Graphical abstract

Keywords: COVID-19, SARS-CoV-2, Mpro, PLpro, Non-covalent inhibitor, Structure-activity relationship (SAR)
Abbreviations: 3CLpro, 3C-like protease or main protease; CoV, coronavirus; COVID-19, coronavirus disease 2019; E protein, envelope protein; EBOV, Ebola virus; Mpro, main protease; M protein, membrane protein; MERS-CoV, Middle East respiratory syndrome coronavirus; N protein, nucleocapsid protein; Nsp, non-structural proteins; NTD, N-terminal domain; ORF, open reading frame; PLpro, papain-like protease; QSAR, Quantitative structure-activity relationship; RdRp, RNA-dependent RNA polymerase; S protein, spike protein; SAR, Structure-activity relationship; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SPCI, Structural and physico-chemical interpretation; WHO, World Health Organization
Highlights
-
•
Prorteases (Mpro and PLpro) are part of the replication machinery of corona virus.
-
•
Mpro and PLpro inhibitors may serve as therapeutic weapons against SARS-CoV-2.
-
•
An exquisite picture of the recent coronavirus protease inhibitors is provided.
-
•
Experimental screening approaches are also highlighted.
-
•
Challenges in the development of effective as well as drug like protease inhibitors is also discussed.
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) brutally perils physical and mental health worldwide. Unavailability of effective anti-viral drug rendering global threat of COVID-19 caused by SARS-CoV-2. In this scenario, viral protease enzymes are crucial targets for drug discovery. This extensive study meticulously focused on two viral proteases such as main protease (Mpro) and papain-like protease (PLpro), those are essential for viral replication. This review provides a detail overview of the targets (Mpro and PLpro) from a structural and medicinal chemistry point of view, together with recently reported protease inhibitors. An insight into the challenges in the development of effective as well as drug like protease inhibitors is discussed. Peptidomimetic and/or covalent coronavirus protease inhibitors possessed potent and selective active site inhibition but compromised in pharmacokinetic parameters to be a drug/drug like molecule. Lead optimization of non-peptidomimetic and/or low molecular weight compounds may be a better option for oral delivery. A masterly combination of adequate pharmacokinetic properties with coronavirus protease activity as well as selectivity will provide potential drug candidates in future. This study is a part of our endeavors which surely dictates medicinal chemistry efforts to discover effective anti-viral agent for this devastating disease.
1. Introduction
In late December 2019, the newly emerged highly contagious novel coronavirus disease 2019 (COVID-19) was identified in Humans.1, 2 The outburst of virus containing a single positive-stranded RNA first found to occur in Wuhan, China and was named as severe acute respiratory syndrome (SARS)-CoV-2 (SARS-CoV-2).3, 4, 5, 6, 7, 8, 9, 10 Worldwide more than millions of cases have been registered.11, 12 According to World Health Organization (WHO), the highly transmissible disease COVID-19 has so far, more than millions confirmed cases and deaths have been reported from 216 countries around the globe.12
Currently, this virus is far more contagious and more catastrophic compared to other flu-viruses with several symptoms like fever, cough, pneumonia, nausea, and fatigue.13 Hence, the World Health Organization was forced to declare a state of global health emergency to organized scientific and medical efforts to quickly develop a cure for patients.14 Presently, there is no specific targeted therapy against this novel virus. Thus, the scientific community is making great efforts to explore diverse mechanisms to restrict the virus replication. As a result, diverse antiviral drugs with similar viral infections were tested on patients. Several drugs like: Remdesivir (designed for the Ebola virus),14 Lopinavir/Ritonavir (designed for the HIV),15 chloroquine and hydroxychloroquine (designed for anti-malarial action)14 and Tocilizumab (designed for rheumatoid arthritis)16 were found to be effective against this deadly virus, but their efficacy still remains controversial.17
The current impact of COVID-19 outbreak and the possibility of forthcoming CoV epidemics prove that there is a need for rapid discovery of anti-COVID-19 drugs. Recent studies revealed that SARS-CoV-2 has a comparable genomic pattern to other corona viruses.18 These viruses mainly comprises of a 5′-untranslated region (UTR), a replicase complex for encoding non-structural proteins (nsps), spike protein (S) gene, envelope protein (E), membrane protein (M) gene, nucleocapsid protein (N) gene, 3′-UTR, and numerous unknown non-structural parts which provide them support against the environmental factors.19, 20, 21, 22 Usually, these viruses harvest several polypeptides which promote proteolytic breakdown to produce 20 additional proteins during their lifecycle. Among them two crucial proteases such as main protease (Mpro) and papain-like protease (PLpro) are vital for virus replication.22, 23, 24 Meanwhile, a tremendous effort has been spent on studying these proteases in order to discover specific inhibitors against this noxious COVID-19.25, 26, 27, 28, 29, 30
Among these the two proteases, the coronavirus Main protease (Mpro) also recognized as 3C like protease (3CLpro) acknowledged great attention for its significant role in enzymatic activity leading to its post-translational processing of replicase polyproteins.26, 27 The Mpro consist of 306 amino acid long and has high structural and sequence resemblance to that of SARS-CoV Mpro.25 SARS-CoV-2 Mpro monomer comprises of three domains (i.e., N-terminal domain-I, N-terminal domain-II, and C-terminal domain-III).25 The Mpro active site consists of two catalytic dyad C145 and H41 (Fig. 1 A–B).
Fig. 1.
Three-dimensional structures of (A) SARS-CoV Mpro (PDB: 3ATW), (B) SARS-CoV-2 Mpro (PDB: 6LU7), (C) SARS-CoV PLpro (PDB: 4OW0), (D) SARS-CoV-2 PLpro (PDB: 6WUU). The amino acids formed catalytic dyad for Mpro and catalytic triad for PLpro are also highlighted.
On the other hand, papain-like protease (PLpro) active site consists of catalytic triad (Fig. 1C–D). PLpro functions by cleaving ISG15, a two-domain Ub-like protein, and Lys48-linked polyUb chains. Hence, their main function lies in the processing of the viral polypeptide into functional proteins, which further deubiquitinize and dampen host anti-viral reactions by hijacking the ubiquitin (Ub) an enzyme playing the pivotal role in host defense mechanism.31 Therefore, the two proteases are equally important for viral lifecycle and are supported by several studies which reveal that most of the coronaviridae genome encrypts two polyproteins, pp1a and pp1ab during their translation stage through ribosomal frame shifting mechanism.32 These polyproteins were further processed into mature non-structural proteins (nsps) by Mpro and PLpro which plays a vital role in the transcription/replication.33 Targeting these may hence institute a valid tactic for antiviral drug design and discovery.
