relevant studies that were not already present in the studies already selected.
From the final papers, we collected first the following details: the DOI; which approach was used (diffusion, MT, T1 relaxometry, T2 relaxometry, or other); which specific MRI measures were compared to histology or equivalent techniques; the magnetic field; the technique used as a reference (histology, immunochemistry, microscopy, electron microscopy); the focus of the study in terms of brain, spinal cord or peripheral nerve; if the subjects were humans or animals, and if the latter which animal; if the tissue under exam was in vivo, in situ or ex vivo, and in the latter case if the tissue was fixed or not; if the tissue was healthy or pathological, and if the latter which pathology; the specific structures examined for correlation purposes; which comparison technique was used (e.g. Pearson correlation, Spearman correlation, linear regression); the number of subjects; the number of ROIs per subject; the male/female ratio; if registration procedures were performed to align MRI and histology; in case of pathological tissue, if control tissue was considered as well; other relevant notes. If before calculating the correlations the data were averaged across subjects, the number of subjects was considered to be one. The same consideration was made for averaging across ROIs. This is because the numbers of subjects and ROIs were used to take into account how many sample points were used when computing the correlation. We set each of those numbers to one for all the studies where the data were averaged respectively across subjects and across ROIs. Finally, in those cases where the number of ROIs or the number of subjects were given as a range rather than specific values, we used the most conservative value and added the related details to the notes.
We then proceeded to collect the quantitative results reported for each measure and for each study in the form of R2. Given that different studies may rely on a different strategy when reporting correlations, we adopted the following reasoning to limit discrepancies across studies while still objectively representing each of them. In case of multiple correlation values reported, for our analysis we selected the ones referring to the whole dataset and the entire brain if available, and considering each ROI in a given subject as a sample if possible; if only correlation values for specific ROIs were reported, the one for the most common reported structure would be chosen. In the case of multiple subjects, if data were provided separately for each group, the correlation for the control group was used. When different comparison methods were reported (e.g. both Pearson and Spearman correlation) or if the MRI data was compared with multiple references (e.g. both histology and immunohistochemistry), the correlations used were chosen on the basis of the following priority orders (from the most preferable to the least): for multiple comparison methods, linear regression, Spearman correlation, Pearson correlation; for multiple reference techniques, electron microscopy, immunohistochemistry, histology. Finally, in any other case where more than one correlation value was available, the most conservative value was used. Any other additional value was in any case mentioned in the notes of the respective study.
Meta-analysis
For the quantitative analysis, we restricted our focus on brain studies and only on the ones providing an indication of both the number of subjects and the number of ROIs. For each study, we computed the sample size as the product between the number of subjects and the number of ROIs per subject. In this way, we were able to compare the reported R2 values across measures taking into account the related number of points actually used for correlation purposes. We note that correlation or regression analyses run on multiple ROIs and subjects represents a repeated measures analysis, for which the degrees of freedom computation can be complex; however, most papers neglected the repeated measures structure of the data and thus the sample size computation here represents a very approximate and optimistic view of the precision of each R2 value.
To estimate the variance of each R2 value, we relied on the correlation properties and the delta method (Lehman, 1999). Let us consider the Pearson’s correlation r of two variables X and Y with population correlation ρ. If r is calculated from N random samples, the sampling variance is (1-ρ2)2/N. Applying the delta method, we then approximated the variance of R2 as 4 R2(1 R2)2/N, assuming R2≈ρ2. As we recognise that some papers computed Spearman correlation, this calculation is again optimistic and may underestimate the sampling variability of the squared Spearman correlation.
To estimate the overall effect size in terms of R2, we have to choose how to model the distribution of true effects given by the data collected from the literature. The two most common approaches are fixed-effects and mixed-effects models. While the underlying mathematical model is the same as the one used for linear regression (more details in the appendix), the assumptions are different: fixed-effects models assume that all the studies share a common effect size, while mixed-effects models assume that the effect size across studies is similar but not identical (Raudenbush, 2009). In our case, as the studies have several factors that influence the R2 values (e.g. histology/microscopy reference, magnetic field strength, pathology model), we expect a distribution of effect sizes due to inter-study differences. This is why we proceeded to fit a mixed-effects model to each measure that was featured in more than two studies. Apart from the effect size distributions, we reported two additional measures, I2 and τ2: the former expresses as a percentage how much of variability in a typical study is due to heterogeneity (i.e. the variation in study outcomes between studies) rather than chance (Higgins and Thompson, 2002), while the latter can be used to calculate the prediction interval (Raudenbush, 2009), which gives the expected range for the measure of interest in future studies. We used forest plots to represent the outcomes, and both the mixed effects estimate of the population estimated R2, with both a 95% confidence and a (larger) 95% prediction interval.
For the explicit purpose of comparing the effect sizes between different MRI measures, we conducted a repeated measures meta-regression on every R2 value recorded. We associated each R2 value with three additional details: (i) the related variance, as done in the measure-specific mixed-effects models; (ii) the related study, used as the random intercept (i.e. random variable) to incorporate potential inter-study variability; and (iii) the related MRI measure, used as the moderator (i.e. categorical variable) to estimate the differences between measures. In this way, the meta-regression leads to R2 intervals for each MRI measure, with the same trend as measure-specific mixed-effects models but with subtle differences. This is because the meta-regression makes two additional assumptions: first, R2 estimates within the same study share the same random effects and second, the between-study variance is the same for all observations. We then used the meta-regression R2 estimates to compute every possible pairwise comparison between MRI measures and to identify significantly different pairs using Tukey's test, while controlling the error rate over all the possible comparisons (Bonferroni correction).
This additional model is necessary, as direct comparisons are not possible with measure-specific analyses. While the repeated measures meta-regression makes direct comparisons straightforward, we reported the main R2 estimates based on the measure-specific mixed-effects models, as they make weaker assumptions.
