Abstract
Decades after identification as essential for protein synthesis, transfer RNAs (tRNAs) have been implicated in various cellular processes beyond translation. tRNA-derived small RNAs (tsRNAs), referred to as tRNA-derived fragments (tRFs) or tRNA-derived, stress-induced RNAs (tiRNAs), are produced by cleavage at different sites from mature or pre-tRNAs. They are classified into six major types representing potentially thousands of unique sequences and have been implicated to play a wide variety of regulatory roles in maintaining normal homeostasis, cancer cell viability, tumorigenesis, ribosome biogenesis, chromatin remodeling, translational regulation, intergenerational inheritance, retrotransposon regulation, and viral replication. However, the detailed mechanisms governing these processes remain unknown. Aberrant expression of tsRNAs is found in various human disease conditions, suggesting that a further understanding of the regulatory role of tsRNAs will assist in identifying novel biomarkers, potential therapeutic targets, and gene-regulatory tools. Here, we highlight the classification, biogenesis, and biological role of tsRNAs in regulatory mechanisms of normal and disease states.
Graphical Abstract
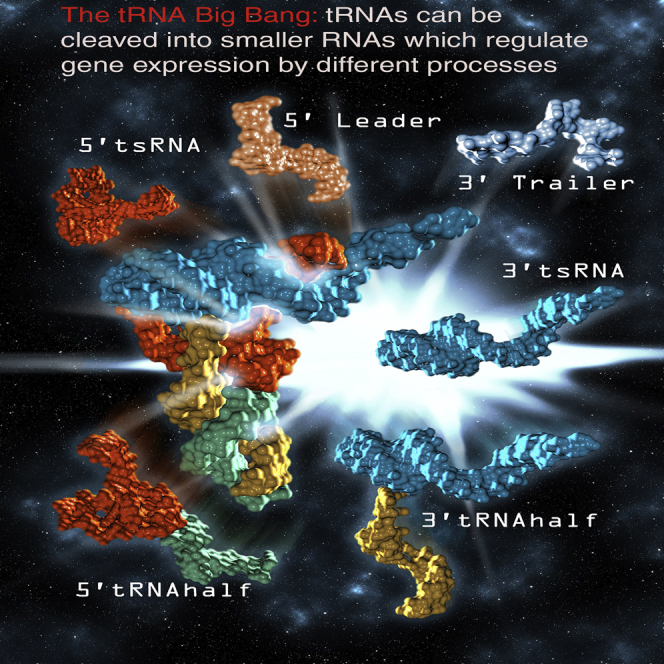
tRNAs can be enzymatically cleaved into different fragments to generate various types of tRNA-derived small RNAs (tsRNAs) that regulate gene expression by various mechanisms that are not well understood. This review describes how tsRNAs are generated and our current understanding of their role(s) in gene regulation in health and disease states.
Main Text
Transfer RNAs (tRNAs) are non-coding RNAs of 76–93 nt in length essential for mRNA translation. They decode genetic information into proteins by transferring amino acids to the growing polypeptide chains that are synthesized on ribosomes.1 tRNAs are processed from pre-tRNAs transcribed by RNA polymerase III (RNA pol III).2 During maturation, pre-tRNAs undergo several modifications, such as the removal of the 5′ leader and 3′ trailer sequences, splicing, post-transcriptional modification, and addition of a CCA sequence at the 3′ end of mature tRNA.
tRNAs are, by far, the most abundant RNA species, and their sequences are highly conserved across species. They are grouped into 64 types according to their anti-codon sequence. More than 500 loci in human genomes have hitherto been identified, and over 400 tRNA genes have unique sequences.3 In addition, there are over 600 tRNA pseudogenes, and tRNA lookalikes in the human nuclear genome and their functional significance remain unknown.3,4
For decades, tRNAs have been mostly considered to play a role in protein synthesis. However, it is becoming increasingly clear that tRNAs have various additional roles involved in cell proliferation, tumor metastasis, and neuronal homeostasis.5 The first tRNA mutations related with human disease were identified in 1990.6,7 These mutations were found in mitochondrial-expressed tRNAs and result in neuromuscular phenotypes. In contrast, no human disease has been associated with a cytoplasmic tRNA mutation, per se, but mutations in genes related to post-transcriptional modification of tRNAs and cytoplasmic tRNA-related protein partners, including splicing factors and aminoacyl tRNA-synthetase (ARSs), are strongly associated with certain human diseases.8 For example, mutations in the tRNA-splicing endonuclease complex (TSEN2, TSEN15, TSEN34, and TSEN54) were identified in PCH (pontocerebellar hypoplasia) type 2 and 4 patients, of which brain tissue showed immature development of the cerebellum and other structural defects.9
tRNA-derived small RNAs (tsRNAs) were first discovered in the urine of cancer patients in the 1970s.10,11 At the time, these RNAs were considered to be degradation products. With the advent of next-generation, high-throughput sequencing (NGS), the sequencing reads corresponding to tsRNAs were often discarded as degradation products or sequencing artifacts. However, just over a decade ago, three groups revealed that tsRNAs have biological roles.12, 13, 14 A growing body of evidence has demonstrated that tsRNA expression is not correlated with cognate tRNA abundance and they are processed at specific cleavage sites of mature and pre-tRNAs.15 Interestingly, tRNAs are differentially expressed in a variety of tissues, cancers, and during normal growth and development, including cellular differentiation.16 The same tRNA isodecoder genes (containing the same anti-codon sequence but with differences in each body sequence) are differentially expressed in several cases,17 which suggests that the differential expression of each tRNA gene may be related to noncanonical tRNA functions (e.g., tsRNA). Thus far, six major types of tsRNAs have been identified in various organisms, from humans to Escherichia. coli (E. coli), and they have been implicated in various diseases with distinct mechanisms. The fact that there are hundreds of tRNA genes that can be cleaved at specific sites to produce many different types and lengths of small RNAs suggests that thousands of unique and de novo non-coding RNAs can potentially be generated in a cell. Together, all of these findings demonstrate that tsRNAs are not merely degradation products but are generated in a precise and regulated manner.
Throughout this review, we will provide an overview of tsRNA classification and biogenesis while delineating known tsRNA-mediated regulatory mechanisms. In addition, we describe potential tsRNA associations with disease-related processes, when known, and how these interactions could empower new disease biomarkers or be manipulated for novel treatments.
Nomenclature and Classification
tsRNAs are also called tRF (tRNA-derived RNA fragment), tDRs (tRNA-derived small RNAs), and tiRNAs (tRNA-derived stress-induced RNAs).12, 13, 14 tsRNAs are differently named according to length and cleavage position on tRNA or pre-tRNA.
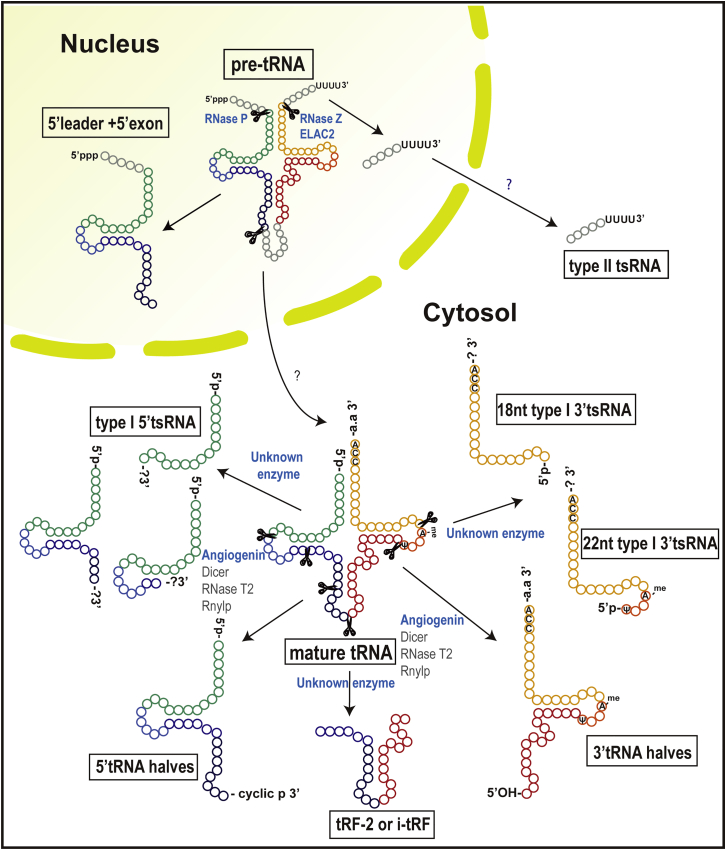
tsRNAs of 18–30 nt in length are categorized as either type I or type II, according to the cleaved position on mature tRNA and pre-tRNA, respectively (Figure 1). Type I tsRNAs are further classified into two subgroups: 5′tsRNA (tRF-5 or 5′tRF) and 3′tsRNA (tRF-3 or 3′tRF), which are derived from the 5′ and 3′ ends of mature tRNA, respectively.14 NGS data show that 5′tsRNAs from HEK293 cells mainly occur at lengths of 14–16 nt (tRF-5a), 22–24 nt (tRF-5b), and 28–30 nt (tRF-5c), which begin at the 5′ termini of mature tRNAs and end at a D-loop (tRF-5a) or at a stem region between the D-loop and the anti-codon loop (tRF-5b and tRF-5c) of a mature tRNA.15 Other lengths of 5′tsRNAs were also identified in prostate cancer cells and Haloferax volcanii,18 indicating that the length of 5′tsRNA might be more heterogeneous than expected, depending on the cell type and species. On the other hand, 3′tsRNAs (3′tRF) are uniformly present in the 18- to 22-nt forms throughout the three domains of life.15 Their 5′ ends reside in the T-loop, and the 3′ end corresponds with the 3′ terminus of mature tRNA, which all ends in CCA. Interestingly, most 22-nt 3′tsRNAs contain an N1-methyl-adenosine (m1A58) at the 58th nt in the T-loop (corresponding to the 19th nt in the tsRNA) and a pseudouridine at or near their 5′ termini,19,20 perhaps because one or both features may play a role in the generation of this type of tsRNA.
Figure 1.
The Classification and Biogenesis of tsRNA
There are more than 6 types of tsRNAs. Each type of tsRNA is processed from mature tRNA or pre-tRNA and has a distinct biogenesis pathway generated by different ribonucleases. All of the involved processing enzymes are indicated; the major processing enzyme is shown in blue. tRNA halves are primarily processed by angiogenin. The major processing enzyme for the tsRNAs derived from mature tRNAs is not well known.