In the 21st century, drug repurposing, screening of databases and designing different inhibitors are the only fastest possibility in terms drug discovery to prevent the catastrophe caused by COVID-19 outbreak. Diverse approaches have also been made in order to get insights into the mechanism of these proteases and to inhibit their functions but still there has been a lot of groundwork to be done for drug discovery and development against these targets. This study, as a part of rational drug design and discovery,10, 34, 35, 36, 37 aims to sketch out the current status of SARS-CoV-2 protease inhibitors based drug discovery. We also try to provide a new insight into coronavirus protease structural biology and discuss the challenges in the development of effective as well as drug like protease inhibitors. The study will offer an initiative to stimulate further research by providing useful guidance to the medicinal chemists for designing of new protease inhibitors effective against COVID-19 in near future.
2. Structural biology of SARS-CoV-2 proteases
CoV is a single-stranded positive sense RNA virus where genome is encapsulated within a membrane envelope.38, 39, 40, 41, 42, 43 The spike glycoprotein of CoV regulates its entry into the host cells.43, 44, 45 Two polyproteins (pp1a and pp1ab) are translated after virion entry into the host cells, which are promptly split by two viral proteases including Mpro and PLpro.46 Further proteolytic cleavage of these two viral polyproteins resulted in sixteen non-structural proteins (nsp1 to nsp16). The PLpro manages the proteolytic cleaving of nsp 1–3, whereas all junctions downstream of nsp4 are cleaved by Mpro.
The Mpro cleaves at no fewer than 11 sites on the large polyprotein 1ab with the recognition sequence of L-Q↓ (S, A, G) (↓ refers the cleavage site).25 The Mpro of SARS-CoV-2 is a 67.6 kDa homodimeric cysteine protease having huge sequence identity with SARS-CoV Mpro (Fig. 1). Mpro of CoV forms a dimer where each monomer consists of N-terminal catalytic region and the C-terminal region. Moreover, the N-terminal residues form a typical chymotrypsin fold while the C-terminal residues form an extra domain. In addition, each protomer containing three domains such as domains I (residues 8–101), II (residues 102–184) and domain III (residues 201–303).22 Domains I and II espouses a double β-barrel fold and the active site is located in a shallow cleft between two antiparallel β-barrels (Fig. 1). Notably, C terminal helical-bundle domain, Domain III, might involve in stabilization of their active homodimer forms. The active site can be further divided into several (sub)sites. Notably, the catalytic dyad formed by H41-C145 is observed at the S1 site (Fig. 2 ). The hydrophobic side chains are found mostly at the S2 and S4 sites. The list of amino acid residues play key role in SARS-CoV-2 Mpro is highlighted in Table 1 .
Fig. 2.
SARS-CoV-2 (A) Mpro and (B) PLpro binding sites.
Table 1.
List of amino acid residues play important role in SARS-CoV-2 Mpro.
| Role in SARS-CoV-2 Mpro | Residue |
|---|---|
| Catalytic dyad | H41, C145 |
| Substrate binding | H41, M49, G143, S144, H163, H164, M165, E166, L167, D187, R188, Q189, T190, A191, Q192 |
| Dimerization | R4, S10, G11, E14, N28, S139, F140, S147, E290, R298 |
Since the sequences of SARS-CoV-2 and SARS-CoV Mpro share 96% of identity and the minimum differences between both enzymes resides at the surface of the proteins. Therefore, inhibitors against SARS-CoV Mpro are expected to inhibit SARS-CoV-2 Mpro.
The ligand-bound X-ray structure of SARS-CoV-2 PLpro was elucidated few days ago.7 A close comparisons of apo SARS-CoV-2 PLpro (PDB: 6W9C) to inhibitor-bound SARS-CoV-2 PLpro (PDB: 6WUU and 6WX4) possessed similar overall structures with little exception of the β14- β15 loop proximal to the catalytic site.7 Catalytic C111 of SARS-CoV-2 PLpro engages in Michael addition to the warhead of inhibitors and rendering the formation of a covalent thio-ether linkage.
The list of amino acid residues play key role in SARS-CoV-2 PLpro is highlighted in Table 2 .
Table 2.
List of amino acid residues play key role in SARS-CoV-2 PLpro.
| Role in SARS-CoV-2 PLpro | Residue |
|---|---|
| Catalytic triad | C111, H272, D286 |
| Substrate binding | Y268, M208, P247, P248, T301, P248, Y264, N267, Q269, L162, C270, G271 and Y273 |
Notably, SARS-CoV-2 PLpro and SARS-CoV PLpro only differ by 54 residues.24 The residues of active site are almost identical. Moreover, the PLpro of SARS-CoV-2 and SARS-CoV demonstrated an almost comparable architecture of S4-S2 site suggesting comparable substrate preferences at positions P4-P2. Interestingly, SARS-CoV-2 PLpro harbors deISGylating activities similar to SARS-CoV PLpro but its potential to hydrolyze K48-linked Ub chains is declined.7 In contrast, SARS-CoV-2 PLpro fails to possess potent interferon-antagonizing and deubiquitinase activities.23
3. Search of protease inhibitors against COVID-19
3.1. Molecular modeling and in silico virtual screening against SARS-CoV-2
Novel coronavirus pandemic caused by SARS-CoV-2 severely threatens public health globally. In its infancy, little knowledge about the exact molecular mechanisms of the disease is obstructing the attempts to develop promising anti-viral drugs.9 Hence, bioinformatics and molecular modeling approaches are the only handy strategy until precise molecular and structural biology is known.
FDA-approved drugs surely claim safe alternatives if it exhibits at least modest activity against SARS-CoV-2. Currently, scientific community are largely focused in the screening of – (i) FDA-approved drug databases, (ii) clinical trials molecules and/or (iii) previously reported coronavirus inhibitors.9 In silico virtual screening (VS) techniques are proficient to explore CoV protease inhibitors.47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86 Yu and co-workers40 reported the computational screening and findings with regard to potential binding luteolin and other natural compounds against Mpro. Notably, luteolin has also been found to bind effectively with other targets (PLpro, Spike protein, and RdRp) of SARS-CoV-2.40, 87, 88
Vast amount of in silico VS studies against SARS-CoV-2 Mpro has been reported over past months.27, 28, 29, 30, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86 As the detailed description on the molecular modeling studies is out of Scope for this current communication, readers interested in learning more about recent molecular modeling studies to identify probable CoV protease inhibitors are directed to mentioned references48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90 and others.
