Abstract
Leukemia diagnosis based on bone marrow cell morphology primarily relies on the manual microscopy of bone marrow smears. However, this method is greatly affected by subjective factors and tends to lead to misdiagnosis. This study proposes using bone marrow cell microscopy images and employs convolutional neural network (CNN) combined with transfer learning to establish an objective, rapid, and accurate method for classification and diagnosis of LKA (AML, ALL, and CML). We collected cell microscopy images of 104 bone marrow smears (including 18 healthy subjects, 53 AML patients, 23 ALL patients, and 18 CML patients). The perfect reflection algorithm and a self-adaptive filter algorithm were first used for preprocessing of bone marrow cell images collected from experiments. Subsequently, 3 CNN frameworks (Inception-V3, ResNet50, and DenseNet121) were used to construct classification models for the raw dataset and preprocessed dataset. Transfer learning was used to improve the prediction accuracy of the model. Results showed that the DenseNet121 model based on the preprocessed dataset provided the best classification results, with a prediction accuracy of 74.8%. The prediction accuracy of the DenseNet121 model that was obtained by transfer learning optimization was 95.3%, which was increased by 20.5%. In this model, the prediction accuracies of the normal groups, AML, ALL, and CML were 90%, 99%, 97%, and 95%, respectively. The results showed that the leukemic cell morphology classification and diagnosis based on CNN combined with transfer learning is feasible. Compared with conventional manual microscopy, this method is more rapid, accurate, and objective.
Keywords: classification, convolutional neural network, diagnosis, leukaemia, transfer learning
1. Introduction
Leukemia (LKA) is a hematopoietic malignancy. In recent years, the prevalence of LKA has increased year by year. In 2018, there were 18.1 million new malignant tumor patients in the world, of which 0.43 million were LKA patients (2.4%), and its incidence was ranked 15th among all malignant tumors. Among the 9.7 million deaths due to malignant tumors, 300,000 deaths are due to LKA (3.2%), and its mortality rate is ranked 10th.[1] In China, the 2003 to 2007 China LKA incidence and mortality report published by the National Cancer Center of China showed that the incidence of LKA was ranked 13th among all cancers and its mortality rate was ranked 9th among all cancers, which was an increase over 1970 and 1990 data.[2] The composition of various types of LKA in descending order is acute myelogenous leukemia (AML), acute lymphoblastic leukemia (ALL), chronic myelocytic leukemia (CML), and chronic lymphocytic leukemia.[3]
The accurate diagnosis, classification, and typing of LKA is a prerequisite for formulating correct and effective treatment regimens. According to the LKA classification and diagnostic criteria formulated by the French-American-British classification, British Committee for Standards in Hematology, and World Health Organization (WHO), cell morphology examination is the basis and core of LKA diagnosis.[4] However, LKA diagnosis by cell morphology based on manual smear reading is a labor- and time-intensive, and highly repetitive job. In addition, because of the complex and variable cell morphology and objective errors introduced during smear preparation and staining, there is a risk of misdiagnosis when subjective manual smear reading is employed. Therefore, there is an urgent need to develop a rapid, accurate, and objective method for cell morphology diagnosis in LKA.
With the development of computers, computer-aided testing/diagnosis is widely used to aid hematologists in analyzing the images of blood cells. These tools can use computer-aided microscopy systems to achieve a more accurate and standard analysis. Image processing-based systems not only increase the accuracy and speed of manual methods but also conserve time, manpower, and costs. The acquisition of images from bone marrow cells under the microscope is the only input for these systems in blood tests. This is an emerging cross-disciplinary technique that integrates digital image processing, computer science, blood smear image processing, and artificial intelligence.
Deep learning, particularly CNN is becoming more widely used in computer-assisted systems as a method to apply artificial intelligence and has obtained good progress in the diagnosis, testing, and classification of different diseases in medical imaging.[5–10] In cell morphology research, CNN can overcome the shortcomings of manual screening, such as high costs, extensive workload, and strong subjectivity, to achieve rapid and accurate cell enumeration and provide cell morphology information. This has provided new ideas for the development of efficient, objective, and automated diagnostic methods for LKA cell morphology. Many researchers have obtained results in this area. Agaian et al[11] used LBP characteristics to achieve classification of 98 peripheral blood microscopy images of normal subjects and ALL subjects and obtained an accuracy of 94%. Moshavash et al[12] fused shapes, colors, and LBP texture characteristics in combination with Support Vector Machines, k-NearestNeighbor, Naive Bayes classifiers, and decision tress to achieve the classification of peripheral blood microscopy images into normal cells and lymphoblasts, and between healthy people and ALL subjects.
To expand the application of deep learning in medical imaging such that existing small sample medical imaging datasets can be fully utilized, the most common method used currently is combining deep learning with transfer learning, in which a model that is trained for 1 problem is simply adjusted to tackle a new problem. In LKA cell morphology diagnosis studies, Shafique et al[13] studied the 3 subtypes of ALL (L1, L2, L3) and normal subjects, and used the AlexNet[14] framework combined with transfer learning to achieve the classification of the 4 subtypes, with an accuracy of 96.06%. Vogado et al[15] established cell image data of peripheral blood smears of healthy leukocytes from healthy adults and ALL cells, and combined transfer learning and CNN to extract image characteristics. Following that, support vector machines, was used for the classification of image characteristics, and a classification accuracy of 99% was achieved. However, these studies are only based on peripheral blood pictures to study a single type of LKA, but not using bone marrow cells that better reflect the pathological conditions.; furthermore, they did not carry out systematic research on the 3 common LKA types (AML, ALL, and CML).