For visual comparisons, we used the Jupyter notebook provided in the supplementary materials. For model fitting, we used the Metafor package, version 2.4–0 (Viechtbauer, 2010).
Acknowledgements
MM was funded by the Wellcome Trust through a Sir Henry Wellcome Postdoctoral Fellowship [213722/Z/18/Z]. TEN was supported by NIH grant R01MH096906.
Appendix 1
Search keywords
(myelin[Title/Abstract] AND ((magnetic[Title/Abstract] AND resonance[Title/Abstract]) OR mr[Title/Abstract] OR mri[Title/Abstract])) AND (histology[Title/Abstract] OR histopathology[Title/Abstract] OR microscopy[Title/Abstract] OR immunohistochemistry[Title/Abstract] OR histological[Title/Abstract] OR histologically[Title/Abstract] OR histologic[Title/Abstract] OR histopathological[Title/Abstract] OR histopathologically[Title/Abstract] OR histopathologic[Title/Abstract]).
Results obtained from the Medline database: 688 (03/06/2020).
Appendix 1—figure 1. PRISMA flowchart for the meta-analysis.
Fixed- and mixed-effects models
While a traditional linear regression model estimates the error variance from residuals, in a fixed effects meta-analysis model, each paper’s response and standard errors, as well as the error variance of the regression model can be directly computed from the supplied response standard deviations. Specifically, for a (non-meta) regression model we have the -th response modeled with covariate values , , where random error has unknown variance . In a fixed-effects meta-analysis, we are given but also , the standard error of , and the regression model has the same form except the variance is known, , and the weighted least squares regression can be computed, estimating beta and its standard error. A mixed-effects meta-analysis accounts for more variance than what can be ascribed to the sampling error of the reported outcome. The regression model has again the same form, except now the variance is , the sum of the reported squared standard error and the unknown between-study variance . Iterative methods are used to estimate and, once estimated, a weighted least squares regression can be computed. The parameter can be interpreted as the variance of noise-free (hypothetical, zero standard error) results from the population of all possible studies. The importance of can also be gauged by , the proportion of variance due to random inter-study differences (i.e. is the proportion attributable to random sampling error of each study) (Higgins and Thompson, 2002).
Abbreviations and mathematical symbols
AD – axial diffusivity
AK – axial kurtosis
AWF – axonal water fraction
FA – fraction anisotropy
ihMTR – inhomogeneous magnetization transfer ratio
k_fm – free water-macromolecular exchange rate
k_mf – macromolecular-free water exchange rate
M0m – macromolecular pool magnetization fraction
MD – mean diffusivity
MK – mean kurtosis
MPF – macromolecular pool fraction
MT – magnetization transfer
MTR – magnetization transfer ratio
MTR-UTE – magnetization transfer ratio (using ultra-short echo time)
MTV – macromolecular tissue volume
MVF-MT – myelin volume fraction (estimated from MT)
MVF-T2 – myelin volume fraction (estimated from T2)
MWF – myelin water fraction
PD – proton density
PN – peripheral nerve
PRISMA – Preferred Reporting Items for Systematic Reviews and Meta-Analyses
QSM – quantitative susceptibility mapping
R1f – free water pool longitudinal relaxation rate
R2* – apparent transverse relaxation rate
RAFF – relaxation along a fictitious field
RD – radial diffusivity
RD-DBSI – radial diffusivity (from diffusion basis spectrum imaging)
RDe – extra-cellular compartment radial diffusivity
RK – radial kurtosis
rSPF – relative semi-solid proton fraction
SC – spinal cord
T1 – longitudinal relaxation time
T1p – adiabatic longitudinal relaxation time
T1sat – longitudinal relaxation time under magnetization transfer irradiation
T2 – transverse relaxation time
T2f – free water pool transverse relaxation time
T2int – transverse relaxation intermediate component
T2m – macromolecular pool transverse relaxation rate
T2p – adiabatic transverse relaxation time
Appendix 1—table 1. Selected studies for qualitative analysis.
| Study | MRI measure(s) | Histology/microscopy measure | Tissue | Condition | Focus |
|---|---|---|---|---|---|
| Schmierer et al., 2004 | T1, MTR | Histology - LFB | Human | Multiple sclerosis | Brain |
| Odrobina et al., 2005 | T1, T2, T2int, MWF, M0m, MTR | Microscopy - Myelin fraction | Animal - Rat | Demyelination - Tellurium | PN |
| Pun et al., 2005 | T1, T2int, MWF | Microscopy - Myelin fraction | Animal - Rat | Demyelination - Tellurium | PN |
| Laule et al., 2006 | MWF | Histology - LFB | Human | Multiple sclerosis | Brain |
| Schmierer et al., 2007a | T1, MTR, MPF, T2m | Histology - LFB | Human | Multiple sclerosis | Brain |
| Schmierer et al., 2007b | FA, MD | Histology - LFB | Human | Multiple sclerosis | Brain |
| Jito et al., 2008 | FA | Microscopy - Myelin sheath area | Animal - Mouse | Healthy | Brain |
| Kozlowski et al., 2008 | MWF, FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Laule et al., 2008 | MWF | Histology - LFB | Human | Multiple sclerosis | Brain |
| Schmierer et al., 2008 | T1, T2, MTR, MPF, MD, FA, AD, RD | Histology - LFB | Human | Multiple sclerosis | Brain |