Since the SARS-CoV-2 Mpro shares about 96% sequence similarity with SARS-CoV Mpro, previously reported SARS-CoV Mpro inhibitors may have huge prospect to show their efficacy against SARS-CoV-2 Mpro also. By May this year, we have endorsed our rational anti-viral drug design efforts through data mining and molecular docking studies.10 In an endeavour, our research team explored the crucial structural fingerprints modulating SARS-CoV PLpro inhibitory activities by the aid of 2D-QSAR, SPCI analysis as well as Monte Carlo optimization based QSAR. Further, QSAR derived virtual screening of some in-house molecules were done which rendered some important hits.
3.2. What efforts are taken to identify COVID-19 protease inhibitors?
In February 2020, the first crystal structure of SARS-CoV-2 virus Mpro (PDB: 6LU7) with covalent inhibitor N3 (Fig. 3 ) was reported by Jin and co-workers.3
Fig. 3.
Structure of SARS-CoV-2 Mpro inhibitors.
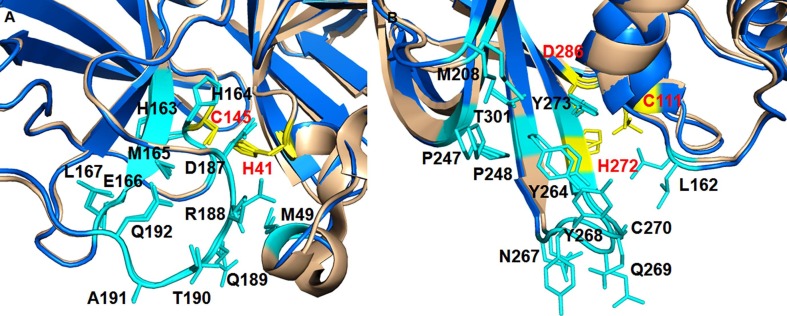
The isobutyl function of N3 embed itself in the hydrophobic S2 site formed by H41, M49, and M169 (Fig. 4 A–B). This study forms the basis of rapid target-based discovery of lead molecules against 2019-nCoV Mpro.
Fig. 4.
(A) Binding modes of SARS-CoV-2 Mpro inhibitors (N3, GC-376, 13b, 11a and 11b) at the active site, (B) Interaction of GC-376 with the amino acid residues, (C) Binding modes of SARS-CoV-2 PLpro inhibitors (VIR250 and VIR251) at the active site, (D) Interaction of VIR250 with the amino acid residues at active site of SARS-CoV-2 PLpro.
After that lot of target-based virtual screening were performed to find out promising protease inhibitors against COVID-19. Jin et al screened more than 10,000 molecules underwent SARS-CoV-2 Mpro structure-based virtual and high-throughput screening, which finally identified six small-molecule inhibitors including two FDA-approved drugs (Disulfiram and Carmofur) and clinical trials compounds (Ebselen, Tideglusib, Shikonin and PX-12) with a IC50 range of 0.67–21.4 μM (Fig. 3). Notably, Ebselen (SARS-CoV-2 Mpro IC50 = 0.67 μM) showed promising anti-viral activity in cell-based assays (EC50 = 4.67 µM).3
Su and co-workers elucidated the SARS-CoV-2 Mpro crystal structure in complex with a non-covalent, non-peptidomimetic inhibitor baicalein (Fig. 5 ) at a resolution of 2.2 Å (PDB: 6M2N).8 Interestingly, baicalein is snugly fitted in the core of the substrate-binding site by interacting catalytic oxyanion loop (residues 138–145), E166, those are critical not only for recognition of substrates but also peptidomimetic inhibitors. Nevertheless, baicalein revealed a unique binding mode in comparison with other covalent or peptidomimetic inhibitors.
Fig. 5.
Structure of SARS-CoV-2 Mpro inhibitors namely Baicalin, Baicalein, Boceprevir, GC-376, calpain inhibitors II and XII.
A comparison analysis of the inhibitor-bound CoV Mpro crystal structures suggested that a peptidomimetic inhibitor portrayed like a ‘sword’, while baicalein set a ‘shield’ near the two catalytic dyads to restrict the binding of the substrate. In addition, baicalein was also found promising in an enzymatic assay against SARS-CoV-2 Mpro (IC50 = 0.94 µM). It also exhibited a dose-dependent inhibition on the replication of SARS-CoV-2 with a half-maximal effective concentrations (EC50 = 1.69 µM).8 The unique binding mode and promising ligand-binding efficiency of baicalein will inspire Researchers for further lead optimization.
Ma et al employed a fluorescence resonance energy transfer (FRET)-based SARS-CoV-2 Mpro enzymatic assay to explore a library of known protease inhibitors.17 Among these, four inhibitors such as boceprevir (HCV NS3/4A protease inhibitor, SARS-CoV-2 Mpro IC50 = 4.13 µM), GC-376 (calpain protease inhibitor, SARS-CoV-2 Mpro IC50 = 0.030 µM), calpain inhibitors II (SARS-CoV-2 Mpro IC50 = 0.97 µM) and XII (SARS-CoV-2 Mpro IC50 = 0.45 µM) exhibited promising SARS-CoV-2 Mpro inhibitory activities (Fig. 5).
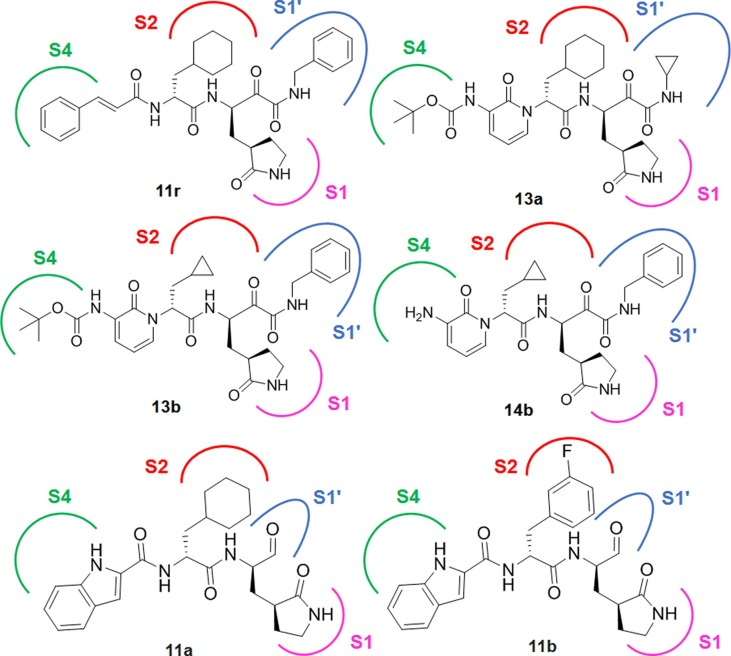
Structure-based design, synthesis and activity assessment by Zhang et al facilitated the development of peptidomimetic α-ketoamides as broad-spectrum inhibitors of beta coronavirus and alpha coronavirus Mpro.4 The most potent compound 11r of this series displayed EC50 of 400 pM against MERS-CoV in virus-infected Huh7 cells (Fig. 6 ). Notably, 11r exhibited broad-spectrum anti-viral activity due to its P2 cyclohexyl moiety which intended to fit the pocket in the enterovirus Mpro.
