Abstract
The regulatory mechanism of long non-coding RNAs (lncRNAs) in trastuzumab resistance is not well established to date. In this research, we identified differentially expressed lncRNA and investigated its regulatory role in trastuzumab resistance of breast cancer. HiSeq sequencing and quantitative real-time PCR were performed to identify the dysregulated lncRNAs. Mass spectrometry, RNA fluorescence in situ hybridization (RNA-FISH), and immunoprecipitation assays were performed to identify the direct interactions between ZNF649-AS1 and other associated targets, such as polypyrimidine tract binding protein 1 (PTBP1) and autophagy related 5 (ATG5). Our results showed that ZNF649-AS1 was more highly expressed in trastuzumab-resistant cells compared to sensitive cells. Increased expression of ZNF649-AS1 was associated with a poorer response and shorter survival time of breast cancer patients. ZNF649-AS1 was upregulated by H3K27ac modification at the presence of trastuzumab treatment, and knockdown of ZNF649-AS1 reversed trastuzumab resistance via modulating ATG5 expression and autophagy. Mechanically, ZNF649-AS1 was associated with PTBP1 protein, which further promoted the transcription activity of the ATG5 gene. In conclusion, we demonstrated that H3K27ac modification-induced upregulation of ZNF649-AS1 could cause autophagy and trastuzumab resistance through associating with PTBP1 and promoting ATG5 transcription.
Keywords: breast cancer, trastuzumab resistance, autophagy, ZNF649-AS1, ATG5
Graphical Abstract
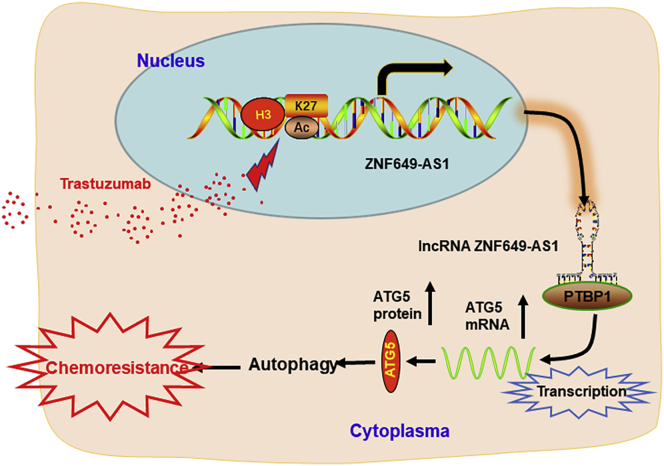
This study reveals that trastuzumab treatment induced lncRNA ZNF649-AS1 via modulation of H3K27ac enrichment at the promoter region. Increased ZNF649-AS1 caused autophagy and trastuzumab resistance via upregulating ATG5 expression through binding with PTBP1. This study provides strong evidence for the essential role of ZNF649-AS1 lncRNA and autophagy in trastuzumab resistance.
Introduction
Human epidermal growth factor receptor 2-positive (HER2+) breast cancer is one of the most common types of breast cancer. HER2 is amplified or overexpressed in 15%–20% of all breast cancer patients.1 Adjuvant trastuzumab currently represents the standard treatment for these patients;2 however, only a fraction of metastatic patients respond to trastuzumab, with approximately 60% developing resistance after the initial response.3 Thus, there is an urgent need to develop effective therapeutic approaches and targets to promote trastuzumab cytotoxicity.
Non-coding RNAs (ncRNAs) have attracted much research interest in recent years due to an accumulation of functional data suggesting that ncRNAs play important roles in cellular proliferation, apoptosis, differentiation, migration, and invasion. ncRNAs have also been found to be dysregulated in several types of human cancers.4 Among the ncRNAs, long ncRNAs (lncRNAs) have many functions, including in tumor progression and in response to therapy.5 The evidence is increasingly indicating that numerous lncRNAs are involved in regulating drug resistance, especially in breast cancer.6 For example, Li et al.7 demonstrated that lncRNA GAS5 suppresses trastuzumab resistance in breast cancer. Zhu et al.8 reported that lncRNA UCA1 induces trastuzumab resistance by sponging miR-18a. Shi et al.9 revealed the critical role of lncRNA ATB in trastuzumab resistance in breast cancer. These studies suggest that lncRNAs may be important regulators in the formation of trastuzumab resistance. However, the specific functions and regulation pathways of these lncRNAs remain to be elucidated.
Autophagy is a highly conserved cellular process that is induced by diverse pathologies and cellular stress conditions, including nutrient deprivation, endoplasmic reticulum stress, and hypoxia.10 Autophagy is known to be involved in the resistance of cancer to multiple chemotherapeutic drugs, including cisplatin,11 paclitaxel,12 and gemcitabine13. Autophagy plays a vital role in regulating cancer chemoresistance, the blocking of which can be used to develop a promising therapeutic strategy for breast cancer treatment.14 By modulating key autophagy-related expression of proteins, lncRNAs play essential roles in regulating autophagy.15 More generally, the contribution of autophagy mediated by lncRNA in cancer therapy and drug resistance has attracted substantial attention in the field of cancer research. In their study, Wang et al.16 reported that lncRNA H19 promotes tamoxifen resistance in breast cancer via autophagy. Therefore, identifying the regulatory relationship between lncRNAs and autophagy in trastuzumab resistance may provide new insights into breast cancer and the development of therapeutic strategies.
In this study, we performed a comprehensive analysis of the lncRNA expression profiles from RNA-sequencing data and confirmed the essential role played by ZNF649-AS1 (19q13.41, ENSG00000268095) in trastuzumab resistance in breast cancer. This finding provides insights into the function and mechanisms underlying the regulation of trastuzumab resistance by ZNF649-AS1.
Results
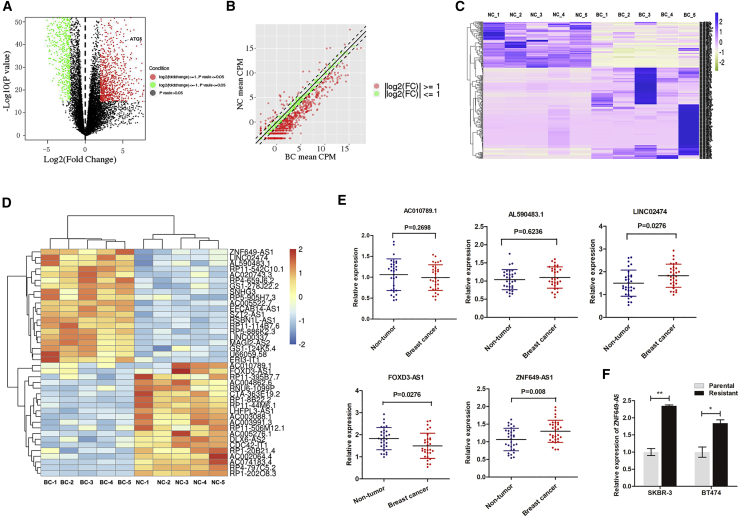
lncRNA ZNF649-AS1 Is Upregulated in Trastuzumab-Resistant Breast Cancer Cells
To identify the lncRNAs that are closely associated with trastuzumab resistance, we performed HiSeq sequencing using the established trastuzumab-resistant cells (Figure S1). Scatter and volcano plots were used to evaluate variations in gene expression between SKBR-3 cells resistant to trastuzumab treatment (SKBR-3-TR cells) and SKBR-3 cells (Figures 1A and 1B). In total, 639 lncRNAs were identified with significant differential expression (p < 0.05 and log2[fold change] > 2) (Figure 1C). To identify the most promising lncRNAs, we focused our analysis on the top 20 most upregulated and 20 downregulated lncRNAs, as shown in the heatmap in Figure 1D. Starting from the lncRNAs with the greatest fold changes, we filtered the appropriate candidate lncRNAs in descending order. Candidates should be plausible for primer design, and only those with consistent levels of expression in cells and tissue samples were selected. Finally, we selected three candidate lncRNAs from the most upregulated group and two from the most downregulated group (Table S2). We tested the expression of the selected genes in 30 paired breast cancer tissues and adjacent non-tumor tissues from HER2+ patients. ZNF649-AS1 (Chr19: 51888025-51900464) was the most upregulated lnRNA, with statistical significance (Figure 1E). Moreover, ZNF649-AS1 was significantly upregulated in SKBR-3-TR cells and BT474 cells resistant to trastuzumab treatment (B7474-TR cells) compared to their parental cells, respectively (Figure 1F).
Figure 1.
lncRNA ZNF649-AS1 Is Upregulated in Trastuzumab-Resistant Breast Cancer Cells
(A and B) Gene expression variations between SKBR-3-TR and SKBR-3 cells were screened by RNA sequencing and presented as a scatterplot (A) and volcano plot (B). NC, negative control, BC, breast cancer. (C) Heatmap was used to show the 639 lncRNAs with significant differential expression (p < 0.05 and log2[fold change] > 2). (D) The top 20 most upregulated and 20 downregulated lncRNAs and shown in a heatmap. (E) The expression levels of AC010789.1, AL590483.1, Linc02474, FOXD3-AS1, and ZNF649-AS1 were detected via quantitative real-time PCR in 30 paired breast cancer tissues and adjacent non-tumor tissues. (F) Quantitative real-time PCR was used for the detection of ZNF649-AS1 in trastuzumab-resistant and parental cells. ∗p < 0.05, ∗∗p < 0.01.
