Abstract
There is growing evidence that implicates epigenetic modification in the pathogenesis of systemic sclerosis (SSc). The complexity of epigenetic regulation and its dynamic nature complicate the investigation of its role in the disease. We will review the current literature for factors that link epigenetics to SSc by discussing DNA methylation, histone acetylation and methylation, and non-coding RNAs (ncRNAs), particularly microRNA changes in endothelial cells, fibroblasts (FBs), and lymphocytes. These three cell types are significantly involved in the early stages and throughout the course of the disease and are particularly vulnerable to epigenetic regulation. The pathogenesis of SSc is likely related to modifications of the epigenome by environmental signals in individuals with a specific genetic makeup. The epigenome is an attractive therapeutic target; however, successful epigenetics-based treatments require a better understanding of the molecular mechanisms controlling the epigenome and its alteration in the disease.
Keywords: Epigenetics, systemic sclerosis, angiogenesis, autoimmune disease, MicroRNAs DNA methylation
Introduction
Systemic sclerosis (SSc) is a chronic, heterogeneous multisystem autoimmune connective tissue disease. It is characterized by three pathological processes: vascular injury and endothelial dysfunction, resulting in vascular intimal proliferation and remodeling, vasoconstriction, and defective angiogenesis; immune dysregulation, resulting in cell-mediated immunity and autoantibodies production; and fibroblast (FB) activation, resulting in excessive collagen and extracellular matrix (ECM) production and accumulation in the skin and other organs (1–4). Of note, vascular manifestations can precede other disease manifestations by several years. The pathogenesis of SSc is still unclear. The disease occurs in a multi-step process involving interaction between genetic and environmental factors in a genetically susceptible individual. This process starts with microvascular endothelial cell dysfunction and overexpression of adhesion molecules and chemokines, attracting diverse types of immune cells, including T cells and activated B cells. These cells release their cytokines when they accumulate in the tissue, stimulating FBs to produce excessive amounts of collagen and other ECM components (5).
SSc is classified as limited cutaneous SSc (lcSSc) when it affects limited portions of skin and has minimal systemic involvement and as diffuse cutaneous SSc (dcSSc) when it affects large portions of skin and involves multiple internal organs. SSc is more prevalent in women with an overall female-to-male ratio of 3:1 or greater and marked ethnic differences (1, 6). There is no clear causative factor for SSc. Genetics plays an important role in the pathogenesis of SSc; however, despite the identification of multiple genetic risk loci such as the major histocompatibility complex (MHC) II (7), which are associated with increased susceptibility to the disease, genetic factors alone do not explain the occurrence of the disease (1–4, 8). This conclusion is supported by the low concordance rate of SSc among monozygotic twins (approximately 4.2%, the lowest among all autoimmune diseases), which is the same as the SSc concordance rate in dizygotic twins (1–6, 9, 10). Thus, it is reasonable to believe that environmental factors play a central role in disease pathogenesis. This view is supported by the geographic clustering of SSc (2). The environmental factors that have been linked to SSc include exposure to silica, viral infections, vinyl chloride, ultraviolet light, and medications such as bleomycin (5, 11). Epigenetic regulation links genetic and environmental factors in the pathogenesis by modifying the gene expression.
Epigenetics is defined as heritable variations in gene expression patterns without alteration in the DNA sequence. These modifications are accomplished through various mechanisms, including DNA methylation, histone acetylation and methylation, and changes in non-coding RNAs (ncRNAs), particularly microRNAs (1). Changes in all three epigenetic modifiers have been described for SSc. Overall, epigenetic mechanisms can alter the chromatin structure in a manner that controls the access to the regulatory sequences, leading to over- or under-expression of the target gene. Epigenetic changes can be swift but reversible and are affected by environmental factors as well as endogenous mediators (1–5, 8, 10, 12–15). The same gene sequence can give rise to multiple cell phenotypes through epigenetic modifications. The cellular inheritance of epigenetic changes during cell replication serves to maintain the particular cellular phenotype, whether this phenotype is normal or pathological. In SSc, epigenetic mechanisms are proposed to be the reason for the persistence of the activated phenotype of SSc FBs over multiple generations (2).