Type II tsRNAs (tRF-1 or 3′U tRF) 16–48 nt in length are processed from a 3′ trailer sequence of pre-tRNA that begins 1 to 2 nt downstream of the 3′ end of the tRNA genomic sequence and ends with a polyuridine sequence (UUUUU, UUCUU, GUCUU, or AUCUU), a RNA pol III termination signal.12,14 Because the positions of the termination signal vary for each tRNA transcript, the length of the type II tsRNAs are more heterogeneous than type I tsRNAs.14,15
tsRNAs of length 30–40 nt are called “tRNA halves” because their lengths are almost half that of mature tRNA.21 They are often referred to as “tiRNA” due to their stress-induced characteristic,13 and some of them are called “SHOT-RNAs” (sex-hormone-dependent tRNA-derived RNA) because they are highly expressed in hormone-dependent cancers.22 They are grouped into 5′tRNA halves (5′tiRNA) and 3′tRNA halves (3′tiRNA) (Figure 1). The 5′ end of the former corresponds with the 5′ end of the mature corresponding tRNA, although the 3′ end is in the anti-codon loop. The latter begins in the anti-codon loop and ends with the 3′ terminus of the mature tRNA (Figure 1).13,22
There are two additional types of tsRNAs that are not included in the classes described above. i-tRF,23 referred to as tRF-2,24 with a variable length is derived from the internal region of mature tRNA straddling the anti-codon region (Figure 1).23,25 The other type begins at the 5′ end of the leader sequences in pre-tRNA and ends in the 3′ terminus of the 5′ exon in the anti-codon loop after removal of introns, which is implicated in neuronal degeneration (Figure 1).26
The information of varied tsRNAs has been retrieved from numerous deep-sequencing analyses and have been made available on public websites: tRFdb (http://genome.bioch.virginia.edu/trfdb) and tRF2Cancer (http://rna.sysu.edu.cn/tRFfinder).27,28
So far, the nomenclature for tsRNA has not been established. In order to intuitively understand which mature or pre-tRNAs generate the indicated tsRNAs in our text, we have added each tRNA name to the term “tsRNA” or “tRNA halves” according to their size and also include the names used in each study. Other tsRNAs not included in the six major subtypes described above will be referred to by the name used in each study.
Biogenesis of tsRNAs
18- to 30-nt tsRNAs
Because type I and II tsRNAs are primarily 22 nt in length and are also known to be 5′ phosphorylated and 3′ hydroxylated,14,29 which are characteristics of microRNAs (miRNAs), early investigation into the biogenesis of tsRNAs was focused on the involvement of Dicer or the microprocessor complex (Drosha/DGCR8), both of which are required for the biogenesis of miRNA.30 tsRNAs (3′ end of Ile tRNA precursor) in mouse embryonic stem cells (mESCs) were initially found to be processed by Dicer, but not DGCR8.31 The Ile tRNA precursor is predicted to form a long hairpin structure as an alternative form, which is considered to be a substrate of Dicer. In human cell lines, 19nt-Gln5′tsRNA and 22nt-GlyGCC3′tsRNA (CU1276) were also reported to be processed by Dicer.32,33 An 18-nt Lys3′tsRNA (PBSncRNA), which is highly expressed in human immunodeficiency virus (HIV)-1-infected MT4 cells, is processed by Dicer.34
In contrast, NGS data analyses from Dicer or Dgcr8 knockout mESCs indicate that type I tsRNAs are not processed using a canonical miRNA pathway.35 Dicer mutants in mouse, D. melanogaster, and S. pombe do not significantly change type I or II tsRNA expression.15 Dicer knockout 293T cells also show no differences in tsRNA expression.36 At most, these data suggest that Dicer and the microprocessor complex may only process a small number of specific tRNAs and/or in a cell-type-specific manner during particular cellular events.
There are many lines of evidence demonstrating that tsRNAs are processed by endonucleases other than the miRNA-processing enzymes. For example, LysTTT3′tsRNA is processed by other ribonucleases, including angiogenin, in an in vitro cleavage assay.35 RNase T2 is responsible for the production of tsRNAs in length of 17–40 nt in plants. Human RNase T2 and yeast Rny1p, a member of the RNase T2 family, are also able to generate 17- to 40-nt tsRNAs.37
Type II tsRNAs are also processed by different endonucleases. RNA pol III transcribes the pre-tRNA gene, which is composed of a 5′ leader, mature tRNA, and 3′ trailer sequence.2,38 Ribonuclease RNase P and RNase Z remove the 5′ leader and 3′ trailer portions from the pre-tRNA sequence in the nucleus, respectively, and a CCA sequence is added at the 3′ end of the mature tRNA.38 Consequently, the released 3′ trailer sequence in the processing of tRNA becomes a type II tsRNA. Haussecker et al. provided the evidence that RNase Z is required for type II tsRNA generation in vitro,14 and Lee et al. identified that ELAC2, a tRNA 3′ endonuclease39 and a prostate cancer susceptibility gene, processes SerTGA type II tsRNA (tRF-1001) from pre-tRNA in a prostate cancer cell line.12 Overall, more work is required to further determine the full detailed mechanism by which type II tsRNAs are processed, the location(s) of processing (i.e., nuclear versus cytoplasmic), and how such processing may be linked to regulating their production.
30- to 40-nt tsRNAs
The biogenesis of tRNA halves 30–40 nt in length is more established than the 17- to 30-nt fragments. tRNA halves were initially found in E. coli and were generated by PrrC nuclease in response to bacteriophage infection.40 Later, it was reported that this cleavage also occurs in Tetrahymena thermophila,21 bacteria,41 fungi,42 and mammalian cells25,43, 44, 45 during various stress conditions, including amino acid or glucose starvation, heat shock, hypoxia, UV irradiation, or heavy metal exposure. However, all stress conditions do not induce the production of tRNA halves, indicating that this cleavage event is not solely caused by reduced translation during stress conditions.13,43,45,46 These stress-induced tRNA halves are called tiRNAs. Two different ribonucleases, angiogenin (a member of RNase A superfamily)13,43 and Rny1p,47 were initially identified to be involved in this stress-induced process in mammalian or yeast cells, respectively. Under normal physiological states, angiogenin does not process tsRNAs because the enzyme is localized to the nucleus or sequestered by assembly with RNH1 (angiogenin inhibitor) in the cytoplasm. Stress condition releases angiogenin from nucleus or RNH1, allowing processing into tRNA halves.46,48,49
Independent of the characteristic stress response, a subset of tRNA halves, including AspGUC and HisGUG tRNA halves, is expressed in estrogen receptor (ER)-positive breast cancer and androgen receptor (AR)-positive prostate cancer cell lines, indicating that regulation of specific tRNA halves is dependent upon sex hormones and their receptors. These SHOT-RNAs are produced by angiogenin-dependent cleavage in the anti-codon loop of the tRNA in response to hormone-signaling pathways.22 3′SHOT-RNAs have an amino acid at the 3′ terminus and do not have introns, suggesting that this type of tRNA half is generated from the mature tRNA.21,22
A number of studies show that angiogenin is not the only ribonuclease that can process tRNA halves. Angiogenin-independent tRNA halves can be formed in the absence of stress,45,46 and the combination of Dicer- and angiogenin is required for processing of certain type of tRNA halves.44 Furthermore, the biogenesis of tRNA halves is modulated by tRNA modifications. As an example, Drosophila DNA methyltransferase DNMT2 methylates mature tRNAsAsp (GTC), Val (AAC), Gly(GCC) and protects them from angiogenin-induced cleavage.50 Additionally, loss of the cytosine-5 RNA methyltransferase, NSUN2, and its anti-cleavage effect on tRNAs results in the accumulation of 5′ tRNA halves by increasing angiogenin-mediated cleavage of tRNAs. This increased cleavage reduces protein translation rate and leads to neuro-developmental disease.51 Finally, RNase T2 in plants and humans can process tRNA halves as well.37 Together, these results suggest there may be multiple pathways that result in the biogenesis of tRNA halves.
The Biological Functions of tsRNAs and Their Role in Disease Processes
There is a growing body of evidence demonstrating that certain types of tsRNAs play a role in a variety of cellular processes. Before explaining these processes, we will first cover the limitations of tsRNA sequencing and quantification and the potential use of tsRNAs as tumor markers.
tsRNAs as Biomarkers in Human Disease
Unique Issues with tsRNA Quantification. One important aspect of tsRNA biology is that the high number of nucleotide modifications38 and sequence similarities in both tRNAs and corresponding tsRNAs, plus the relative abundance of closely related tsRNAs and various length isoforms, can result in mis-quantification for a given tsRNA species. For example, a chemical modification that interferes with reverse transcription may result in an overestimation of the smaller isoform and underestimation of the longer isoform of a respective tsRNA in NGS analysis. Other means of quantification between size variants can be distinguished using northern blots and, as such, may be at odds with sequencing-based quantification.52 However, northern blot quantification can suffer from cross-hybridization to a number of tsRNAs that have high sequence homologies.
As a result, newer forms of NGS are being developed to overcome these limitations.53, 54, 55 Cozen et al.53 and Zheng et al.54 separately developed a novel library preparation method that removes methyl groups from tRNA. They used engineered E. coli AlkB to demethylate N1-methyladenosine (m1A), N3-methylcytidine (m3C), and N1-methylguanosine (m1G).53,54 Their methods prevent the inhibitory effects of methylation but did not resolve the potential effects of other types of modified nucleotides during sequencing. A better solution for tsRNA sequencing might be nanopore sequencing, also recently developed, which detects ionic current through nanopores and recognizes the change in current when RNA or DNA passes through the nanopores.56 Because there is no cDNA synthesis step, nanopore sequencing might be the best-existing method to resolve the inhibitory effects of modified nucleotides on reverse transcription during sequencing. However, a higher error rate and inadequate character for short biomolecules need to be resolved.
To date, the modified nucleotides in mature tRNAs likely make it difficult to discover biomarkers from tsRNAs in a high-throughput manner. However, type II tsRNA (derived from 3′ end of pre-tRNA) have high potential to be used as biomarkers with current sequencing technology because pre-tRNAs are the least-modified form of tRNA.
The Characteristic Expression of Type II tsRNAs in Cancer. The first identified tsRNA identified to play a role in cancer was SerTGA type II tsRNA (tRF-1001). This tsRNA is highly expressed in several cancer cell lines, including prostate and colon cancers. It is highly associated with the proliferation of colon cancer cells (HCT-116) and is required for the cell cycle transition from the G2 to M phase.12
After this initial finding, Balatti et al.57 screened type II tsRNA expression using custom tsRNA microarray chips and revealed that many type II tsRNAs are differentially expressed in different tumors, including chronic lymphocytic leukemia (CLL) and lung, breast, ovarian, or colorectal cancers. In both CLL and lung cancer, HisGTG1 type II tsRNA (Ts-46), ArgTCG4 type II tsRNA (ts-47), ThrCGT15 type II tsRNA (ts-49), SerGCT-4-3-type II tsRNA (ts-53), and ThrAGT-1-1 type II tsRNA (ts-101) are downregulated although PseudoCTT51 type II tsRNA (ts-4) is upregulated. In breast cancer, IleAAT154 type II tsRNA (ts-66) is upregulated, whereas ts-86 is downregulated. In ovarian cancer, ten tsRNAs are up or downregulated. SerGCT-4-3-type II tsRNA (ts-53) and ThrAGT-1-1 type II tsRNA (ts-101) are also downregulated in adenoma, but not in adenocarcinoma. AspGTC48 type II tsRNA (ts-3) is upregulated in CLL and ovarian cancer. These differential expression patterns suggest that tsRNAs could be potentially tumor-promoting or suppressing genes. tsRNAs are highly regulated in HRAS, KRAS, PIK3CA-activated breast cancer cell lines, and MYC-activated lymphocytes, also suggesting that tsRNAs can be important molecules in tumorigenesis.57
The regulation of type II tsRNA biogenesis has only recently started to be revealed. Farina et al.58 determined that four type II tsRNAs (ts-19, ts-29, ts-46, and ts-112) expression levels were responsive to the RUNX1 tumor suppressor expression and that ts-112 was inversely correlated with RUNX1 and possessed oncogenic activity. This finding suggests that expression of some type II tsRNAs might be regulated by tumor-suppressor genes or oncogenes, which could also explain differential expression of tsRNAs in cancer cells.
