Abstract
This study aimed to identify significantly altered long non-coding RNAs (lncRNAs), microRNAs (miRNAs), mRNAs, pathways in preeclampsia (PE), and to investigate their targeted relationships and biological functions.
GSE96985 from Gene Expression Omnibus database was extracted, involving 3 PE and 4 normal tissues. After the differential expression analysis of miRNAs, lncRNAs, and mRNAs using the limma package, protein-protein interaction (PPI) network and module analyses were performed for differentially expressed mRNAs (dif-mRNAs). Combined with the miRanda and miRWalk tools, a regulatory relationship between dif-miRNAs and dif-mRNAs/lncRNAs (dif-mRNAs/dif-lncRNAs) was predicted. Finally, mRNA-miRNA-lncRNA regulatory network construction was performed using Cytoscape software.
A total of 511 dif-mRNAs were screened in PE. The top 5 nodes in the PPI networks included up-regulated complement component 3 (C3), C-X-C motif chemokine ligand 8 (CXCL8), and fibronectin 1 (FN1). Three significant network modules were identified for dif-mRNAs. C3 and CXCL8 were identified in module A, and FN1 was identified in module C. A disintegrin and metalloproteinase with thrombospondin motifs 6 (ADAMTS6) was down-regulated by the miR-210-3p. Therefore, lnc-CTD-2383M3.1 functions as a competing endogenous RNA in ADAMTS6 expression regulation by competitively binding to miR-210-3p during the regulation process of PE.
C3, CXCL8, FN1, and ADAMTS6 might be involved in the development of PE. The lnc-CTD-2383M3.1-miR-210-3p-ADAMTS6 axis might be a potential regulatory mechanism in PE.
Keywords: ADAMTS6, C3, ceRNACXCL8, FN1
1. Introduction
Preeclampsia (PE) is a common pregnancy-associated disorder and is one of the main causes of pregnancy-related maternal and neonatal morbidity with clinical characteristics of pregnancy-induced hypertension and proteinuria.[1] Every year, approximately 76,000 women and 500,000 babies die from PE, which is one of the major causes of maternal mortality.[2] PE is an unpredictable multifaceted syndrome that has been associated with a high risk of cardiovascular complications and diabetes mellitus. PE can rapidly develop into the life-threatening condition, eclampsia.[3,4] However, few effective treatments are currently available for PE patients in clinic. The only available cure for PE is delivery of the fetus and placenta, which reduces proinflammatory substances in the maternal cardiovasculature.[5,6] The cause of PE remains elusive and numerous research studies have been carried out to investigate it.[7] With the emergence of clinical signs and symptoms of PE in the third trimester of pregnancy, identification of early diagnosis/prognosis bio-markers has become a relevant research focus.
In the past few years, numerous studies have been performed on the biomarkers involved in PE. Vascular endothelial growth factor has been considered a down-regulated gene that plays an essential role in the abnormality of placenta and vascular dysfunction of PE.[8] Furthermore, the elements involved in the activity of placental unfolded protein response, including activating transcription factor-6, phosphorylated inositol-requiring transmembrane kinase/endoribonuclease 1α, transcription factor X-box-binding protein-1, and glucose-regulated proteins GRP78 and GRP94, have a higher expression in PE patients compared to normotensive controls.[9] Previous studies have shown that microRNAs (miRNAs) had a crucial role in PE. MiR-141 is one of the most abundant miRNAs in placental plasma during pregnancy.[10] Recently, miR-141 has been found to have a crucial function during intercellular communication between the fetal trophoblast and maternal immune cells. It can be used as a marker of potential diseases like PE during pregnancy.[11] Non-coding RNAs account for 90% of the human genome and are the largest group of RNA transcripts. In particular, long non-coding RNAs (lncRNAs) can potentially sponge their target miRNAs and mediate post-transcriptional regulation of target mRNAs by impairing target miRNA ability.[12]
In the present study, a microarray dataset for PE was downloaded from a public database. Then, differentially expressed mRNAs (dif-mRNAs), differentially expressed miRNAs (dif-miRNAs), and differentially expressed lncRNAs (dif-lncRNAs) between PE and control tissues were investigated using enrichment analysis. To further explore the mechanism of key mRNAs, lncRNAs, and miRNAs in PE, protein-protein interaction (PPI) network and regulatory network analyses were performed. This study might provide novel therapeutic targets for PE.