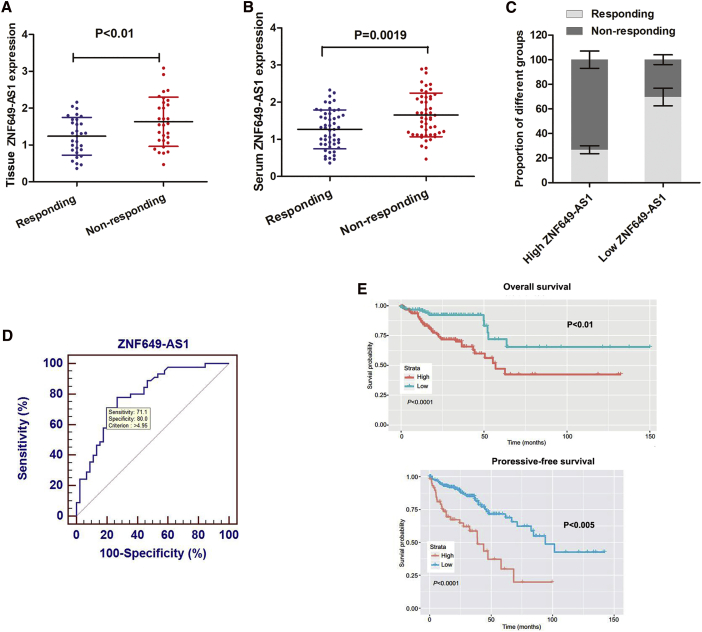
lncRNA ZNF649-AS1 Is Associated with Poor Response to Trastuzumab Treatment
To verify whether ZNF649-AS1 is associated with trastuzumab treatment, we detected its expression in 60 breast cancer tissues from patients receiving trastuzumab treatment (30 responding and 30 non-responding patients). As shown in Figure 2A, ZNF649-AS1 is upregulated in non-responding patients compared to responding patients. Predictive biomarkers should be blood based, as blood samples can be easily obtained and allow for the monitoring of cancer progression. Thus, 90 serum samples from patients receiving trastuzumab treatment, including 45 responding and 45 non-responding patients, were used to determine the expression of ZNF649-AS1. ZNF649-AS1 was found to be dramatically upregulated in non-responding patients compared to responding patients (Figure 2B). Moreover, the receiver operator characteristic (ROC) curve showed a relatively high diagnostic value of ZNF649-AS1 expression, with an area under the curve (AUC) of 0.794, and a diagnostic sensitivity and specificity of 71.1% and 80.0%, respectively (Figure 2C). By stratifying patients into high or low ZNF649-AS1 expression groups, using the stratification criteria (1.95) obtained from the ROC curve, we found that the proportion of patients responding to trastuzumab therapy was much lower in the high ZNF649-AS1-expressing group than in low ZNF649-AS1-expressing group (Figure 2D). More importantly, Kaplan-Meier survival analysis indicated that high ZNF649-AS1 expression was associated with poor overall survival and progression-free survival (Figure 2E). Cox regression analysis indicated that levels of ZNF649-AS1 in the serum and TNM stage served as independent predictive factors for overall survival (OS) and progression-free survival (PFS) (Table S3).
Figure 2.
lncRNA ZNF649-AS1 Is Associated with Poor Response to Trastuzumab Treatment
(A) Quantitative real-time PCR showed that ZNF649-AS1 was significantly upregulated in 30 trastuzumab-resistant tissues compared with 30 responding tissues. (B) Serum ZNF649-AS1 was determined in 45 resistant and 45 responding patients. (C) Statistical analysis showed that the proportion of responding patients in the high-ZNF649-AS1 group was significantly lower in than that in low-ZNF649-AS1 group. (D) ROC curve was established to evaluate the potential of ZNF649-AS1 expression in discriminating responding and non-responding patients. (E) Kaplan-Meier survival analysis proved that high ZNF649-AS1 expression is associated with poorer overall survival (upper panel) and progressive-free survival (lower panel).
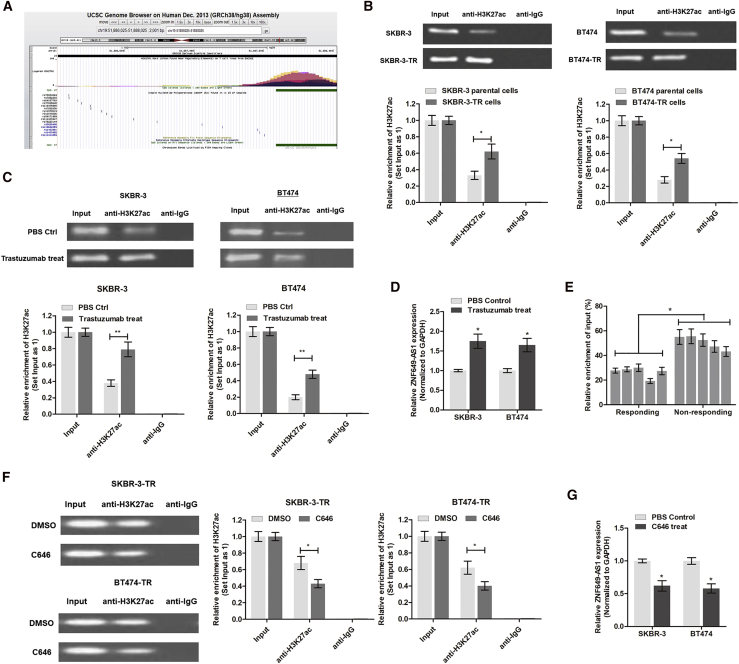
Trastuzumab Increases ZNF649-AS1 Expression by Inducing H3K27 Acetylation
Next, we investigated the underlying mechanism of ZNF649-AS1 upregulation in trastuzumab-resistant cells. By analyzing the transcriptional modification regions on the UCSC Genome Browser (http://genome.ucsc.edu/), we found that there was an enriched H3K27ac binding area upstream of the ZNF649-AS1 promoter region in seven cell lines (Figure 3A). We then performed a chromatin immunoprecipitation (ChIP) assay using anti-H3K27ac antibody and found that H3K27ac was enriched at the ZNF649-AS1 promoter region. Moreover, the enrichment level of H3K27ac was significantly increased in SKBR-3-TR and BT474-TR cells compared to their parental cells, respectively (Figure 3B). To further confirm the effects of trastuzumab treatment, we treated SKBR-3 and BT474 cells with trastuzumab for 48 h. As shown in Figures 3C and 3D, the levels of H3K27ac enrichment and ZNF649-AS1 were increased in trastuzumab-treated cells compared to the control cells. Consistently, H3K27ac enrichment was also increased in breast cancer tissues from trastuzumab-resistant patients compared to those from chemo-sensitive patients (Figure 3E). We then treated cells with C646, a well-known acetyltransferase inhibitor, and found that H3K27ac enrichment and ZNF649-AS1 expression were dramatically suppressed (Figures 3F and 3G). Collectively, our results suggest that trastuzumab treatment increases the levels of ZNF649-AS1 by inducing H3K27ac enrichment at the promoter region. However, further study is needed to elucidate a detailed regulatory mechanism.
Figure 3.
Trastuzumab Increases ZNF649-AS1 Expression by Inducing H3K27
(A) The H3K27ac enriched area at the promoter of ZNF649-AS1 was predicted online at the UCSC Genome Browser (http://genome.ucsc.edu). (B) RIP assay using anti-H3K27ac antibody showed that H3K27ac was enriched at the promoter region of ZNF649-AS1. Moreover, the enriched level was significantly increased in trastuzumab-resistant cells in contrast to parental sensitive cells. (C) RIP assay showed that trastuzumab treatment increased the enrichment of H3K27ac at the ZNF649-AS1 promoter in SKBR-3 and BT474 cells. (D) Quantitative real-time PCR was performed to measure the differential expression of ZNF649-AS1 in trastuzumab-treated cells and controlled cells. (E) ChIP sequencing (ChIP-seq) was performed in tissues from responding and non-responding patients. (F) The acetyltransferase inhibitor C646 significantly suppressed the binding level of H3K27ac. (G) C646 treatment significantly inhibited the expression of ZNF649-AS1 in SKBR-3 and BT474 cells. ∗p < 0.05, ∗∗p < 0.01.