Next, we will discuss specific functional roles for tsRNAs that have been identified as potential targets for cancer detection and therapeutics.
Extracellular tsRNAs. Although there are certain limitations to measure tsRNA levels correctly, there have been a number of attempts to identify extracellular tsRNAs. tsRNAs in extracellular vesicles (EVs) were initially detected from murine immune cells59 and then identified to originate after secretion by T cells. Activating T cells released EVs enriched with tsRNAs, suggesting that T cells may selectively shed tsRNAs that might repress immune activation.60 Recently, a subset of tsRNAs was recently detected in EVs from liver cancer patients,61 suggesting that tsRNAs could be promising biomarkers for cancer diagnosis.
Like all research on extracellular RNAs, the role of tsRNAs outside of cells is unresolved. It is possible that specific tsRNAs in serum or EVs could be used as biomarkers in specific disease states. Any possible functionality to their release from the cell and ultimate fate needs to be determined with additional studies.62,63
Gene Silencing—Do tsRNAs Interact with the miRNA Machinery?
In early investigations, tsRNAs were considered as a different type or class of miRNAs, and it remained unclear whether tsRNAs interact with various miRNA components, such as the Ago proteins, a central element of the RNA-induced silencing complex (RISC). Nevertheless, there remains conflicting evidence for and against the argument that tsRNA interacts with miRNA machinery. In part, some studies in support use supraphysiologic levels of tsRNA mimics to establish binding to various RNA-binding proteins, an approach that can be fraught with artifact. In addition, tsRNA-mediated functional knockdown of mRNAs containing tsRNA complementary targets is generally not as robust as with similarly designed miRNA targets. Finally, Ago proteins mostly require double-stranded RNA for loading,64 making it mechanistically difficult to envision how some of the tsRNAs would become functionally attached to these proteins.
In this section, we demonstrate the conflicting evidence for and against tsRNA-Ago association and various examples of tsRNA-mediated gene silencing processing.
Ago Associations
GlyTCC3′tsRNA (Cand23), GlyGCC3′tsRNA (Cand33 or CU1276), and type II-SerTGAtsRNA (Cand45 or tRF-1001) are more abundantly associated with Ago 3 and 4 rather than Ago 1 and 2 determined by RNA-immunoprecipitation assay with overexpressed Agos.14,33 This preference toward specific Ago families was also substantiated by two different NGS-based analyses, PAR-CLIP (photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation) and CLASH (crosslinking, ligation, and sequencing of hybrids), both of which delineated that type I tsRNAs prefer Ago 1, 3, and 4 proteins to Ago 2, although most type II tsRNAs did not bind to any Ago proteins.15 The preference for different Ago proteins was also affected by age in flies, and the target prediction results of these age-dependent tsRNAs suggest the potential roles of tsRNAs in development and neuronal functions associated with age.65 Recently, the luciferase assay with overexpressed LeuAAG3′tsRNA, CysGCA3′tsRNA, and LeuTAA3′tsRNA showed that 18-nt 3′tsRNAs repress gene expression in a Dicer-independent but Ago-dependent manner. Furthermore, they were associated with the GW182 protein, which represses translation of target mRNAs and degrades them.36
In contrast to the findings above, there are also studies suggesting that tsRNAs are poorly or not at all associated with Ago families. Cole et al.32 revealed that Gln3′tsRNA was poorly associated with Ago1 and Ago2 proteins by determining that the majority of this tsRNA was enriched in fractions lacking Ago2 proteins in HeLa cells. Hasler et al.66 showed that the lupus autoantigen La protein, which is required for tRNA processing and folding,67 prevents tsRNAs from being incorporated into Ago 1–4 proteins, thus protecting the miRNA pathway from tsRNA incorporation into the Ago proteins. However, they also revealed that Ile-typeII-tsRNA transports to the cytoplasm for Dicer processing and Ago protein loading via exportin (Xpo5),66 suggesting that the interaction with Agos might vary by individual tsRNAs. Kim et al.52 also determined that LeuCAG5′ and 3′tsRNA, AspGTC3′tsRNA, and SerGCT-typeII-tsRNA (Cand45 or tRF-1001 in another study) were not associated with endogenous Ago 1–3 proteins, which was different from previously published results.35
Specific Examples of Gene Silencing
The first validated target of tsRNA-mediated gene-silencing activity was in HIV. The 18-nt LysUUU3′tsRNA (3′ end of tRNAlys3) is complementary to the HIV primer binding site (PBS) and associates with the Ago2 protein. It was induced from HIV-infected MT4 T cells and remarkably reduced HIV-1 RNA copies, indicating that this tsRNA might potentially inhibit HIV-1 replication using RNAi machinery and be a novel molecular target for modulating HIV replication (Figure 2A).34 Like LysUUU3′tsRNA, HisGTG3′tsRNA and LeuCAG3′tsRNA were also found to be associated with the Ago2 proteins when Ago2 was overexpressed in HeLa cells.35
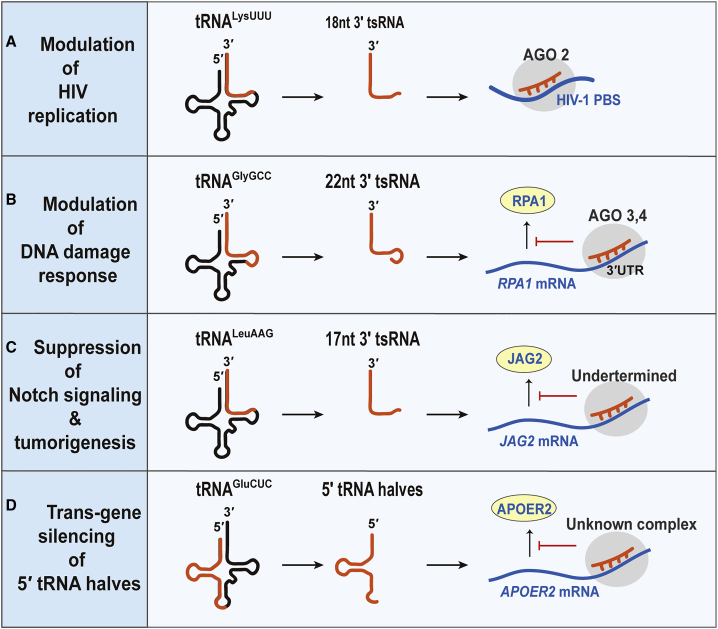
Figure 2.
Gene-Silencing Mechanism
(A) Modulation of HIV replication. LysUUU 3′tsRNA binds to the HIV-1 primer-binding site (PBS) associated with the Ago2 protein. (B) Modulation of DNA damage response is shown. GlyGCC 3′tsRNA modulates the molecular response to DNA damage by repressing endogenous RPA1. (C) Suppression of Notch signaling and tumorigenesis is shown. LeuAAG 3′tsRNA (tRF/miR-1280 fragment) binds to the JAG2 mRNA 3′ UTR and induces its degradation, which inhibits the activation of the Notch pathway in colorectal cancer cells. (D) Trans-gene silencing of 5′ tRNA halves is shown. 5′GluCUC tRNA halves recognize the 3′ UTR of apolipoprotein E receptor 2 (APOER2) mRNA and suppresses its expression.
GlyGCC3′tsRNA (CU1276) also has trans-gene-silencing activity, even though it is skewed toward interactions with Ago1, 3, and 4 rather than Ago2. The GlyGCC3′tsRNA is highly expressed in normal germinal center B cells but was markedly reduced in germinal-center-derived lymphomas. Overexpressed GlyGCC3′tsRNA suppresses lymphoma cell viability and modulates the molecular response to DNA damage by repressing RPA1 expression, which stabilizes single-stranded DNA intermediates during DNA replication or stress (Figure 2B).33
17-nt tRF/miR-1280 is known to be processed from both LeuAAG mature tRNA and a miRNA precursor. Furthermore, this tRF/miR-1280 suppresses Notch signaling by repressing Notch ligand JAG2 expression, resulting in reduced tumor formation and metastasis, as well as suppression of the stem-cell-like phenotype of colorectal cancer cells (Figure 2C).68 It remains to be determined which Ago protein is involved in the gene-silencing activity of this tsRNA.
Recently, Luo et al.69 found that 5′tsRNAs inhibit global translation by suppressing the synthesis of translation-machinery-related genes, including ribosomal proteins, via interaction with the Ago2 protein. Kuscu et al.36 also found that the 18-nt 3′tsRNA represses endogenous target genes associated with Ago and GW182 proteins involved in miRNA-mediated translation repression and target mRNA degradation.
30- to 40-nt tRNA halves have been implicated in viral replication by gene-silencing mechanisms as well. Wang et al.70 identified a subset of 5′tRNA halves (termed tRFs in their study) that were enriched in human respiratory syncytial virus (RSV)-infected human airway epithelial cells, in which GluCUC, GlyCCC, LysCUU, and CysGCA5′tRNA halves (tRF5-GluCUC, GlyCCC, LysCUU, and CysGCA) promoted viral replication. These tsRNAs are generated by angiogenin and repress gene expression through base pairing with the 3′ UTR in a target mRNA. Distinct from miRNA-mediated regulation, the 3′ end region of tRNA halves is important for selecting target sites of tRNA halves, suggesting they might interact with other protein complexes, other than RISC.70,71 RSV-induced GluCTC5′tRNA halves downregulate the viral replication repressor host gene APOER2 and promote viral replication,72 which can be a potential target for inhibiting RSV replication (Figure 2D). Together, these studies begin to illuminate the functional role of tsRNA species in cancer and infections, which appear to be diverse in their mechanisms and likely targetable for therapeutic interventions.
Piwi-Dependent Gene Regulation
Some tsRNAs of similar length to PIWI-interacting RNAs (piRNAs) of 26–31 nt in length have been found to associate with PIWI proteins. piRNAs are generally expressed in the germline and repress transposable elements (TEs). In addition, piRNAs can affect histone modification and DNA methylation in somatic cells.73 Of interest, an association of PIWI protein and tsRNAs has also been observed in Tetrahymena. 3′tsRNAs are associated with Tetrahymena Piwi 12 (Twi12) protein,29 which is essential for growth.74 3′tsRNAs-associated Twi12 protein is assembled with the Xrn2 and Tan1 proteins, resulting in stimulation of Xrn2 exonuclease activity in the nucleus, which is required for ribosomal RNA processing (Figure 3A).75 These interactions add to the growing body of evidence that tsRNA plays a substantiative role in essential biological processes.
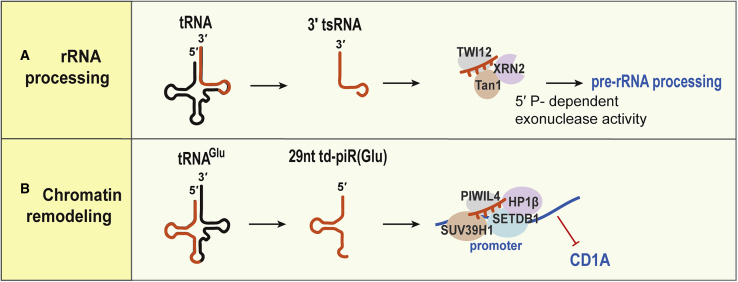
Figure 3.