Fig. 6.
Structure of SARS-CoV-2 Mpro inhibitors (11r, 13a, 13b, 14b, 11a and 11b).
In another study, the same group modified the chemical structure of 11r by replacing the hydrophobic cinnamoyl moiety by comparatively less hydrophobic Boc group and concealing the P3-P2 amide bond within a six member pyridone ring.25 This led to the development of 13a (Fig. 6) with improved solubility in plasma and also reduced binding to plasma proteins, however, the SARS-CoV-2 Mpro inhibition was compromised (13a: SARS-CoV-2 Mpro IC50 = 2.39 µM vs 11r: SARS-CoV-2 Mpro IC50 = 0.18 µM). Further replacement P2 cyclohexyl moiety of 13a with cyclopropyl resulted an increase in the anti-viral property against SARS-CoV-2 Mpro (13b: IC50 = 0.67 µM, Fig. 6). This molecule 13b also showed potency against Mpro of SARS-CoV (IC50 = 0.90 µM) and MERS-CoV (IC50 = 0.58 μM). Furthermore, the X-ray crystal structure of 13b-bound SARS-CoV-2 Mpro conferred that the carbonyl oxygen of pyridone in the P3-P2 position formed hydrogen bond with the main-chain amide of E166. Despite the protecting Boc group on P3 unable to occupy the canonical S4 site of Mpro the protease, it was found at close enough to P168 and subsequently, directed outward by more than 2 Å relative to apo-Mpro structure of SARS-CoV-2. Further removal of Boc group in compound 14b (Fig. 6) led to fall in the inhibitory action, suggesting that hydrophobicity and bulkiness of Boc group would be important to cross the cellular membrane.25
Dai et al elucidated two crystal structures of SARS-CoV-2 Mpro in complex with two indole based covalent inhibitors (Fig. 6) at a high resolution.21 The indole ring of 11a at P3 occupied the solvent exposed S4 site to form a 2.6-Å hydrogen bond with E166 along with hydrophobic interactions with side chains of residues P168 and Q189. Since the S2 site of CoV Mpro withholds bulky P2 fragment, the cyclohexyl moiety of 11a is buried snugly into the S2 pocket of SARS-CoV-2 Mpro and stacking with the imidazole ring of H41. It also interacts with the side chains of M49, Y54, M165, D187 and R188. In contrast, fluophenyl function of 11b at P2 undergoes a significant downward rotation and form additional hydrogen bond Q189 that is likely to enhance Mpro inhibitory activity. Notably, the aldehyde functions of both 11a and 11b act as a warhead in P1 to form a covalent bond with cysteine residue. Moreover, the (S)-γ-lactam ring immerses into the S1 site of CoV Mpro to form several hydrogen bonds with H163, F140 and E166. Both of these inhibitors displayed excellent Mpro inhibitory activities (11a: SARS-CoV-2 Mpro IC50 = 0.053 µM; 11b: SARS-CoV-2 Mpro IC50 = 0.040 µM) along with good PK properties in vivo.21 Hence, from the above studies3, 17, 21, 25 it may be observed that S2 site in SARS-CoV-2 Mpro can board a broad range of hydrophobic substitutions. The isobutyl, cyclopropyl, cyclohexyl and 3-FPh moieties of inhibitors embed themselves in the hydrophobic S2 site formed by H41, M49, and M169.17
Despite huge research efforts on SARS-CoV-2 Mpro inhibitors, proteomic and structural biology works on SARS-CoV-2 PLpro and its inhibitor have been very few. Nevertheless, Rut and co-workers utilised HyCoSuL (Hybrid Combinatorial Substrate Library) to scrutinize substrate specificity of SARS-CoV-2 PLpro enzymes.7 Two irreversible inhibitors namely VIR250 and VIR251 having high degree of PLpro selectivity over other proteases were identified (Fig. 7 ). The same study first time reported the inhibitor-bound crystal structures of SARS-CoV-2 PLpro. Altogether, these crystal structures in complex with VIR250 (PDB: 6WUU) and VIR251 (PDB: 6WX4) provide a basic for rapid rational drug design against SARS-CoV-2 PLpro (Fig. 4 C–D).
Fig. 7.
Structure of SARS-CoV-2 PLpro inhibitors (VIR250 and VIR251) and naphthyl based SARS-CoV-2 PLpro inhibitors.
We have reported the SARs and QSAR of naphthalene based SARS-CoV PLpro inhibitors.10 Very recently, Freitas and co-workers reported these naphthalene based derivatives against SARS-CoV-2 PLpro and SARS-CoV-2 replication.24 Since the residues in the BL2 loop of PLpro are almost identical between SARS-CoV-2 and SARS-CoV-1, this probably facilitated the binding of naphthalene based derivatives. GRL-0617 (IC50 of 2.4 μM and 600 nM against SARS-CoV-2 PLpro and SARS-CoV-1 PLpro, respectively), compound 6 (IC50 of 5 μM and 2.6 μM against SARS-CoV-2 PLpro and SARS-CoV-1 PLpro, respectively) exhibited excellent potency against SARS-CoV-2 replication (Fig 7). In addition, the lead compounds, 77247723 and 6577871 exhibited IC50 values of 23.5 μM and 100.7 μM, respectively against SARS-CoV-2 PLpro. Previously they (77247723 and 6577871) displayed 20 μM and 59 μM SARS-CoV PLpro IC50 values, respectively.