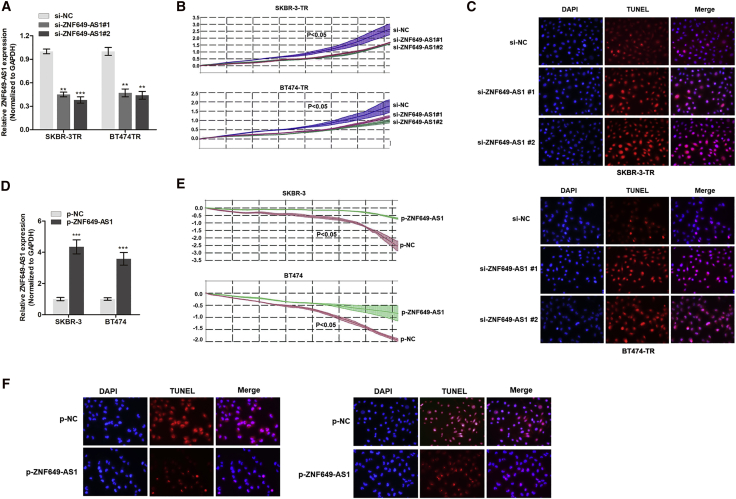
Silencing lncRNA ZNF649-AS1 Reverses Trastuzumab Resistance of Breast Cancer Cells
To identify the role of ZNF649-AS1 in trastuzumab resistance, we silenced ZNF649-AS1 expression using specific inhibitors. As shown in Figure 4A, we observed a significant reduction in the expression of ZNF649-AS1 in cells transfected with small interfering RNA (siRNA) against (si-)ZNF649-AS1#1 or si-ZNF649-AS1#2 compared to the control cells. Real-time cellular analysis (RTCA) revealed that the knockdown of ZNF649-AS1 significantly suppressed cell viability upon trastuzumab treatment (Figure 4B). Flow cytometry showed that the silencing of ZNF649-AS1 did not affect the cell cycle mode (data not shown), suggesting that ZNF649-AS1 may influence cell viability independent of its regulation of cell proliferation or the cell cycle. Thereafter, we assessed its role in apoptosis. The results of a TUNEL (terminal deoxynucleotidyltransferase-mediated deoxyuridine triphosphate nick end labeling) assay showed that cell apoptosis was increased after ZNF649-AS1 knockdown (Figure 4C), which was consistent with the flow cytometry results (Figure S1).
Figure 4.
Silencing lncRNA ZNF649-AS1 Reverses Trastuzumab Resistance of Breast Cancer Cells
(A) ZNF649-AS1 was silenced by two specific small interfering oligonucleotides in SKBR-3-TR and BT474-TR cells. (B) RTCA of cell viability in breast cancer cells silenced with ZNF649-AS1. (C) TUNEL assay showed that silence of ZNF649-AS1 promoted cell apoptosis compared to control cells. (D) Quantitative real-time PCR verified the upregulation of ZNF649-AS1 after transfection of ZNF649-AS1-loaded plasmids in SKBR-3 and BT474 cells. (E and F) Overexpression of ZNF649-AS1 increased cell viability (E) and suppressed apoptosis (F) in cells treated with trastuzumab. ∗∗p < 0.01, ∗∗∗p < 0.001, compared to control groups.
Next, we investigated the influence of ZNF649-AS1 overexpression in trastuzumab resistance in SKBR-3 and BT474 parental cells. As shown in Figure 4D, ZNF649-AS1 was overexpressed in these cells after transfection of ZNF649-AS1-sequenced plasmids. By treating SKBR-3 and BT474 cells with trastuzumab (3 μg/mL for 48 h), we found an enhanced ZNF649-AS1-induced resistance to trastuzumab compared to the control (Figure 4E). Furthermore, ZNF649-AS1 was found to suppress the apoptosis of SKBR-3 and BT474 cells (Figure 4F).
lncRNA ZNF649-AS1 Is Essential for Trastuzumab-Induced Autophagy
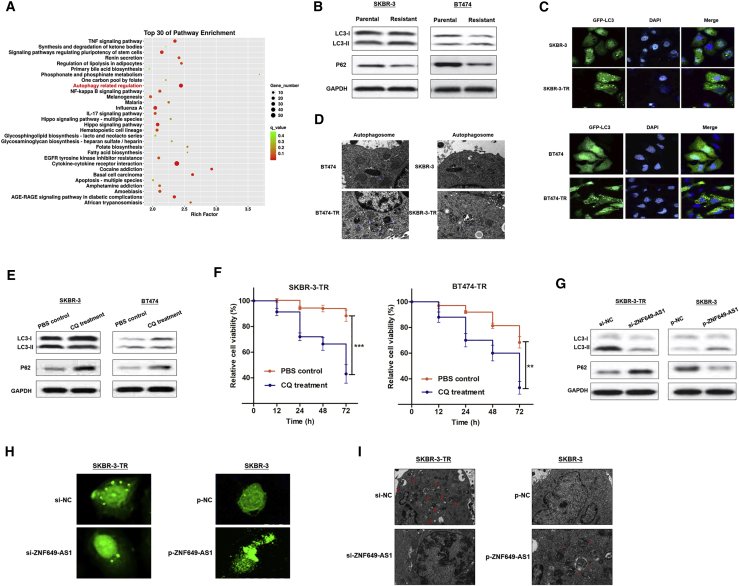
Using Gene Ontology (GO) analysis, we hypothesized that autophagy may be involved in the function of ZNF649-AS1 (Figure 5A). To validate the hypothesis, we measured the levels of LC3 and p62, which are widely used as autophagy markers.17,18 Remarkably, trastuzumab-resistant cells showed higher LC3-II and lower p62 protein levels than did the corresponding parental cells, suggesting that autophagic flux was induced when chemoresistance occurred (Figure 5B). Consistently, trastuzumab-resistant cells showed an increased formation of LC3 puncta (Figure 5C) and autophagosomes (Figure 5D). To verify whether autophagy is essential for trastuzumab resistance, we treated trastuzumab-resistant cells with chloroquine (CQ), a well-known autophagy inhibitor.19 Our results showed that CQ treatment suppressed autophagy and reversed the chemoresistance status (Figures 5E and 5F).
Figure 5.
Trastuzumab Treatment Induced Autophagy via lncRNA ZNF649-AS1
(A) GO pathway analysis showed that ZNF649-AS1 is associated with autophagy regulation. (B) Western blot showed that trastuzumab-resistant cells induced increased the proportion of LC3II/LC3I and decreased p62 expression. (C and D) Trastuzumab-resistant cells expressed increased LC3 puncta (C) and autophagosomes formation (D). (E) CQ treatment suppressed LC3II/LC3I proportion and increased p62 expression. (F) CCK-8 assay revealed that CQ treatment suppressed cell viability. (G) Western blot showed that knockdown of ZNF649-AS1 suppressed autophagy activity. (H and I) GFP staining suggested that ZNF649-AS1 positively regulated LC3 puncta (H) and autophagosomes formation (I). ∗∗p < 0.01, ∗∗∗p < 0.001.
Next, we investigated the role of ZNF649-AS1 in autophagic activity. The knockdown of ZNF649-AS1 resulted in reduced levels of LC3-II and enhanced levels of p62 in SKBR-3-TR. In contrast, in SKBR-3 cells with ZNF649-AS1 overexpression, the level of LC3-II was increased, while the level of p62 was decreased (Figure 5G). Silencing of ZNF649-AS1 in SKBR-3-TR cells reduced the numbers of LC3 puncta and blocked the formation of autophagosomes, whereas the overexpression of ZNF649-AS1 led to an increase in the number of LC3 puncta and autophagosomes in SKBR-3 cells (Figures 5H and 5I). Taken together, our results indicate that trastuzumab resistance induces enhanced autophagy activity, and that the inhibition of ZNF649-AS1 could reverse autophagy, thereby inducing chemosensitivity.
ZNF649-AS1 Induces Trastuzumab Resistance and Autophagy by Upregulating ATG5
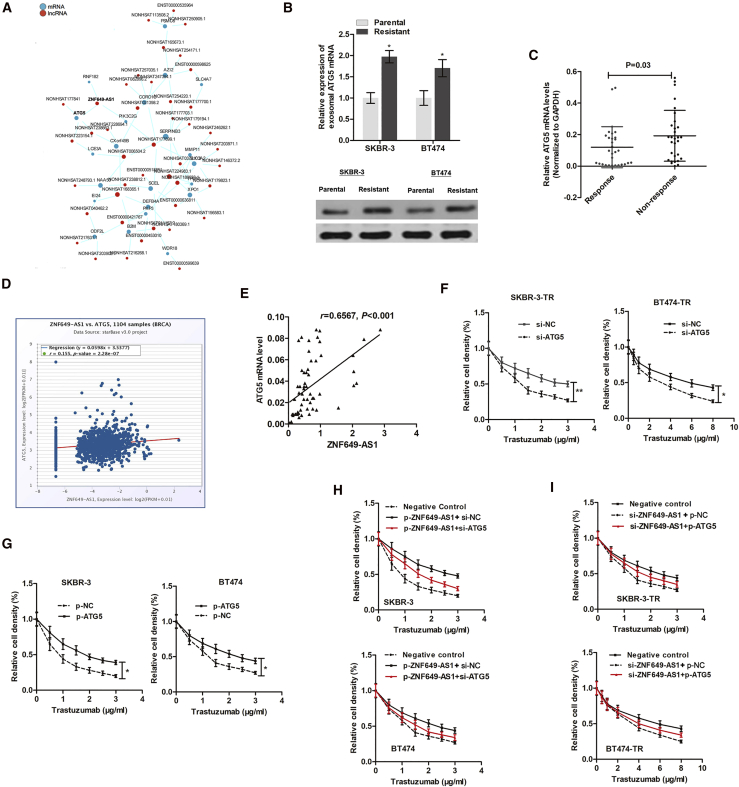
To find the genes responsible for ZNF649-AS1-induced autophagy and trastuzumab resistance, we analyzed the co-expression network of ZNF649-AS1 and target mRNAs (Figure 6A). Interestingly, we identified an autophagy-related mRNA, autophagy related 5 (ATG5), that was targeted by ZNF649-AS1. ATG5 was upregulated in SKBR-3-TR and BT474-TR cells compared to parental cells at both the transcript and protein levels (Figure 6B). In addition, ATG5 was upregulated in the tissues of patients showing resistance to trastuzumab compared to patients responding to therapy (Figure 6C). According to the data obtained from the Pan-Cancer Atlas, ZNF649-AS1 was slightly positively associated with ATG5 expression (Figure 6D). This correlation was further confirmed in 60 tissues from HER2+ patients receiving trastuzumab treatment (Figure 6E). Next, we sought to determine whether ATG5 is essential for ZNF649-AS1-regulated trastuzumab resistance. By performing a series of gain- or loss-of-function experiments, we found that the inhibition of ATG5 attenuated chemoresistance of trastuzumab-resistant cells (Figure 6F), while increasing ATG5 promoted trastuzumab resistance in parental cells (Figure 6G). Moreover, the suppression of ATG5 abolished the trastuzumab resistance induced by ZNF649-AS1 in sensitive cells (Figure 6H), whereas enhanced ATG5 abrogated the ZNF649-AS1 knockdown-induced chemosensitivity of resistant cells (Figure 6I).
