Abstract
Introduction
: The recent pandemic outbreak of SARS-CoV-2 has been associated with a lethal atypical pneumonia, making COVID-19 an urgent public health issue with an increasing rate of mortality and morbidity. There are currently no vaccines or therapeutics available for COVID-19, which is causing an urgent search for a new drug to combat the COVID-19 pandemic. The lipid membrane alternation efficiency of small antimicrobial lipopeptides enables them to block viral membrane fusion to the host cell. Lipopeptides could serve as potential antiviral agents, by interacting or competing with viral fusion proteins.
Methods
: This study screened seven different lipopeptides (tsushimycin, daptomycin, surfactin, bacillomycin, iturin, srfTE, and LPD-12) and docked them individually against the spike (S)-glycoprotein of SARS-CoV-2.
Results
: Based on the maximum docked score and minimum atomic contact energy, LPD-12 (–1137.38 kcal) was the appropriate molecule for proper binding with the S-glycoprotein of SARS-CoV-2 and thus significantly interrupted its affinity of binding with angiotensin-converting enzyme-2 (ACE2), which is the only receptor molecule found to be facilitating disease development. The results confirmed a strong binding affinity of LPD-12 with ACE2, with a binding free energy of –1621.62 kcal, which could also reciprocally prevent the binding of S-protein.
Conclustion
: It can be concluded that LPD-12 may act as a potential therapeutic drug, by reducing the entry of SARS-CoV-2 to the human cells via the ACE2 receptor and related infections.
Keywords: COVID-19, Lipopeptide, Novel coronavirus, Docking, Fusion protein
1. Introduction
A previously unknown and potentially lethal coronavirus SARS-CoV-2 was reported in December 2019 in Wuhan, Hubei province of China [1]. SARS-CoV-2 was associated with an ongoing outbreak of lethal atypical pneumonia named COVID-19 and its possible origin was suggested to be from bats [2]. SARS-CoV-2 infections are now pandemic and were declared a public health emergency of international concern by the World Health Organization. MERS-CoV, SARS-CoV and SARS-CoV-2 are the three highly pathogenic zoonotic coronaviruses that all belong to β-coronavirus [3]. Apart from the highly pathogenic zoonotic pathogens, MERS-CoV, SARS-CoV and SARS-CoV-2, which cause severe disease in humans, four low pathogenic coronaviruses are endemic in humans and cause disease with mild symptoms: HCoV-HKU1, HCoV-NL63, HCoV-OC43, and HCoV-229E [3]. To date there are no antivirals, vaccines or therapeutic strategies approved against any of the seven human-infecting coronaviruses. All of the coronaviruses that have caused pandemics were finally controlled by conventional control measures like travel restrictions, patient isolation and strategic social distancing.
The first instance of the viral infection is mediated by a homo-trimers form of trans-membrane spike (S)-glycoprotein in the surface of SARS-CoV-2 [4]. Coronaviruses use distinct binding receptors of the host cells that are recognized by specific domains present in the S1 subunit of different viral species. SARS-CoV and SARS-CoV-2 bind to angiotensin-converting enzyme 2 (ACE2) and enter target host cells using the SB domain of the S1 subunit of the S-glycoprotein [5]. Therefore, coronavirus entry into the susceptible host cells is a complex process involving specific receptor binding and host protease-mediated proteolytic cleavage of the S-glycoprotein.
Conventional antiviral drugs have been designed to target viral proteins and host cell elements that are involved in the infection process. However, the first step of viral infection needs the involvement of lipid membranes of both virus and host cells that are conserved in nature. Small antimicrobial peptides have been reported to disrupt cell membranes of various pathogens, including bacteria, fungi and cancer cells [6]. Surfactins are cyclic lipopeptides naturally produced by various strains of Bacillus subtilis and reported to have a broad spectrum of bioactivities [7]. Surfactin has shown antiviral activity against several enveloped viruses such as herpes-simplex-virus (HSV-1, HSV-2), vesicular stomatitis virus, simian immunodeficiency virus, Newcastle disease virus, and porcine epidemic diarrhea virus [8], [9], [10]. The antiviral activity of surfactin is due to the inhibition of membrane fusion events between the virus and host cells [11]. Entry of the Influenza A viruses (IAV) has been shown to restrict by inhibiting the membrane fusion event by blocking or interacting with the HA2 subunit of hemagglutinin [12]. Another study confirmed that coiled-coil interaction of MERS-CoV and IAVs spike proteins to host cell membranes were inhibited by short α-helical lipopeptides, thus showing the potential of lipopeptides being used as broad-spectrum antiviral agents. In the focus of the same concern, the present study suggested small lipopeptides as potential tools to combat SARS-CoV-2 infections.