| Wu et al., 2008 | T2 | Histology - LFB | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Zaaraoui et al., 2008 | MTR | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Takagi et al., 2009 | FA, AD | EM - Myelin thickness | Animal - Rat | Degeneration - Contusive injury | PN |
| Wang et al., 2009 | FA, RD | Histology - LFB | Animal - Rat | Ischemia - Induced hypoxia | Brain |
| Zhang et al., 2009 | RD | Histology - LFB | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Schmierer et al., 2010 | MTR, T2 | Histology - LFB | Human | Multiple sclerosis | Brain |
| Fatemi et al., 2011 | MTR | Immunohistochemistry - MBP | Animal - Mouse | Ischemia - Induced hypoxia | Brain |
| Laule et al., 2011 | MWF | Immunohistochemistry - MBP | Human | Multiple sclerosis | Brain |
| Underhill et al., 2011 | MPF | Histology - LFB | Animal - Mouse | Healthy | Brain |
| Chandran et al., 2012 | FA, RD | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Tardif et al., 2012 | T1, T2, MTR, PD | Immunohistochemistry - MBP | Human | Multiple sclerosis | Brain |
| Fjær et al., 2013 | MTR | Immunohistochemistry - PLP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Harkins et al., 2013 | MWF, MPF | Microscopy - Myelin fraction | Animal - Rat | Edema - Hexaclorophene | SC |
| Janve et al., 2013 | MPF, R1a, k_ba, FA, RD, MD, AD | Histology - LFB | Animal - Rat | Demyelination - Lipopolysaccharide | Brain |
| Thiessen et al., 2013 | MPF, R1f, k_fm, k_mf, T2f, T2m, MD, RD, AD, FA, T1, T2 | EM - Myelin thickness | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Kozlowski et al., 2014 | MWF | Immunohistochemistry - MBP | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Wang et al., 2014 | RD, RD-DBSI | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Autoimmune encephalomyelitis | SC |
| Fjær et al., 2015 | MTR | Immunohistochemistry - PLP | Animal - Mouse | Demyelination - Autoimmune encephalomyelitis | Brain |
| Seehaus et al., 2015 | FA, RD, MD | Histology - Silver | Human | Healthy | Brain |
| Turati et al., 2015 | MPF | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Wang et al., 2015 | RD-DBSI | Histology - LFB | Human | Multiple sclerosis | SC |
| Aojula et al., 2016 | FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Rat | Hydrocephalus | Brain |
| Hakkarainen et al., 2016 | T1, T2, MTR, T1p, T2p, RAFF | Histology - Gold chloride | Animal - Rat | Healthy | Brain |
| Jelescu et al., 2016 | RD, RK, AWF, Rde, T2, MTR | EM - Myelin fraction | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Kelm et al., 2016 | MD, RD, MK, RK, AWF | EM - Myelin fraction | Animal - Mouse | Demyelination - Knockout | Brain |
| Reeves et al., 2016 | T1, T2 | Immunohistochemistry - MBP | Human | Epilepsy | Brain |
| Tu et al., 2016 | FA, AD, RD, MD, MTR | Immunohistochemistry - MBP | Animal - Rat | Traumatic brain injury | Brain |
| Chang et al., 2017 | FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Mouse | Healthy | Brain |
| Chen et al., 2017 | MWF | EM - Myelin fraction | Animal - Rat | Injury - Dorsal columnar transection | SC |
| Khodanovich et al., 2017 | MPF | Histology - LFB | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Lehto et al., 2017a | RAFF, MTR, T1sat, FA, MD, AD, RD | Histology - Gold chloride | Animal - Rat | Demyelination - Lipopolysaccharide | Brain |
| Lehto et al., 2017b | MTR | Histology - Gold chloride | Animal - Rat | Traumatic brain injury | Brain |
| van Tilborg et al., 2018 | FA | Immunohistochemistry - MBP | Animal - Rat | White matter injury | Brain |
| Beckmann et al., 2018 | MTR | Histology - LFB | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Berman et al., 2018 | MTV | EM - Myelin fraction | Animal - Mouse | Demyelination - Knockout | Brain |
| Hametner et al., 2018 | R2*, T1, QSM | Histology - LFB | Human | Vascular diseases | Brain |
| Praet et al., 2018 | MK, RK, AK, FA, MD, RD, AD | Immunohistochemistry - MBP | Animal - Mouse | Amyloidosis | Brain |
| Wendel et al., 2018 | FA, AD, RD, MD | Immunohistochemistry - MBP | Animal - Mouse | Traumatic brain injury | Brain |
| West et al., 2018 | MPF, MWF, MVF-T2, MVF-MT | EM - Myelin fraction | Animal - Mouse | Demyelination - Knockout | Brain |
| Yano et al., 2018 | FA, RD, MD | Immunohistochemistry - PLP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Abe et al., 2019 | FA, RD, AD | Microscopy - Myelin thickness | Animal - Mouse | Optogenetic stimulation | Brain |
| Duhamel et al., 2019 | ihMTR, MTR | Microscopy - Fluorescence | Animal - Mouse | Healthy | Brain |
| Khodanovich et al., 2019 | MPF | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Mollink et al., 2019 | FA | Immunohistochemistry - MBP | Human | Amyotrophic lateral sclerosis | Brain |
| Peters et al., 2019 | FA, MD | Histology - LFB | Human | Tuberous sclerosis complex | Brain |
| Pol et al., 2019 | QSM, FA, MD | Histology - Solochrome | Animal - Mouse | Healthy | Brain |
| Soustelle et al., 2019 | MPF, RD, MWF, rSPF | Immunohistochemistry - MBP | Animal - Mouse | Demyelination - Cuprizone | Brain |
| Guglielmetti et al., 2020 | MTR, MTR-UTE | Immunohistochemistry - MBP | Animal - Mouse | Healthy | Brain |
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Matteo Mancini, Email: ingmatteomancini@gmail.com.
Saad Jbabdi, University of Oxford, United Kingdom.
Chris I Baker, National Institute of Mental Health, National Institutes of Health, United States.
Funding Information
This paper was supported by the following grants:
Wellcome Trust 213722/Z/18/Z to Matteo Mancini.