Consequently, bone marrow cell microscopy images of healthy subjects and LKA patients were used as the study subjects in this study, and CNN was combined with transfer learning to carry out bone marrow cell morphology classification and diagnosis of common types of LKA in China.
2. Materials and methods
2.1. Sample collection
In this study, the microscopy images of bone marrow smears were directly obtained from the departments of hematology of the PLA No. 74 Hospital, Guangdong Second Provincial General Hospital, and Zhujiang Hospital of Southern Medical University. There were 104 subjects, of which bone marrow smears were taken from 18 healthy subjects, 53 AML patients, 23 ALL patients, and 18 CML patients. Table 1 lists the detailed information of bone marrow smear samples.
Table 1.
Bone marrow smear sample information statistics table.
| Samples | Age | Male | Female | Total |
| Healthy | 18–40 | 12 | 8 | 18 |
| AML | 13–73 | 30 | 23 | 53 |
| ALL | 3–58 | 13 | 11 | 23 |
| CML | 21–68 | 10 | 8 | 10 |
| Total | 65 | 39 | 104 |
To avoid changes in bone marrow cell count and morphology caused by clinical treatment from affecting this study, all samples used were bone marrow smears of patients taken during initial diagnosis. Hematologists with 15 years or more of experience were responsible for obtaining the clinical diagnosis results, based on the French-American-British classification criteria and WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues and in combination with various morphologic, immunologic, cytogenetic, and molecular biologic classification markers.
The acquisition of microscopy images from bone marrow smear samples was achieved through the constructed image acquisition device, and its major components included the CX40 microscope from SDPTOP Co., Ltd., which has a magnification of 100 times, and the KMC-630H CCD camera manufactured by Guangzhou Koster Scientific instrument Co., Ltd. After the microscope imaging system acquired bone marrow smear microscopy images that were clear and showed uniform cell distribution, they were ultimately stored as color images with a resolution of 1920∗1200.
2.2. Convolutional neural network (CNN)
CNN is a deep learning network structure that was first proposed by Hubel and Wiesel and is widely used in pattern classification. Compared with conventional image classification, deep learning does not require the manual extraction of image characteristics and can extract abstract high-level semantic features from input images, which effectively increases the efficiency of feature learning and feature extraction. In addition, deep learning has shown significant results in natural image classification[16,17] and other tasks (e.g., image segmentation).[18] To develop models with high accuracy that are closer to results from human experts, we employed 3 of the most promising frameworks.
GoogLeNet[19] is a new deep learning structure that was proposed by Szegedy in 2014 and was the winner of the 2014 ImageNet Large Scale Visual Recognition Challenge. GoogLeNet is slightly different from AlexNet, which is purely dependent on a deepening network structure to improve network performance. In GoogLeNet, the Inception structure was introduced while the network was deepened (22 layers) to replace the conventional model of simple combined convolutional activation. The Inception structure module contains 1 × 1 convolution kernels, 3 × 3 convolution kernels, 5 × 5 convolution kernels, and one 3 × 3 bottom sampling layer. This structure increases the breadth of single convolutional layers and increases the expressivity of the network.
ResNet[20] was proposed by He et al from Microsoft Research in 2015 and is a residual learning framework with advantages, such as easy optimization and low computational load. ResNet mainly uses residual blocks to solve the problems of network degeneration and vanishing gradient to train networks with deeper layers and was awarded the 2015 ImageNet Large Scale Visual Recognition Challenge award. In the residual structure, a shortcut is also present for direct output of input information in addition to the conventional route. This will protect information integrity and limit the learning needs of the entire network to just the input and output residuals, which greatly simplifies learning difficulty.
DenseNet[21] was proposed by Huang et al to solve CNN network structures with vanishing gradient problems and surpassed the best result of ResNet in 2016. This is a CNN with dense connections, and the core concept is “skip connection”. In this network, the input of each layer is the combined output of all preceding layers. In addition, the feature map learned by that layer is directly transmitted to all the following layers as output, avoiding information loss during layer-to-layer transmission and the vanishing gradient problem.
2.3. Transfer learning
Conventional data mining and machine learning algorithms use previously collected statistical models trained by labeled or unlabeled data for future data prediction, and when labeled data is too small they use large amounts of unlabeled data and small amounts of labeled data to establish a good classifier. Transfer learning allows the regions, tasks, and distribution used for training and testing to be different. Figure 1 shows the differences in the learning process between conventional learning and transfer learning technologies. With conventional machine learning technology, learning is accomplished by task from the start. Meanwhile, with transfer learning technology, knowledge from other tasks is transferred to the current task, requiring less data for learning and adaptation to the target task.
Figure 1.