Figure 6.
ATG5 Is Essential for ZNF649-AS1-Induced Trastuzumab Resistance and Autophagy
(A) The co-expression network of ZNF649-AS1 and target mRNAs was analyzed by using the RNA sequencing data. (B) ATG5 was upregulated in trastuzumab-resistant cells in both transcript and protein levels. (C) ATG5 mRNA expression was increased in 30 non-responding tissues and 30 responding tissues. (D) ZNF649-AS1 was slightly positively associated with ATG5 expression according to The Cancer Genome Atlas (TCGA) database. (E) The positive correlation was further validated in 60 tissue samples. (F and G) Inhibition of ATG5 attenuated chemoresistance of trastuzumab-resistant cells (F) while it increased ATG5-promoted trastuzumab resistance in parental cells (G). (H and I) Suppression of ATG5 abolished the trastuzumab resistance induced by ZNF649-AS1 in sensitive cells (H), whereas enhanced ATG5 abrogated the ZNF649-AS1 knockdown-induced chemosensitivity of resistant cells (I). ∗p < 0.05.
ZNF649-AS1 Activates the Transcription of ATG5 by Recruiting PTBP1
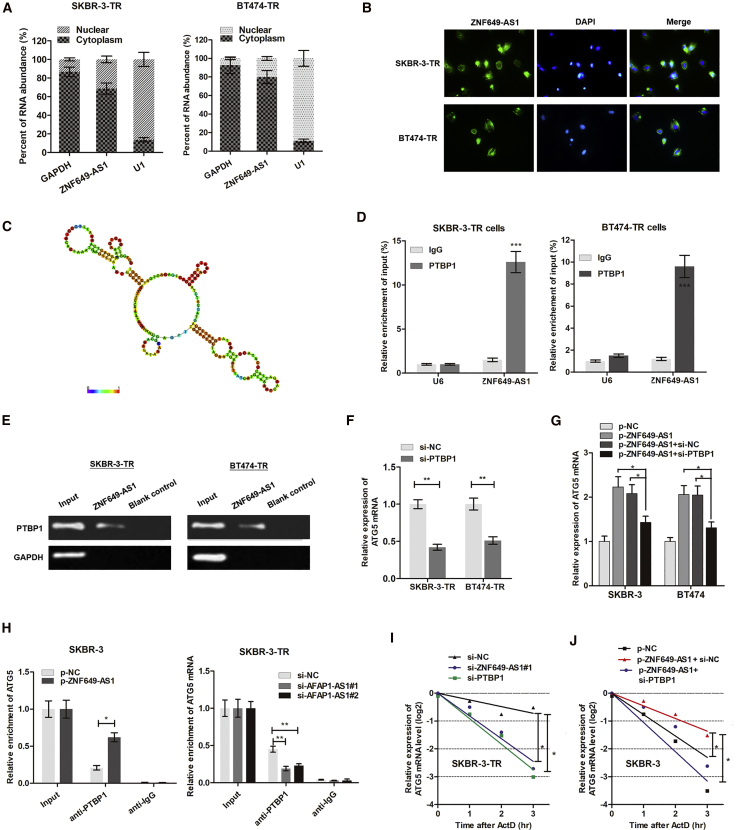
We used cellular fractionation polymerase chain reaction (PCR) to find that ZNF649-AS1 was mainly distributed in the cytoplasm of breast cancer cells (Figure 7A). RNA fluorescence in situ hybridization (RNA-FISH) confirmed this conclusion (Figure 7B), suggesting that ZNF649-AS1 regulates downstream pathways at the post-transcriptional level. Based on an online minimum free energy (MFE) evaluation (http://rna.tbi.univie.ac.at/), we predicted that the ZNF649-AS1 transcript at the 981–1190 nt loci formed stem-loop structures (Figure 7C), which is essential for the association with targeted RNA-binding proteins. To verify the proteins associated with ZNF649-AS1, RNA pull-down followed by mass spectrometry was performed. As a result, several potential ZNF649-AS1-interacting proteins were identified (Table S4), among which we found polypyrimidine tract binding protein 1 (PTBP1), which may participate in localization, translation initiation, and mRNA stability. By designing a ZNF649-AS1 probe and performing an RNA pull-down assay, we found that PTBP1 protein was enriched by ZNF649-AS1 (Figure 7D). Moreover, an RNA immunoprecipitation (RIP) assay verified that ZNF649-AS1 was precipitated by PTBP1 antibody (Figure 7E). These findings suggest that ZNF649-AS1 is associated with PTBP1 protein to play critical bio-functions.
Figure 7.
ZNF649-AS1 Activates the Transcription of ATG5 via Recruiting PTBP1
(A) Cellular fractionation PCR showed that ZNF649-AS1 was mainly distributed in the cytoplasm section. (B) RNA fluorescence in situ hybridization (RNA-FISH) verified that ZNF649-AS1 was mainly distributed in cytoplasm of SKBR-3-TR and BT474-TR cells. (C) Prediction of 981–1190 nt ZNF649-AS1 structure was based on minimum free energy (MFE) and partition function (http://rna.tbi.univie.ac.at/). (D) RNA pull-down assay showed that PTBP1 protein was directly enriched by ZNF649-AS1 probe in breast cancer cells. (E) RIP was performed using anti-PTBP1 and control IgG antibodies, followed by quantitative real-time PCR to examine the enrichment of ZNF649-AS1 and U6. U6 served as a negative control. (F) ATG5 mRNA was silenced by knockdown of PTBP1. (G) Quantitative real-time PCR showed that ZNF649-AS1 upregulated ATG5 expression; however, this upregulation was significantly reversed by co-transfection of si-PTBP1. (H) The endogenous PTBP1 binding to ATG5 mRNA was increased by overexpression of ZNF649-AS1 in SKBR-3 cells, while it was decreased by knockdown of ZNF649-AS1 in SKBR-3-TR cells. (I and J) SKBR-3-TR (I) and SKBR-3 (J) cells silenced or overexpressed with ZNF649-AS1 were cultured with 2 μg/mL actinomycin D (ActD) for 1–3 h, then subjected to qPCR analysis of ATG5. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
We sought to prove that ZNF649-AS1 increases ATG5 mRNA stability by binding with PTBP1. Figure 7F shows that the silencing of PTBP1 downregulated the ATG5 mRNA levels. Moreover, the silencing of PTBP1 abrogated the ZNF649-AS1-induced increase of ATG5 mRNA (Figure 7G). To determine the role of PTBP1, we performed a RIP assay. The overexpression of ZNF649-AS1 increased endogenous PTBP1 binding to ATG5 in SKBR-3 cells, while the knockdown of ZNF649-AS1 exerted the opposite effect in SKBR-3-TR cells (Figure 7H). In addition, we treated trastuzumab-resistant cells with actinomycin D (ActD), which is widely used for the experimental blockage of the transcriptional process, and found that silencing ZNF649-AS1 or PTBP1 caused an increased degradation of ATG5 mRNA (Figure 7I). In addition, the overexpression of ZNF649-AS1 increased its half-life, while the knockdown of PTBP1 reduced the half-life back to normal levels (Figure 7J). As such, our results proved that ZNF649-AS1-bound PTBP1 increased the expression levels of ATG5 by enhancing the stability of mRNA.