National Institutes of Health R01MH096906 to Thomas E Nichols.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Data curation, Formal analysis, Visualization, Methodology, Writing - original draft, Writing - review and editing.
Visualization, Writing - review and editing.
Conceptualization, Writing - review and editing.
Conceptualization, Writing - review and editing.
Conceptualization, Methodology, Writing - review and editing.
Conceptualization, Methodology, Writing - review and editing.
Additional files
In order to execute the notebook, the Python (3.7) and R (3.6) interpreters are required, as well as the R packages metafor (2.4) and multcomp (1.4), and the following Python packages: numpy (1.18.4); pandas (0.25.3); plotly (4.8.1); rpy2 (3.3.4); xlrd (1.2.0). The notebook assumes that the spreadsheet is in the same path as the notebook itself. More details are provided here: https://github.com/matteomancini/myelin-meta-analysis (Mancini, 2020; copy archived at swh:1:rev:17ca8673c9e15c54ad0b814248b69232b63c3a38).
Data availability
All the data collected from the selected studies for this meta-analysis are provided in the spreadsheet file Source data 1.
References
- Abe Y, Komaki Y, Seki F, Shibata S, Okano H, Tanaka KF. Correlative study using structural MRI and super-resolution microscopy to detect structural alterations induced by long-term optogenetic stimulation of striatal medium spiny neurons. Neurochemistry International. 2019;125:163–174. doi: 10.1016/j.neuint.2019.02.017. [DOI] [PubMed] [Google Scholar]
- Aojula A, Botfield H, McAllister JP, Gonzalez AM, Abdullah O, Logan A, Sinclair A. Diffusion tensor imaging with direct cytopathological validation: characterisation of decorin treatment in experimental juvenile communicating hydrocephalus. Fluids and Barriers of the CNS. 2016;13:9. doi: 10.1186/s12987-016-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR in Biomedicine. 2002;15:435–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- Beaulieu C. CHAPTER 6 - The Biological Basis of Diffusion Anisotropy. In: Johansen-Berg H, Behrens T. E. J, editors. Diffusion MRI. Academic Press; 2009. pp. 105–126. [Google Scholar]
- Beckmann N, Giorgetti E, Neuhaus A, Zurbruegg S, Accart N, Smith P, Perdoux J, Perrot L, Nash M, Desrayaud S, Wipfli P, Frieauff W, Shimshek DR. Brain region-specific enhancement of remyelination and prevention of demyelination by the CSF1R kinase inhibitor BLZ945. Acta Neuropathologica Communications. 2018;6:9. doi: 10.1186/s40478-018-0510-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman S, West KL, Does MD, Yeatman JD, Mezer AA. Evaluating g-ratio weighted changes in the corpus callosum as a function of age and sex. NeuroImage. 2018;182:304–313. doi: 10.1016/j.neuroimage.2017.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cercignani M, Dowell NG, Tofts PS. Quantitative MRI of the Brain. CRC Press; 2018. [Google Scholar]
- Chandran P, Upadhyay J, Markosyan S, Lisowski A, Buck W, Chin CL, Fox G, Luo F, Day M. Magnetic resonance imaging and histological evidence for the blockade of cuprizone-induced demyelination in C57BL/6 mice. Neuroscience. 2012;202:446–453. doi: 10.1016/j.neuroscience.2011.10.051. [DOI] [PubMed] [Google Scholar]
- Chang EH, Argyelan M, Aggarwal M, Chandon TS, Karlsgodt KH, Mori S, Malhotra AK. Diffusion tensor imaging measures of white matter compared to myelin basic protein immunofluorescence in tissue cleared intact brains. Data in Brief. 2017;10:438–443. doi: 10.1016/j.dib.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HS, Holmes N, Liu J, Tetzlaff W, Kozlowski P. Validating myelin water imaging with transmission electron microscopy in a rat spinal cord injury model. NeuroImage. 2017;153:122–130. doi: 10.1016/j.neuroimage.2017.03.065. [DOI] [PubMed] [Google Scholar]
- Cohen-Adad J. Microstructural imaging in the spinal cord and validation strategies. NeuroImage. 2018;182:169–183. doi: 10.1016/j.neuroimage.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Cohen-Adad J, Wheeler-Kingshott CA. Quantitative MRI of the Spinal Cord. Academic Press; 2014. [Google Scholar]
- Does MD. Inferring brain tissue composition and microstructure via MR relaxometry. NeuroImage. 2018;182:136–148. doi: 10.1016/j.neuroimage.2017.12.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Does MD, Gore JC. Rapid acquisition transverse relaxometric imaging. Journal of Magnetic Resonance. 2000;147:116–120. doi: 10.1006/jmre.2000.2168. [DOI] [PubMed] [Google Scholar]
- Du J, Takahashi AM, Bydder M, Chung CB, Bydder GM. Ultrashort TE imaging with off-resonance saturation contrast (UTE-OSC) Magnetic Resonance in Medicine. 2009;62:527–531. doi: 10.1002/mrm.22007. [DOI] [PubMed] [Google Scholar]
- Duhamel G, Prevost VH, Cayre M, Hertanu A, Mchinda S, Carvalho VN, Varma G, Durbec P, Alsop DC, Girard OM. Validating the sensitivity of inhomogeneous magnetization transfer (ihMT) MRI to myelin with fluorescence microscopy. NeuroImage. 2019;199:289–303. doi: 10.1016/j.neuroimage.2019.05.061. [DOI] [PubMed] [Google Scholar]