PIWI Protein-Dependent Gene Regulation
(A) rRNA processing regulation. Twi12-bound 3′ tsRNAs assemble the nuclear exonuclease Xrn2 and Tan1, which is required for cellular ribosomal RNA processing. (B) Chromatin remodeling regulation is shown. tRNA-Glu-derived piRNA (td-piR(Glu)) is associated with PIWIL4, leading to recruitment of SETDB1, SUV39H1, and heterochromatin protein 1β to the CD1A promoter region, and facilitates H3K9 methylation. The transcription of CD1A is consequently inhibited.
Another study by Zhang et al.76 identified a new piRNA, td-piR(Glu), derived from the 5′ end of tRNA-Glu that is highly expressed in human monocytes rather than in differentiated dendritic cells. td-piR(Glu) is 29 nt in length and 2′-O methylated at the 3′ terminus, which is characteristic of a piRNA. PIWIL4 protein interacts with the td-piR(Glu) and recruits SETDB1, SUV39H1, and heterochromatin protein 1β to the CD1A promoter, which leads to H3K9 methylation and inhibition of CD1A transcription in monocytes. This result suggests that tsRNAs can play a role in piRNA regulation of chromatin remodeling in somatic cells (Figure 3B). The authors also revealed that angiogenin, which processes tRNA halves from mature tRNAs, did not affect the processing of the td-piR(Glu). Instead, during the differentiation of monocytes into dendritic cells, cytokine interleukin-4 (IL-4) is likely to be involved in the generation of td-piRs.76 An investigation into the generation and modification of tRNA-derived piRNAs might shed light on piRNA biogenesis and function in somatic cells.
SerGCT-4-3-typeII-tsRNA (ts-4521 or later called ts-101) and ThrAGT-1-1-typeII-tsRNA (ts-3676, later called ts-53) were originally identified as miRNAs (miR-4521 and miR-3676, respectively). They bind to overexpressed Ago1 and Ago2 proteins as well as overexpressed Piwi-like protein (PiwiL2), suggesting that these tsRNAs might play a role as a piwi RNA.77
Regulation of Transposons
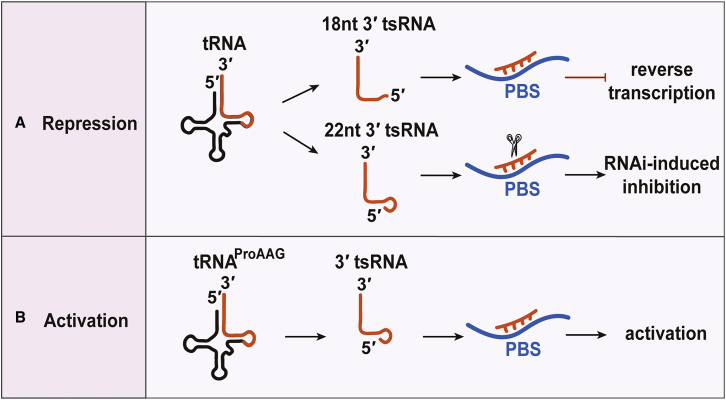
TEs are called mobile genomic DNAs, and their transposition activities are usually suppressed by epigenetic markers, like DNA methylation and histone modification.78 TEs are also regulated by small RNAs, including piRNAs, at the level of post-transcription and/or pre-transcription during epigenetic marker reprogramming.73 Most epigenetic markers and piRNAs disappear during preimplantation embryo development, in which other pathways or short RNAs are needed to protect the genome.79 The possibility that tsRNAs can be newly identified regulators of TE was raised due to their perfect sequence complementarity with the PBS sequence contained in the long terminal repeat (LTR) of retrotransposons (also known as endogenous retroviruses [ERVs]). Recently, Schorn et al.80 found that 18- and 22-nt 3′tsRNAs (CCA 3′tRF) accumulated in Setdb1−/−, but not in Dnmt-1−/− mESCs. Dnmt-1 mediates DNA methylation, resulting in inhibition of most non-LTR retrotransposons, whereas Setdb1 mediates histone H3K9 trimethylation (H3K9me3), leading to inhibition of most LTR retrotransposons.78 This finding implies that tsRNAs might be regulators of the expression of the LTR retrotransposon, perhaps through their perfect sequence complementarity. In vitro assay results demonstrate that the 18-nt LysUUU3′tsRNA (tRF-Lys-AAA) inhibits retroviral cDNA synthesis by competing with mature tRNAs for binding to the PBS. Another mechanism proposed was that the 22-nt LysUUU3′tsRNA induced RNAi silencing activity, resulting in the inhibition of coding-competent ERVs (Figure 4A).80 Contrary to the inhibitory effect of these tsRNAs on retrotransposons, ProAGG3′tsRNA (tRF-3019), which is perfectly complementary to the PBS of human T cell leukemia virus type 1 (HTLV-1), primes HTLV-1 reverse transcriptase in an in vitro assay. This finding suggests that ProAGG3′tsRNA might be required for HTLV-1 reverse transcription (Figure 4B).81
Figure 4.
The Regulation of LTR-Retrotransposon (ERV)
(A) Repression. 18-nt 3′tsRNAs block ERV reverse transcription by competing with intact tRNAs for PBS; 22-nt 3′tsRNAs exert post-transcriptional silencing of retroviral protein production by targeting its mRNA. (B) Activation is shown. The ProAAG3′tsRNA (tRF-3019) is capable of priming HTLV-1 reverse transcriptase. The ProAAG3′tsRNA may play an important role in priming HTLV-1 reverse transcription and thus represents a novel target to control HTLV-1 infection.
Thus, the miRNA and piRNA-like activities of some tsRNAs have been described in the context of their individual biological roles, across several cellular processes. Next, we will discuss some of the more-unique functions ascribed to tsRNAs.
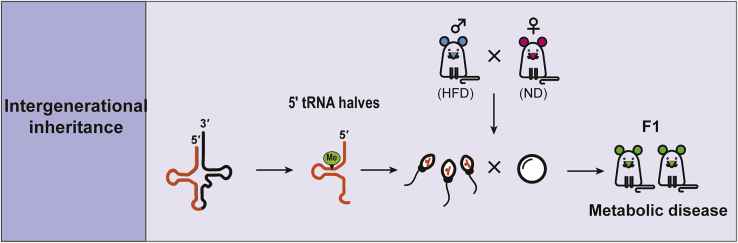
Intergenerational Inheritance and Metabolic Disease
One of the unique functions of tsRNAs is related to epigenetic inheritance. The mechanism of non-genetic inheritance, including histone modifications, DNA methylation, and RNA transcription, remains largely undetermined.82 The metabolic disorders in offspring resulting from a paternal diet-induced metabolic change83 strongly suggests that sperm contain information related to intergenerational non-genetic inheritance. Peng et al.84 identified distinct expression patterns of small non-coding RNAs in adult testis, mature sperm, and uteri. Although most of the small RNAs in adult testis are piwi RNAs, the majority in mature sperm are 5′tRNA halves, suggesting that tRNA halves might play an important role in parental hereditary information. Because mammalian PIWI proteins are not expressed in mature sperm,85 5′tRNA halves in mature sperm might not be considered to have a piRNA function requiring PIWI proteins. Recently, two different groups revealed that 5′tRNA halves displayed significant differences in mature sperm from mice fed a high-fat diet (HFD)86 or low-protein diet.87
Chen et al.86 determined that male offspring from mice fed a HFD or sperm from normal mice whose zygotes were injected with tsRNAs isolated from HFD sperm had impaired glucose tolerance as a result of suppressed metabolic-pathway-related genes. These genes were more downregulated in both the eight-cell embryos and blastocysts than any other stage in preimplantation mouse development. Their results suggest that sperm 5′tRNA halves affect gene expression in early embryogenesis, leading to a metabolic disorder. Sperm 5′tRNA halves preferentially match promoter regions rather than coding sequences in 1,134 genes, of which 922 are expressed in eight-cell embryos, suggesting that sperm 5′tRNA halves might play a role in transcriptional regulation (Figure 5).86 Later, Yang et al.88 determined 5′tRNA halves were highly expressed in eight-cell embryos, supporting the concept that 5′tRNA halves might play a role in early embryogenesis.
Figure 5.
Intergenerational Inheritance
5′tRNA halves in sperm transmits paternal diet-induced metabolic change to progeny, inducing metabolic disorders and addictive behavior. The tsRNAs were loaded into maturing sperm via epididymosomes. HFD, high-fat diet; ND, normal diet.
Further studies show that sperm 5′tRNA halves isolated from mice fed an HFD had more m2G and m5C modifications,86 which might contribute to RNA stability89 or RNA-mediated transgenerational epigenetic inheritance.90 Zhang et al.91 determined that a deletion in the Dnmt2 (mouse tRNA methyltransferase) gene inhibited the formation of these modifications and abolished sperm 5′tRNA half-mediated intergenerational inheritance. They also showed that Dnmt2-regulated m5C affects the secondary structure and biological properties of sperm 5′tRNA halves, as well as rRNA-derived small RNAs (Figure 5). Their results suggest that modification of sperm non-coding RNAs is important for parental epigenetic inheritance.
Other studies focused on epigenetic inheritance conducted by Sharma et al.87 identified that 5′tRNA halves derived from GluCYC, GlyCCC, GlyGCC, GlyTCC, LysCTT, and HisGTG tRNA were 2- to 3-fold upregulated in cauda sperm in low-protein-diet-fed mice. These 5′tRNA halves are not abundant in the epididymis. Inhibition of GlyCCC 5′tRNA halves repressed genes regulated by the LTR of endogenous retroelement MERVL in both embryonic stem cells and embryos, suggesting that low-protein-diet-induced sperm 5′tRNA halves play a role in regulating retrotransposon activity or expression.
The role of tRNA halves in intergenerational inheritance is also supported by the recent evidence that paternal exercise alters mouse small non-coding RNAs in sperm and reduces anxiety traits from male offspring. Intriguingly, paternal exercise highly increased 299 5′tRNA halves and a few miRNAs, although 3′tsRNAs of 16 and 22 nt in length were significantly reduced.92 The reason for the reduced 3′tsRNAs and their mode of regulation remain to be determined.
Despite discrepancies regarding the mechanism by which tRNA halves regulate metabolic changes in offspring from each group, the results provide strong evidence that sperm 5′tRNA halves are distinct from stress-induced 5′tRNA halves (tiRNAs) and play a role in intergenerational inheritance. The detailed mechanism by which they affect metabolic status in offspring requires further investigation.
Global Translation Regulation
One of the most studied tsRNA functions is translation regulation. Translation is complex and highly regulated at many levels. The process is generally divided into three basic steps: initiation; elongation; and termination. The regulation of translation initiation has been well studied. The eukaryotic initiation factor 4F (eIF4F) complex is composed of eIF4A, 4E, and 4G. EIF4F binds to the 7-methylguanosince cap on mRNAs and recruits the 43S ribosomal pre-initiation complex (40S ribosomal subunit and the eIF2–GTP–Met-tRNAiMet ternary complex), which scans in the 5′ to 3′ direction to reach the start codon. During this initiation step, eIF4E is regulated by MYC, mitogen-activated protein kinase (MAPK)-interacting serine/threonine kinase (MNK), and the mTOR complex 1 (mTORC1). Elongation is also known to be regulated by mTORC1 via ribosomal protein S6 kinase (Figure 6).93
Figure 6.