Knockdown of ZNF649-AS1 Reverses Trastuzumab Resistance and Autophagy In Vivo
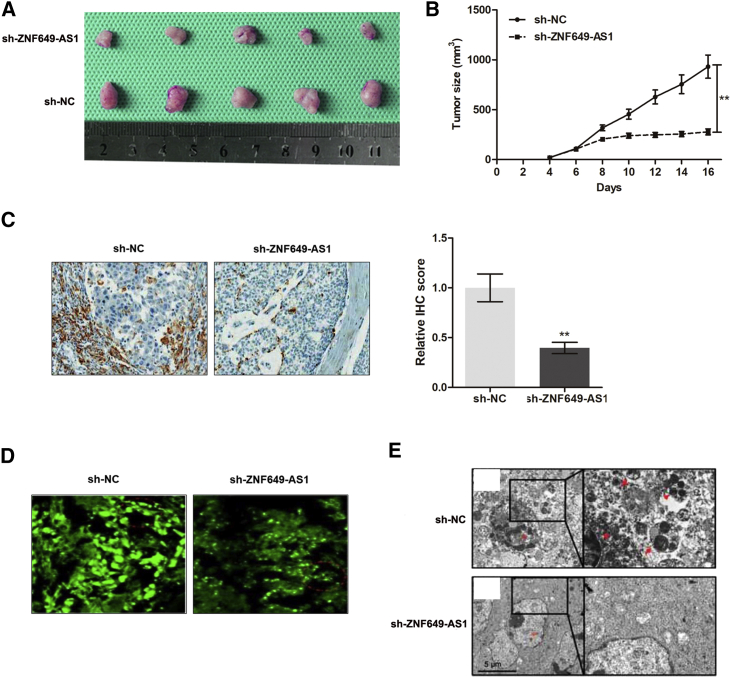
Based on our in vitro observations, we sought to validate our data by establishing xenografts in BALB/c nude mice models. We generated xenografts by subcutaneously injecting SKBR-3-TR cells stably infected with short hairpin RNA (shRNA) targeting (sh-)ZNF649-AS1 or sh-negative control (NC), followed by intraperitoneal treatment with trastuzumab (described in Materials and Methods). By stripping the tumors from the nude mice, we established xenografts for the different groups after 20 days of treatment (Figure 8A). The tumors that formed in the sh-ZNF649-AS1 group were found to be substantially smaller than those in the sh-NC group (Figure 8B). By conducting an immunohistochemistry (IHC) assay, we found that the tumors formed from sh-ZNF649-AS1-infected cells exhibited decreased expression levels of ATG5 compared to tumors formed from the control cells (Figure 8C). Moreover, lower levels of LC3 puncta (Figure 8D) and autophagosome (Figure 8E) formation were observed in the mice from the sh-ZNF649-AS1 group.
Figure 8.
Knockdown of ZNF649-AS1 Reverses Trastuzumab Resistance and Autophagy In Vivo
(A) Representative images of the xenograft after intraperitoneal injection with once every two days 3 mg/kg trastuzumab treatment for 3 weeks in sh-NC or sh-ZNF649-AS1 cell groups. (B) Tumor sizes were calculated every 2 days. (C) Immunohistochemistry analysis revealed that the tumors developed from sh-ZNF649-AS1 cells displayed lower ATG5 staining than did the sh-NC group. (D) GFP staining showed that LC3 puncta was suppressed in ZNF649-AS1-silenced xenograft tissues compared to controlled tissues. (E) The formed autophagosomes were significantly decreased in tumors developed from ZNF649-AS1-silenced cells compared to controlled tissues. ∗∗p < 0.01.
Discussion
It has now become widely accepted that mammalian genomes encode numerous lncRNAs.20 The dysregulation of some of these lncRNAs has been found in various types of cancers at various stages, including cancer initiation, progression, and chemoresistance, in breast cancer.21 However, the functions and mechanisms of these lncRNAs in resistance to treatment, such as trastuzumab resistance, are not yet fully understood. Based on our lncRNA sequencing data, we identified an antisense lncRNA, ZNF649-AS1, which was highly expressed in trastuzumab-resistant cells compared to the corresponding parental cells. Gain- and loss-of-function assays showed that the knockdown of ZNF649-AS1 reverses trastuzumab resistance. Experimental analysis revealed that the silencing of ZNF649-AS1 suppressed autophagy, thereby reversing trastuzumab resistance of breast cancer. ATG5 was verified as a functional target of ZNF649-AS1 during autophagy and chemoresistance. Mechanistically, ZNF649-AS1 enhanced the translation of ATG5 mRNA by binding with PTBP1, thereby inducing autophagy and trastuzumab resistance (Figure 9).
Figure 9.
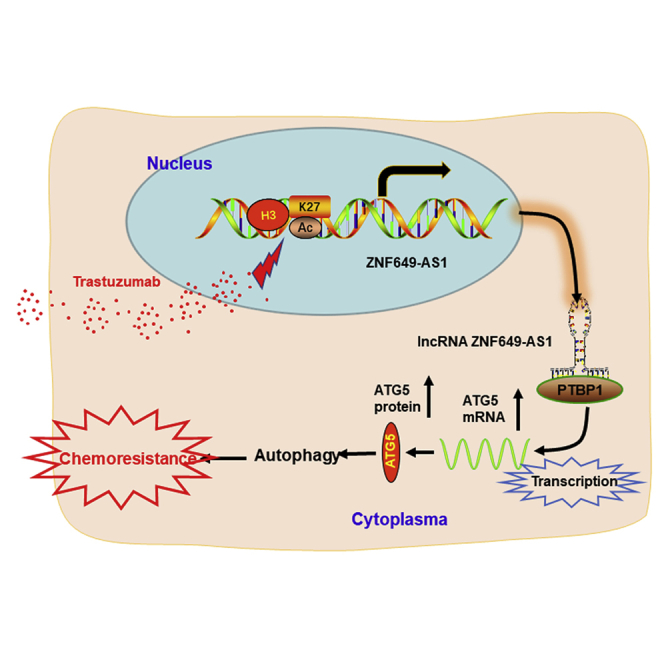
A Scheme of the Proposed Mechanisms
Trastuzumab treatment increases the H3K27ac enrichment at the ZNF649-AS1 promoter region, inducing upregulation of ZNF649-AS1, which thereby increases ATG5 expression by guiding PTBP1 to activate its transcription. Then, increased ATG5 protein induced autophagy and trastuzumab resistance.
More effective therapies using trastuzumab emtansine are now available.22 Unfortunately, resistance to trastuzumab and other therapies that target the HER2 pathway is still found in cancer patients, who cannot benefit from a trastuzumab-based drug regimen. In addition, the identification of patients who could benefit from HER2-directed therapies is greatly hindered by a lack of biomarkers for predicting the therapeutic response of patients.23 This is despite the extensive retrospective analysis of phase III clinical trial data for the potential roles of related receptor tyrosine kinases (RTKs) (epidermal growth factor receptor [EGFR], HER3, and insulin growth factor 1 receptor [IGF-1R]), ligands (EGF and transforming growth factor α [TGF-α]), or phosphatidylinositol 3-kinase (PI3K) pathway alterations. The expression of HER2 itself is currently the only biomarker available to guide treatment decisions within the HER2+ patient cohort.24 As a result, there are currently numerous studies focusing on identifying potential pathways and predictive biomarkers for these patients.25,26
Functionally, the dysregulation of lncRNAs is closely associated with aberrant biological behaviors of human cancers.27 Also, note that some lncRNAs oriented in the antisense direction with respect to a protein coding loci in the opposite strand usually act as regulators in many pathological processes.28 Previous studies have shown that natural antisense transcripts play critical roles in various physiological and pathological processes by regulating gene promoter activation and transcription.29 The cDNA of ZNF649 is 3,176 bp and encodes a protein of 505 aa in the nuclei. It has been reported that ZNF649 is expressed in most of the examined human adult and embryonic tissues.30 ZNF649 is a transcription suppressor when fused to GAL-4 DNA-binding domain and co-transfected with VP-16. In this study, we identified its antisense transcript, ZNF649-AS1, as an upregulated lncRNA during trastuzumab resistance. By analyzing the transcriptional modification region using the UCSC Genome Browser (http://genome.ucsc.edu/), we found an enriched H3K27ac binding area upstream of the ZNF649-AS1 promoter region. Moreover, we found that ZNF649-AS1 was transcriptionally activated via trastuzumab treatment-induced acetylation. Although the role of ZNF649-AS1 in cancer has not yet been elucidated, our findings confirm that its function in trastuzumab resistance is meaningful.
Autophagy is known to be involved in the maintenance of tumor cell survival under a variety of adverse conditions, including nutrient deficiency, chemotherapy, and radiation treatment.31 Accumulating evidence suggests that the regulation of autophagic activity could enhance the action of many antitumor agents, including oxaliplatin, cisplatin, doxorubicin, and 5-fluorouracil (5-FU). Thus, autophagy has been proposed as a potential drug target to reverse drug resistance. Previous reports have shown that a series of lncRNAs regulate drug resistance by modulating autophagy.32,33 Notably, we found that trastuzumab-resistant cells showed increased levels of autophagic activity compared to their corresponding parental cells, and further confirmed ZNF649-AS1 as a vital autophagy-regulating lncRNA by inducing autophagy to enhance trastuzumab resistance. The molecular mechanism of autophagy involves several conserved autophagy-related genes (ATGs), which have multiple functions in various physiological contexts. Among these genes, ATG5 protein conjugated with ATG12 and ATG8 (LC3) has been previously found to be involved in the early stages of autophagosome formation.34 ATG5 is important for development, cell differentiation, and homeostasis maintenance, including early neonatal starvation.35 In our study, we demonstrated that ZNF649-AS1 regulates the expression of ATG5 at the post-transcriptional level, indicating the great value of the ZNF49-AS1-ATG5 regulatory pathway in autophagy and trastuzumab resistance.
To determine the underlying mechanism by which ZNF649-AS1 regulates the ATG5 protein levels, we screened ZNF649-AS1-interacting proteins and identified PTBP1. PTB was discovered as a splicing regulator, perhaps because of its large number of targets and its efficient crosslinking to RNA with short-wave UV light.36 The PTB locus on human chromosome 19 (PTBP1 gene) produces several spliced isoforms. In the present study, we found that PTBP1 was associated with ZNF649-AS1 and may act as an adaptor protein that cooperates with PTBP1 to bind to the ATG5 gene. By performing a RIP assay, we revealed that PTBP1 interacts directly with ATG5 mRNA, and that ZNF649-AS1 increased this association level. Moreover, ZNF649-AS1 showed no effect on the ATG5 protein stability, which further supported our hypothesis.