- Dula AN, Gochberg DF, Valentine HL, Valentine WM, Does MD. Multiexponential T2, magnetization transfer, and quantitative histology in white matter tracts of rat spinal cord. Magnetic Resonance in Medicine. 2010;63:902–909. doi: 10.1002/mrm.22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi A, Wilson MA, Phillips AW, McMahon MT, Zhang J, Smith SA, Arauz EJ, Falahati S, Gummadavelli A, Bodagala H, Mori S, Johnston MV. In vivo magnetization transfer MRI shows dysmyelination in an ischemic mouse model of periventricular leukomalacia. Journal of Cerebral Blood Flow & Metabolism. 2011;31:2009–2018. doi: 10.1038/jcbfm.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg DA, Oshio K. GRASE (gradient- and spin-echo) MR imaging: a new fast clinical imaging technique. Radiology. 1991;181:597–602. doi: 10.1148/radiology.181.2.1924811. [DOI] [PubMed] [Google Scholar]
- Fields RD. A new mechanism of nervous system plasticity: activity-dependent myelination. Nature Reviews Neuroscience. 2015;16:756–767. doi: 10.1038/nrn4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjær S, Bo L, Lundervold A, Myhr KM, Pavlin T, Torkildsen O, Wergeland S. Deep gray matter demyelination detected by magnetization transfer ratio in the cuprizone model. PLOS ONE. 2013;8:e84162. doi: 10.1371/journal.pone.0084162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjær S, Bo L, Myhr KM, Torkildsen Ø, Wergeland S. Magnetization transfer ratio does not correlate to myelin content in the brain in the MOG-EAE mouse model. Neurochemistry International. 2015;83-84:28–40. doi: 10.1016/j.neuint.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Guglielmetti C, Boucneau T, Cao P, Van der Linden A, Larson PEZ, Chaumeil MM. Longitudinal evaluation of demyelinated lesions in a multiple sclerosis model using ultrashort Echo time magnetization transfer (UTE-MT) imaging. NeuroImage. 2020;208:116415. doi: 10.1016/j.neuroimage.2019.116415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakkarainen H, Sierra A, Mangia S, Garwood M, Michaeli S, Gröhn O, Liimatainen T. MRI relaxation in the presence of fictitious fields correlates with myelin content in normal rat brain. Magnetic Resonance in Medicine. 2016;75:161–168. doi: 10.1002/mrm.25590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hametner S, Endmayr V, Deistung A, Palmrich P, Prihoda M, Haimburger E, Menard C, Feng X, Haider T, Leisser M, Köck U, Kaider A, Höftberger R, Robinson S, Reichenbach JR, Lassmann H, Traxler H, Trattnig S, Grabner G. The influence of brain iron and myelin on magnetic susceptibility and effective transverse relaxation - A biochemical and histological validation study. NeuroImage. 2018;179:117–133. doi: 10.1016/j.neuroimage.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Harkins KD, Dula AN, Does MD. Effect of intercompartmental water exchange on the apparent myelin water fraction in multiexponential T2 measurements of rat spinal cord. Magnetic Resonance in Medicine. 2012;67:793–800. doi: 10.1002/mrm.23053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harkins KD, Valentine WM, Gochberg DF, Does MD. In-vivo multi-exponential T2, magnetization transfer and quantitative histology in a rat model of intramyelinic edema. NeuroImage: Clinical. 2013;2:810–817. doi: 10.1016/j.nicl.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Statistics in Medicine. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Höftberger R, Lassmann H. Chapter 19 - Inflammatory demyelinating diseases of the central nervous system. In: Kovacs G. G, Alafuzoff I, editors. Handbook of Clinical Neurology. Elsevier; 2018. pp. 263–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janve VA, Zu Z, Yao SY, Li K, Zhang FL, Wilson KJ, Ou X, Does MD, Subramaniam S, Gochberg DF. The radial diffusivity and magnetization transfer pool size ratio are sensitive markers for demyelination in a rat model of type III multiple sclerosis (MS) lesions. NeuroImage. 2013;74:298–305. doi: 10.1016/j.neuroimage.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelescu IO, Zurek M, Winters KV, Veraart J, Rajaratnam A, Kim NS, Babb JS, Shepherd TM, Novikov DS, Kim SG, Fieremans E. In vivo quantification of demyelination and recovery using compartment-specific diffusion MRI metrics validated by electron microscopy. NeuroImage. 2016;132:104–114. doi: 10.1016/j.neuroimage.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jito J, Nakasu S, Ito R, Fukami T, Morikawa S, Inubushi T. Maturational changes in diffusion anisotropy in the rat corpus callosum: comparison with quantitative histological evaluation. Journal of Magnetic Resonance Imaging. 2008;28:847–854. doi: 10.1002/jmri.21496. [DOI] [PubMed] [Google Scholar]
- Kelm ND, West KL, Carson RP, Gochberg DF, Ess KC, Does MD. Evaluation of diffusion kurtosis imaging in ex vivo hypomyelinated mouse brains. NeuroImage. 2016;124:612–626. doi: 10.1016/j.neuroimage.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodanovich MY, Sorokina IV, Glazacheva VY, Akulov AE, Nemirovich-Danchenko NM, Romashchenko AV, Tolstikova TG, Mustafina LR, Yarnykh VL. Histological validation of fast macromolecular proton fraction mapping as a quantitative myelin imaging method in the cuprizone demyelination model. Scientific Reports. 2017;7:46686. doi: 10.1038/srep46686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodanovich M, Pishchelko A, Glazacheva V, Pan E, Akulov A, Svetlik M, Tyumentseva Y, Anan’ina T, Yarnykh V. Quantitative imaging of white and gray matter remyelination in the cuprizone demyelination model using the macromolecular proton fraction. Cells. 2019;8:1204. doi: 10.3390/cells8101204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski P, Raj D, Liu J, Lam C, Yung AC, Tetzlaff W. Characterizing white matter damage in rat spinal cord with quantitative MRI and histology. Journal of Neurotrauma. 2008;25:653–676. doi: 10.1089/neu.2007.0462. [DOI] [PubMed] [Google Scholar]