2. Materials and Methods
2.1. Molecular docking
Molecular docking was performed to monitor the interaction of the surface spike glycoprotein (S protein) of SARS-CoV-2 (PDB ID: 5 × 5B) with seven different lipopeptides (tsushimycin, daptomycin, surfactin, bacillomycin, iturin, srfte, and LPD-12) using PATCHDOCK, and images were visualized using PyMOL. Protein-Ligand Interaction Profiler (PLIP) was used to understand the point of contact between the receptor and ligand. Based on the better efficacy of binding, the ligand was chosen. After the first stage of screening, the compound showing greater affinity of binding was docked with ACE2 (PDB1R42), where ACE2 acted as the receptor and lipopeptide as the ligand. The resultant docked complex was again docked with SARS-CoV-2, which ultimately hindered the binding of SARS-CoV-2 with ACE2 by forming a barrier in-between.
3. Results and Discussion
With the potential of lipopeptides to inhibit viral membrane fusion with the host, this study screened available crystal structures of potential lipopeptides. Interestingly, a few potentially interacting lipopeptides with strong binding affinity to SARS-CoV-2 spike (S)-glycoprotein were found (Table 1 ). These lipopeptides may serve as potential drugs to combat the COVID-19 pandemic; however, further in vitro and in vivo validation is required. Further, these results provide a relatively broad scope for development of lipopeptide-based drug development to treat COVID-19 and related viral diseases. The docking results of all tested lipopeptides and their interaction residues, along with bond distances, are described in detail in Figure S1 and Table S1.
Table 1.
Determination of binding energy from the docking analysis of different lipopeptides with spike glycoprotein (S-protein) of SARS-CoV-2 (PDB ID: 5 × 5B).
| INTERACTION | BINDING FREE ENERGY (kcal) |
|---|---|
| Tsushimycin (1W3M) | –1055.60 |
| Daptomycin (1XT7) | –550.98 |
| Surfactin (2NPV) | –539.61 |
| Bacillomycin (2IGZ) | –493.44 |
| Iturin (2IHO) | –311.53 |
| srfTE (1JMK) | –343.45 |
| LPD12 (3CAY) | –1137.38 |
In silico analysis of seven of the most abundant lipopeptides with the S-protein of SARS-CoV-2 revealed that LDP-12 (Lipopeptide detergent-12) was the best one among them (Table 1). The interaction between LPD-12 and S-protein was higher than S-protein with its receptor ACE2. The binding free energy was calculated as –1137.38 kcal, which was the best among all the tested lipopeptides and significantly higher than the –866.21 kcal of S-protein and ACE2 complex (Table 1). The binding free energy of LPD-12 and S-protein interaction was –1137.38 kcal, which suggests that LDP-12 has more affinity to bind with S-protein. Again, a similar type of docking was performed with ACE2, to see how strongly it binds with LPD-12 and helps in hindering the entry of virus within the body (Figure S2). In silico docking of ACE2 with LPD-12 clearly depicted strong binding energy of –1621.62 kcal.