To understand the role of epigenetics in DNA modification, it is helpful to review the chromosome structure (16). Each chromosome is composed of two chromatids, and each chromatid is composed of compressed and folded chromatin. Chromatin is composed of condensed nucleosomes. Each nucleosome comprises eight histone proteins and 146 nucleotides wrapped around them. The chromatin structure can be modified into different conformations through interactions among the histones. The degree of chromatin condensation is regulated by epigenetic mechanisms. Chromatin can exist in two forms: euchromatin, which is the decondensed and relaxed form, and heterochromatin, which is the condensed form. For the chromatin to be transcriptionally active, it has to be decondensed to allow transcription factors to reach the DNA promoter region, which lies upstream of the coding region (3, 15).
DNA methylation
DNA methylation is the process by which a -CH3 (methyl) group, derived from S-adenosyl-L-methionine, covalently binds to position 5 in the cytosine ring within the CpG dinucleotides in the DNA. This process is catalyzed by DNA methyltransferases (DNMTs), namely DNMT1, DNMT3a, and DNMT3b. Even DNMT3L belongs to this group; however, it has no catalytic activity. Nonetheless, it stimulates de novo methylation of cytosine by DNMT3a and DNMT3b (15). DNMT3a and DNMT3b play important roles in the de novo methylation and generation of specific DNA methylation patterns. DNMT3a has a significant role in genomic imprinting during gametogenesis (17–19), whereas DNMT3b has an important role in embryonic development. DNMT1 is responsible for the maintenance of DNA methylation patterns during replication, ensuring faithful inheritance of epigenetic changes during replication and mitotic divisions (20). Therefore, any abnormalities or changes in the inherited methylation patterns can be attributed to DNMT1 dysfunction (21). DNA methylation is the proposed mechanism for genomic imprinting and X chromosome inactivation (21, 22).
DNA methylation of the cytosine in the CpG sites marks the heterochromatin closed structure, resulting in transcriptional silencing by direct physical interference with the binding of transcriptional factors or by binding to methyl-CpG-binding proteins such as MeCP2 and methylated DNA-binding domain (MBD)-containing proteins (23). DNA methylation can be reversed by a demethylation process that can be active or passive. Active demethylation is mediated by 10–11 translocation oxidases and passive demethylation occurs through replication in the absence of DNMT1 activity (2). The demethylation of CpG islands in the gene promoter region results in increased transcriptional activity.
DNA methylation plays a significant role in SSc pathogenesis as described above in the three key cells associated with disease pathogenesis—FBs, endothelial cells, and lymphocytes. These cells mediate the major SSc manifestations of fibrosis, vasculopathy, and immune dysregulation.
Fibroblasts (FBs)
Fibrosis is the result of excessive production of collagen and ECM components, and defective remodeling of ECM. Previous work in our lab demonstrated that the transcription factor Friend leukemia integration 1 (Fli1), encoded by the FLI1 gene and a potent negative regulator of collagen transcription, is downregulated in SSc skin and FBs. Further examination of the FLI1 gene displayed substantial methylation of CpG sites in the promoter region in SSc FBs (24, 25). The downregulation of Fli1 and the overexpression of collagen were reversed in vitro when the SSc FBs were treated with 5-azacytidine, an inhibitor of DNMT1 and a universal demethylating agent (1). Moreover, MBD-1, MBD-2, and MeCP-2 were significantly elevated in SSc FBs compared with healthy FBs (1). Genome-wide DNA methylation investigations confirmed hypomethylation and overexpression of two collagen genes (COL23A1 and COL4A2) in dcSSc FBs and lcSSc FBs compared with control FBs, in addition to hypomethylation of several other collagen genes in each subset separately (26). Moreover, TNXB was shown to be hypomethylated in dcSSc FBs and lcSSc FBs (26). TNXB encodes a member of the tenascin family of ECM glycoproteins, which are involved in matrix maturation (2). These observations indicate that the methylation status of the collagen and ECM-protein encoding genes is intimately involved in ECM accumulation in SSc.
Endothelial cells
One of the major functions of microvascular endothelial cells (MVECs) is to control the vascular tone by maintaining a healthy balance between vasoconstrictors and vasodilators. Nitric oxide (NO) is one of the most important vasodilators and is synthesized by endothelial nitric oxide synthase (eNOS) that is encoded by the gene NOS3. NO has vasodilatory, antithrombotic, antiplatelet, and antioxidant properties (27). Significant downregulation of NOS3 expression was observed in SSc MVECs. In addition, substantial methylation of the CpG sites in the promoter region of NOS3 was identified in association with gene repression. The addition of 5-azacytidine reversed CpG sites methylation, leading to normalization of NOS3 expression. This observation may explain the propensity for vasospasm and platelet activation in SSc (1, 28). The bone morphogenic protein receptor II (BMPRII) promoter region was found to be intensely methylated in SSc MVECs (29). BMPRII is a member of the transforming growth factor-β (TGF-β) superfamily that coordinates cell proliferation, differentiation, and survival. Signaling through BMPRII in MVECs results in apoptosis resistance and promotes the survival of MVECs. Downregulation of BMPRII expression was observed in SSc MVECs with enhanced response to apoptotic signals, including growth factor withdrawal (29). The heightened response to apoptosis induction was reversed with exposure to 5-azacytidine. This mechanism may enhance endothelial apoptosis in SSc.