- Kozlowski P, Rosicka P, Liu J, Yung AC, Tetzlaff W. In vivo longitudinal myelin water imaging in rat spinal cord following dorsal column transection injury. Magnetic Resonance Imaging. 2014;32:250–258. doi: 10.1016/j.mri.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule C, Leung E, Lis DK, Traboulsee AL, Paty DW, MacKay AL, Moore GR. Myelin water imaging in multiple sclerosis: quantitative correlations with histopathology. Multiple Sclerosis Journal. 2006;12:747–753. doi: 10.1177/1352458506070928. [DOI] [PubMed] [Google Scholar]
- Laule C, Vavasour IM, Kolind SH, Li DK, Traboulsee TL, Moore GR, MacKay AL. Magnetic resonance imaging of myelin. Neurotherapeutics. 2007;4:460–484. doi: 10.1016/j.nurt.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laule C, Kozlowski P, Leung E, Li DK, Mackay AL, Moore GR. Myelin water imaging of multiple sclerosis at 7 T: correlations with histopathology. NeuroImage. 2008;40:1575–1580. doi: 10.1016/j.neuroimage.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Laule C, Vavasour IM, Leung E, Li DK, Kozlowski P, Traboulsee AL, Oger J, Mackay AL, Moore GR. Pathological basis of diffusely abnormal white matter: insights from magnetic resonance imaging and histology. Multiple Sclerosis Journal. 2011;17:144–150. doi: 10.1177/1352458510384008. [DOI] [PubMed] [Google Scholar]
- Laule C, Moore GRW. Myelin water imaging to detect demyelination and remyelination and its validation in pathology. Brain Pathology. 2018;28:750–764. doi: 10.1111/bpa.12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman EL. Elements of Large-Sample Theory. Springer; 1999. [Google Scholar]
- Lehto LJ, Albors AA, Sierra A, Tolppanen L, Eberly LE, Mangia S, Nurmi A, Michaeli S, Gröhn O. Lysophosphatidyl choline induced demyelination in rat probed by relaxation along a fictitious field in high rank rotating frame. Frontiers in Neuroscience. 2017a;11:433. doi: 10.3389/fnins.2017.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto LJ, Sierra A, Gröhn O. Magnetization transfer SWIFT MRI consistently detects histologically verified myelin loss in the thalamocortical pathway after a traumatic brain injury in rat. NMR in Biomedicine. 2017b;30:e3678. doi: 10.1002/nbm.3678. [DOI] [PubMed] [Google Scholar]
- MacKay A, Whittall K, Adler J, Li D, Paty D, Graeb D. In vivo visualization of myelin water in brain by magnetic resonance. Magnetic Resonance in Medicine. 1994;31:673–677. doi: 10.1002/mrm.1910310614. [DOI] [PubMed] [Google Scholar]
- MacKay AL, Laule C. Magnetic resonance of myelin water: an in vivo Marker for Myelin. Brain Plasticity. 2016;2:71–91. doi: 10.3233/BPL-160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M. myelin-meta-analysis. 17ca867GitHub. 2020 https://github.com/matteomancini/myelin-meta-analysis
- Mollink J, Hiemstra M, Miller KL, Huszar IN, Jenkinson M, Raaphorst J, Wiesmann M, Ansorge O, Pallebage-Gamarallage M, van Cappellen van Walsum AM. White matter changes in the perforant path area in patients with amyotrophic lateral sclerosis. Neuropathology and Applied Neurobiology. 2019;45:570–585. doi: 10.1111/nan.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nave KA, Werner HB. Myelination of the nervous system: mechanisms and functions. Annual Review of Cell and Developmental Biology. 2014;30:503–533. doi: 10.1146/annurev-cellbio-100913-013101. [DOI] [PubMed] [Google Scholar]
- Odrobina EE, Lam TY, Pun T, Midha R, Stanisz GJ. MR properties of excised neural tissue following experimentally induced demyelination. NMR in Biomedicine. 2005;18:277–284. doi: 10.1002/nbm.951. [DOI] [PubMed] [Google Scholar]
- Peters JM, Struyven RR, Prohl AK, Vasung L, Stajduhar A, Taquet M, Bushman JJ, Lidov H, Singh JM, Scherrer B, Madsen JR, Prabhu SP, Sahin M, Afacan O, Warfield SK. White matter mean diffusivity correlates with myelination in tuberous sclerosis complex. Annals of Clinical and Translational Neurology. 2019;6:1178–1190. doi: 10.1002/acn3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiet A, Adanyeguh I, Aigrot MS, Poirion E, Nait-Oumesmar B, Santin M, Stankoff B. Ultrahigh field imaging of myelin disease models: toward specific markers of myelin integrity? Journal of Comparative Neurology. 2019;527:2179–2189. doi: 10.1002/cne.24598. [DOI] [PubMed] [Google Scholar]
- Pol S, Sveinsson M, Sudyn M, Babek N, Siebert D, Bertolino N, Modica CM, Preda M, Schweser F, Zivadinov R. Teriflunomide's Effect on Glia in Experimental Demyelinating Disease: A Neuroimaging and Histologic Study. Journal of Neuroimaging. 2019;29:52–61. doi: 10.1111/jon.12561. [DOI] [PubMed] [Google Scholar]