2. Materials and methods
2.1. Data source
Microarray dataset GSE96985 (miRNA sequencing platform: GPL20712 Agilent-070156 Human miRNA [miRNA version], lncRNA and mRNA sequencing platform: GPL22120 Agilent-078298 human competing endogenous RNA (ceRNA) array V1.0 4X180K [Probe Name Version]) from the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database was downloaded. The database included 3 PE patients and 4 normal tissues. GSE96985 is a public dataset. Therefore neither ethics committee approval nor patient informed consent was required for analyzing GSE96985 data.
2.2. Differential expression analysis
The original data were preprocessed using the limma package.[13,14] Data preprocessing included background correction, normalization, and concentration prediction. Gene expression values were calculated by mapping probes to symbols utilizing the microarray dataset and annotation files from the chip platform. The average value was defined as the value of gene expression when multiple probes were matched to one symbol.
The dif-mRNAs, dif-miRNAs, and dif-lncRNAs between PE patients and normal tissues were analyzed using a typical Bayesian method and corrected by the Benjamini/Hochberg tests. The heat maps for dif-mRNAs, dif-miRNAs, and dif-lncRNAs were plotted using the R package in pheatmap software (version: 1.0.10, https://cran.r-project.org/web/packages/pheatmap/index.html).[15] The dif-miRNAs were defined as miRNAs with |log fold change (FC)| > 1 and P value <.05. The dif-lncRNAs and dif-mRNAs were defined as lncRNAs or mRNAs with |log2 (FC)| > 2 and P values <.05.
2.3. PPI network and module analyses for dif-mRNA
Using the STRING database (version: 10.0, http://www.string-db.org/),[16] interactions between dif-mRNAs were retrieved. A PPI score = 0.4 was defined as the median confidence. Cytoscape software[17] (version: 3.2.0, http://www.cytoscape.org) was used to construct the PPI network based on obtained interactions. Significant network modules were analyzed using Molecular Complex Detection (MCODE) plugin (version: 1.4.2, http://apps.cytoscape.org/apps/MCODE)[18] in Cytoscape software. Modules with scores ≥ 8 were considered to be significant.
2.4. Pathway enrichment analysis
Based on the clusterprofiler tool in R package[19] (version: 2.4.3, https://bioconductor.org/packages/3.2/bioc/html/clusterProfilter.html), the Kyoto Encyclopedia of Genes and Genomes (KEGG)[20] pathway enrichment analysis was performed for dif-mRNAs, mRNAs in significant modules, and those in the lncRNA-miRNA-mRNA network. Related genes involved in the ceRNA net were also evaluated using the KEGG pathway enrichment analyses. Significantly enriched pathways with P values <.05 and involving no less than two mRNAs were selected.
2.5. Regulatory network analysis of miRNA with mRNA/lncRNA
The dif-miRNA and dif-lncRNA pairs were predicted using a score ≥150 and energy < −20 based on the miRanda tool (version: 3.3a, https://omictools.com/miranda-tool).[21]
The miRWalk 2.0[22] (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/) was used to predict the targeted mRNAs for dif-miRNAs in the above relationship. The mRNA-dif-miRNA pairs were selected according to the condition that the pairs appeared at least 5 times among the following databases: miRWalk, miRanda, miRDB, miRMap, Pictar2, RNA22, and Targetscan. The mRNAs-lncRNA pairs were finally selected among the acquired regulation pairs of mRNAs-lncRNAs with the condition that mRNAs belong to the dif-mRNA group.
2.6. LncRNA-mRNA coexpression network analysis
The dif-mRNAs among dif-mRNA-dif-miRNA pairs and dif-lncRNAs among dif-lncRNA-dif-miRNA pairs were selected to construct relative pairs of dif-mRNA-dif-lncRNAs. The Pearson correlation coefficient for dif-lncRNA-dif-mRNAs was calculated. The threshold values for significantly correlated dif-lncRNA-dif-mRNA pairs were R-value for top 100 with a false discovery rate <0.05.
2.7. LncRNA-miRNA-mRNA integration analysis
The Cytoscape tool was used to construct a complex network of lncRNA-miRNA-mRNAs by integrating the obtained lncRNA-mRNA, miRNA-mRNA, and miRNA-lncRNA pairs.