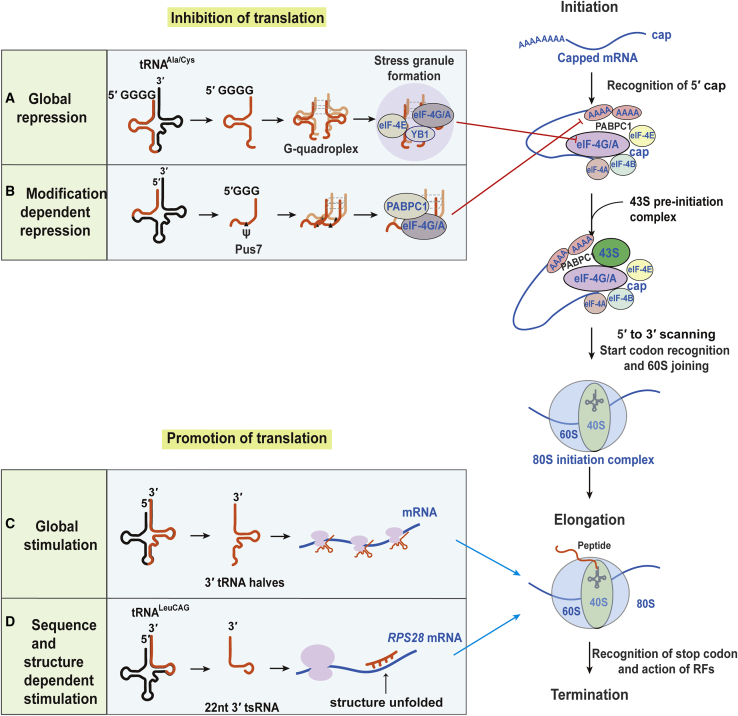
Translational Regulation
A number of tsRNAs are involved in translation regulation. Multiple mechanisms are involved. The tsRNAs can inhibit translation or promote the translation of cellular mRNAs. The 3 major translation steps are shown on the right. (A) Global repression is shown. 5′ tsRNAs with terminal oligo-G (TOG) motifs form an RNA G-quadruplex (RG4) structure, which displaces translational initiation factors (eukaryotic initiation factor [eIF]-4A, F, and G) from the m7G-capped mRNAs and sequesters them in stress granules, leading to global translational inhibition. (B) Modification-dependent repression is shown. Shorter forms of TOG-5′ tsRNAs (18 nt) forming RG4 show different binding affinities with translational initiation factors, depending on whether they are modified at the C8 position, where Pus7 converts cytidine to pseudouridine. The 18-nt U8-TOG-5′tsRNAs inhibit translation, although the 18-nt C8-TOG-5′tsRNAs do not. (C) Global stimulation is shown. tRNAThr 3′half is produced during stress conditions in Trypanosoma brucei. tRNAThr halves are associated with ribosomes and polysomes, which stimulate translation by facilitating mRNA loading during stress recovery once starvation conditions. (D) Sequence- and structure-dependent stimulation is shown. The 22-nt LeuCAG3′tsRNA binds to the mRNA of small ribosome protein (RPS28/15) mRNAs and unfolds the duplexed RNA structure at the target site, which increases the translation of the RPS28/15 protein. RPS28 is essential for ribosome biogenesis.
Studies in recent years have shown that tsRNAs may fine-tune global translation or the translation of a specific subset of mRNAs under different cellular conditions. These events can involve either inhibition or stimulation of the translational processes that regulate cellular events, such as ribosome biogenesis, cellular proliferation, and/or apoptosis.
The first identified function of tsRNAs was global translation inhibition under stress in plant and animal cells.13,94 tRNA cleavage events under stress conditions were earlier regarded as a cellular process to reduce the abundance of mature tRNAs. However, even with the formation of tRNA halves, the mature tRNA pool did not significantly change.13,21,45 Instead, tRNA halves were found to play an independent role in global translation inhibition. In plants, Zhang et al.94 identified that tRNA halves or larger sizes of tRNA fragments were enriched in the phloem sap (PS) of pumpkin plants and inhibited global translation in vitro. In mammals, Yamasaki et al.13 identified that tRNA halves were induced under oxidative stress, heat shock, or UV irradiation and angiogenin is required for the generation of tRNA halves.
Translation is inhibited by the formation of cytoplasmic stress granules (SGs) under various environmental stresses. There are two distinct pathways that result in inducing SG formation. One is to phosphorylate eukaryotic translation initiation factor 2α (eIF2α) to reduce ternary complexes, leading to the SGs formation. The other is an eIF2α phosphorylation-independent SG formation pathway.95,96
Intriguingly, 5′tRNA halves, but not 3′tRNA halves, induce the eIF2α phosphorylation-independent formation of SGs.13 The Anderson lab determined that selected tRNA halves (Ala5′tRNA halves and Cys5′tRNA halves), bearing terminal oligonucleotide (TOG) motifs (four to five guanine residues) at their 5′ termini, form intermolecular RNA G-quadruplexes (RG4) to displace the translation initiation factor eIF4F complex from mRNAs.97,98 Additionally, these tRNA halves were associated with the YB-1 (YBX) protein to facilitate the assembly of SGs (Figure 6A).97,99, 100, 101 However, the YB-1 protein is dispensable for displacing translation initiation factors from capped mRNAs,101 indicating that other mechanisms are required for displacing translation initiation factors. Anderson’s group further investigated the mechanism by which RG4 displace translation initiation factors from mRNAs and found that RG4 binds to eIF4G to impair 40S ribosome scanning on mRNAs, leading to the formation of eIF2α-independent stress granules.102
Guzzi et al.103 identified that global translation is dependent upon the presence of a pseudouridine on tsRNAs in embryonic and hematopoietic stem cells. 18-nt TOG-bearing 5′tsRNAs with a pseudouridine molecule at the 8th position bind to polyadenylate-binding protein 1 (PABC1), displacing PABPC1 and translation initiation factors from mRNAs, leading to global translation inhibition; conversely, tsRNAs without pseudouridine did not affect translation (Figure 6B).
Gebetsberger et al.18 identified that fourteen different 5′tsRNAs of 20–44 nt in length were stress-dependent, ribosome-associated ncRNAs in Haloferax volcanii. In particular, 26-nt ValGAC5′tsRNA (ValtRF) binds to the polysome and 30S subunit during high pH stress and suppresses global translation by inhibiting peptide bond formation in vitro. A further study revealed that ValGAC5′tsRNA displaces mRNA from the translation initiation complex.104 tsRNAs were also shown to interact with ribosome-associated aminoacyl-tRNA synthetases in yeast and inhibited in vitro translation through effects on tRNA aminoacylation.105
Recently, ribosome-associated translation stimulation was also reported. Fricker et al.106 analyzed the small non-coding RNA interactome of ribosomes in Trypanosoma brucei during different growth conditions and life stages and found that Thr3′tRNA half (tRNAThr 3′half) was markedly induced under nutrient deprivation. During the recovery phase from starvation conditions, Thr3′tRNA half bound to both ribosomes and polysomes to enhance translation by facilitating mRNA loading onto actively translating ribosomes (Figure 6C).
Keam et al.107 performed immunoprecipitation and SILAC (stable isotope labeling with amino acids in cell culture) mass spectroscopy and revealed that 19-nt Gln5′tsRNA (5′ tRF Gln19) was associated with the human multisynthetase complex, which increases ribosomal and poly(A)-binding protein translation.
Ribosome Biogenesis and Enhanced Translation
Different from the global translation regulation, tsRNAs can regulate mRNA translation in a sequence-specific manner. Kim et al.52 found that a specific tsRNA derived from the 3′ end of tRNALeu(CAG) could selectively enhance the translation of specific RPS mRNAs. LeuCAG3′tsRNA binds to double-stranded regions in ribosomal protein S28 and S15 (RPS28 and RPS15) mRNAs in a sequence-specific manner and enhances their translation by altering the secondary structure of the target region from human cancer cells. RPS28 is especially required for 18S ribosomal RNA processing.108 Therefore, inhibition of LeuCAG3′tsRNA reduced the abundance of RPS28 and RPS15 proteins and, in turn, impaired ribosome biogenesis, leading to apoptosis in HeLa, HEK293, and HCT-116 cells. This inhibition also suppressed human hepatocellular carcinoma growth in a patient-derived orthotopic xenograft model in mice (Figure 6D).52 The target site of LeuCAG3′tsRNA is conserved in many vertebrate species. LeuCAG3′tsRNA-regulated mouse RPS28 translation occurs in a similar manner in humans, suggesting that LeuCAG3′tsRNA-regulated translation might be conserved in vertebrates.109 Kim et al.109 also determined that LeuCAG3′tsRNA regulates translation after initiation in humans and mice. These studies still raise questions about which tsRNAs play a role in translation in a manner similar to LeuCAG3′tsRNA.
This work gives an additional layer to the complexity of translational regulation and provides a potential therapeutic target for cancer treatment. Below, we show another tsRNA may act as a tumor suppressor.
Destabilization of Oncogenic Transcripts
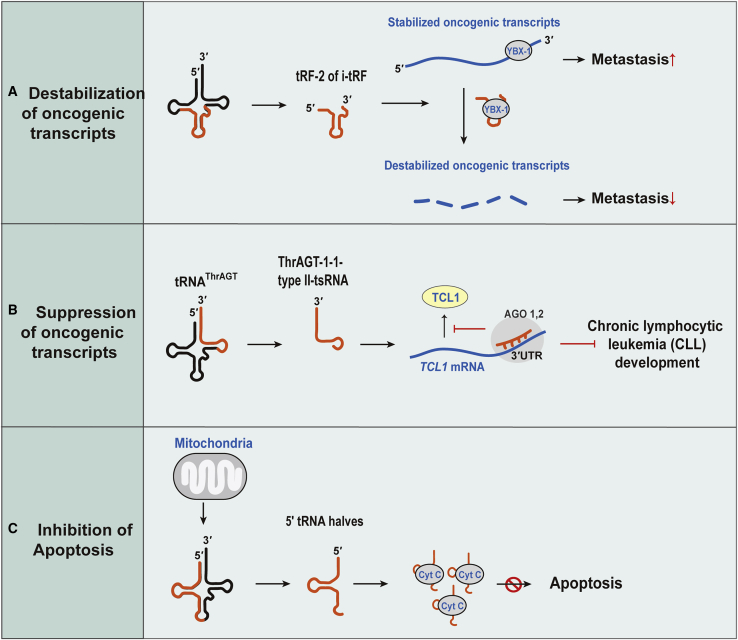
Goodarzi et al.110 identified that a subset of internal tsRNAs (termed as tRF-2 or i-tRF) are induced in breast cancer cells under hypoxic conditions. They are derived from GluYTC, AspGTC, GlyTCC, and TyrGTA mature tRNAs. Their cleavage position and length vary, but mostly anti-codon regions are included in these tsRNAs. A common sequence motif (SCUBYC) from these tsRNAs is similar to the target sequences of the YB-1 protein, which binds to the 3′ UTR of multiple oncogenic gene transcripts to stabilize them. The induced internal tsRNAs competitively displace target oncogenic transcripts from the YB-1 protein and decrease the stability of those transcripts, resulting in suppression of the metastatic progression of breast cancer. They are present at significantly lower levels in highly metastatic breast cancer cells, and more YB-1 protein is known to be present in metastatic cancer cells, implying that highly metastatic cells have a mechanism to evade the suppressive function of the internal tsRNAs (Figure 7A). Although tiRNAs are associated with the YB-1 protein via their TOG motif, internal tsRNAs do not have a TOG motif.110 These studies revealed a novel mechanism for suppression of tumors by tsRNAs, and their biogenesis warrants future investigation.
Figure 7.