- Praet J, Manyakov NV, Muchene L, Mai Z, Terzopoulos V, de Backer S, Torremans A, Guns PJ, Van De Casteele T, Bottelbergs A, Van Broeck B, Sijbers J, Smeets D, Shkedy Z, Bijnens L, Pemberton DJ, Schmidt ME, Van der Linden A, Verhoye M. Diffusion kurtosis imaging allows the early detection and longitudinal follow-up of amyloid-β-induced pathology. Alzheimer's Research & Therapy. 2018;10:8. doi: 10.1186/s13195-017-0329-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasloski T, Rauscher A, MacKay AL, Hodgson M, Vavasour IM, Laule C, Mädler B. Rapid whole cerebrum myelin water imaging using a 3D GRASE sequence. NeuroImage. 2012;63:533–539. doi: 10.1016/j.neuroimage.2012.06.064. [DOI] [PubMed] [Google Scholar]
- Pun TW, Odrobina E, Xu QG, Lam TY, Munro CA, Midha R, Stanisz GJ. Histological and magnetic resonance analysis of sciatic nerves in the tellurium model of neuropathy. Journal of the Peripheral Nervous System : JPNS. 2005;10:38–46. doi: 10.1111/j.1085-9489.2005.10107.x. [DOI] [PubMed] [Google Scholar]
- Raudenbush SW. Analyzing effect sizes: Random-effects models. In: Hedges L. V, editor. The Handbook of Research Synthesis and Meta-Analysis, Russell Sage Foundation; 2009. pp. 295–315. [Google Scholar]
- Reeves C, Tachrount M, Thomas D, Michalak Z, Liu J, Ellis M, Diehl B, Miserocchi A, McEvoy AW, Eriksson S, Yousry T, Thom M. Combined ex vivo 9.4 T MRI and Quantitative Histopathological Study in Normal and Pathological Neocortical Resections in Focal Epilepsy. Brain Pathology. 2016;26:319–333. doi: 10.1111/bpa.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney WD, Johnson G, Li X, Cohen ER, Kim SG, Ugurbil K, Springer CS. Magnetic field and tissue dependencies of human brain longitudinal 1h2o relaxation in vivo. Magnetic Resonance in Medicine. 2007;57:308–318. doi: 10.1002/mrm.21122. [DOI] [PubMed] [Google Scholar]
- Sampaio-Baptista C, Johansen-Berg H. White matter plasticity in the adult brain. Neuron. 2017;96:1239–1251. doi: 10.1016/j.neuron.2017.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer K, Scaravilli F, Altmann DR, Barker GJ, Miller DH. Magnetization transfer ratio and myelin in postmortem multiple sclerosis brain. Annals of Neurology. 2004;56:407–415. doi: 10.1002/ana.20202. [DOI] [PubMed] [Google Scholar]
- Schmierer K, Tozer DJ, Scaravilli F, Altmann DR, Barker GJ, Tofts PS, Miller DH. Quantitative magnetization transfer imaging in postmortem multiple sclerosis brain. Journal of Magnetic Resonance Imaging. 2007a;26:41–51. doi: 10.1002/jmri.20984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer K, Wheeler-Kingshott CA, Boulby PA, Scaravilli F, Altmann DR, Barker GJ, Tofts PS, Miller DH. Diffusion tensor imaging of post mortem multiple sclerosis brain. NeuroImage. 2007b;35:467–477. doi: 10.1016/j.neuroimage.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer K, Wheeler-Kingshott CA, Tozer DJ, Boulby PA, Parkes HG, Yousry TA, Scaravilli F, Barker GJ, Tofts PS, Miller DH. Quantitative magnetic resonance of postmortem multiple sclerosis brain before and after fixation. Magnetic Resonance in Medicine. 2008;59:268–277. doi: 10.1002/mrm.21487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer K, Parkes HG, So PW, An SF, Brandner S, Ordidge RJ, Yousry TA, Miller DH. High field (9.4 tesla) magnetic resonance imaging of cortical grey matter lesions in multiple sclerosis. Brain. 2010;133:858–867. doi: 10.1093/brain/awp335. [DOI] [PubMed] [Google Scholar]
- Seehaus A, Roebroeck A, Bastiani M, Fonseca L, Bratzke H, Lori N, Vilanova A, Goebel R, Galuske R. Histological validation of high-resolution DTI in human post mortem tissue. Frontiers in Neuroanatomy. 2015;9:98. doi: 10.3389/fnana.2015.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled JG. Modelling and interpretation of magnetization transfer imaging in the brain. NeuroImage. 2018;182:128–135. doi: 10.1016/j.neuroimage.2017.11.065. [DOI] [PubMed] [Google Scholar]
- Soustelle L, Antal MC, Lamy J, Rousseau F, Armspach JP, Loureiro de Sousa P. Correlations of quantitative MRI metrics with myelin basic protein (MBP) staining in a murine model of demyelination. NMR in Biomedicine. 2019;32:e4116. doi: 10.1002/nbm.4116. [DOI] [PubMed] [Google Scholar]
- Stanisz GJ, Kecojevic A, Bronskill MJ, Henkelman RM. Characterizing white matter with magnetization transfer and T(2) Magnetic Resonance in Medicine. 1999;42:1128–1136. doi: 10.1002/(SICI)1522-2594(199912)42:6<1128::AID-MRM18>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Stanisz GJ, Webb S, Munro CA, Pun T, Midha R. MR properties of excised neural tissue following experimentally induced inflammation. Magnetic Resonance in Medicine. 2004;51:473–479. doi: 10.1002/mrm.20008. [DOI] [PubMed] [Google Scholar]
- Takagi T, Nakamura M, Yamada M, Hikishima K, Momoshima S, Fujiyoshi K, Shibata S, Okano HJ, Toyama Y, Okano H. Visualization of peripheral nerve degeneration and regeneration: monitoring with diffusion tensor tractography. NeuroImage. 2009;44:884–892. doi: 10.1016/j.neuroimage.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Tardif CL, Bedell BJ, Eskildsen SF, Collins DL, Pike GB. Quantitative magnetic resonance imaging of cortical multiple sclerosis pathology. Multiple Sclerosis International. 2012;2012:1–13. doi: 10.1155/2012/742018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiessen JD, Zhang Y, Zhang H, Wang L, Buist R, Del Bigio MR, Kong J, Li X-M, Martin M. Quantitative MRI and ultrastructural examination of the cuprizone mouse model of demyelination. NMR in Biomedicine. 2013;26:1562–1581. doi: 10.1002/nbm.2992. [DOI] [PubMed] [Google Scholar]