Recent studies have revealed the therapeutic potential of lncRNAs for use in cancer treatment, designing specific silencing molecular or overexpression vectors to knock down or upregulate specific oncogenic lncRNAs.37 Additionally, silencing nucleic acids against specific molecular targets may serve as new-generation therapeutic drugs to overcome an existing resistance.38 Therefore, the inhibition of trastuzumab resistance via the precise control of key genes represents a potential therapeutic method.
We would point out several limitations of this study. First, HiSeq sequencing analysis was only performed for SKBR-3-TR/SKBR-3 cells. In future, it may be more powerful if BT474-TR/BT474 cells are included in the sequencing analysis. However, we confirmed the upregulation of ZNF649-AS1 in both cell lines during the validation phase. Second, although we preliminarily proved the role of ZNF649-AS1/PTBP1/ATG5 in autophagy and trastuzumab resistance, the precise regulatory model and interactions with other cellular factors in the tumor microenvironment will require further study to be fully elucidated.
In conclusion, we found that ZNF649-AS1 plays a crucial role in trastuzumab resistance in breast cancer by inducing the direct suppression of ATG5 expression, which results in the inhibition of autophagy. Our results provide insight into ncRNA-regulated therapeutic effects in cancer therapy and a basis for the development of more efficient strategies to reverse trastuzumab resistance in patients with breast cancer.
Materials and Methods
Patient Samples
Tissue samples from 90 HER2+ patients who had previously received trastuzumab treatment were enrolled in this study. Another independent cohort of serum samples obtained from nine HER2+ patients was included to investigate the predictive role of serum ZNF649-AS1. Cancerous tissues were collected from January 2012 to August 2014 and snap-frozen in liquid nitrogen at ‒80°C. Patients who had previously received radiotherapy and chemotherapy before surgical treatment were excluded. For the remaining patients, their general clinical information and detailed pathological records were collected. Written informed consent was obtained from all patients. Our study protocol was approved by the Research Scientific Ethics Committee of The First Affiliated Hospital of Zhengzhou University and Hainan General Hospital. All participants signed informed consent prior to using the tissues and serum samples for scientific research. The overall survival rates of the enrolled patients were followed up over a median period of 65 months. OS was calculated from the date of surgery to the date of mortality or the final follow-up. PFS was calculated from the date of surgery to the date of first recurrence or the final follow-up.
Cell Culture and Treatment
Human HER2+ breast cancer cell lines SKBR-3 and BT474 were purchased from American Type Culture Collection (Manassas, VA, USA) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies, Grand Island, NY, USA) in humidified air at 37°C with 5% CO2. The cell lines were authenticated by short tandem repeat (STR) profiling. Trastuzumab (Herceptin) was purchased from Roche (Basel, Switzerland) and used by dissolving in phosphate-buffered saline (PBS). SKBR-3-TR and BT474-TR cells were obtained by establishing xenografts followed by four courses of trastuzumab treatment, as previously described.39
Vector Construction and Cell Transfection
si-PTBP1 and si-ZNF649-AS1#1 and si-ZNF649-AS1#2 were synthesized and purchased from GenePharma (Shanghai, China). Negative control siRNA was purchased from Invitrogen (catalog no. 12935-110; Carlsbad, CA, USA). ZNF549-AS1 cDNA or negative control cDNA was enlarged and cloned into a pcDNA3.1 expression vector, denoted as p-ZNF649-AS1 and p-NC. sh-ZNF649-AS1 or sh-NC was synthesized and packaged into lentivirus by GeneChem (Shanghai, China). The transfection of sh-ZNF649-AS1 or sh-NC into human HEK293T cells with lentiviral packaging vectors was performed to yield high-titer lentiviruses, also manufactured and verified by GeneChem. Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was used for in vitro transfection with a final concentration of 100 nM according to the manufacturer’s instructions. For stable infections, SKBR-3-TR and BT474-TR cells were transduced with sh-ZNF649-AS1 or sh-NC lentiviruses in the presence of Polybrene (10 μg/mL) (MilliporeSigma, Shanghai, China) at a multiplicity of infection (MOI) of 30 for 72 h. A selection procedure was then carried out for 5–7 days using blasticidin (15 μg/mL) (MilliporeSigma, Shanghai, China). Thereafter, clear-edge healthy multicell colonies were collected, re-plated in six-well plates containing fresh culture medium without blasticidin, and allowed to proliferate for three to five passages. The sequences of the oligonucleotides are provided in Table S1.
Quantitative Real-Time PCR Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and treated with DNase I (Thermo Fisher Scientific, Waltham, MA, USA), followed by conversion into cDNA using random primers and RevertAid Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Thermo Fisher Scientific). The cDNA templates were amplified by real-time PCR using the SYBR Green PCR kit (TaKaRa, Tokyo, Japan). Quantitative real-time PCR was conducted on an Applied Biosystems 7500 sequence detection system (Applied Biosystems, Thermo Fisher Scientific) using the following thermal cycling conditions: 95°C for 30 s, followed by 45 cycles of 95°C for 5 s and 60°C for 30 s. The final extension was 72°C for 5 min. Details on the specific primers used are provided in Table S1. The experiments were repeated three times and the relative expression of RNA was calculated using the 2−ΔΔCt method.40
Expression Profile Analysis of lncRNAs
SKBR-3-TR and SKBR-3 cells were used to screen the differentially expressed lncRNAs. Five samples were enrolled for both groups. The extraction of total RNA, sequencing platform, data analysis, and the construction of the cDNA library were performed as previously described.39 For sequencing, the raw reads were preprocessed by filtering out rRNA reads, sequencing adapters, short-fragment reads, and other low-quality reads. TopHat v2.0.9 was used to map the cleaned reads to the human GRCh38 reference genome with two mismatches. For known gene models, Cufflinks v2.1.1 was run with a reference annotation to generate fragments per kilobase of transcript per million mapped reads (FPKM) values after genome mapping. Cuffdiff was used to evaluate differentially expressed genes. The differentially expressed genes were selected using the following filter criteria: p ≤ 0.05 and fold change ≥ 2.
Cell Viability Analysis by RTCA and CCK-8 Assays
The cell proliferation ability was evaluated by RTCA on xCELLigence (ACEA Biosciences, CA, USA), according to the manufacturer’s instructions. Each well of E-plates was filled with 50 μL of culture media, and background impedance measurement was performed for 10 min. Cell suspensions at a cell density of 1 × 104 cells/well were then seeded onto the E-plate and incubated for 30 min at room temperature to allow for an initial cell adhesion at the bottom of each well, followed by incubation at 37°C. RTCA software was used for the analysis of the cell index values.
Cell viability was assessed using Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan), according to the manufacturer’s instructions. A total of 5,000 cells with corresponding treatment were seeded onto 96-well culture plates. At a specific time point, CCK-8 reagent was added to the wells and incubated for 2 h at 37°C. Lastly, absorbance was measured at 450 nm using an Infinite M200 spectrophotometer (Tecan, Switzerland).
Cell Cycle and Apoptosis Analysis by Flow Cytometry
Cells were fixed in absolute ethyl alcohol at ‒20°C overnight, washed twice with PBS, and resuspended in propidium iodide (PI) staining solution containing 0.1 mg/mL RNase A, 50 μg/mL PI, and 0.2% Triton X-100. Cell cycle distribution was analyzed using bivariate flow cytometry on a LSRFortessa flow cytometry platform.
To evaluate the cell apoptosis, 2 × 105 cells were harvested and resuspended in 500 μL of annexin V binding buffer, and then incubated with 5 μL of annexin V for 10 min and 5 μL of 7-aminoactinomycin D (7-AAD) for 15 min by flow cytometry, according to the manufacturer’s instructions.
RIP and ChIP
RIP was implemented using a Magna RIP RNA-binding protein immunoprecipitation (IP) kit (Millipore, Cambridge, MA, USA), according to the manufacturer’s instructions. After harvesting the cells in IP lysis buffer and mechanical shearing using a homogenizer, antibodies against PTBP1 (catalog no. ab61193; Abcam, Cambridge, MA, USA) and immunoglobulin G (IgG) (catalog no. 12-371; EMD Millipore) were added and cultured with the cell extract overnight under 4°C. Streptavidin-coated magnetic beads were then added and incubated for 2 h. The isolated and purified RNAs in which ZNF649-AS1 may be enriched were then analyzed using quantitative real-time PCR.
A ChIP assay was carried out using a Simple ChIP enzymatic ChIP kit (Cell Signaling Technology, CA, USA). The cross-linked cell lines were cultured in 4% formaldehyde for 30 min at room temperature, followed by lysis in radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific, Waltham, MA, USA). The lysates were ultrasonicated to obtain DNA fragments that were 200 to 1,000 bp in length. IP was conducted by incubating with specific antibodies against H3K27ac antibody (catalog no. ab4729; Abcam) or the negative control IgG antibody (catalog no. 12-371; EMD Millipore) overnight at 4°C. After rinsing and elution, the de-cross-linked chromatin samples were retrieved and quantitated using quantitative real-time PCR, with IgG antibody as the negative control. All experimental procedures were performed at least in triplicate.