The Biological Role of tsRNA in Cancer Progression and Apoptosis
(A) Destabilization of oncogenic transcripts. YBX1 binds to oncogenic transcripts and protects them from degradation. Internal tsRNAs interact with YBX1, leading to sequestration of YBX1. As a result, oncogenic transcripts are degraded. (B) Suppression of oncogenic transcripts is shown. ThrAGT-1-1-typeII-tsRNA binds to TCL1 mRNA and post-transcriptionally represses its protein product, leading to inhibition of CLL development. (C) Inhibition of apoptosis is shown. tsRNA binds to cytochrome C (Cyt C) and inhibits apoptosome formation, thus promoting cell survival.
Balatti et al.111 determined that ThrAGT-1-1-typeII-tsRNA suppresses a T cell leukemia/lymphoma 1 (TCL1) gene, a key oncogene in the development of aggressive CLL, through binding to its 3′ UTR like a miRNA. Loss of ThrAGT-1-1-typeII-tsRNA upregulates TCL1 expression, leading to CLL progression (Figure 7B).
Nonetheless, here and unlike the studies by Kim et al.52 showing downregulation of a LeuCAG3′tsRNA may be a suitable cancer target, these tsRNAs would have to be added back or upregulated if they were to be potential cancer targets. Potential therapeutic strategies of tsRNA addition or downregulation could be similar to strategies used to regulate tumor suppressor versus proto-oncogene, respectively.
tsRNAs and Apoptosis
Another activity of tsRNA that might be useful for cancer treatment is related to the induction of apoptosis. Saikia et al.112,113 revealed that angiogenin induced tRNA halves from mouse embryonic fibroblast (MEF) cells after interaction with released cytochrome c (Cyt c) in a ribonucleoprotein (Cyt c-RNP) complex. Because Cyt c is a component of the apoptosome, these tRNA halves inhibit apoptosome assembly and ultimately lead to the protection of cells from apoptosis under hyperosmotic stress (Figure 7C). They also found that angiogenin treatment protected primary cortical neurons under hyperosmotic stress from apoptosis. Their results suggest a subset of tRNA halves play a role in cell viability using a function distinct from translational inhibition under different stress conditions, and the mechanisms by which their roles are regulated need to be determined.
Intriguingly, full-length mature tRNAs also inhibit apoptosis through direct binding to Cyt c, followed by the suppression of apoptosome formation in the absence of stress,114 suggesting that full-length mature tRNAs and tRNA halves play a role in protecting cells from apoptosis under normal or stress conditions, respectively. Why do cells require two different molecules for cytoprotection from apoptosis? What is the role of mature tRNA in cytoprotection during stress? How is binding to Cyt c or inhibition of global translation regulated under stress? New efforts to answer these questions may contribute to our understanding of how to protect cells from apoptosis.
In addition to the above findings, new studies have shed some light on how higher concentrations of tsRNAs might be related to apoptosis in cancer cells. Zhou et al.115 determined that tsRNA-26576 activated cellular multiplication and migration but suppressed cellular apoptosis in breast cancer cells. La Ferlita et al.116 identified around 300 tsRNAs in the National Cancer Institute cancer cell lines (NCI-60) and The Cancer Genome Atlas (TCGA) databases, analyzed their expression profiles, and developed a public database, tRFexplorer. Veneziano et al.117 determined 964 type I tsRNAs were upregulated more than 2-fold, although 701 type I tsRNAs were downregulated at least 2-fold in CLL compared to normal B cells. These data suggest that type I tsRNAs could be used for biomarkers for cancer diagnosis, including CLL, and might affect apoptosis in cancer cells.
At this point in time, the role of differentially expressed tsRNA in various cancers is not well understood, and further study is needed to establish the value of each in its particular context for future anti-cancer therapies.
tsRNAs in Neurodegenerative Disease
The last tsRNA function to be discussed is related to neurodegenerative diseases. Hanada et al.26 identified that a distinct class of tsRNAs is processed from intron-containing pre-tRNAs. Tyrosine pre-tRNA-derived fragments accumulate in kinase-dead CLP1 (Clp1K/K) MEF cells, the spinal cords of C57BL/6 Clp1K/K neonatal mice, and 4-month-old CBA/J Clp1K/K mice. CLP1 was the first identified mammalian RNA kinase, which phosphorylates the 5′ hydroxyl termini of RNAs and is implicated in tRNA, mRNA, and siRNA maturation.118,119 CLP1 is associated with the tRNA splicing endonuclease (TSEN) complex, which removes the intron within the anti-codon loop of a limited number of pre-tRNAs, producing 5′ and 3′ tRNA exon halves.119,120 A kinase-dead CLP1 causes impairment of pre-tRNA splicing and instead results in the accumulation of a 41- to 46-nt 5′ leader sequence followed by 5′ exon fragments and 3′ exon fragments from tyrosine pre-tRNA, which are localized in the nucleus. These tsRNAs sensitize cells to p53-dependent cell death under oxidative stress but did not affect translation. Even though these tsRNAs are induced under oxidative stress, their localization in the nucleus and angiogenin-independent processing suggests that their biological roles are different from angiogenin-induced tRNA halves.26
Schaffer at al.121 determined that a Clp1 mutation in patients led to progressive brain atrophy and observed an accumulation of intron-containing pre-tRNAs and a depletion of corresponding mature tRNAs, including TyrGTA, IleTAT, and LeuCAA tRNA from these patients-derived neurons. Intriguingly, 5′ exon fragments did not affect cell viability. Instead, the 5′ phosphorylated 3′ exon fragment allowed CLP1 mutant cells to survive under oxidative stress, although the 5′ hydroxylated 3′ exon fragment did not.121 Together, these studies suggest that tsRNAs or tRNA processing are highly correlated with neuronal development and neurodegeneration.
Closing Remarks
Since the first evidence for a functional role of tsRNAs in gene regulation was found a decade ago, the aberrant expression of cytoplasmic tsRNAs has been reported to contribute to various human disease states. Indeed, the wide diversity of tsRNAs represents a novel group of non-coding RNAs and appears to have distinct functions for regulating gene expression or affecting cell physiology. With additional work, the validity and importance of the various tsRNA-described functions will become more apparent. Nevertheless, it seems logical to ask whether there is a direct connection between the canonical function of tRNAs in protein translation and the generation of functional tsRNAs across the genera. Might the diversity and/or redundancy of the tRNA genes in organisms be connected to the function of the various tRNAs and/or tsRNAs? Interestingly, a common theme to many of the current studies discussed in this review suggests that tsRNAs likely play various roles to fine-tuning protein synthesis through differing mechanisms.
Roughly 30%–50% of cellular energy is consumed in protein synthesis,122,123 and the coordination in producing appropriate stoichiometric amounts of the structural RNAs and required protein components is complex. Thus, multi-faceted coordination through feedback between the various protein synthetic components (e.g., tsRNAs) would make sense.
tsRNA modifications also needed to be thoroughly investigated because they are highly correlated with tRNA stability, tsRNA biogenesis, and its function.50,51,91,92,103 Currently, the purification methods of endogenous tsRNAs based on their modification state have just started to be developed.124,125 More-advanced, modification-based tsRNA purification technology will be important for understanding the role of modification on tsRNA and developing tsRNAs as therapeutic drugs. Nevertheless, we have just started to scratch the surface in our understanding of the biological role of these non-coding RNAs. In the next decade, we are likely to see (1) improvement in tsRNA quantification methods and hence better expression profiles during normal growth and development, as well as pathogenic states; (2) a better understanding of tsRNA biogenesis and mechanisms regulating their cellular concentrations; (3) more mechanistic insights into how the tsRNAs regulate gene expression, perhaps at the level of global translation versus selective translation; and (4) insights into the potential of tsRNAs as diagnostic and/or therapeutic targets for various disease states (Figure 8).
Figure 8.
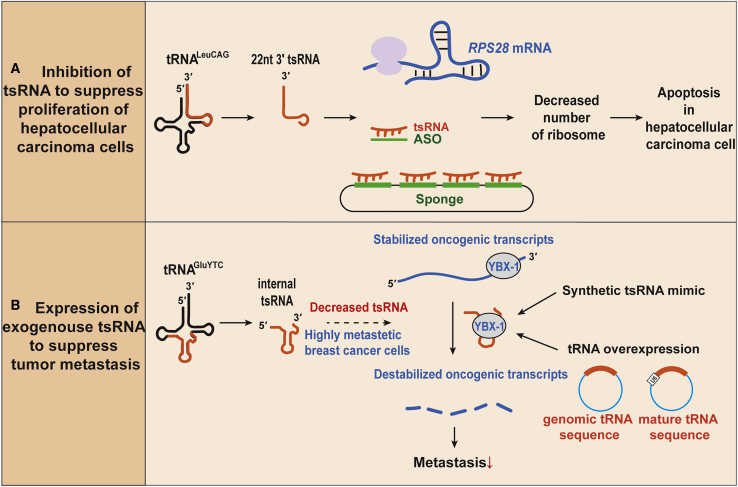
Two Different Strategies Using tsRNAs as Anti-cancer Targets
(A) Inhibition of tsRNA to suppress proliferation of hepatocellular carcinoma. The LeuCAG3′tsRNA directly binds and enhances translation of RPS28 transcripts by the unfolding of their secondary structures. As a result, increased LeuCAG3′tsRNA upregulates the number of ribosomes, promoting proliferation of hepatocellular carcinoma cells. Antisense oligonucleotides (ASOs) complementary to the tsRNA or possibly tsRNA sponges act as competitive inhibitors and “soak up” the endogenous tsRNA. Inhibition of LeuCAG3′tsRNA results in apoptosis and can suppress hepatocellular carcinoma growth. (B) Addition of exogenous tsRNAs derived from tRNAGluYTC to suppress tumor metastasis is shown. These tsRNAs destabilize oncogenic transcripts by competing for binding with the YBX1 protein. In highly metastatic breast cancer cells, the tsRNAs are suppressed, which induces metastatic progression. Addition of synthetic tsRNAs or overexpression of tsRNAs from an tRNA overexpression plasmid can destabilize oncogenic transcripts, leading to inhibition of metastasis.
Author Contributions
H.K.K. and M.A.K. wrote and edited the manuscript. J.-H.Y. prepared the figures.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the National Institutes of Health R01-114483 (M.A.K.) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MIST) (grant no. 2020R1A2C1005942; H.K.K.). We thank Calvin Stephens for critical reading of the manuscript. Mary O’Reilly designed the graphical abstract.
Contributor Information
Hak Kyun Kim, Email: hakyun@cau.ac.kr.
Mark A. Kay, Email: markay@stanford.edu.