Therefore, it may be postulated that previously designed naphthalene based SARS-CoV PLpro inhibitors follows the similar trend of structure–activity relationship against SARS-CoV-2 PLpro also.24
3.3. Challenges in drug discovery efforts for SARS-CoV-2 protease inhibitors
The corona virus protease inhibitors discovery effort targeting Mpro and PLpro have presenting a substantial challenge owing to poor pharmacokinetic properties of peptidomimetic/macromolecular compounds and low inhibitory potency of non-peptidomimetic and/or compounds having low molecular weight.91, 92 To be an effective drug/drug candidate, a molecule must have ability not only to reach its desire target in the body in sufficient concentration but also to possess expected biological responses. Drug discovery and development markedly depends on assessment of absorption, distribution, metabolism and excretion (ADME) characteristics. Notably, the macromolecule approach of developing SARS-CoV-2 Mpro as well as PLpro inhibitors has been advantageous over the low molecular weight compounds in terms of inhibitory potency and selectivity. In fact the former can occupy the different parts of the active sites of the Mpro and PLpro enzyme to manifest potential inhibitory property. We have collected recently published different SARS-CoV-2 Mpro inhibitors and analyzed for their drug likeliness and other physicochemical properties. From the analysis we have seen that most of these compounds fail to pass the drug-likeness criteria (Fig. 8 A–D).
Fig. 8.
(A) Radar plot of four prototype compounds N3, 13b, Disulfiram, Carmofur after calculating ADME data (http://www.swissadme.ch/) suggesting the drug-likeness. Pink area represents the optimal range of each property. LIPO = Lipophilicity, SIZE = Molecular weight, POLAR = Polarity, INSOLU = Solubility, INSATU = Saturation, FLEX = Flexibility; (B) Correlation of SARS-CoV-2 Mpro inhibitory activity with eleven molecular descriptors (N = 25); (C) The correlation matrix of the physic-chemical and structural properties along with SARS-CoV-2 Mpro pIC50 (N = 25) at significant p-statics; (D) The correlation matrix of the ADME properties along with SARS-CoV-2 Mpro pIC50 (N = 25) at significant p-statics.
Molecules including Baicalein, Disulfiram, Carmofur (Fig. 8 A), Ebselen, Tideglusib, Shikonin and PX-12 coherently passed the drug likeness to be suitable drug but adding challenge to achieve potency and selectivity against coronavirus protease. A similar trend is seen with the SARS-CoV-2 PLpro inhibitors also. Hence, this should be addressed in Mpro and PLpro protease-based drug discovery which could help harness the therapeutic potential against COVID-19.
Meanwhile, a pool of 2D and fingerprint descriptors for these twenty five SARS-CoV-2 Mpro inhibitors (those having exact biological endpoint from Table 3 ) was calculated to frisk the linear relationships. However, the similar analysis has not been possible for the SARS-CoV-2 PLpro inhibitors due to the insufficient number of reported compounds. The correlation of SARS-CoV-2 Mpro inhibitory activity with eleven molecular descriptors (N = 25) at significant p-statics is graphically represented in Fig. 8 B.
Table 3.
Reported SARS-CoV-2 Mpro inhibitors and biological properties.
| Entry | Name | SARS-CoV-2 Mpro IC50 (µM) | pIC50 |
|---|---|---|---|
| 1 | N3 | – | – |
| 2 | Ebselen | 0.67 | 6.174 |
| 3 | Disulfiram | 9.35 | 5.029 |
| 4 | Tideglusib | 1.55 | 5.810 |
| 5 | Carmofur | 1.82 | 5.740 |
| 6 | Shikonin | 15.75 | 4.803 |
| 7 | PX-12 | 21.39 | 4.670 |
| 8 | Cinanserin | 124.93 | 3.903 |
| 9 | 11a | 0.053 | 7.276 |
| 10 | 11b | 0.04 | 7.398 |
| 11 | 11r | 0.18 | 6.745 |
| 12 | 13a | 2.39 | 5.622 |
| 13 | 13b | 0.67 | 6.174 |
| 14 | 14b | – | – |
| 15 | Simeprevir | 13.74 | 4.862 |
| 16 | Boceprevir | 4.13 | 5.384 |
| 17 | Narlaprevir | 5.73 | 5.242 |
| 18 | MG-132 | 3.9 | 5.409 |
| 19 | Calpeptin | 10.69 | 4.971 |
| 20 | Calpain inhibitor III | >20 | – |
| 21 | Calpain inhibitor VI | >20 | – |
| 22 | Calpain inhibitor I (ALLN) | 8.6 | 5.066 |
| 23 | MG-115 | 3.14 | 5.503 |
| 24 | Calpain inhibitor II (ALLM) | 0.97 | 6.013 |
| 25 | Calpain inhibitor XII | 0.45 | 6.347 |
| 26 | PSI | 10.38 | 4.984 |
| 27 | GC-376 | 0.03 | 7.523 |
| 28 | Rupintrivir | >20 | – |
| 29 | Camostat | >20 | – |
| 30 | Lopinavir | >20 | – |
| 31 | Ritonavir | >20 | – |
| 32 | Baicalin | 6.41 | 5.193 |
| 33 | Baicalein | 0.94 | 6.027 |
A positive coefficient of the eight descriptors referred to the favourable effect on the SARS-CoV-2 Mpro inhibitory potential of the studied compounds, while the negative coefficient of three descriptors, i.e., Fraction Csp3, WLOGP, MLOGP, exhibited detrimental effect on the inhibitory properties. The correlation matrix of the physico-chemical and structural properties along with SARS-CoV-2 Mpro pIC50 (N = 25) at significant p-statics is depicted in Fig. 8 C.
Commonly, among these 25 derivatives, several molecules carrying hydrogen bond donor (HBD) > 3 and total polar surface area (TPSA) > 150 were poorly active. Besides, molecules with TPSA < 90 and HBD < 3 frequently possessed lower Mpro inhibitory potency. Regarding the influence of lipophilicity in Mpro activity, highly hydrophobic (WLOGP > 2.9 and MLOGP > 1.6) derivatives were ill-active against Mpro. Interestingly, in this dataset, the least active compound namely cinanserin possessed a lower molecular weight (MW = 340.48), lesser hydrogen bond donor (HBD) and acceptor (HBA) groups (HBD = 1 and HBA = 2), lower total polar surface area (TPSA = 57.64) as well as high hydrophobic nature (WLOGP = 4.08 and MLOGP = 3.76) compared to the most potent Mpro inhibitor (GC-376: MW = 507.53, HBD = 4, HBA = 8, TPSA = 182.34, WLOGP = 0.75 and MLOGP = −0.08).