Recent work demonstrated significant genome-wide DNA methylation anomaly in SSc MVECs, characterized by differential methylation of 2,455 CpG sites, representing 1,301 genes. The hypermethylated genes identified in SSc MVECs were NOS1, DNMT3A, DNMT3B, HDAC4, and ANGPT2. Gene ontology analysis demonstrated enrichment of genes involved in angiogenesis (30).
Lymphocytes
Both CD4+ and CD8+ cells participate in the pathogenesis of SSc. CD8+ cells infiltrate the tissue at an early phase of the inflammatory response, followed by CD4+ predominance when fibrosis is evident (31). CD4+ T cells in SSc are characterized by global hypomethylation with decreased expression and activity of DNMT1 (32). The global hypomethylation of DNA in CD4+ T cells may lead to the reactivation of endoparasitic sequences, such as LINE-1 retrotransposable elements that contribute to autoimmune activities (33). In animal models of autoimmunity, defects in the extracellular signal-regulated kinase signaling pathway in CD4+ T cells were reported (34, 35).
A significant reduction in the number of regulatory T cells (Tregs) was observed in SSc (36). Tregs are CD4+ cells with immunosuppressive activity aimed at maintaining self-tolerance, regulating immune responses, and averting autoimmunity (37, 38). The reduced number of Tregs in SSc is linked to the gene methylation status of forkhead box P3 (FOXP3) transcription factor (39). FOXP3 is a lineage-specifying factor with an important role in the differentiation and regulation of Tregs. The all-trans retinoic acid (ATRA), which is an active metabolite of vitamin A, was found to improve skin manifestations of SSc and decrease DNA methylation at the promoter region of FOXP3 gene, leading to increased levels of mRNA and protein of FOXP3 and the percentage and number of SSc Tregs. The addition of 5-azacytidine to SSc CD4+ T cells resulted in a similar effect (40). This study suggested a potential role for ATRA as a therapeutic agent in SSc and explained the epigenetic mechanism for its effect.
The maturation of B cells involves the interaction between CD40 on B cells and CD40 ligand (CD40L, also known as CD154) on CD4+ T cells. The CD40L gene is located on the inactivated X chromosome. Hypomethylation of CD40L promoter leads to the overexpression of the ligand, resulting in extensive interaction with CD40 on B cells and additional B cell activation (41, 42).
Another co-stimulatory molecule that is expressed on both B and T cells is CD70. The promoter region of CD70 in SSc CD4+ T cells was found to be hypomethylated, leading to its overexpression (41, 43). Similarly, CD11a was found to be overexpressed in SSc CD4+ T cells and its gene promoter region was found to be hypomethylated (44).
Other examples of hypomethylated genes include ACTA (45), CTNNA3, CTNND2, COL1A1, COL6A3, COL12A1, PDGF-C, TNXB, PAX9, ADAM12, and ITGA9 (46). Examples of hypermethylated genes are C8ORF4 (47), KLF5 (48), SOX2OT (46), DKK1, SFRP1 (49), and RORC1 and RORC2 (50). Table 1 summarizes the biological consequences of these modifications.
Table 1.
Summary of epigenetic modifications in SSc.