References
- 1.Rodnina M.V., Wintermeyer W. The ribosome as a molecular machine: the mechanism of tRNA-mRNA movement in translocation. Biochem. Soc. Trans. 2011;39:658–662. doi: 10.1042/BST0390658. [DOI] [PubMed] [Google Scholar]
- 2.Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- 3.Chan P.P., Lowe T.M. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016;44(D1):D184–D189. doi: 10.1093/nar/gkv1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Telonis A.G., Loher P., Kirino Y., Rigoutsos I. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front. Genet. 2014;5:344. doi: 10.3389/fgene.2014.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schimmel P. The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018;19:45–58. doi: 10.1038/nrm.2017.77. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi Y., Momoi M.Y., Tominaga K., Momoi T., Nihei K., Yanagisawa M., Kagawa Y., Ohta S. A point mutation in the mitochondrial tRNA(Leu)(UUR) gene in MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes) Biochem. Biophys. Res. Commun. 1990;173:816–822. doi: 10.1016/s0006-291x(05)80860-5. [DOI] [PubMed] [Google Scholar]
- 7.Shoffner J.M., Lott M.T., Lezza A.M., Seibel P., Ballinger S.W., Wallace D.C. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell. 1990;61:931–937. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 8.Abbott J.A., Francklyn C.S., Robey-Bond S.M. Transfer RNA and human disease. Front. Genet. 2014;5:158. doi: 10.3389/fgene.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Budde B.S., Namavar Y., Barth P.G., Poll-The B.T., Nürnberg G., Becker C., van Ruissen F., Weterman M.A., Fluiter K., te Beek E.T. tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat. Genet. 2008;40:1113–1118. doi: 10.1038/ng.204. [DOI] [PubMed] [Google Scholar]
- 10.Borek E., Baliga B.S., Gehrke C.W., Kuo C.W., Belman S., Troll W., Waalkes T.P. High turnover rate of transfer RNA in tumor tissue. Cancer Res. 1977;37:3362–3366. [PubMed] [Google Scholar]
- 11.Speer J., Gehrke C.W., Kuo K.C., Waalkes T.P., Borek E. tRNA breakdown products as markers for cancer. Cancer. 1979;44:2120–2123. doi: 10.1002/1097-0142(197912)44:6<2120::aid-cncr2820440623>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y.S., Shibata Y., Malhotra A., Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamasaki S., Ivanov P., Hu G.-F., Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 2009;185:35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haussecker D., Huang Y., Lau A., Parameswaran P., Fire A.Z., Kay M.A. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar P., Anaya J., Mudunuri S.B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gingold H., Tehler D., Christoffersen N.R., Nielsen M.M., Asmar F., Kooistra S.M., Christophersen N.S., Christensen L.L., Borre M., Sørensen K.D. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Dittmar K.A., Goodenbour J.M., Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebetsberger J., Zywicki M., Künzi A., Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozenski J., Crain P.F., McCloskey J.A. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson J.T., Droogmans L. Fine-Tuning of RNA Functions by Modification and Editing, H. Grosjean. Springer; 2005. Biosynthesis and function of 1-methyladenosine in transfer RNA; pp. 121–139. [Google Scholar]
- 21.Lee S.R., Collins K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005;280:42744–42749. doi: 10.1074/jbc.M510356200. [DOI] [PubMed] [Google Scholar]
- 22.Honda S., Loher P., Shigematsu M., Palazzo J.P., Suzuki R., Imoto I., Rigoutsos I., Kirino Y. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. USA. 2015;112:E3816–E3825. doi: 10.1073/pnas.1510077112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telonis A.G., Loher P., Honda S., Jing Y., Palazzo J., Kirino Y., Rigoutsos I. Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies. Oncotarget. 2015;6:24797–24822. doi: 10.18632/oncotarget.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar P., Kuscu C., Dutta A. Biogenesis and function of transfer RNA-related fragments (tRFs) Trends Biochem. Sci. 2016;41:679–689. doi: 10.1016/j.tibs.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodarzi H., Liu X., Nguyen H.C.B., Zhang S., Fish L., Tavazoie S.F. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell. 2015;161:790–802. doi: 10.1016/j.cell.2015.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanada T., Weitzer S., Mair B., Bernreuther C., Wainger B.J., Ichida J., Hanada R., Orthofer M., Cronin S.J., Komnenovic V. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature. 2013;495:474–480. doi: 10.1038/nature11923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar P., Mudunuri S.B., Anaya J., Dutta A. tRFdb: a database for transfer RNA fragments. Nucleic Acids Res. 2015;43:D141–D145. doi: 10.1093/nar/gku1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng L.-L., Xu W.-L., Liu S., Sun W.-J., Li J.-H., Wu J., Yang J.H., Qu L.H. tRF2Cancer: a web server to detect tRNA-derived small RNA fragments (tRFs) and their expression in multiple cancers. Nucleic Acids Res. 2016;44(W1):W185–W193. doi: 10.1093/nar/gkw414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couvillion M.T., Sachidanandam R., Collins K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010;24:2742–2747. doi: 10.1101/gad.1996210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 31.Babiarz J.E., Ruby J.G., Wang Y., Bartel D.P., Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole C., Sobala A., Lu C., Thatcher S.R., Bowman A., Brown J.W.S., Green P.J., Barton G.J., Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maute R.L., Schneider C., Sumazin P., Holmes A., Califano A., Basso K., Dalla-Favera R. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc. Natl. Acad. Sci. USA. 2013;110:1404–1409. doi: 10.1073/pnas.1206761110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeung M.L., Bennasser Y., Watashi K., Le S.-Y., Houzet L., Jeang K.-T. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 2009;37:6575–6586. doi: 10.1093/nar/gkp707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Z., Ender C., Meister G., Moore P.S., Chang Y., John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res. 2012;40:6787–6799. doi: 10.1093/nar/gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuscu C., Kumar P., Kiran M., Su Z., Malik A., Dutta A. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA. 2018;24:1093–1105. doi: 10.1261/rna.066126.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Megel C., Hummel G., Lalande S., Ubrig E., Cognat V., Morelle G., Salinas-Giegé T., Duchêne A.M., Maréchal-Drouard L. Plant RNases T2, but not Dicer-like proteins, are major players of tRNA-derived fragments biogenesis. Nucleic Acids Res. 2019;47:941–952. doi: 10.1093/nar/gky1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phizicky E.M., Hopper A.K. tRNA biology charges to the front. Genes Dev. 2010;24:1832–1860. doi: 10.1101/gad.1956510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takaku H., Minagawa A., Takagi M., Nashimoto M. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res. 2003;31:2272–2278. doi: 10.1093/nar/gkg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levitz R., Chapman D., Amitsur M., Green R., Snyder L., Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haiser H.J., Karginov F.V., Hannon G.J., Elliot M.A. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jöchl C., Rederstorff M., Hertel J., Stadler P.F., Hofacker I.L., Schrettl M., Haas H., Hüttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res. 2008;36:2677–2689. doi: 10.1093/nar/gkn123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu H., Feng J., Liu Q., Sun F., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 2009;583:437–442. doi: 10.1016/j.febslet.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 44.Liu S., Chen Y., Ren Y., Zhou J., Ren J., Lee I., Bao X. A tRNA-derived RNA fragment plays an important role in the mechanism of arsenite-induced cellular responses. Sci. Rep. 2018;8:16838–16839. doi: 10.1038/s41598-018-34899-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson D.M., Lu C., Green P.J., Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson D.M., Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Thompson D.M., Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shapiro R., Vallee B.L. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc. Natl. Acad. Sci. USA. 1987;84:2238–2241. doi: 10.1073/pnas.84.8.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuji T., Sun Y., Kishimoto K., Olson K.A., Liu S., Hirukawa S., Hu G.F. Angiogenin is translocated to the nucleus of HeLa cells and is involved in ribosomal RNA transcription and cell proliferation. Cancer Res. 2005;65:1352–1360. doi: 10.1158/0008-5472.CAN-04-2058. [DOI] [PubMed] [Google Scholar]
- 50.Schaefer M., Pollex T., Hanna K., Tuorto F., Meusburger M., Helm M., Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., Lukk M., Lombard P., Treps L., Popis M. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014;33:2020–2039. doi: 10.15252/embj.201489282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim H.K., Fuchs G., Wang S., Wei W., Zhang Y., Park H., Roy-Chaudhuri B., Li P., Xu J., Chu K. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature. 2017;552:57–62. doi: 10.1038/nature25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cozen A.E., Quartley E., Holmes A.D., Hrabeta-Robinson E., Phizicky E.M., Lowe T.M. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat. Methods. 2015;12:879–884. doi: 10.1038/nmeth.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng G., Qin Y., Clark W.C., Dai Q., Yi C., He C., Lambowitz A.M., Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maguire S., Lohman G.J.S., Guan S. A low-bias and sensitive small RNA library preparation method using randomized splint ligation. Nucleic Acids Res. 2020;48:e80. doi: 10.1093/nar/gkaa480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kono N., Arakawa K. Nanopore sequencing: review of potential applications in functional genomics. Dev. Growth Differ. 2019;61:316–326. doi: 10.1111/dgd.12608. [DOI] [PubMed] [Google Scholar]
- 57.Balatti V., Nigita G., Veneziano D., Drusco A., Stein G.S., Messier T.L., Farina N.H., Lian J.B., Tomasello L., Liu C.G. tsRNA signatures in cancer. Proc. Natl. Acad. Sci. USA. 2017;114:8071–8076. doi: 10.1073/pnas.1706908114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farina N.H., Scalia S., Adams C.E., Hong D., Fritz A.J., Messier T.L., Balatti V., Veneziano D., Lian J.B., Croce C.M. Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer. J. Cell. Physiol. 2020;235:5318–5327. doi: 10.1002/jcp.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nolte-‘t Hoen E.N.M., Buermans H.P.J., Waasdorp M., Stoorvogel W., Wauben M.H.M., ’t Hoen P.A.C. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiou N.-T., Kageyama R., Ansel K.M. Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 2018;25:3356–3370.e4. doi: 10.1016/j.celrep.2018.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu L., Li J., Gong Y., Wu Q., Tan S., Sun D., Xu X., Zuo Y., Zhao Y., Wei Y.-Q. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol. Cancer. 2019;18:74. doi: 10.1186/s12943-019-1000-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dhahbi J.M., Spindler S.R., Atamna H., Yamakawa A., Boffelli D., Mote P., Martin D.I. 5′ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics. 2013;14:298. doi: 10.1186/1471-2164-14-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y., Zhang Y., Shi J., Zhang H., Cao Z., Gao X., Ren W., Ning Y., Ning L., Cao Y. Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 2014;6:172–174. doi: 10.1093/jmcb/mjt052. [DOI] [PubMed] [Google Scholar]
- 64.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 65.Karaiskos S., Naqvi A.S., Swanson K.E., Grigoriev A. Age-driven modulation of tRNA-derived fragments in Drosophila and their potential targets. Biol. Direct. 2015;10:51. doi: 10.1186/s13062-015-0081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hasler D., Lehmann G., Murakawa Y., Klironomos F., Jakob L., Grässer F.A., Rajewsky N., Landthaler M., Meister G. The lupus autoantigen La prevents mis-channeling of tRNA fragments into the human microRNA pathway. Mol. Cell. 2016;63:110–124. doi: 10.1016/j.molcel.2016.05.026. [DOI] [PubMed] [Google Scholar]
- 67.Bayfield M.A., Maraia R.J. Precursor-product discrimination by La protein during tRNA metabolism. Nat. Struct. Mol. Biol. 2009;16:430–437. doi: 10.1038/nsmb.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang B., Yang H., Cheng X., Wang D., Fu S., Shen W., Zhang Q., Zhang L., Xue Z., Li Y. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 2017;77:3194–3206. doi: 10.1158/0008-5472.CAN-16-3146. [DOI] [PubMed] [Google Scholar]