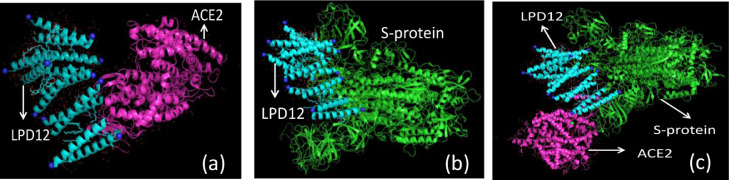
LPD-12 and ACE2 complex again interacted with the S-protein of SARS-CoV-2 (Figure 1 ). The binding free energy of the SARS-CoV-2 complex with the ACE2-LPD-12 complex was calculated as –755.66 kcal. In this docking, the binding free energy between LPD-12-ACE2 complexes with SARS-CoV-2 was dramatically reduced due to the strong binding affinity of LPD-12 with ACE2. ACE2 has always been prone to binding with S-protein. As LPD-12 came into play between the two molecules, the affinity of binding of ACE2 was much reduced. The bond distance was also calculated between the SARS-CoV-2 with ACE2, SARS-CoV-2 with LPD-12 and LPD-12 with ACE2, showing maximum binding affinity between the two different atoms present in each chain (Table S1). In the S-protein of SARS-CoV-2 and ACE2 docking, it was clearly seen that the bond distance between C:ILE 1097 of S-protein and A:ASP 615 of ACE2 was measured as 6.645 Å, whereas in S-protein-LPD-12-ACE2 docking, the similar bond distance was increased to 108.690 Å, which showed that the binding site changed where LPD-12 came into play. Similarly, in SARS-CoV-2 and LPD-12 docking, the bond distance between B:PRO 547 of SARS-CoV-2 and H:ALA 25 of LPD-12 was measured as 3.387 Å, which was the same in SARS-CoV-2/LPD-12/ACE2 complex docking. This data signifies that the docking position of the S-protein with LPD-12 remains unchanged. Again, in LPD-12-ACE2 docking, the bond distance was measured between L:TYR 12 of LPD-12 and A:ASP 615 of ACE2 as 5.555Å, which was also similar in SARS-CoV-2-LPD-12-ACE2 docking. This highlights that there was no change in the bonding position of ACE2 and LPD-12, whereas a change occurred for the S-protein and ACE2 complex. Moreover, the complex formation between S-protein and ACE2 was significantly reduced in the presence of LPD-12.
Figure 1.
Docked image of LPD12/ACE2/ S-protein of SARS-CoV-2 complex viewed in PyMOL. (a) Protein-ligand interaction site between LPD-12 with S-protein of SARS CoV-2; (b) protein-ligand interaction site between LPD-12 with ACE2; and (c) LPD-12-ACE2 complex with S-protein.
S-glycoprotein contains two functional subunits – S1 and S2 – which are responsible for binding to the host cell receptor and fusion of the viral and host cell membrane, respectively [5]. The S-glycoprotein is cleaved by host proteases at the interface of S1/S2 subunits, which remain non-covalently bound in the prefusion conformation before infection [13]. The distal S1-subunit with receptor-binding domain(s) plays a role in stabilization of the prefusion conformation of the membrane-anchored S2-subunit that has fusion machinery [5]. Further, proteolytic cleavage at the S2’ site (located immediately upstream to the fusion peptide in the S2 domain) within the S2 subunit is needed for the fusion of viral and host cell membranes, which includes extensive irreversible conformational changes [13]. Lipopeptides are well known for their antimicrobial activity with the mechanism of membrane disruption [7,14,15]. Lipopeptide detergents (LPDs) are a class of amphiphiles, which are designed for the structural study of the integral membrane proteins [16]. LPDs are designed as α-helices, which may self-assemble into cylindrical micelles to provide a closely mimicking environment of acyl chain packing-like lipid bilayers. LPD-12 is good enough to mimic the lipid bilayer width. LPD-12 monomers successfully adopt lipid bilayer conformation and are associated into cylindrical octamers [17]. LPD-12 proved to be a very well behaving lipopeptide detergent with acetyl-A(O12)AEAAEKAAKYAAEAAEKAAKA(O12)A-amide sequence, where O12 refers to modified ornithine residues.