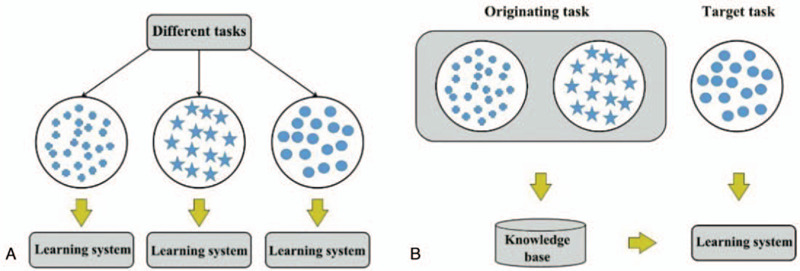
Model learning process: (a) traditional machine learning; (b) migration learning.
The most commonly used strategy for applying transfer learning in CNN networks is to first pre-train a CNN model on a large-scale dataset (usually an ImageNet dataset), and subsequently, to use these pre-trained model parameters for classification of the new dataset. Specifically, the methods for combining CNN and transfer learning can be divided into 2.
-
(1)
Consider the pre-trained model as a fixed feature extractor. The parameters of all convolutional layers in the pre-trained model were directly copied (i.e., parameter transfer) to all convolutional layers in the target domain model before they are frozen, that is, parameter updating is no longer carried out during training, and parameter training only involves the final fully connected layer to complete recognition tasks in the target domain. Figure 2(a) shows the process diagram. Therefore, CNN can be considered as a feature extractor during this process.[22] As this feature extractor has achieved good classification results in the ImageNet dataset, it can be used to achieve simple and more expressive feature vectors from most images.
-
(2)
Fine tuning of the pre-trained model. First, the parameters of all convolutional layers in the pre-trained model are transferred to all convolutional layers in the target domain model and are used as the initial values of the parameters. Subsequently, random initialization of the parameters in the fully connected layer is carried out. Finally, the parameters in the target domain are updated through back propagation to achieve recognition tasks in the target domain.[16] As the deep CNN model contains a million parameters, when there is insufficient data in the target domain, problems may occur, such as the inability of the algorithm to converge or model overfitting. When the parameters from the convolutional layer of the pre-trained model are transferred to the target domain model, complete transfer or partial transfer can be selected. In conclusion, the transfer parameters can be suitably adjusted and revised according to the target task. Figure 2(b) shows the process diagram.
Figure 2.
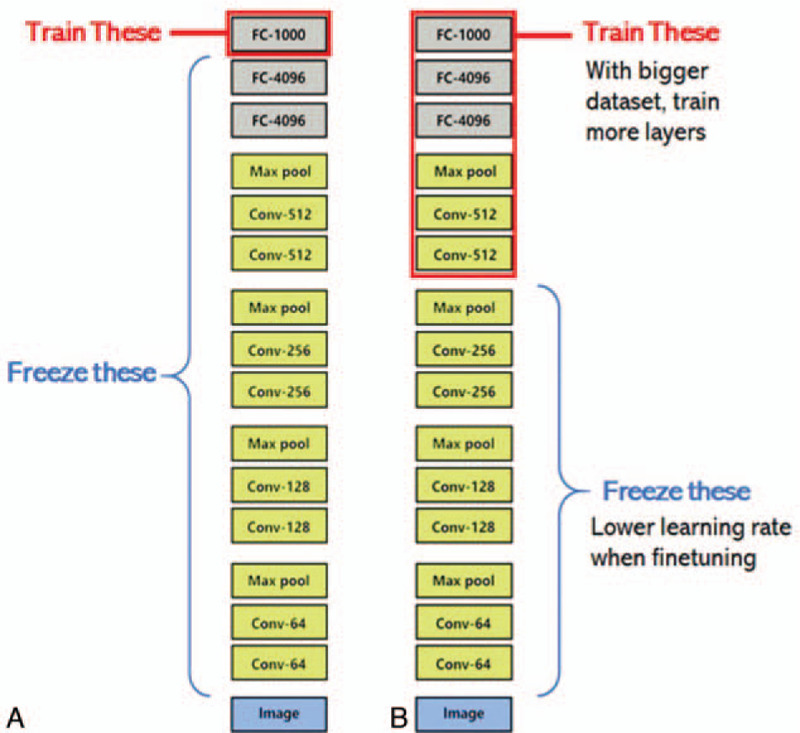
Schematic diagram of migration learning combined with convolutional neural network: (a) using the pre-trained model as a fixed feature extractor; (b) fine-tuning the pre-trained model.
2.4. Model evaluation
Model evaluation is used to evaluate the parameter space and feature extraction results from different models. Classification accuracy refers to the ratio of the number of statistical samples that are correctly identified to the total number of samples when the prediction set is used to test the constructed model in the classification model. The closer the accuracy is to 1, the better the model classification result. In this experiment, the accuracy and the confusion matrix are used for the evaluation of the multi-classification model performance, and the calculation formula is as follows:
| (1) |
| (2) |
In the equation, TP represents the number of positive samples from the pre-trained set that were correctly classified by the model, FN represents the number of positive samples from the pre-trained set that were wrongly classified by the model, FP represents the number of negative samples from the pre-trained set that were wrongly classified by the model, and TN represents the number of negative samples from the pre-trained set that were correctly classified by the model.
2.5. Ethics approval and consent to participate
The studies involving human LKA were approved by the Ethics Committee of Jinan University. The studies using bone marrow smears samples were approved by the biological and medical Ethics Committee of No.74 Group Army Hospital, and written-informed consent was obtained from each participant.