3. Results
3.1. Differential expression analysis
According to the screening criteria, there were 511 dif-mRNAs (364 up-regulated and 147 down-regulated), 251 dif-lncRNAs (93 up-regulated and 158 down-regulated), and 36 dif-miRNAs (14 up-regulated and 22 down-regulated) in PE tissues compared with normal tissues.
The bidirectional clustering heat maps for dif-mRNAs, dif-lncRNAs, and dif-miRNAs are shown in Figure 1. The difference analysis can significantly separate the 3 PE patients from the 4 normal tissues on the level of mRNA, lncRNA, and miRNA. Different analysis results were reliable.
Figure 1.

Bidirectional clustering heat map for mRNAs, dif-lncRNAs, and dif-miRNAs. (A) Heat map of differentially expressed mRNAs from PE and normal tissues (|logfold change (FC)| > 1, adjusted P < .05). (B) Heat map of differentially expressed miRNAs from PE and normal tissues (|logfold change (FC)| > 2, adjusted P < .05). (C) Heat map of differentially expressed lncRNAs from PE and normal tissues (|logfold change (FC)| > 2, adjusted P < .05).
3.2. Enrichment analysis
Enrichment analysis for the up- and down-regulated mRNAs was conducted. The up-regulated mRNAs were implicated in 36 pathways. The top ten pathways are shown in Figure 2A. Down-regulated mRNAs were implicated in 6 pathways (Fig. 2B).
Figure 2.

Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for differentially expressed mRNAs (dif-mRNAs). (A) Top ten up-regulated dif-mRNAs. (B) Down-regulated dif-mRNAs.
3.3. PPI network and module analyses
The PPI network included 229 nodes and 605 interactions (Fig. 3). Three significant models were separately identified from the PPI network (Fig. 3). Model A had 17 nodes and 136 interactions, model B had 10 nodes and 45 interactions, and model C had 9 nodes and 36 interactions. Complement component 3 (C3), C-X-C motif chemokine ligand 8 (CXCL8), and fibronectin 1 (FN1) were identified in the modules and among the top 5 nodes (C3 and CXCL8 were identified in module A and FN1 was identified in module C). The KEGG enrichment analysis was conducted for the mRNAs in significant modules. The top 10 results are shown in Figure 4.
Figure 3.
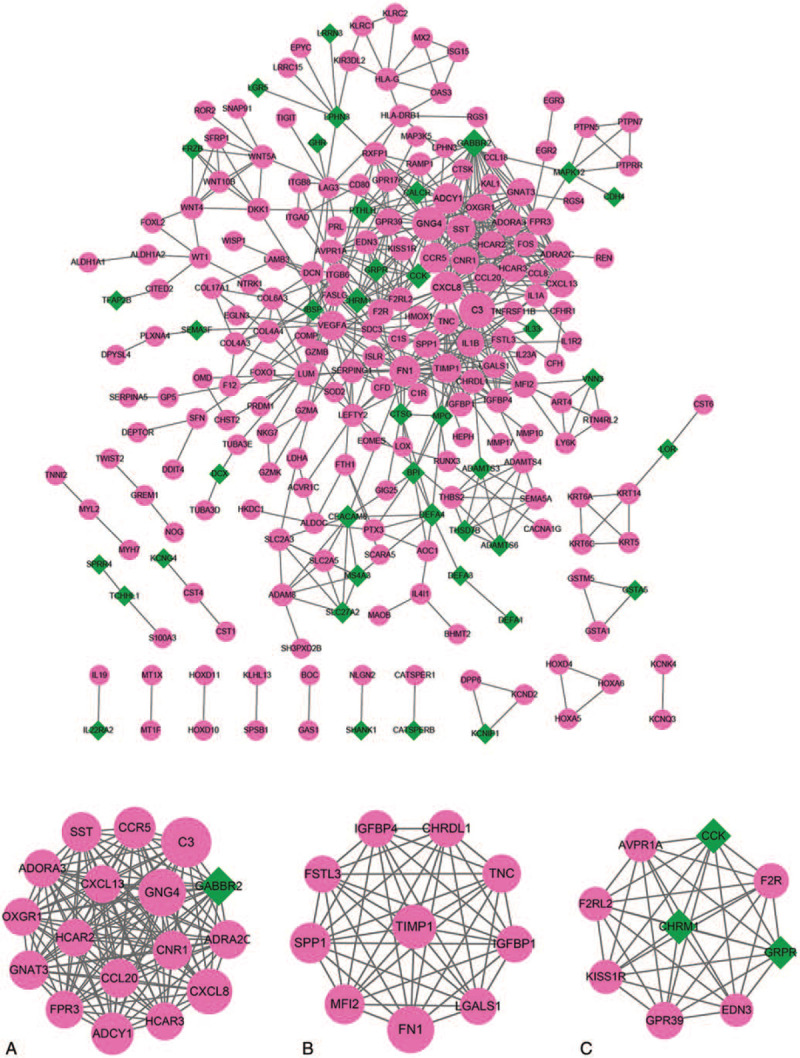
Protein-protein interaction (PPI) network for differentially expressed mRNAs and modules A, B, and C identified from PPI network for dif-mRNAs. Red circles represent up-regulated mRNAs, green diamonds represent down-regulated mRNAs. Node size and degree are proportional.