LPD-12 is designed to have a high propensity of α-helices formation, which might interact more efficiently with integral membrane lipids and disintegrate the lipid membrane. Interestingly, a recent study showed that α-helical lipopeptides efficiently targeted viral membrane fusion proteins, suggesting it as a potentially broad-spectrum antiviral therapy. By considering this fact, LPD-12 could be used as a docking ligand against SARS-CoV-2 S-protein, due to its strong binding efficiency. LPD-12 was found to alter the binding of S-protein with ACE2 and reduce their affinity by blocking the target site. These results suggest that LPD-12 may be used as a potential therapeutic agent to treat COVID-19; however, further experimental validation is needed.
Declarations
Funding: No funding.
Competing Interests: Nil.
Ethical Approval: Nil.
Dr. Po-Ren Hsueh
References
- 1.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature. 2020 doi: 10.1101/2020.01.22.914952. [DOI] [Google Scholar]
- 3.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The Proximal Origin of SARS-CoV-2. Nat Med. 2020;25:450–452. doi: 10.2106/JBJS.F.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vankadari Naveen, Jacqueline A. Wilce. Emerging WuHan (COVID-19) coronavirus: glycan shield and structure prediction of spike glycoprotein and its interaction with human CD26. Emerg Microbes Infect. 2020;9:601–604. doi: 10.1080/22221751.2020.1739565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fjell CD, Hiss JA, Hancock REW, Schneider G. Designing antimicrobial peptides: Form follows function. Nat Rev Drug Discov. 2012 doi: 10.1038/nrd3591. [DOI] [PubMed] [Google Scholar]
- 7.Mandal SM, Barbosa AEAD, Franco OL. Lipopeptides in microbial infection control : Scope and reality for industry. Biotechnol Adv. 2013;31:338–345. doi: 10.1016/j.biotechadv.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Vollenbroich D, Pauli G, Özel M, Vater J. Antimycoplasma properties and application in cell culture of surfactin, a lipopeptide antibiotic from Bacillus subtilis. Appl Environ Microbiol. 1997 doi: 10.1128/aem.63.1.44-49.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X, Lu Z, Zhao H, Bie X, Lü FX, Yang S. Antiviral activity of antimicrobial lipopeptide from Bacillus subtilis fmbj against Pseudorabies Virus, Porcine Parvovirus, Newcastle Disease Virus and Infectious Bursal Disease Virus in vitro. Int J Pept Res Ther. 2006 doi: 10.1007/s10989-006-9041-4. [DOI] [Google Scholar]
- 10.Yuan L, Zhang S, Peng J, Li Y, Yang Q. Synthetic surfactin analogues have improved anti-PEDV properties. PLoS One. 2019 doi: 10.1371/journal.pone.0215227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan L, Zhang S, Wang Y, Li Y, Wang X, Yang Q. Surfactin Inhibits Membrane Fusion during Invasion of Epithelial Cells by Enveloped Viruses. J Virol. 2018 doi: 10.1128/jvi.00809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu W, Wang J, Lin D, Chen L, Xie X, Shen X. Super short membrane-active lipopeptides inhibiting the entry of influenza A virus. Biochim Biophys Acta - Biomembr. 2015 doi: 10.1016/j.bbamem.2015.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Mille JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy A, Mahata D, Paul D, Korpole S, Franco OL, Mandal SM. Purification, biochemical characterization and self-assembled structure of a fengycin-like antifungal peptide from bacillus thuringiensis strain SM. Front Microbiol. 2013 doi: 10.3389/fmicb.2013.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mandal SM, Sharma S, Pinnaka AK, Kumari A, Korpole S. Isolation and characterization of diverse antimicrobial lipopeptides produced by Citrobacter and Enterobacter. BMC Microbiol. 2013 doi: 10.1186/1471-2180-13-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGregor CL, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A. Lipopeptide detergents designed for the structural study of membrane proteins. Nat Biotechnol. 2003 doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- 17.Ho DN, Pomroy NC, Cuesta-Seijo JA, Privé GG. Crystal structure of a self-assembling lipopeptide detergent at 1.20 Å. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0801941105. [DOI] [PMC free article] [PubMed] [Google Scholar]