RNA Pull-Down and Mass Spectrometry
ZNF649-AS1 was transcribed using T7 RNA polymerase in vitro (Ambion Life), followed by purification with an RNeasy Plus mini kit (QIAGEN) and treatment with DNase I (QIAGEN). The cell lysates were freshly prepared using a ProteaPrep zwitterionic cell lysis kit, mass spec grade (Protea), using lysis buffer supplemented with anti-RNase, protease/phosphatase inhibitor cocktail, panobinostat, and methylstat. The beads were washed briefly three times and boiled in SDS buffer. The retrieved protein was separated using electrophoresis, then silver stained and analyzed by mass spectrometry (ekspert nanoLC; AB Sciex TripleTOF 5600+). Data were analyzed using ProteinPilot software (https://pdf.medicalexpo.com/pdf/sciex/proteinpilot-software/79612-124319.html). The retrieved protein PTBP1 was validated using a western blot.
Nucleocytoplasmic Separation
Nuclear and cytosolic fractions were separated using the PARIS kit (Am1921; Thermo Fisher Scientific, USA), according to the manufacturer’s instructions. The expression levels of GAPDH, U1, and ZNF649-AS1 in the cytoplasm or nucleus of breast cancer cells were then detected using quantitative real-time PCR.
RNA-FISH
GFP-labeled ZNF649-AS1 probes were obtained from RiboBio. Hybridizations were carried out using a FISH kit (RiboBio), according to the manufacturer’s instructions. Briefly, 4% paraformaldehyde was used to fix SKBR-3-TR and BT474-TR cells, followed by treatment with 0.5% Triton X-100. The cells were then cultured with specific probes overnight. All of the fluorescence images were captured using a Nikon A1Si laser scanning confocal microscope (Nikon Instruments, Japan). The sequence used for the ZNF649-AS1 probe was as follows: 5′-ATTCCTTTATTTTATGGGATGTTCTGTAGGGAGTT-3′.
Autophagy Detection
A total of 2 × 104 cells per well were seeded into six-well plates and cultured in complete medium. Autophagy was detected using monomeric red fluorescent protein (mRFP)-GFP-LC3 adenoviral vectors purchased from HanBio Technology (HanBio, Shanghai, China), and observed using a confocal microscope (Zeiss LSM 710; Zeiss, Oberkochen, Germany) using a ×63 lens. The acquisition setting was set between samples and experiments as follows: XY resolution, 1,024 × 1,024 pixels; pinhole adjusted to 1.1 μm of Z-thickness; increments between stack images were 1 μm; laser power and gain were set for each antibody. Autophagic flux was determined by calculating the number of GFP and mRFP dots (dots/cell were counted).
IHC Analysis and Scoring Methods
Paraffin-embedded sections of tumor tissues from nude mice were placed in an incubator maintained at 60°C for 2 h, followed by immersion in xylene. Different concentrations of ethanol (including 70%, 85%, 95%, and 100%) and deionized water were used to hydrate the samples. The samples were then immersed in citrate buffer solution (0.01 mol/L [pH 6.0]) and heated at a temperature between 95°C and 100°C for 30 min. After washing with PBS, the samples were incubated with 0.5% Triton X-100 for 30 min. This fraction was then stained using the biotin-streptavidin horseradish peroxidase (HRP) detection system (ZSGB, China), followed by incubation with primary antibody targeting ATG5 (1:200) (ab108327; Abcam) overnight at 4°C. The presence of brown chromogen was taken to be indicative of positive immunoreactivity.
The immunostaining intensity of each sample was graded as follows: negative, 0; weak, 1; moderate, 2; or strong, 3. The proportion of positively stained cells was assessed as a percentage. The score was then calculated as the intensity score multiplied by the percentage of cells stained: score = intensity × % of positive cells. Images were visualized using a Nikon Eclipse Ti (Fukasawa, Japan) microscope and processed using Nikon software.
In Vivo Nude Mouse Model
Tumor xenografts were established using female BALB/c nude mice (4–6 weeks old) purchased from the Model Animal Research Center of Nanjing University (Nanjing, China). Equal numbers of SKBR-3-TR and SKBR-3 cells (3 × 106) were subcutaneously injected into each mouse to establish the breast cancer xenograft model. After 1 week, the mice received an intraperitoneal injection of trastuzumab (3 mg/kg) once every 2 days for 20 days. On day 28, the animals were euthanized and their tumors were removed. The diameters of the tumors were recorded once every 2 days starting from day 12 using a caliper. Tumor volume was calculated as follows: (length × width)2 × 0.5. The mice were sacrificed after 30 days, and the weight of neoplasm was measured immediately after resection. Tissues were fixed to obtain pathological slides fixed with 4% paraformaldehyde, followed by IHC staining for ATG5. Animal experiments were approved by the Institutional Review Board of The First Affiliated Hospital of Zhengzhou University (Henan, China).
Bioinformatics Analysis
The putative modification at the promoter of the ZNF649-AS1 gene was predicted using the UCSC Genome Browser (http://genome.ucsc.edu). Based on MFE and partition function, the stem-loop structure of ZNF649-AS1 was established using the ViennaRNA Web Services (http://rna.tbi.univie.ac.at/).
Western Blotting Assay
The cells from all of the groups were collected, washed with 3 mL of pre-cooled PBS, placed on ice, and boiled in SDS-sample buffer. The proteins samples were resolved by electrophoresis using 10% SDS-PAGE, then transferred onto polyvinylidene fluoride (PVDF) membranes. Membranes were blocked for 1 h in 5% skim milk at room temperature, followed by incubation with primary rabbit antibodies against PTBP1 (1:1,000) (ab133734; Abcam), ATG5 (1:1,000) (ab228668; Abcam), mouse antibodies against P62 (1:1,000) (ab56416; Abcam), LC3B (1:1,000) (ab48394; Abcam), and GAPDH (1:5,000) (ab8245; Abcam) at 4°C overnight. Membranes were then incubated with the corresponding secondary antibody at 37°C for 1 h. Lastly, the PVDF membranes were developed using ECL chemiluminescent reagent, and protein bands were visualized using a Bio-Rad Gel Doc XR+ system (Bio-Rad, Hercules, CA, USA).
Statistical Analysis
All experiments were performed in triplicate. Data are presented as the mean ± standard deviation (SD). Comparison between two groups was performed using a Student’s t test. One-way analysis of variance (ANOVA) was used to compare multiple groups (more than two). A Fisher’s exact test was used to evaluate the differences in proportions between different groups. Statistical analyses were performed using GraphPad Prism (v5.01) (GraphPad, San Diego, CA, USA). p < 0.05 was considered as statistically significant. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
M.H. and H.D. acquired the data and created a draft of the manuscript; M.H., H.D., X.Q., H.C., F.W., X.L., N.H., X.Y., Y.Y., D.D., J.H., and W.W. collected clinical samples and performed the in vitro and in vivo assays; X.Q., F.Z., and J.H. analyzed and interpreted the data and performed statistical analysis; H.D. reviewed the manuscript, figures, and tables. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81960475, 81702557, 81600377), and by the “Nanhai series” talent cultivation program of Hainan Province. We thank Prof. Manran Liu (The Key Laboratory of Laboratory Medical Diagnostics, Chinese Ministry of Education, Chongqing Medical University, Chongqing 400016, China) for critical technical support.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2020.07.019.
Contributor Information
Mingli Han, Email: minglihan@126.com.
Huaying Dong, Email: dr_dhy@163.com.