| Gene/Pathway | Epigenetic modification | Cell type | Consequences | Reference number |
|---|---|---|---|---|
| I-DNA methylation | ||||
| NOS3 | Hypermethylation | MVECs | Reduced NOS activity in MVECs Increased expression of proinflammatory and vasospastic genes | (20) |
| FL1 | Hypermethylation | FB | Overexpression of collagen genes in SSc FBs | (24) |
| DNA demethylase | Downregulation | MVECs | Hypermethylation | (24) |
| MBD1 | Overexpression | FB | Recruitment of HDACs, resulting in unfavorable chromatin structure | (24) |
| DNMT1 | Overexpression | FB, MVECs | Increased expression of DNMT1 could contribute to hypermethylation of certain genes such as FLI1 | (24, 25) |
| BMPRII | Hypermethylation | MVECs | Failure of the inhibitory mechanism for cell proliferation and induction of apoptosis. | (29) |
| DNMT1 | Downregulation | CD4+ T cells | Global hypomethylation in CD4+ T cells | (32) |
| FOXP3 | Hypermethylation | CD4+ T cells | Decreased FOXP3 expression leads to quantitative defects in Tregs that may contribute to autoimmunity | (39) |
| CD40L | Hypomethylation | CD4+ T cells | May contribute to female susceptibility in SSc | (41) |
| CD70 (TNFSF7) | Hypomethylation | CD4+ T cells | Co-stimulatory molecule, role is not clear in SSc | (43) |
| CD11a | Hypomethylation | CD4+ T cells | Overexpression of CD11a leads to increased proliferation of CD4+ T cells and increased production of IgG antibodies by B cells | (44) |
| ACTA | Hypomethylation | Human alveolar FB | Lung FBs exhibit significantly low levels of DNA methylation of the ACTA promoter, not clear if demethylation of ACTA is a prerequisite for FB activation | (45) |
| CTNNA3, CTNND2 | Hypomethylation | FBb | Involved in the Wnt/b-catenin pathway activation | (46) |
| COL1A1, COL6A3, COL12A1 | Hypomethylation | FBb | May contribute to collagen overexpression | (46) |
| SOX2OT | Hypermethylation | FBa | Encodes lnc-RNAs | (46) |
| PDGF-C | Hypomethylation | FBa | PDGF-C is a profibrotic factor, contributes to FB activation and their transformation to myofibroblasts | (46) |
| CDH11 | Hypomethylation | FBa | Overexpression of CDH11 leads to increased cadherin-11 that facilitates myofibroblast differentiation | (46) |
| TNXB | Hypomethylation | FBb | May contribute to matrix maturation by increasing the expression of tenascin family of glycoproteins | (46) |
| PAX9 | Hypomethylation | FBb | Overexpression of pro-α 2 chain of type I collagen | (46) |
| ADAM12 | Hypomethylation | FBb | Contributes to fibrosis through activating the TGF-β signaling pathway | (46) |
| ITGA9 | Hypomethylation | FBb | Overexpression of ITAG9 leads to TGF-β upregulation | (46) |
| C8ORF4 | Hypermethylation | Lung FB | Decreased expression of C8ORF4 may contribute to the deceased capacity of fibrotic lung FBs to produce COX-2 and PEG2 | (47) |
| KLF5 | Hypermethylation | FBa | Downregulation of KLF5 and FLI1 works synergistically to enhance the expression of connective tissue growth factors | (48) |
| DKK1, SFRP1 | Hypermethylation | FBb/Becha,b | Decreased expression of DKK1 and SFRP1 results in aberrant Wnt signaling | (49) |
| RORC1, RORC2 | Hypermethylation | PBMCsa | Hypermethylation of RORC1 and RORC2 correlated with inflammatory marker and Scl-70+ | (50) |
| II-Histone modification | ||||
| H3, H4 acetylation | Reduction | FB | Unfavorable chromatin structure for target gene expression | (24) |
| FLI1 | H3, H4 Hypoacetylation | FB | Repression of FLI1 leads to overexpression of collagen genes | (24) |
| KLF5 | H3, H4 Hypoacetylation | FB | Downregulation of FLI1 and KLF5 works synergistically to enhance the expression of connective tissue growth factors and promote fibrosis | (48) |
| FRA2 | H3K27me3 inhibition | FB | Inhibition of H3K27me3 induces the expression of the profibrotic transcription factor Fra-2 | (53) |
| VEGF, FGF2, DNMT1, DNMT3A, MECP2 | H3K27me3 inhibition | FB | Inhibition of H3K27me3 results in downregulation of pro-angiogenic genes and genes involved in DNA methylation | (56) |
| TGF-β | Unclear | FB | HDAC inhibitor prevents SSc-related tissue fibrosis by reducing collagen I and fibronectin in dermal SSc FBs | (57) |
| Global H4 acetylation, H3K methylation | Increased H4 acetylation, decreased H3K methylation | B cells | Favor target gene expression in B cells and activation of genes in the immune system and antibody production | (66) |