3. Results
All algorithm processing was achieved using Python 3.5, and the hardware used was a 3 GHz Intel Core i5 CPU and 11 GB NVIDIA GTX1080Ti.
3.1. Dataset division
Scanning of all bone marrow smears was carried out to obtain 1322 bone marrow cell images, including 380 images from healthy subjects, 400 from AML patients, 302 from ALL patients, and 240 from CML patients. The bone marrow image dataset was constructed based on the clinical diagnosis results of the microscopy images (as shown in Fig. 3), and this was divided by a 3:1 ratio into a training set and prediction set, of which there were 991 and 331, respectively, as presented in Table 2.
Figure 3.
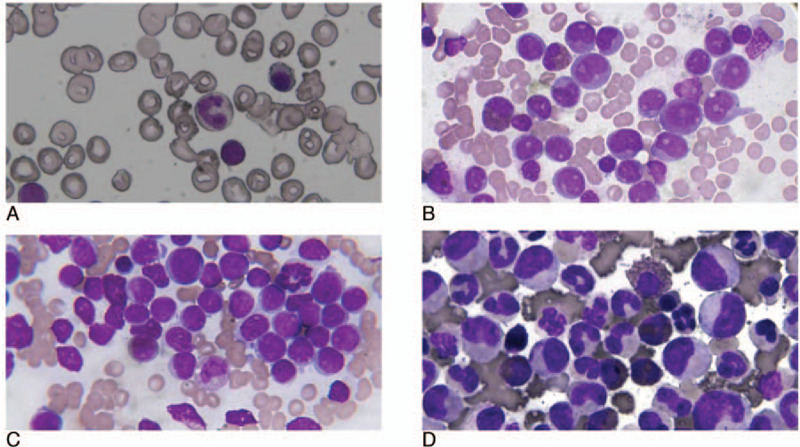
Dataset image dataset: (a) normal group; (b) acute myelogenous leukemia (c) acute lymphoblastic leukemia; (d) chronic myelocytic leukemia.
Table 2.
Data set partition results.
| Samples | Healthy | AML | ALL | CML | Total |
| Data set | 380 | 400 | 302 | 240 | 1322 |
| Train set | 285 | 300 | 226 | 180 | 991 |
| Test set | 95 | 100 | 76 | 60 | 331 |
3.2. Image preprocessing
The background of bone marrow cell images acquired from the microscope is complex and includes erythrocytes, platelets, and debris. During clinical diagnosis, manual microscopy examination is carried out as soon as possible after the staining of bone marrow smears, as the storage interval does not exceed 24 hours. In this experiment, the collected bone marrow smear samples are greatly affected by non-uniform staining, long storage duration, and storage environment. Therefore, preprocessing of microscopy images in the dataset was first carried out to enhance the target cell image. This was achieved by employing the perfect reflection algorithm and self-adaptive filters during preprocessing (as shown in Fig. 4).
Figure 4.
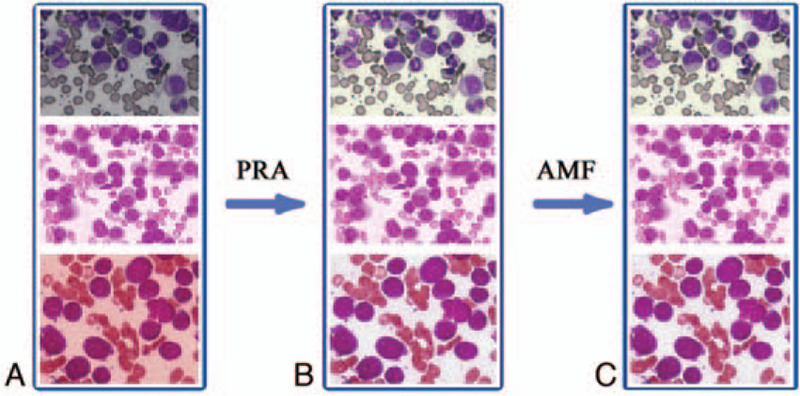
Dataset microscopic image preprocessing results (a) Original image (b) perfect reflection algorithm (c) perfect reflection algorithm and adaptive median filtering.
3.3. Model classification results
To maintain the rational utilization of CPU resources, the batch size was set to 8, and cross entropy was used as the loss function. The specific hyperparameter settings are presented in Table 3.
Table 3.
Hyperparameter setting of convolutional neural network model.
| Hyperparameter | Settings |
| Optimizer | SGD (Stochastic Gradient Descent) |
| Pooling method | Max-pooling |
| Activation function | ReLU (Rectifier Linear Unit) |
| Loss function | Cross-Entropy |
| Batch-size | 8 |
| Learning rate | 0.001 |
| Momentum | 0.5 |
3.4. Effects of different CNN frameworks on classification results
To examine the effects of different CNN frameworks on model performance, we tested 3 frameworks (GoogLeNet, ResNet, and DenseNet) on raw images and preprocessed images. During this experiment, the specific networks used were the: Inception-V3, ResNet50, and DenseNet121 structures. Table 3 presents the specific hyperparameter settings of all models. The number of iterations (epoch) was set to 50, and accuracy was used to measure the classification accuracy of the model. Table 4 and Table 5 present the model construction results when the raw dataset and the preprocessed dataset were used, respectively.
Table 4.