Supplemental Information
References
- 1.Menendez J.A., Vellon L., Lupu R. Antitumoral actions of the anti-obesity drug orlistat (Xenical™) in breast cancer cells: blockade of cell cycle progression, promotion of apoptotic cell death and PEA3-mediated transcriptional repression of Her2/neu (erb B-2) oncogene. Ann. Oncol. 2005;16:1253–1267. doi: 10.1093/annonc/mdi239. [DOI] [PubMed] [Google Scholar]
- 2.Daniels B., Kiely B.E., Lord S.J., Houssami N., Lu C.Y., Ward R.L., Pearson S.A. Long-term survival in trastuzumab-treated patients with HER2-positive metastatic breast cancer: real-world outcomes and treatment patterns in a whole-of-population Australian cohort (2001–2016) Breast Cancer Res. Treat. 2018;171:151–159. doi: 10.1007/s10549-018-4804-0. [DOI] [PubMed] [Google Scholar]
- 3.Adamczyk A., Kruczak A., Harazin-Lechowska A., Ambicka A., Grela-Wojewoda A., Domagała-Haduch M., Janecka-Widła A., Majchrzyk K., Cichocka A., Ryś J., Niemiec J. Relationship between HER2 gene status and selected potential biological features related to trastuzumab resistance and its influence on survival of breast cancer patients undergoing trastuzumab adjuvant treatment. OncoTargets Ther. 2018;11:4525–4535. doi: 10.2147/OTT.S166983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kołat D., Hammouz R., Bednarek A.K., Płuciennik E. Exosomes as carriers transporting long non-coding RNAs: molecular characteristics and their function in cancer (Review) Mol. Med. Rep. 2019;20:851–862. doi: 10.3892/mmr.2019.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang W.T., Han C., Sun Y.M., Chen T.Q., Chen Y.Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019;12:55. doi: 10.1186/s13045-019-0748-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malih S., Saidijam M., Malih N. A brief review on long noncoding RNAs: a new paradigm in breast cancer pathogenesis, diagnosis and therapy. Tumour Biol. 2016;37:1479–1485. doi: 10.1007/s13277-015-4572-y. [DOI] [PubMed] [Google Scholar]
- 7.Li W., Zhai L., Wang H., Liu C., Zhang J., Chen W., Wei Q. Downregulation of lncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7:27778–27786. doi: 10.18632/oncotarget.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu H.Y., Bai W.D., Ye X.M., Yang A.G., Jia L.T. Long non-coding RNA UCA1 desensitizes breast cancer cells to trastuzumab by impeding miR-18a repression of Yes-associated protein 1. Biochem. Biophys. Res. Commun. 2018;496:1308–1313. doi: 10.1016/j.bbrc.2018.02.006. [DOI] [PubMed] [Google Scholar]
- 9.Shi S.J., Wang L.J., Yu B., Li Y.H., Jin Y., Bai X.Z. lncRNA-ATB promotes trastuzumab resistance and invasion-metastasis cascade in breast cancer. Oncotarget. 2015;6:11652–11663. doi: 10.18632/oncotarget.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueno T., Masuda N., Kamigaki S., Morimoto T., Saji S., Imoto S., Sasano H., Toi M. Differential involvement of autophagy and apoptosis in response to chemoendocrine and endocrine therapy in breast cancer: JBCRG-07TR. Int. J. Mol. Sci. 2019;20:984. doi: 10.3390/ijms20040984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou F., Yang X., Zhao H., Liu Y., Feng Y., An R., Lv X., Li J., Chen B. Down-regulation of OGT promotes cisplatin resistance by inducing autophagy in ovarian cancer. Theranostics. 2018;8:5200–5212. doi: 10.7150/thno.27806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan Y., Wang K., Li Q., Zou Y., Chen B., Gong Q., Ho H.I., Yin T., Zhang F., Lu Y. The novel autophagy inhibitor alpha-hederin promoted paclitaxel cytotoxicity by increasing reactive oxygen species accumulation in non-small cell lung cancer cells. Int. J. Mol. Sci. 2018;19:3221. doi: 10.3390/ijms19103221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma T., Chen W., Zhi X., Liu H., Zhou Y., Chen B.W., Hu L., Shen J., Zheng X., Zhang S. USP9X inhibition improves gemcitabine sensitivity in pancreatic cancer by inhibiting autophagy. Cancer Lett. 2018;436:129–138. doi: 10.1016/j.canlet.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Yu T., Guo F., Yu Y., Sun T., Ma D., Han J., Qian Y., Kryczek I., Sun D., Nagarsheth N. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563.e16. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam Khan M.Z., Tam S.Y., Law H.K.W. Autophagy-modulating long non-coding RNAs (lncRNAs) and their molecular events in cancer. Front. Genet. 2019;9:750. doi: 10.3389/fgene.2018.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J., Xie S., Yang J., Xiong H., Jia Y., Zhou Y., Chen Y., Ying X., Chen C., Ye C. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 2019;12:81. doi: 10.1186/s13045-019-0747-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 18.Shvets E., Elazar Z. Autophagy-independent incorporation of GFP-LC3 into protein aggregates is dependent on its interaction with p62/SQSTM1. Autophagy. 2008;4:1054–1056. doi: 10.4161/auto.6823. [DOI] [PubMed] [Google Scholar]
- 19.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merry C.R., Niland C., Khalil A.M. Diverse functions and mechanisms of mammalian long noncoding RNAs. Methods Mol. Biol. 2015;1206:1–14. doi: 10.1007/978-1-4939-1369-5_1. [DOI] [PubMed] [Google Scholar]
- 21.Huang P., Li F., Li L., You Y., Luo S., Dong Z., Gao Q., Wu S., Brünner N., Stenvang J. lncRNA profile study reveals the mRNAs and lncRNAs associated with docetaxel resistance in breast cancer cells. Sci. Rep. 2018;8:17970. doi: 10.1038/s41598-018-36231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verma S., Miles D., Gianni L., Krop I.E., Welslau M., Baselga J., Pegram M., Oh D.Y., Diéras V., Guardino E., EMILIA Study Group Trastuzumab emtansine for HER2-positive advanced breast cancer. N. Engl. J. Med. 2012;367:1783–1791. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez E.A., de Haas S.L., Eiermann W., Barrios C.H., Toi M., Im Y.H., Conte P.F., Martin M., Pienkowski T., Pivot X.B. Relationship between tumor biomarkers and efficacy in MARIANNE, a phase III study of trastuzumab emtansine ± pertuzumab versus trastuzumab plus taxane in HER2-positive advanced breast cancer. BMC Cancer. 2019;19:517. doi: 10.1186/s12885-019-5687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shetty P., Patil V.S., Mohan R., D’souza L.C., Bargale A., Patil B.R., Dinesh U.S., Haridas V., Kulkarni S.P. Annexin A2 and its downstream IL-6 and HB-EGF as secretory biomarkers in the differential diagnosis of Her-2 negative breast cancer. Ann. Clin. Biochem. 2017;54:463–471. doi: 10.1177/0004563216665867. [DOI] [PubMed] [Google Scholar]
- 25.Montemurro F., Prat A., Rossi V., Valabrega G., Sperinde J., Peraldo-Neia C., Donadio M., Galván P., Sapino A., Aglietta M. Potential biomarkers of long-term benefit from single-agent trastuzumab or lapatinib in HER2-positive metastatic breast cancer. Mol. Oncol. 2014;8:20–26. doi: 10.1016/j.molonc.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponde N., Bradbury I., Lambertini M., Ewer M., Campbell C., Ameels H., Zardavas D., Di Cosimo S., Baselga J., Huober J. Cardiac biomarkers for early detection and prediction of trastuzumab and/or lapatinib-induced cardiotoxicity in patients with HER2-positive early-stage breast cancer: a NeoALTTO sub-study (BIG 1-06) Breast Cancer Res. Treat. 2018;168:631–638. doi: 10.1007/s10549-017-4628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riddihough G. A very focused function for lncRNAs. Science. 2017;355:35–37. doi: 10.1126/science.355.6320.35-k. [DOI] [PubMed] [Google Scholar]
- 28.Jadaliha M., Gholamalamdari O., Tang W., Zhang Y., Petracovici A., Hao Q., Tariq A., Kim T.G., Holton S.E., Singh D.K. A natural antisense lncRNA controls breast cancer progression by promoting tumor suppressor gene mRNA stability. PLoS Genet. 2018;14:e1007802. doi: 10.1371/journal.pgen.1007802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pian L., Wen X., Kang L., Li Z., Nie Y., Du Z., Yu D., Zhou L., Jia L., Chen N. Targeting the IGF1R pathway in breast cancer using antisense lncRNA-mediated promoter cis competition. Mol. Ther. Nucleic Acids. 2018;12:105–117. doi: 10.1016/j.omtn.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H., Yuan W., Wang Y., Zhu C., Liu B., Wang Y., Yang D., Li Y., Wang C., Wu X., Liu M. ZNF649, a novel Kruppel type zinc-finger protein, functions as a transcriptional suppressor. Biochem. Biophys. Res. Commun. 2005;333:206–215. doi: 10.1016/j.bbrc.2005.05.101. [DOI] [PubMed] [Google Scholar]
- 31.Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X., Zhou Y., Yang L., Ma Y., Peng X., Yang S., Li H., Liu J. lncRNA NEAT1 promotes autophagy via regulating miR-204/ATG3 and enhanced cell resistance to sorafenib in hepatocellular carcinoma. J. Cell. Physiol. 2020;235:3402–3413. doi: 10.1002/jcp.29230. [DOI] [PubMed] [Google Scholar]
- 33.Xin L., Zhou Q., Yuan Y.W., Zhou L.Q., Liu L., Li S.H., Liu C. METase/lncRNA HULC/FoxM1 reduced cisplatin resistance in gastric cancer by suppressing autophagy. J. Cancer Res. Clin. Oncol. 2019;145:2507–2517. doi: 10.1007/s00432-019-03015-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Bars R., Marion J., Le Borgne R., Satiat-Jeunemaitre B., Bianchi M.W. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 2014;5:4121. doi: 10.1038/ncomms5121. [DOI] [PubMed] [Google Scholar]
- 35.Rai S., Arasteh M., Jefferson M., Pearson T., Wang Y., Zhang W., Bicsak B., Divekar D., Powell P.P., Naumann R. The ATG5-binding and coiled coil domains of ATG16L1 maintain autophagy and tissue homeostasis in mice independently of the WD domain required for LC3-associated phagocytosis. Autophagy. 2019;15:599–612. doi: 10.1080/15548627.2018.1534507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keppetipola N., Sharma S., Li Q., Black D.L. Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 2012;47:360–378. doi: 10.3109/10409238.2012.691456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velagapudi S.P., Cameron M.D., Haga C.L., Rosenberg L.H., Lafitte M., Duckett D.R., Phinney D.G., Disney M.D. Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl. Acad. Sci. USA. 2016;113:5898–5903. doi: 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Disney M.D., Angelbello A.J. Rational design of small molecules targeting oncogenic noncoding RNAs from sequence. Acc. Chem. Res. 2016;49:2698–2704. doi: 10.1021/acs.accounts.6b00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dong H., Hu J., Zou K., Ye M., Chen Y., Wu C., Chen X., Han M. Activation of lncRNA TINCR by H3K27 acetylation promotes trastuzumab resistance and epithelial-mesenchymal transition by targeting microRNA-125b in breast cancer. Mol. Cancer. 2019;18:3. doi: 10.1186/s12943-018-0931-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Tuomi J.M., Voorbraak F., Jones D.L., Ruijter J.M. Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods. 2010;50:313–322. doi: 10.1016/j.ymeth.2010.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.