Moreover, from the properties such as, blood–brain-barrier permeability (BBB permiant), inhibition capability of CYP1A2 (CYP1A2 inhibitor), CYP2C19 (CYP2C19 inhibitor) as well as permeability coefficient (log Kp (cm/s)) of these molecules can be negatively correlated with their Mpro inhibitory potency (Fig. 8 D). Noticeably, most of this dataset molecule with a log Kp (cm/s) value ≥ −6.0 exhibited lesser to-poorly active nature against Mpro. Hence, this should be addressed in Mpro and PLpro protease-based drug discovery which could help harness the therapeutic potential against COVID-19.
4. Conclusion and future perspective
COVID-19 disease is few months old. Until precise molecular and structural biology underlying SARS-CoV-2 replication are available, bioinformatics and multi-target molecular modeling driven in vitro anti-viral study as well as repurposing of previous SARS-CoV protease inhibitors are the handy strategies.
From the SARs, it may be postulated that peptidomimetic and/or covalent coronavirus protease inhibitors possessed potent and selective active-site inhibition. However, these inhibitors exhibited poor absorption, distribution, metabolism, and excretion as well as toxicology parameters to be a drug/drug like molecule.92 Consequently, repurposing/new protease inhibitors discovery efforts of peptidomimetic compounds have presenting a substantial challenge owing to poor pharmacokinetic properties. On the other hand, non-peptidomimetic and/or compounds having low molecular weight coherently passed the drug likeness to be suitable drug but adding challenge to achieve potency and selectivity against coronavirus protease. Hence, lead optimization of non-peptidomimetic and/or low molecular weight compounds should be focused. In this scenario, fragment based drug design (FBDD) approaches can play a significant role in designing and developing potential protease inhibitors. The effective strategy for drug discovery of potential protease inhibitors may consist of following steps: step 1: identification of low molecular weight compounds as protease inhibitors; step 2: identification of good fragments from peptidomimetic compounds by different experimental and computational methods; step 3: incorporating of good fragments during the lead optimization of the low molecular weight compounds; step 4: final optimization of these hybrid molecules for satisfactory pharmacokinetic and pharmacodynamics properties. A masterly combination of adequate pharmacokinetic properties with coronavirus protease activity as well as selectivity will provide strong drug candidates in future.
Based on recent mechanistic and structural data on other viral proteases including HIV, we can anticipate or rather suggest to target the allosteric sites of coronavirus proteases as strategies-based drug discovery tool.92 This effort may soon emerge as frontiers in SARS-CoV-2 Mpro and PLpro drug discovery to triumph the battle against COVID-19.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Financial assistance from the Council of Scientific and Industrial Research (CSIR), New Delhi, India in the form of a Senior Research Fellowship (SRF) [FILE NO.: 09/096(0967)/2019-EMR-I, Dated: 01-04-2019] to Sk. Abdul Amin is thankfully acknowledged. Suvankar Banerjee and Tarun Jha are thankful for the financial support from RUSA 2.0 of UGC, New Delhi, India to Jadavpur University, Kolkata, India. We are very much thankful to the Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India and Department of Pharmaceutical Sciences, Dr. Harisingh Gour University, India for providing the research facilities.
Biographies
Sk. Abdul Amin is a Senior Research Fellow at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India. His research area includes design and synthesis of small molecules with anti-cancer and anti-viral properties, computational chemistry, and large-scale structure-activity relationship analysis. He has published sixty seven research/review articles in different reputed peer-reviewed journals and four book chapters. His SCOPUS h-index is 16 (till October, 2020). Apart from that he is a heritage enthusiast and travel writer. His interests are history through the lens of Art, culture, and religion. He enjoys a good conversation on science, regional history, contemporary art and books.
Suvankar Banerjee is a Research Scholar at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India. He is working under the guidance of Tarun Jha. His research area includes design of small molecules with anti-cancer and anti-viral properties. He has published nine articles in different reputed peer-reviewed journals.
Kalyan Ghosh is a student of Master of Pharmacy (M. Pharm.) at Dr. Harisingh Gour University, Sagar, India. He is working under the guidance of Shovanlal Gayen. His research area includes molecular modeling and drug design. He has published four research articles in the different reputed peer-reviewed journals.
Shovanlal Gayen, is an Assistant Professor at Department of Pharmaceutical Sciences, Dr. Harisingh Gour University, Sagar, India. He is actively involved in different drug design and discovery projects. He has published more than ninety research articles in different reputed peer-reviewed journals and has filed two Indian patents.
Tarun Jha, a Professor at Department of Pharmaceutical Technology, Jadavpur University, Kolkata, India, has supervised 16 Ph.D. students and guided nine research projects funded by different organizations. He has published more than 165 research articles in different reputed peer-reviewed journals. His research area includes design and synthesis of anti-cancer small molecules. Prof. Jha is a member of the Academic Advisory Committee of National Board of Accreditation (NBA), New Delhi, India.
References
- 1.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.Y. Coronaviruses—drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15:327. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z., Du X., Xu Y. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Lin D., Kusov Y. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem. 2020;63:4562–4578. doi: 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh A.K., Brindisi M., Shahabi D., Chapman M.E., Mesecar A.D. Drug development and medicinal chemistry efforts toward SARS-coronavirus and Covid-19 therapeutics. ChemMedChem. 2020;15:907–932. doi: 10.1002/cmdc.202000223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gil C., Ginex T., Maestro I. COVID-19: Drug targets and potential treatments. J Med Chem. 2020 doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 7.Rut W., Lv Z., Zmudzinski M. Activity profiling and structures of inhibitor-bound SARS-CoV-2-PLpro protease provides a framework for anti-COVID-19 drug design. BioRxiv. 2020 doi: 10.1101/2020.04.29.068890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su H., Yao S., Zhao W., Li M., Liu J., Shang W. Discovery of baicalin and baicalein as novel, natural product inhibitors of SARS-CoV-2 3CL protease in vitro. BioRxiv. 2020 doi: 10.1101/2020.04.13.038687. [DOI] [Google Scholar]
- 9.Amin S.A., Jha T. Fight against novel coronavirus: a perspective of medicinal chemists. Eur J Med Chem. 2020;201 doi: 10.1016/j.ejmech.2020.112559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amin S.A., Ghosh K., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Monte Carlo based QSAR, virtual screening and molecular docking study of some in-house molecules as papain-like protease (PLpro) inhibitors. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1780946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization. WHO Director-General's opening remarks at the media briefing on COVID-19-11 March 2020: https://www.who.int/dg/speeches/detail/who-director-general-s-openingremarks-at-the-media-briefing-on-covid-19 (accessed Oct 13, 2020).