Model classification results of the original data set.
| Models | Accuracy of train data | Accuracy of prediction data |
| Inception-V3 | 96.5% | 64.3% |
| ResNet50 | 98.4% | 66.2% |
| DenseNet121 | 98.9% | 70.6% |
Table 5.
Model classification results of preprocessed data sets.
| Models | Accuracy of train data | Accuracy of prediction data |
| Inception-V3 | 97.1% | 60.6% |
| ResNet50 | 98.7% | 69.3% |
| DenseNet121 | 99.2% | 74.8% |
From Table 4, we can see that the accuracy of the Inception-V3, ResNet50, and DenseNet121 prediction datasets was 64.3%, 66.2%, and 70.6%, respectively. When the raw dataset constructed models were used to test the 3 frameworks, the DenseNet121 model obtained the highest classification accuracy, with a prediction accuracy of 70.6%, which was 6.3% higher than the Inception-V3, which had the lowest classification accuracy.
From Table 5, we can see that after preprocessing, the accuracy of the Inception-V3, ResNet50, and DenseNet121 prediction datasets was 60.6%, 69.3%, and 74.8%, respectively. When the preprocessed dataset was used to construct the 3 models, the DenseNet121 model shows the highest classification accuracy, with a prediction accuracy of 74.8%.
However, image preprocessing shows limited improvement on the model classification accuracy, and the current classification accuracy of the 3 models still has room for improvement. Therefore, the next step mainly employs transfer learning to improve model accuracy.
3.5. Effects of transfer learning on classification results
The use of transfer learning in CNN mainly refers to the transfer of parameters from the pre-trained CNN network to the target CNN model and is an optimization model that can prevent overfitting from occurring. Transfer learning is mainly used in small sample set modeling in CNN, as introducing transfer learning can enable rapid convergence for the model and save computation time and resources.
To examine the effects of transfer learning on CNN model performance, we used DenseNet 121 as the basic model, and 2 transfer learning strategies were employed for model construction of the preprocessed dataset. These 2 strategies include:
-
1)
Using the pre-trained model for feature extraction, that is, transferring the parameters of the convolutional layer in the pre-trained model to the target model and not updating the parameters. During iteration, training was only carried out on parameters in the fully connected layer. The model constructed using this method is known as CNN-1.
-
2)
Fine tuning of the pre-trained model, that is, transferring the parameters of the convolutional layer in the pre-trained model to the target model as initialization parameters and carrying out parameter updating during every iteration. The model constructed using this method is known as CNN-2. Subsequently, a comparative analysis of the 2 types of parameter transfer models and 2 models with random initialization of parameters was conducted. Table 6 presents the model classification results of the datasets when different modeling methods were used.
Table 6.
Model classification results of different modeling methods on the data set.
| Methods | Model | Accuracy of train set | Accuracy of test set | Time |
| Non | CNN | 99.2% | 74.8% | 45 min |
| Transfer learning | CNN-1 | 99.4% | 84.9% | 8 min |
| CNN-2 | 99.7% | 95.3% | 20 min |
From the results listed in Table 6, the time required for training in CNN, CNN-1, and CNN-2 was 45, 8, and 20 minutes, respectively. The convergence speed is extremely fast for the CNN model, which is combined with transfer learning. The CNN-2 model, which was constructed through fine tuning, required half the time as that of the CNN model. The CNN-2 model conducted training of randomly initialized parameters and could adapt very well to the target classification task with a predicted classification accuracy of 95.3%, which is better than the performance of the other 2 models. The model with the second-best prediction accuracy is the CNN-1 model, which is based on feature extraction and has a prediction accuracy of 84.9%.
Figure 5 shows the confusion matrix of the fine-tuning model CNN-2. Each cell shows the proportion of each type of predicted image. The horizontal axis and vertical axis show the class labels of actual and predicted values. The diagonal values correspond to the true positive rate for each class, while the remaining entries show the probability of wrong classification. The prediction accuracy of bone marrow cell microscopy images for AML, ALL, CML, and healthy subjects was 90%, 99%, 97%, and 95%, respectively. Among these groups, the prediction accuracy for AML is low. This is because 8% of bone marrow cell microscopy images were wrongly classified as ALL, while 2% of images were wrongly classified as healthy.
Figure 5.
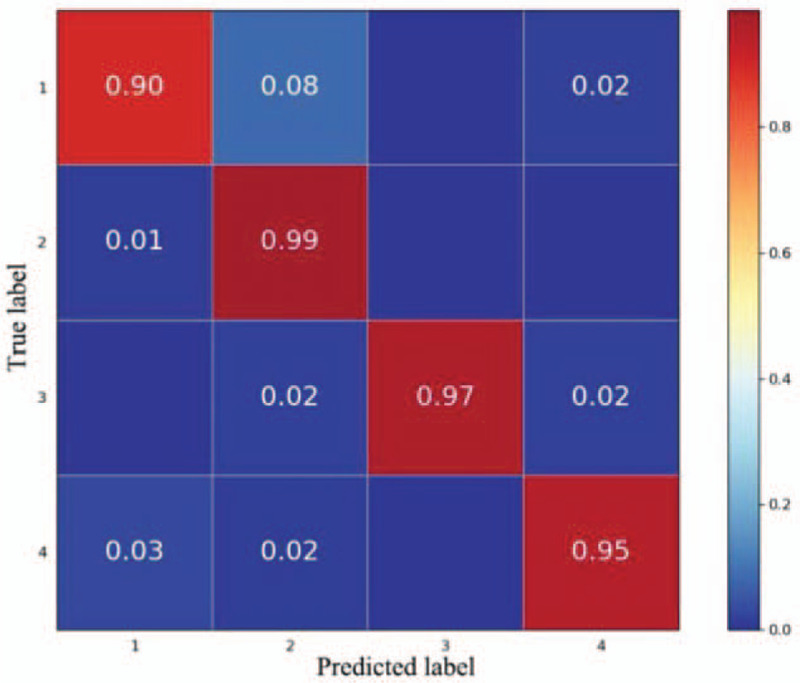
Confusion matrix based on the pre-processing data set fine-tuning model convolutional neural network-2 (1. acute myelogenous leukemia; 2. acute lymphoblastic leukemia; 3. chronic myelocytic leukemia; 4. Normal).