Figure 4.
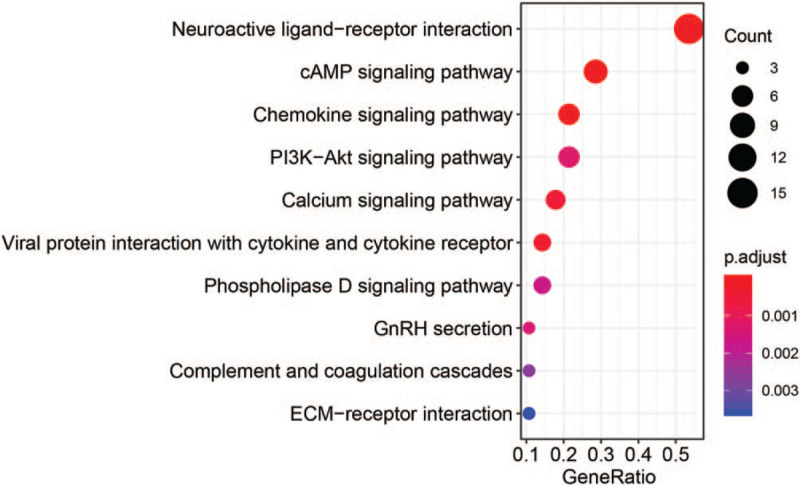
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for differentially expressed mRNAs involved in modules (top ten listed).
3.4. Regulatory predictive analysis
The miRanda tool was used to predict the regulatory relationship for all dif-miRNAs and dif-lncRNAs. A total of 71 lncRNA-miRNA interaction pairs were obtained, including 24 dif-miRNAs and 45 dif-lncRNAs. To predict the targeted mRNAs for the above dif-miRNA, the miRWalk2.0 database was utilized. A total of 439 miRNA-mRNA interaction pairs were identified, including 22 dif-miRNAs and 186 dif-mRNAs.
Furthermore, the 22 dif-miRNA-regulated mRNAs and lncRNAs were screened for co-expression. The top 100 positively correlated lncRNA-mRNA pairs were obtained based on the screening threshold.
3.5. Regulatory network analysis
LncRNA-miRNA-mRNA pairs were further sorted according to the information available for all the pairs. A total of 605 interaction pairs were obtained, including 100 lncRNA-mRNA co-expressed, 439 miRNA-mRNA interaction, and 65 lncRNA-miRNA interaction pairs. The lncRNA-miRNA-mRNA network contained 9 up-regulated miRNAs, 13 down-regulated miRNAs, 137 up-regulated mRNAs, 49 down-regulated mRNAs, 16 up-regulated lncRNAs, and 11 down-regulated lncRNAs (Fig. 5). The lnc-CTD-2383M3.1 might function as a ceRNA in regulating disintegrin and metalloproteinase with thrombospondin motifs 6 (ADAMTS6) expression in PE by competitively binding to miR-210-3p (Fig. 5). The KEGG pathway enrichment analysis was carried out for all dif-mRNAs in the network (Fig. 6).
Figure 5.