- 69.Luo S., He F., Luo J., Dou S., Wang Y., Guo A., Lu J. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res. 2018;46:5250–5268. doi: 10.1093/nar/gky189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Q., Lee I., Ren J., Ajay S.S., Lee Y.S., Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 2013;21:368–379. doi: 10.1038/mt.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou J., Liu S., Chen Y., Fu Y., Silver A.J., Hill M.S., Lee I., Lee Y.S., Bao X. Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J. Gen. Virol. 2017;98:1600–1610. doi: 10.1099/jgv.0.000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng J., Ptashkin R.N., Chen Y., Cheng Z., Liu G., Phan T., Deng X., Zhou J., Lee I., Lee Y.S., Bao X. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 2015;23:1622–1629. doi: 10.1038/mt.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Siomi M.C., Sato K., Pezic D., Aravin A.A. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 2011;12:246–258. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 74.Couvillion M.T., Lee S.R., Hogstad B., Malone C.D., Tonkin L.A., Sachidanandam R., Hannon G.J., Collins K. Sequence, biogenesis, and function of diverse small RNA classes bound to the Piwi family proteins of Tetrahymena thermophila. Genes Dev. 2009;23:2016–2032. doi: 10.1101/gad.1821209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Couvillion M.T., Bounova G., Purdom E., Speed T.P., Collins K. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol. Cell. 2012;48:509–520. doi: 10.1016/j.molcel.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang X., He X., Liu C., Liu J., Hu Q., Pan T., Duan X., Liu B., Zhang Y., Chen J. IL-4 inhibits the biogenesis of an epigenetically suppressive PIWI-interacting RNA to upregulate CD1a molecules on monocytes/dendritic cells. J. Immunol. 2016;196:1591–1603. doi: 10.4049/jimmunol.1500805. [DOI] [PubMed] [Google Scholar]
- 77.Pekarsky Y., Balatti V., Palamarchuk A., Rizzotto L., Veneziano D., Nigita G., Rassenti L.Z., Pass H.I., Kipps T.J., Liu C.G., Croce C.M. Dysregulation of a family of short noncoding RNAs, tsRNAs, in human cancer. Proc. Natl. Acad. Sci. USA. 2016;113:5071–5076. doi: 10.1073/pnas.1604266113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Slotkin R.K., Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 79.Zhang Y., Shi J., Chen Q. tsRNAs: new players in mammalian retrotransposon control. Cell Res. 2017;27:1307–1308. doi: 10.1038/cr.2017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schorn A.J., Gutbrod M.J., LeBlanc C., Martienssen R. LTR-retrotransposon control by tRNA-derived small RNAs. Cell. 2017;170:61–71.e11. doi: 10.1016/j.cell.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ruggero K., Guffanti A., Corradin A., Sharma V.K., De Bellis G., Corti G., Grassi A., Zanovello P., Bronte V., Ciminale V., D’Agostino D.M. Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J. Virol. 2014;88:3612–3622. doi: 10.1128/JVI.02823-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heard E., Martienssen R.A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014;157:95–109. doi: 10.1016/j.cell.2014.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rando O.J., Simmons R.A. I’m eating for two: parental dietary effects on offspring metabolism. Cell. 2015;161:93–105. doi: 10.1016/j.cell.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peng H., Shi J., Zhang Y., Zhang H., Liao S., Li W., Lei L., Han C., Ning L., Cao Y. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thomson T., Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 87.Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Q., Lin J., Liu M., Li R., Tian B., Zhang X., Xu B., Liu M., Zhang X., Li Y. Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci. Adv. 2016;2:e1501482. doi: 10.1126/sciadv.1501482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 90.Kiani J., Grandjean V., Liebers R., Tuorto F., Ghanbarian H., Lyko F., Cuzin F., Rassoulzadegan M. RNA-mediated epigenetic heredity requires the cytosine methyltransferase Dnmt2. PLoS Genet. 2013;9:e1003498. doi: 10.1371/journal.pgen.1003498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang Y., Zhang X., Shi J., Tuorto F., Li X., Liu Y., Liebers R., Zhang L., Qu Y., Qian J. Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat. Cell Biol. 2018;20:535–540. doi: 10.1038/s41556-018-0087-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Short A.K., Yeshurun S., Powell R., Perreau V.M., Fox A., Kim J.H., Pang T.Y., Hannan A.J. Exercise alters mouse sperm small noncoding RNAs and induces a transgenerational modification of male offspring conditioned fear and anxiety. Transl. Psychiatry. 2017;7:e1114. doi: 10.1038/tp.2017.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Truitt M.L., Ruggero D. New frontiers in translational control of the cancer genome. Nat. Rev. Cancer. 2016;16:288–304. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang S., Sun L., Kragler F. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol. 2009;150:378–387. doi: 10.1104/pp.108.134767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dang Y., Kedersha N., Low W.-K., Romo D., Gorospe M., Kaufman R., Anderson P., Liu J.O. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 2006;281:32870–32878. doi: 10.1074/jbc.M606149200. [DOI] [PubMed] [Google Scholar]
- 96.Farny N.G., Kedersha N.L., Silver P.A. Metazoan stress granule assembly is mediated by P-eIF2alpha-dependent and -independent mechanisms. RNA. 2009;15:1814–1821. doi: 10.1261/rna.1684009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ivanov P., Emara M.M., Villen J., Gygi S.P., Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell. 2011;43:613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lyons S.M., Gudanis D., Coyne S.M., Gdaniec Z., Ivanov P. Identification of functional tetramolecular RNA G-quadruplexes derived from transfer RNAs. Nat. Commun. 2017;8:1127. doi: 10.1038/s41467-017-01278-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Emara M.M., Ivanov P., Hickman T., Dawra N., Tisdale S., Kedersha N., Hu G.F., Anderson P. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ivanov P., O’Day E., Emara M.M., Wagner G., Lieberman J., Anderson P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc. Natl. Acad. Sci. USA. 2014;111:18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lyons S.M., Achorn C., Kedersha N.L., Anderson P.J., Ivanov P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res. 2016;44:6949–6960. doi: 10.1093/nar/gkw418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lyons S.M., Kharel P., Akiyama Y., Ojha S., Dave D., Tsvetkov V., Merrick W., Ivanov P., Anderson P. eIF4G has intrinsic G-quadruplex binding activity that is required for tiRNA function. Nucleic Acids Res. 2020;48:6223–6233. doi: 10.1093/nar/gkaa336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Guzzi N., Cieśla M., Ngoc P.C.T., Lang S., Arora S., Dimitriou M., Pimková K., Sommarin M.N.E., Munita R., Lubas M. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell. 2018;173:1204–1216.e26. doi: 10.1016/j.cell.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 104.Gebetsberger J., Wyss L., Mleczko A.M., Reuther J., Polacek N. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol. 2017;14:1364–1373. doi: 10.1080/15476286.2016.1257470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mleczko A.M., Celichowski P., Bąkowska-Żywicka K. Transfer RNA-derived fragments target and regulate ribosome-associated aminoacyl-transfer RNA synthetases. Biochim. Biophys. Acta Gene Regul. Mech. 2018;1861:647–656. doi: 10.1016/j.bbagrm.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 106.Fricker R., Brogli R., Luidalepp H., Wyss L., Fasnacht M., Joss O., Zywicki M., Helm M., Schneider A., Cristodero M., Polacek N. A tRNA half modulates translation as stress response in Trypanosoma brucei. Nat. Commun. 2019;10:118. doi: 10.1038/s41467-018-07949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Keam S.P., Sobala A., Have S.T., Hutvagner G. tRNA-derived RNA fragments associate with human multisynthetase complex (MSC) and modulate ribosomal protein translation. J. Proteome Res. 2017;16:413–420. doi: 10.1021/acs.jproteome.6b00267. [DOI] [PubMed] [Google Scholar]
- 108.Robledo S., Idol R.A., Crimmins D.L., Ladenson J.H., Mason P.J., Bessler M. The role of human ribosomal proteins in the maturation of rRNA and ribosome production. RNA. 2008;14:1918–1929. doi: 10.1261/rna.1132008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim H.K., Xu J., Chu K., Park H., Jang H., Li P., Valdmanis P.N., Zhang Q.C., Kay M.A. A tRNA-derived small RNA regulates ribosomal protein S28 protein levels after translation initiation in humans and mice. Cell Rep. 2019;29:3816–3824.e4. doi: 10.1016/j.celrep.2019.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goodarzi H., Nguyen H.C.B., Zhang S., Dill B.D., Molina H., Tavazoie S.F. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell. 2016;165:1416–1427. doi: 10.1016/j.cell.2016.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Balatti V., Rizzotto L., Miller C., Palamarchuk A., Fadda P., Pandolfo R., Rassenti L.Z., Hertlein E., Ruppert A.S., Lozanski A. TCL1 targeting miR-3676 is codeleted with tumor protein p53 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2015;112:2169–2174. doi: 10.1073/pnas.1500010112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Saikia M., Krokowski D., Guan B.-J., Ivanov P., Parisien M., Hu G.-F., Anderson P., Pan T., Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J. Biol. Chem. 2012;287:42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saikia M., Jobava R., Parisien M., Putnam A., Krokowski D., Gao X.-H., Guan B.J., Yuan Y., Jankowsky E., Feng Z. Angiogenin-cleaved tRNA halves interact with cytochrome c, protecting cells from apoptosis during osmotic stress. Mol. Cell. Biol. 2014;34:2450–2463. doi: 10.1128/MCB.00136-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mei Y., Yong J., Liu H., Shi Y., Meinkoth J., Dreyfuss G., Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol. Cell. 2010;37:668–678. doi: 10.1016/j.molcel.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou J., Wan F., Wang Y., Long J., Zhu X. Small RNA sequencing reveals a novel tsRNA-26576 mediating tumorigenesis of breast cancer. Cancer Manag. Res. 2019;11:3945–3956. doi: 10.2147/CMAR.S199281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.La Ferlita A., Alaimo S., Veneziano D., Nigita G., Balatti V., Croce C.M., Ferro A., Pulvirenti A. Identification of tRNA-derived ncRNAs in TCGA and NCI-60 panel cell lines and development of the public database tRFexplorer. Database (Oxford) 2019;2019:baz115. doi: 10.1093/database/baz115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Veneziano D., Tomasello L., Balatti V., Palamarchuk A., Rassenti L.Z., Kipps T.J., Pekarsky Y., Croce C.M. Dysregulation of different classes of tRNA fragments in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2019;116:24252–24258. doi: 10.1073/pnas.1913695116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weitzer S., Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 119.Weitzer S., Hanada T., Penninger J.M., Martinez J. CLP1 as a novel player in linking tRNA splicing to neurodegenerative disorders. Wiley Interdiscip. Rev. RNA. 2015;6:47–63. doi: 10.1002/wrna.1255. [DOI] [PubMed] [Google Scholar]
- 120.Trotta C.R., Miao F., Arn E.A., Stevens S.W., Ho C.K., Rauhut R., Abelson J.N. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 121.Schaffer A.E., Eggens V.R.C., Caglayan A.O., Reuter M.S., Scott E., Coufal N.G., Silhavy J.L., Xue Y., Kayserili H., Yasuno K. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell. 2014;157:651–663. doi: 10.1016/j.cell.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Buttgereit F., Brand M.D. A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 1995;312:163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Russell J.B., Cook G.M. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Drino A., Oberbauer V., Troger C., Janisiw E., Anrather D., Hartl M., Kaiser S., Kellner S., Schaefer M.R. Production and purification of endogenously modified tRNA-derived small RNAs. RNA Biol. 2020;17:1104–1115. doi: 10.1080/15476286.2020.1733798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Akiyama Y., Kharel P., Abe T., Anderson P., Ivanov P. Isolation and initial structure-functional characterization of endogenous tRNA-derived stress-induced RNAs. RNA Biol. 2020;17:1116–1124. doi: 10.1080/15476286.2020.1732702. [DOI] [PMC free article] [PubMed] [Google Scholar]