- 12.https://www.who.int/emergencies/diseases/novel-coronavirus-2019 (accessed Oct 13, 2020).
- 13.Hall D.C., Ji H.F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect Dis. 2020;101646 doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M., Cao R., Zhang L. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao B., Wang Y., Wen D. A trial of Lopinavir-Ritonavir in adults hospitalized with severe Covid-19. N Engl J Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu X., Han M., Li T. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci USA. 2020;117:10970–10975. doi: 10.1073/pnas.2005615117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma C., Sacco M.D., Hurst B. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. BioRxiv. 2020 doi: 10.1101/2020.04.20.051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pillaiyar T., Meenakshisundaram S., Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today. 2020;25:668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y., Yan L., Huang Y. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dai W., Zhang B., Su H. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020 doi: 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goyal B., Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb Sci. 2020 doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 23.Yuen C.K., Lam J.Y., Wong W.M. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infec. 2020 doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freitas B.T., Durie I.A., Murray J. Characterization and noncovalent inhibition of the deubiquitinase and deISGylase activity of SARS-CoV-2 papain-like protease. ACS Infect Dis. 2020 doi: 10.1021/acsinfecdis.0c00168. [DOI] [PubMed] [Google Scholar]
- 25.Zhang L., Lin D., Sun X. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peele K.A., Chandrasai P., Srihansa T. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inform Med Unlocked. 2020 doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Das S., Sarmah S., Lyndem S., Roy A.S. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Umesh, Kundu D., Selvaraj C., Singh S.K., Dubey V.K. Identification of new anti-nCoV drug chemical compounds from Indian spices exploiting SARS-CoV-2 main protease as target. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1763202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): an in-silico screening of alkaloids and terpenoids from African medicinal plants. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar D., Kumari K., Jayaraj A. Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1752310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keng C.T., Åkerström S., Leung C.S. SARS coronavirus 8b reduces viral replication by down-regulating E via an ubiquitin-independent proteasome pathway. Microbesinfect. 2011;13:179–188. doi: 10.1016/j.micinf.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiel V., Ivanov K.A., Putics A. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 33.Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3CLpro of the COVID-19 via structure-based ligand design and molecular modeling. Chem Phys Lett. 2020;18 doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adhikari N., Baidya S.K., Saha A., Jha T. Structural insight into the viral 3Clike protease inhibitors: comparative SAR, QSAR approaches. In: Gupta S.P., editor. Viral Proteases and their inhibitors. Academic Press; USA: 2017. pp. 317–402. Chapter 11. [Google Scholar]
- 35.Amin S.A., Adhikari N., Jha T. Design of aminopeptidase N inhibitors as anti-cancer agents. J Med Chem. 2018;61:6468–6490. doi: 10.1021/acs.jmedchem.7b00782. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh K., Amin S.A., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: Exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J Mol Struct. 2020;1224 doi: 10.1016/j.molstruc.2020.129026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banerjee S., Amin S.A., Baidya S.K., Adhikari N., Jha T. Exploring the structural aspects of ureido-amino acid-based APN inhibitors: a validated comparative multi-QSAR modelling study. SAR QSAR Env Res. 2020;31:325–345. doi: 10.1080/1062936X.2020.1734080. [DOI] [PubMed] [Google Scholar]
- 38.Petushkova A.I., Zamyatnin A.A., Jr. Papain-Like proteases as coronaviral drug targets: Current Inhibitors, opportunities, and limitations. Pharmaceuticals (Basel) 2020;13:E277. doi: 10.3390/ph13100277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;16 doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu R., Chen L., Lan R., Shen R., Li P. Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:1–8. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shang J., Ye G., Shi K. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020:1–4. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lan J., Ge J., Yu J. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020:1–6. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 45.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Báez-Santos Y.M., John S.E., Mesecar A.D. The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 2015;115:21–38. doi: 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiménez-Alberto A., Ribas-Aparicio R.M., Aparicio-Ozores G., Castelán-Vega J.A. Virtual screening of approved drugs as potential SARS-CoV-2 main protease inhibitors. Comput Biolog Chem. 2020;88 doi: 10.1016/j.compbiolchem.2020.107325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kumar V., Roy K. Development of a simple, interpretable and easily transferable QSAR model for quick screening antiviral databases in search of novel 3C-like protease (3CLpro) enzyme inhibitors against SARS-CoV diseases. SAR QSAR Environ Res. 2020:1–16. doi: 10.1080/1062936X.2020.1776388. [DOI] [PubMed] [Google Scholar]
- 49.Havranek B., Islam S.M. An in silico approach for identification of novel inhibitors as potential therapeutics targeting COVID-19 main protease. J Biomol Struct Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1776158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao K., Nguyen D.D., Chen J., Wang R., Wei G. Repositioning of 8565 existing drugs for COVID-19. J Phys Chem Lett. 2020 doi: 10.1021/acs.jpclett.0c01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ngo S.T., QuynhAnh Pham N., Thi Le L., Pham D.H., Vu V.V. Computational determination of potential inhibitors of SARS-CoV-2 main protease. J Chem Inf Model. 2020 doi: 10.1021/acs.jcim.0c00491. [DOI] [PubMed] [Google Scholar]
- 52.Wang J. Fast Identification of possible drug treatment of coronavirus disease-19 (COVID-19) through computational drug repurposing study. J Chem Inf Model. 2020 doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang R., Hozumi Y., Yin C., Wei G. Decoding SARS-CoV-2 transmission, evolution and ramification on COVID-19 diagnosis, vaccine, and medicine. J Chem Inf Model. 2020 doi: 10.1021/acs.jcim.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Khafaji K., AL-Duhaidahawi L.D., TaskinTok T. Using Integrated computational approaches to identify safe and rapid treatment for SARS -CoV-2. J. Biomol. Struct. Dyn. 2020 doi: 10.1080/07391102.2020.1764392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reiner Ž., Hatamipour M., Banach M. Statins and the COVID-19 main protease: in silico evidence on direct interaction. Arch Med Sci. 2020;16:490–496. doi: 10.5114/aoms.2020.94655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from Tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2020:1–3. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Joshi T., Joshi T., Sharma P. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. Eur Rev Med Pharmacol Sci. 2020;24:4529–4536. doi: 10.26355/eurrev_202004_21036. [DOI] [PubMed] [Google Scholar]