- Tu T-W, Williams RA, Lescher JD, Jikaria N, Turtzo LC, Frank JA. Radiological-pathological correlation of diffusion tensor and magnetization transfer imaging in a closed head traumatic brain injury model. Annals of Neurology. 2016;79:907–920. doi: 10.1002/ana.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turati L, Moscatelli M, Mastropietro A, Dowell NG, Zucca I, Erbetta A, Cordiglieri C, Brenna G, Bianchi B, Mantegazza R, Cercignani M, Baggi F, Minati L. In vivo quantitative magnetization transfer imaging correlates with histology during de- and remyelination in cuprizone-treated mice. NMR in Biomedicine. 2015;28:327–337. doi: 10.1002/nbm.3253. [DOI] [PubMed] [Google Scholar]
- Turner R. Myelin and modeling: bootstrapping cortical microcircuits. Frontiers in Neural Circuits. 2019;13:34. doi: 10.3389/fncir.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill HR, Rostomily RC, Mikheev AM, Yuan C, Yarnykh VL. Fast bound pool fraction imaging of the in vivo rat brain: association with myelin content and validation in the C6 glioma model. NeuroImage. 2011;54:2052–2065. doi: 10.1016/j.neuroimage.2010.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Tilborg E, Achterberg EJM, van Kammen CM, van der Toorn A, Groenendaal F, Dijkhuizen RM, Heijnen CJ, Vanderschuren L, Benders M, Nijboer CHA. Combined fetal inflammation and postnatal hypoxia causes myelin deficits and autism-like behavior in a rat model of diffuse white matter injury. Glia. 2018;66:78–93. doi: 10.1002/glia.23216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma G, Duhamel G, de Bazelaire C, Alsop DC. Magnetization transfer from Inhomogeneously broadened lines: a potential marker for myelin. Magnetic Resonance in Medicine. 2015;73:614–622. doi: 10.1002/mrm.25174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36:i03. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- Wang S, Wu EX, Cai K, Lau HF, Cheung PT, Khong PL. Mild hypoxic-ischemic injury in the neonatal rat brain: longitudinal evaluation of white matter using diffusion tensor MR imaging. American Journal of Neuroradiology. 2009;30:1907–1913. doi: 10.3174/ajnr.A1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cusick MF, Wang Y, Sun P, Libbey JE, Trinkaus K, Fujinami RS, Song SK. Diffusion basis spectrum imaging detects and distinguishes coexisting subclinical inflammation, demyelination and axonal injury in experimental autoimmune encephalomyelitis mice. NMR in Biomedicine. 2014;27:843–852. doi: 10.1002/nbm.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun P, Wang Q, Trinkaus K, Schmidt RE, Naismith RT, Cross AH, Song SK. Differentiation and quantification of inflammation, demyelination and axon injury or loss in multiple sclerosis. Brain. 2015;138:1223–1238. doi: 10.1093/brain/awv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Cao P, Bischof A, Henry RG, Larson PEZ, Liu C. MRI gradient-echo phase contrast of the brain at ultra-short TE with off-resonance saturation. NeuroImage. 2018;175:1–11. doi: 10.1016/j.neuroimage.2018.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel KM, Lee JB, Affeldt BM, Hamer M, Harahap-Carrillo IS, Pardo AC, Obenaus A. Corpus callosum vasculature predicts white matter microstructure abnormalities after pediatric mild traumatic brain injury. Journal of Neurotrauma. 2018;23:e5670. doi: 10.1089/neu.2018.5670. [DOI] [PubMed] [Google Scholar]
- West KL, Kelm ND, Carson RP, Gochberg DF, Ess KC, Does MD. Myelin volume fraction imaging with MRI. NeuroImage. 2018;182:511–521. doi: 10.1016/j.neuroimage.2016.12.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu QZ, Yang Q, Cate HS, Kemper D, Binder M, Wang HX, Fang K, Quick MJ, Marriott M, Kilpatrick TJ, Egan GF. MRI identification of the rostral-caudal pattern of pathology within the corpus callosum in the cuprizone mouse model. Journal of Magnetic Resonance Imaging : JMRI. 2008;27:446–453. doi: 10.1002/jmri.21111. [DOI] [PubMed] [Google Scholar]
- Yano R, Hata J, Abe Y, Seki F, Yoshida K, Komaki Y, Okano H, Tanaka KF. Quantitative temporal changes in DTI values coupled with histological properties in cuprizone-induced demyelination and remyelination. Neurochemistry International. 2018;119:151–158. doi: 10.1016/j.neuint.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Yarnykh VL. Fast macromolecular proton fraction mapping from a single off-resonance magnetization transfer measurement. Magnetic Resonance in Medicine. 2012;68:166–178. doi: 10.1002/mrm.23224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaaraoui W, Deloire M, Merle M, Girard C, Raffard G, Biran M, Inglese M, Petry KG, Gonen O, Brochet B, Franconi J-M, Dousset V. Monitoring demyelination and remyelination by magnetization transfer imaging in the mouse brain at 9.4 T. Magnetic Resonance Materials in Physics, Biology and Medicine. 2008;21:357–362. doi: 10.1007/s10334-008-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Jones M, DeBoy CA, Reich DS, Farrell JA, Hoffman PN, Griffin JW, Sheikh KA, Miller MI, Mori S, Calabresi PA. Diffusion tensor magnetic resonance imaging of wallerian degeneration in rat spinal cord after dorsal root axotomy. Journal of Neuroscience. 2009;29:3160–3171. doi: 10.1523/JNEUROSCI.3941-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]