The mRNA-miRNA-lncRNA regulatory network. Red circles and green diamonds represent up- and down-regulated mRNAs, respectively. Yellow hexagons and blue quadrilaterals represent up- and down-regulated lncRNAs, respectively. Red triangles and dark green arrows represent up- and down-regulated miRNAs, respectively.
Figure 6.
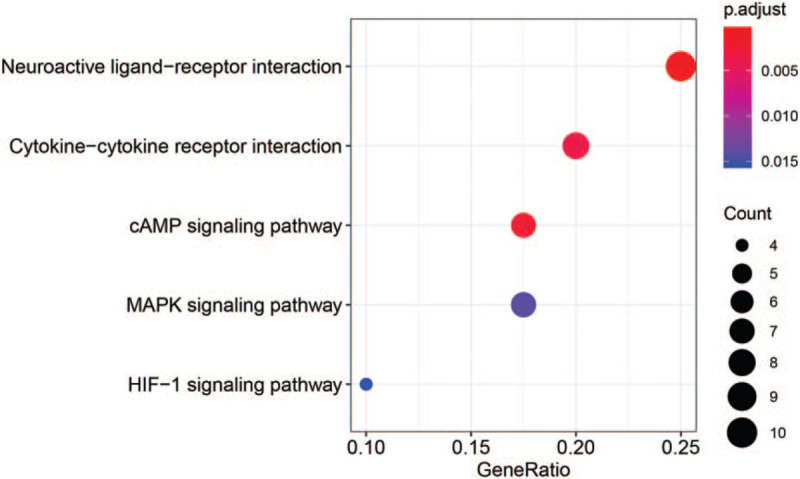
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enriched for differentially expressed mRNAs involved in the mRNA-miRNA-lncRNA regulatory network.
4. Discussion
In the present study, 511 dif-mRNAs (364 up-regulated and 147 down-regulated), 251 dif-lncRNAs (93 up-regulated and 158 down-regulated), and 36 dif-miRNAs (14 up-regulated and 22 down-regulated) in PE tissues were screened. In the dif-mRNA PPI network, 3 (module A, B, and C) significant network modules were identified for dif-mRNAs. In addition, C3, CXCL8, and FN1 were identified in the modules and among the top 5 nodes (C3 and CXCL8 were identified in module A, FN1 was identified in module C). In the regulatory network, miR-210-3p targeted ADAMTS6 and showed a negative correlation. The lnc-CTD-2383M3.1 was predicted to be involved in the ceRNA network of ADAMTS6 and miR-210-3p.
Pregnancy is an immunological process in which the immune system should allow the fetus to thrive and protect both the mother and fetus from pathogens. Even though PE is a vascular disease, immunological mechanisms have been implicated.[23] Previous studies have suggested that inadequate control of the maternal complement system contributes to the etiology of PE.[24,25] The complement system belongs to the immune system, which can be activated by lectin. Studies have shown that complement C3 is the central component of all activation pathways.[26] Indeed, the abundance of C3 in human serum indicated that C3 is a key component of immunity in the process of discriminating between self and non-self.[27] Studies have shown that in complement inhibitor-deficient mice, activation of the complement alternative pathway is the key mechanism for reproductive failure.[28] Recently, it has been shown that an alternative complement pathway is activated in human pregnancies with severe PE.[29] Eculizumab is a targeted inhibitor of complement protein C5 that was able to successfully treat a patient suffering from the HELLP syndrome, which is a life-threatening complication of PE. This demonstrated that the complement system could provide a promising target for drug development in severe PE.[30] Due to regulation of protective haplotype, the extravillous trophoblasts (EVTs) invading the maternal tissue during placentation do not encounter a vigorous complement attack of C3 activation.[31] Concurrently, with an increased levels of factor B in early pregnancy, the predisposing haplotype might result in an increased level of complement activation. This indicates that a complement attack of the EVT invasion would consequently damage the placental function, resulting in an increased occurrence of severe PE.[29,32] In the present study, C3 was significantly up-regulated in PE. The KEGG pathway enrichment analysis showed that C3 was involved in the complement and coagulation cascade pathway. Thus, C3 might play a significant role in PE.