- 58.Choudhury C. Fragment tailoring strategy to design novel chemical entities as potential binders of novel corona virus main protease. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1771424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shamsi A., Mohammad T., Anwar S. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible therapeutic implication in COVID-19 therapy. Biosci Rep. 2020;40 doi: 10.1042/BSR20201256/881803/bsr-2020-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gurung A.B., Ali M.A., Lee J., Farah M.A., Al-Anazi K.M. Unravelling lead antiviral phytochemicals for the inhibition of SARS-CoV-2 Mpro enzyme through in silico approach. Life Sci. 2020;255 doi: 10.1016/j.lfs.2020.117831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsuji M. Potential anti-SARS-CoV-2 drug candidates identified through virtual screening of the ChEMBL database for compounds that target the main coronavirus protease. FEBS Openbio. 2020 doi: 10.1002/2211-5463.12875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer A., Sellner M., Neranjan S., Smieško M., Lill M.A. Potential inhibitors for novel coronavirus protease identified by virtual screening of 606 million compounds. Int J Mol Sci. 2020;21:3626. doi: 10.3390/ijms21103626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gimeno A., Mestres-Truyol J., Ojeda-Montes M.J. Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int J Mol Sci. 2020;21:3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lobo-Galo N., Terrazas-López M., Martínez-Martínez A., Díaz-Sánchez Á.G. FDA-approved thiol-reacting drugs that potentially bind into the SARS-CoV-2 main protease, essential for viral replication. J Biomol Struct Dyn. 2020:1–2. doi: 10.1080/07391102.2020.1764393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liang J., Pitsillou E., Karagiannis C. Interaction of the prototypical α-ketoamide inhibitor with the SARS-CoV-2 main protease active site in silico: molecular dynamic simulations highlight the stability of the ligand-protein complex. Comput Biol Chem. 2020;107292 doi: 10.1016/j.compbiolchem.2020.107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mittal L., Kumari A., Srivastava M., Singh M., Asthana S. Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. J Biomol Struct Dyn. 2020:1–26. doi: 10.1080/07391102.2020.1768151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu C., Liu Y., Yang Y. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ren X., Shao X.X., Li X.X. Identifying potential treatments of COVID-19 from Traditional Chinese Medicine (TCM) by using a data-driven approach. J Ethnopharmacol. 2020;258 doi: 10.1016/j.jep.2020.112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Calligari P., Bobone S., Ricci G., Bocedi A. Molecular investigation of SARS–CoV-2 proteins and their interactions with antiviral drugs. Viruses. 2020;12:445. doi: 10.3390/v12040445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang J. Fast identification of possible drug treatment of coronavirus disease -19 (COVID-19) through computational drug repurposing study. J Chem Inf Model. 2020 doi: 10.1021/acs.jcim.0c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Z.J., Wu W.Y., Hou J.J. Active constituents and mechanisms of Respiratory Detox Shot, a traditional Chinese medicine prescription, for COVID-19 control and prevention: network-molecular docking-LC–MSE analysis. J Integr Med. 2020 doi: 10.1016/j.joim.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elmezayen A.D., Al-Obaidi A., Şahin A.T., Yelekçi K. Drug repurposing for coronavirus (COVID-19): in-silico screening of known drugs against coronavirus 3CL hydrolase and protease enzymes. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1758791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khan S.A., Zia K., Ashraf S., Uddin R., Ul-Haq Z. Identification of chymotrypsin-like protease inhibitors of SARS-CoV-2 Via integrated computational approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1751298. [DOI] [PubMed] [Google Scholar]
- 74.Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251 doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pant S., Singh M., Ravichandiran V., Murty U.S., Srivastava H.K. Peptide-like and small-molecule inhibitors against Covid-19.J. Biomol Struct Dyn. 2020:1. doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen YW, Yiu CP B, Wong KY. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates [version 1; peer review: 3 approved] F1000 Res 2020;9:129. DOI: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed]
- 77.Zhang D.H., Wu K.L., Zhang X., Deng S.Q., Peng B. In silico screening of Chinese herbal medicines with the potential to directly inhibit 2019 novel coronavirus. J Integr Med. 2020;18:152–158. doi: 10.1016/j.joim.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Qamar M.T.ul., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Anal. 2020 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hall D.C., Jr., Ji H.F. A search for medications to treat COVID-19 via in silico molecular docking models of the SARS-CoV-2 spike glycoprotein and 3CL protease. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ton A.T., Gentile F., Hsing M., Ban F., Cherkasov A. Rapid identification of potential inhibitors of SARS-CoV-2 main protease by deep docking of 1.3 billion compounds. Mol Inf. 2020;39:2000028. doi: 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Macchiagodena M., Pagliai M., Procacci P. Identification of potential binders of the main protease 3CLpro of the COVID-19 via structure-based ligand design and molecular modeling. Chem Phys Lett. 2020;750 doi: 10.1016/j.cplett.2020.137489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mothay D., Ramesh K.V. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. Virus Dis. 2020 doi: 10.1007/s13337-020-00585-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Islam R., Parves M.R., Paul A.S. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Abu-Saleh A.A.A., Awad I.E., Yadav A., Poirier R.A. Discovery of potent inhibitors for SARS-CoV-2's main protease by ligand-based/structure-based virtual screening, MD simulations, and binding energy calculations. Phys Chem Chem Phys. 2020 doi: 10.1039/d0cp04326e. [DOI] [PubMed] [Google Scholar]
- 87.Russo M., Moccia S., Spagnuolo C., Tedesco I., Russo G.L. Roles of flavonoids against coronavirus infection. Chem Biol Interact. 2020;328 doi: 10.1016/j.cbi.2020.109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang F., Li Y., Leung E.L. A review of therapeutic agents and Chinese herbal medicines against SARS-COV-2 (COVID-19) Pharmacol Res. 2020;58 doi: 10.1016/j.phrs.2020.104929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Q., Kang C. Progress in developing inhibitors of SARS-CoV-2 3C-like protease. Microorganisms. 2020;8:1250. doi: 10.3390/microorganisms8081250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020;30 doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 92.Drag M., Salvesen G.S. Emerging principles in protease-based drug discovery. Nat Rev Drug Discov. 2010;9:690–701. doi: 10.1038/nrd3053. [DOI] [PMC free article] [PubMed] [Google Scholar]