4. Discussion
This study sought to design an intelligence-assisted diagnosis method based on combining CNN and transfer learning to replace the manual interpretation of bone marrow cell morphology. We achieved this by constructing bone marrow cell microscopy image datasets for AML, ALL, CML, and healthy subjects and used 3 different CNN frameworks (GoogleNet, ResNet, and DenseNet) to construct classification models and carry out comparative analysis. Simplified image preprocessing combined with transfer learning was used to improve the classification accuracy of the model and achieve classification of myelograms from AML, ALL, CML, and healthy subjects. Results showed that this method can automatically learn differences in bone marrow cell image features between different types and achieve effective interpretation. This process is fast and efficient and produces objective and reliable results.
In recent years, the feasibility of using microscopy images of bone marrow or peripheral blood smears for automatic recognition to achieve LKA diagnosis has been validated in many studies. Most studies employ cell segmentation to carry out targeted feature extraction from target cells. However, cell segmentation will result in errors in cell shape,[11] thereby affecting feature extraction. In addition, the algorithm structure is complex, time-consuming, and expensive. In this study, we employed the perfect reflection algorithm and self-adaptive filters to carry out the preprocessing of bone marrow cell images, retaining all the features of bone marrow cells while removing most of the background. This conserves time and cost and simplifies the process.
In contrast to conventional machine learning methods, CNN possesses automatic feature learning and feature expression capabilities. Therefore, it has rapidly become the first choice for analyzing medical images.[23–25] However, the superior performance of CNN is dependent on a structural framework that is composed of many parameters. This means that training the CNN requires a large amount of computational resources and much time and costs, as well as a large sample of high-quality training data.[26] To overcome this limitation of CNN, the method that is usually used for a small sample medical image dataset is data enhancement and transfer learning. At present, related studies that employ transfer learning for blood images were studies regarding a single type of LKA,[13,15] and these studies used the microscopy images of peripheral blood smears for LKA classification and diagnosis. In addition, there are few studies involving microscopy images of bone marrow cells. However, compared with peripheral blood, bone marrow smear examinations can intuitively reflect the synthesis, maturation, and release of nucleated cells in the bone marrow, and the morphology of pathological cells; as such, it is a basic method for diagnosing LKA.[27]
Therefore, microscopy images of bone marrow cells were used as study subjects in this study, and the combination of CNN and transfer learning was used for the cell morphology diagnosis of 3 types of LKA (AML, ALL, and CML), which are common in China, to achieve effective classification of multiple diseases. We examined the effects of image preprocessing, different CNN frameworks, and transfer learning on model classification results in the experiment. The comparison of the results in this study is shown in Figure 6:
Figure 6.
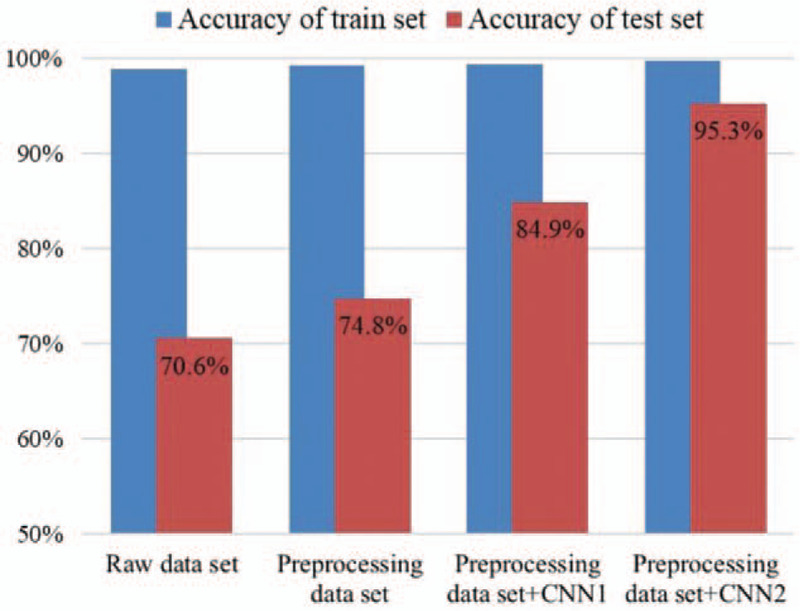
Comparison of modeling results based on DenseNet framework.