Studies have shown that PE pathogenesis is involved in insufficient trophoblast invasion, causing dysfunction in the endothelial system and an increased inflammatory response.[33–35] In PE cases, insufficient invasion of the trophoblast leads to insufficient remodeling of the spiral artery, which might result in hypoxia-reperfusion injury and oxidative stress in the placenta.[36] CXCL8, a C-X-C motif chemokine ligand, is present in the decidua and trophoblast, which also express CXCL8 receptors.[37] CXCL8-stimulated migration and invasion of the trophoblast cells is aided by the increasing expression of matrix metalloproteinase (MMP)-2 and -9.[38] FN might contribute to the maturation of the trophoblast cells, which is an important component of trophoblastic basement membranes.[39,40] Studies have shown that an inadequate trophoblast invasion might induce the pathogenesis of PE. FN1 is primarily synthesized by endothelial cells and has been reported to be involved in regulatory functions of cell interactions. FN1 circulating values correspond to endothelial damage. In addition, PE is considered to be a pathology of the endothelial cells.[41] FN tissue remodeling is an important process during embryonic development and wound healing. In the present study, CXCL8 and FN1 were significantly up-regulated in PE. The KEGG pathway enrichment analysis showed that CXCL8 was involved in the pathway of viral protein interaction with the cytokine and cytokine receptor FN1 was involved in the PI3K-Akt signaling pathway. Therefore, CXCL8 and FN1 might be involved in the development of PE.
The present study also found that ADAMTS6 expression was down-regulated by the miR-210-3p. Lnc-CTD-2383M3.1 might serve as a ceRNA in this process. ADAMTS genes are members of MMP, which play an important role in degradation and repair processes of extracellular matrix (ECM). ADAMTS roles in PE might include invasion regulation of the spiral artery and ECM arrangement of the placenta.[42,43] An increasing number of studies have found that miR-210-3p is mainly expressed in the villous and tunica adventitia trophoblastic layers.[44,45] MiR-210-3p is involved in the process of placenta. Through the mitogen-activated protein kinase-dependent mechanism, elevated levels of miR-210-3p decreased the invasion of EVT.[46] Hence, it was suggested that lnc-CTD-2383M3.1 acted as a ceRNA in regulating ADAMTS6 expression by competitively binding to miR-210-3p in the progression of PE.
5. Conclusion
In conclusion, a series of bioinformatics analyses was conducted in PE patients. The results revealed that C3, CXCL8, and FN1 might be implicated in the pathogenesis of PE. In addition, lnc-CTD-2383M3.1 might function as a ceRNA in ADAMTS6 expression regulation in PE by competitively binding to miR-210-3p. These results must be validated and supported by further experimental studies.
Author contributions
Conceptualization: Ruo-an Jiang.
Data curation: Feifei Zhou.
Formal analysis: Ruo-an Jiang, Ting Wang, Jing He.
Investigation: Dong Xu.
Methodology: Ruo-an Jiang, Ting Wang, Yingsha Yao.
Resources: Feifei Zhou.
Supervision: Dong Xu.
Writing – original draft: Ruo-an Jiang.
Writing – review & editing: Ruo-an Jiang, Ting Wang, Feifei Zhou, Yingsha Yao, Jing He, Dong Xu.
Footnotes
Abbreviations: ADAMTS6 = A disintegrin and metalloproteinase with thrombospondin motifs 6, C3 = complement component 3, ceRNA = competing endogenous RNA, CXCL8 = C-X-C motif chemokine ligand 8, dif-lncRNAs = differentially expressed long non-coding RNAs, dif-miRNAs = differentially expressed microRNAs, dif-mRNAs = differentially expressed mRNAs, ECM = extracellular matrix, EVTs = extravillous trophoblasts, FN1 = fibronectin 1, GEO = Gene Expression Omnibus, KEGG = Kyoto Encyclopedia of Genes and Genomes, lncRNAs = long non-coding RNAs, miRNAs = microRNAs, MMP2 = matrix metalloproteinase-2, PE = preeclampsia, PPI = protein-protein interaction.
How to cite this article: Jiang R, Wang T, Zhou F, Yao Y, He J, Xu D. Bioinformatics-based identification of miRNA-, lncRNA-, and mRNA-associated ceRNA networks and potential biomarkers for preeclampsia. Medicine. 2020;99:45(e22985).