-
1)
The classification results of the bone marrow cell microscopy image dataset in this study by DenseNet121 were better than those of Inception-V3 and ResNet50. After simple color correction and noise-reduction filtering, the classification accuracy of the model was increased. This shows that image preprocessing can increase the classification accuracy of the model. Therefore, in medical image classification tasks, particularly when the effects of force majeure factors, such as acquisition equipment, lighting, and noise, are greater, it is necessary to carry out image preprocessing.
-
2)
The fine-tuning pre-trained model method shows good adaptivity to the experiment dataset, and a classification accuracy of 95.3% was obtained, of which the prediction accuracy for AML, ALL, CML, and healthy subjects was 90%, 99%, 97%, and 95%, respectively. This shows that combining CNN and transfer learning is a feasible and efficient method for the classification of small sample datasets.
As training is only required for the last fully connected layer in the feature extraction method, the time spent is extremely low, which the fine-tuning model CNN-2 cannot match. However, the ImageNet database only contains a few lymphocyte and lymphoblast images; thus, there are still large differences with the complete set of bone marrow cell microscopy images. Therefore, the prediction results of the CNN-1 model are not as good as those of the fine-tuning model CNN-2. If the target database and ImageNet database have high similarity, the feature extraction method can rapidly converge, and good classification accuracy can be obtained. From the confusion matrix of the best model, CNN-2, we observed that the prediction accuracy for AML is low. This is because 8% of bone marrow cell microscopy images were wrongly classified as ALL, while 2% of images were wrongly classified as healthy. This misclassification is because the microscopy images of AML bone marrow cells contain many immature granulocytes and monocytes, and ALL bone marrow cell microscopy images contain many immature lymphocytes.[28,29] Meanwhile, although the bone marrow cell microscopy images of CML patients and healthy subjects mainly contain mature cells, immature cells have a high nucleus-to-cytoplasm rate, and the area of the cell nucleus is larger than that of mature cells.[30] Differentiating between immature granulocytes and lymphocytes is dependent on some fine differences, such as the texture and degree of staining of the cell nucleus. Therefore, AML and ALL images are more prone to misclassification than other types of LKA.
Even though the diagnostic methods for LKA are rapidly changing, diagnosis based on bone marrow cell morphology is still indispensable. In this study, we attempt to provide a feasible, objective, and reliable assisted diagnosis method that can replace the manual interpretation of bone marrow cell morphology. Results showed that this method can identify subtle morphological changes that cannot be identified by the naked eye and avoid errors due to manual interpretation, which significantly increases diagnostic accuracy. This avoids objective influencing factors caused by manual smear reading. Therefore, this method can be used to achieve standardization of bone marrow smear diagnosis. Bone marrow cell morphology examinations include peripheral blood smears, bone marrow smears, bone marrow imprints, and bone marrow sections. Each method has its pros and cons. Bone marrow sections are optimal for evaluating hyperplasia of nucleated cells, however obtaining materials for bone marrow smears is simplest and most convenient. These 4 methods are all morphological diagnosis methods. Therefore, the method used in this study can be used for the classification diagnosis of peripheral blood smears, bone marrow imprints, and bone marrow sections.
We carried out classification research on common types of LKA in this study but did not include types of LKA with low incidence in China, such as chronic lymphocytic leukemia. In future studies, we will carry out morphological studies on bone marrow cells from LKA patients that were diagnosed using morphologic, immunologic, cytogenetic and molecular biologic classification diagnostic criteria (e.g., WHO criteria) to further examine the precise typing and classification of CNN in LKA.
In summary, we constructed a new classification and diagnosis method based on leukemic bone marrow cell morphology in this study. This method is different from conventional manual interpretation. By combining CNN and transfer learning, a classification model for bone marrow cell images was constructed, and this was used for 4 types of classification: healthy subjects and 3 types of LKA. The results are fast, objective, reliable, and can avoid errors, misdiagnosis, and misjudgment due to human factors. Hence, this method can be applied for the morphological diagnosis of other similar types of LKA.
Author contributions
Conceptualization: Furong Huang, Peiwen Guang, Fucui Li, Weimin Zhang, Wendong Huang.
Formal analysis: Furong Huang, Peiwen Guang, Fucui Li.
Investigation: Furong Huang.
Resources: Weimin Zhang, Wendong Huang.
Supervision: Weimin Zhang, Wendong Huang.
Validation: Fucui Li, Xuewen Liu.
Writing – original draft: Furong Huang, Peiwen Guang, Fucui Li, Weimin Zhang.
Writing – review & editing: Weimin Zhang, Wendong Huang.
Footnotes
Abbreviations: ALL = acute lymphoblastic leukemia, AML = acute myelogenous leukemia, CML = chronic myelocytic leukemia, CNN = convolutional neural network, LKA = Leukemia, WHO = World Health Organization.
How to cite this article: Huang F, Guang P, Li F, Liu X, Zhang W, Huang W. AML, ALL, and CML classification and diagnosis based on bone marrow cell morphology combined with convolutional neural network: a STARD compliant diagnosis research. Medicine. 2020;99:45(e23154).
WH and WZ have contributed equally to this work. So WH is a corresponding author too.
This work was supported by the National Natural Science Foundation of China (61975069); Natural Science Foundation of Guangdong Province, China (2018A0303131000).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
ALL = acute lymphoblastic leukemia, AML = acute myelogenous leukemia, CML = chronic myelocytic leukemia.
ALL = acute lymphoblastic leukemia, AML = acute myelogenous leukemia, CML = chronic myelocytic leukemia.
CNN = convolutional neural network.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Chen W, Shan B, Zheng R, et al. Analysis of incidence and mortality of leukemia in registration areas of China from 2003 to 2007. Tumor 2012;32:251–5. [Google Scholar]
- [3].Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- [4].Gisslinger H, Jeryczynski G, Gisslinger B, et al. Clinical impact of bone marrow morphology for the diagnosis of essential thrombocythemia: comparison between the BCSH and the WHO criteria. Leukemia 2016;30:1126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Li Q, Cai WD, Wang XG, et al. Medical image classification with convolutional neural network. I C Cont Automat Rob 2014;844–8. [Google Scholar]
- [6].Scotti F. Automatic morphological analysis for acute leukemia identification in peripheral blood microscope images. Proceedings of the 2005 IEEE International Conference on Computational Intelligence for Measurement Systems and Applications. 2005; 96-101. [Google Scholar]
- [7].Hubel DH, Wiesel TN. Receptive fields and functional architecture of monkey striate cortex. J Physiol 1968;195:215–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Fukushima K. Neocognitron: a self-organizing neural network model for a mechanism of pattern recognition unaffected by shift in position. Biol Cybern 1980;36:193–202. [DOI] [PubMed] [Google Scholar]
- [9].Lu Y, Yu QY, Gao YX, et al. Identification of metastatic lymph nodes in MR imaging with faster region-based convolutional neural networks. Cancer Res 2018;78:5135–43. [DOI] [PubMed] [Google Scholar]
- [10].Torkaman A, Charkari NM, Aghaeipour M, et al. A recommender system for detection of leukemia based on cooperative game. Med C Contr Automat 2009;1126–30. [Google Scholar]
- [11].Agaian S, Madhukar M, Chronopoulos AT. A new acute leukaemia-automated classification system. Comp M Bio Bio E-IV 2018;6:303–14. [Google Scholar]
- [12].Moshavash Z, Danyali H, Helfroush MS. An automatic and robust decision support system for accurate acute leukemia diagnosis from blood microscopic images. J Digit Imaging 2018;31:702–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shafique S, Tehsin S. Acute lymphoblastic leukemia detection and classification of its subtypes using pretrained deep convolutional neural networks. Technol Cancer Res T 2018;17:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun Acm 2017;60:84–90. [Google Scholar]
- [15].Vogado LHS, Veras RMS, Araujo FHD, et al. Leukemia diagnosis in blood slides using transfer learning in CNNs and SVM for classification. Eng Appl Artif Intell 2018;72:415–22. [Google Scholar]
- [16].Shin HC, Roth HR, Gao MC, et al. Deep convolutional neural networks for computer-aided detection: CNN Architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 2016;35:1285–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Araujo T, Aresta G, Castro E, et al. Classification of breast cancer histology images using convolutional neural networks. PLoS One 2017;12:e0177544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- [19].Szegedy C, Liu W, Jia YQ, et al. Going deeper with convolutions. Proc CVPR IEEE 2015;1–9. [Google Scholar]
- [20].He KM, Zhang XY, Ren SQ, et al. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2016;770–8. [Google Scholar]
- [21].Huang G, Liu Z, van der Maaten L, et al. Densely connected convolutional networks. 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2017) 2017;2261–9. [Google Scholar]
- [22].Yosinski J, Clune J, Bengio Y, et al. How transferable are features in deep neural networks ? Adv Neur In 2014;27:3320–8. [Google Scholar]
- [23].Frid-Adar M, Diamant I, Klang E, et al. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018;321:321–31. [Google Scholar]
- [24].Khatami A, Babaie M, Tizhoosh HR, et al. A sequential search-space shrinking using CNN transfer learning and a Radon projection pool for medical image retrieval. Expert Syst Appl 2018;100:224–33. [Google Scholar]
- [25].Lim HN, Mashor MY, Supardi NZ, Hassan R. Color and morphological based techniques on white blood cells segmentation. Proceedings 2015 2nd International Conference on Biomedical Engineering (Icobe 2015). 2015. [Google Scholar]
- [26].LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- [27].Thiyagarajan P, Suresh TN, Anjanappa R, et al. Bone-marrow spectrum in a tertiary care hospital: clinical indications, peripheral smear correlation and diagnostic value. Med J DY Patil Univ 2015;8:490–4. [Google Scholar]
- [28].Rapin N, Bagger FO, Jendholm J, et al. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood 2014;123:894–904. [DOI] [PubMed] [Google Scholar]
- [29].Shalapour S, Eckert C, Seeger K, et al. Leukemia-associated genetic aberrations in mesenchymal stem cells of children with acute lymphoblastic leukemia. J Mol Med 2010;88:249–65. [DOI] [PubMed] [Google Scholar]
- [30].Ghane N, Vard A, Talebi A, et al. Classification of chronic myeloid leukemia cell subtypes based on microscopic image analysis. EXCLI J 2019;18:382–404. [DOI] [PMC free article] [PubMed] [Google Scholar]