The authors have no funding and conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Gathiram P, Moodley J. Pre-eclampsia: its pathogenesis and pathophysiolgy. Cardiovasc J Afr 2016;27:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Poon LC, Shennan A, Hyett JA, et al. The International Federation of Gynecology and Obstetrics (FIGO) initiative on pre-eclampsia: a pragmatic guide for first-trimester screening and prevention. Int J Gynaecol Obstet 2019;145:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Romero R, Chaiworapongsa T. Preeclampsia: a link between trophoblast dysregulation and an antiangiogenic state. J Clin Invest 2013;123:2775–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Staff AC, Redman CW, Williams D, et al. Pregnancy and long-term maternal cardiovascular health: progress through harmonization of research cohorts and biobanks. Hypertension 2016;67:251–60. [DOI] [PubMed] [Google Scholar]
- [5].Staff AC. The two-stage placental model of preeclampsia: an update. J Reprod Immunol 2019;134-135:1–0. [DOI] [PubMed] [Google Scholar]
- [6].Brown MA, Magee LA, Kenny LC, et al. Hypertensive disorders of pregnancy: ISSHP classification, diagnosis, and management recommendations for international practice. Hypertension 2018;72:24–43. [DOI] [PubMed] [Google Scholar]
- [7].Zhao G, Zhou X, Chen S, et al. Differential expression of microRNAs in decidua-derived mesenchymal stem cells from patients with pre-eclampsia. J Biomed Sci 2014;21:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Selda Demircan S, Mert Kü, Firuzan Ka?ar Dg, et al. VEGF, PIGF and HIF-1α in placentas of early- and late-onset pre-eclamptic patients. Gynecolog Endocrinol 2013;29:797–800. [DOI] [PubMed] [Google Scholar]
- [9].Wa YH, Daniel A, Tim CS, et al. Differential activation of placental unfolded protein response pathways implies heterogeneity in causation of early- and late-onset pre-eclampsia. J Pathol 2015;234:262–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chatterjee P, Weaver LE, Doersch KM, et al. Placental Toll-like receptor 3 and Toll-like receptor 7/8 activation contributes to preeclampsia in humans and mice. PloS One 2012;7:e41884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ospina-Prieto S, Chaiwangyen W, Herrmann J, et al. MicroRNA-141 is upregulated in preeclamptic placentae and regulates trophoblast invasion and intercellular communication. Transl Res 2016;172:61–72. [DOI] [PubMed] [Google Scholar]
- [12].Arun K, Arunkumar G, Bennet D, et al. Comprehensive analysis of aberrantly expressed lncRNAs and construction of ceRNA network in gastric cancer. Oncotarget 2018;9:18386–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet 2000;25:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Smyth GK. limma: Linear Models for Microarray Data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, (eds). Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Statistics for Biology and Health . Springer, New York, NY. 2005. [DOI] [Google Scholar]
- [15].Wu Y, Zhang L, Zhang Y, et al. Bioinformatics analysis to screen for critical genes between survived and non-survived patients with sepsis. Mol Med Rep 2018;18:3737–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2010;39: suppl_1: D561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bandettini WP, Kellman P, Mancini C, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson 2012;14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol 2003;5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Dweep H, Gretz N. miRWalk2. 0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. [DOI] [PubMed] [Google Scholar]
- [23].Pijnenborg R, Vercruysse L, Hanssens M. The uterine spiral arteries in human pregnancy: facts and controversies. Placenta 2006;27:939–58. [DOI] [PubMed] [Google Scholar]
- [24].M Camille H, Rumer KK, Anita K, et al. Maternal and fetal alternative complement pathway activation in early severe preeclampsia. Am J Reprod Immunol 2013;71:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Boij R, Svensson J, Nilssonekdahl K, et al. Biomarkers of coagulation, inflammation, and angiogenesis are independently associated with preeclampsia. Am J Reprod Immunol 2012;94:109–109. [DOI] [PubMed] [Google Scholar]
- [26].Nonaka M, Kimura A. Genomic view of the evolution of the complement system. Immunogenetics 2006;58:701–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Meri S. Self-nonself discrimination by the complement system. Febs Letters 2016;590:2418–34. [DOI] [PubMed] [Google Scholar]
- [28].Mao D, Wu X, Deppong C, et al. Negligible role of antibodies and C5 in pregnancy loss associated exclusively with C3-dependent mechanisms through complement alternative pathway. Immunity 2003;19:813–22. [DOI] [PubMed] [Google Scholar]
- [29].Hoffman MC, Rumer KK, Kramer A, et al. Maternal and fetal alternative complement pathway activation in early severe preeclampsia. Am J Reprod Immunol 2014;71:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Burwick R, Feinberg B. Eculizumab for the treatment of preeclampsia/HELLP syndrome. Placenta 2013;34:201–3. [DOI] [PubMed] [Google Scholar]
- [31].Lokki AI, Heikkinen-Eloranta J, Jarva H, et al. Complement activation and regulation in preeclamptic placenta. Front Immunol 2014;5:312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Velickovic I, Dalloul M, Wong KA, et al. Complement factor B activation in patients with preeclampsia. J Reprod Immunol 2015;109:94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Goldman-Wohl D, Yagel S. Regulation of trophoblast invasion: from normal implantation to pre-eclampsia. Mol Cell Endocrinol 2002;187:233–8. [DOI] [PubMed] [Google Scholar]
- [34].Young Nam K, Dae Shim L, Dae Hoon J, et al. The relationship of the level of circulating antiangiogenic factors to the clinical manifestations of preeclampsia. Prenat Diagn 2010;29:464–70. [DOI] [PubMed] [Google Scholar]
- [35].Liu X, Dai LI, Zhou R. Association between preeclampsia and the CXC chemokine family (Review). Exp Ther Med 2015;9:1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ana Sofia CS, Ananth K. Angiogenic factors in preeclampsia and related disorders. Cold Spring Harb Perspect Med 2012;2:705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jovanović M, Stefanoska I, Radojcić L, et al. Interleukin-8 (CXCL8) stimulates trophoblast cell migration and invasion by increasing levels of matrix metalloproteinase (MMP)2 and MMP9 and integrins alpha5 and beta1. Reproduction 2010;139:789–98. [DOI] [PubMed] [Google Scholar]
- [38].Arenberg DA, Kunkel SL, Polverini PJ, et al. Inhibition of interleukin-8 reduces tumorigenesis of human non-small cell lung cancer in SCID mice. J Clin Invest 1996;97:2792–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sekiguchi K, Klos AM, Hirohashi S, et al. Human tissue fibronectin: expression of different isotypes in the adult and fetal tissues. Biochem Biophys Res Commun 1986;141:1012–7. [DOI] [PubMed] [Google Scholar]
- [40].Virtanen I, Laitinen L, Vartio T. Differential expression of the extra domain-containing form of cellular fibronectin in human placentas at different stages of maturation. Histochemistry 1988;90:25–30. [DOI] [PubMed] [Google Scholar]
- [41].Saleh AA, Bottoms SF, Farag AM, et al. Markers for endothelial injury, clotting and platelet activation in preeclampsia. Arch Gynecol Obstet 1992;251:105–10. [DOI] [PubMed] [Google Scholar]
- [42].Gökdemir İE, Evliyaoğlu ÖBÇ. The role of ADAMTS genes in preeclampsia. v Turk J Obstet Gynecol 2016;13:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kandemir Sİ, Taşkin Iİ, Pektanç G, et al. Altered expression of ADAMTSs and HAPLNs in preeclamptic placenta. Erciyes Med J 2018;40:87–92. [Google Scholar]
- [44].Lee D-C, Romero R, Kim J-S, et al. miR-210 targets iron-sulfur cluster scaffold homologue in human trophoblast cell lines: siderosis of interstitial trophoblasts as a novel pathology of preterm preeclampsia and small-for-gestational-age pregnancies. Am J Pathol 2011;179:590–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ishibashi O, Ohkuchi A, Ali MM, et al. Hydroxysteroid (17-() dehydrogenase 1 is dysregulated by miR-210 and miR-518c that are aberrantly expressed in preeclamptic placentas: a novel marker for predicting preeclampsia. Hypertension 2012;59:265–73. [DOI] [PubMed] [Google Scholar]
- [46].Anton L, Olarerin-George AO, Schwartz N, et al. miR-210 inhibits trophoblast invasion and is a serum biomarker for preeclampsia. Am J Pathol 2013;183:1437–45. [DOI] [PMC free article] [PubMed] [Google Scholar]