| WIF1 | Histone hypoacetylation | FB | Oxidative DNA damage induced by SSc autoreactive antibodies leads to histone deacetylase 3 (HDAC3) recruitment, hypoacetylation of the WIF-1 promoter, inhibiting its expression and enabling Wnt activation | (68) |
| NR4A1 | H3, H4 hyperacetylation | FB | The antifibrotic effect of NR4A1 is inhibited by AKT-and HDAC-dependent mechanisms under persistent activation of TGF-β | (69) |
| COL1A2 | H4 hyperacetylation | FB | TGF-β signaling enhances both p300 recruitment and histone H4 acetylation at the COL1A2 (collagen, type I, α2) locus | (60) |
| III-miRNAs anomaly | ||||
| miR-29 | Downregulation | FB | Antifibrotic factor, putative target is type I collagen | (80, 89) |
| miR let-7a | Downregulation | FB, serum | Putative target is type I collagen | (83) |
| miR-196a | Downregulation | FB, serum, hair shafts | Putative target is type I collagen | (84, 85) |
| miR-150 | Downregulation | FB, serum | Induction of integrin β3 | (86) |
| miR-129-5p | Downregulation | FB | Putative target is type I collagen | (87) |
| miR-21 | Overexpression | FB | Profibrotic factor, targets SMAD7 | (89) |
| miR-31 | Overexpression | Skin, FB | Putative target is type I collagen | (89) |
| miR-145 | Downregulation | Skin, FB | Putative target is SMAD3 | (89) |
| miR-146 | Overexpression | Skin, FB | Putative target is SMAD4 | (89) |
| miR-503 | Overexpression | Skin, FB | Putative target is SMAD7 | (89) |
| miR-7 | Overexpressed in SSc, down regulated in lcSSc | FB, skin, sera | Target type I collagen | (91, 92) |
| miR-142 | Overexpression | Serum | Regulation of integrin α-V expression | (95) |
| miR-92-a | Overexpression | FB, serum | Inhibits MMP-1 | (96) |
| miR-483-5p | Overexpression | FB, serum, MVECs | Promotes fibrosis, increases transcription of COL4A1 and COL4A2 in F-Bs and endothelial cells | (97) |
| miR-152 | Downregulation | MVECs | Overexpression of DNMT1 | (98) |
All cell types are human in origin, unless otherwise specified. adiffuse SSc. blimited SSc.
ACTA: actin, alpha 2, smooth muscle, aorta; BMPRII: bone morphogenetic protein type II receptor; COX-2: prostaglandin-endoperoxide synthase 2; DNA: deoxyribonucleic acid; DNMT1: DNA (cytosine-5-)-methyltransferase 1; ECM: extracellular matrix; FLI1: Friend leukemia integration 1; FB: fibroblast; H3K27me3: trimethylation of histone H3 on lysine 27; HDACs: histone deacetylases; IgG: immunoglobulin G; MBD1: methyl-CpG-binding domain protein 1; MeCP2: methyl-CpG-binding protein 2; MMP-1: matrix metalloproteinase 1; MVEC: microvascular endothelial cell; NOS: nitric oxide synthase; PGE2: prostaglandin E2; PBMC: peripheral blood mononuclear cell; RNA: ribonucleic acid; RORC: RAR-related orphan receptor C; SMAD: intracellular proteins that transduce extracellular signals from TGF-β ligands; SSc: systemic sclerosis; TGF-β: transforming growth factor beta; Tregs: regulatory T cells.
It is interesting to note hyper- and hypo-methylation patterns in different cells that are likely to contribute to SSc pathogenesis. The epigenetic modification differs depending on the cell type (hypermethylation in FBs and MVECs and hyper- and hypo-methylation in CD4+ T cells). Mapping all the patterns of epigenetic modifications in cells is essential to completely understand the role of DNA methylation in the pathogenesis of SSc.
Histone modification
Histones are an essential part of the eukaryotic nucleosomes and are the key building blocks of chromatin (16). There are five different types of histones, which are divided into two main groups: core histones (H2A, H2B, H3, and H4) and linker histones (H1 and H5) (9). Post-translational modifications of histones occur on their N-terminal domains. These modifications include methylation, acetylation, phosphorylation, citrullination, ubiquitination, and sumoylation (2). The most studied modifications are histone acetylation and methylation. Histone acetylation results from the transfer of an acetyl group from acetyl coenzyme A to the N-terminal domain of lysines on histone H3 or H4 (4, 51, 52). This process is regulated by histone acetyltransferases (HATs) such as P300/CBP, PCAF, and MYST; histone deacetylases (HDACs) such as class 1, 2, and 4 HDACs; and sirtuins such as SIRT1-7, which is also known as class 3 HDAC (4). Histone acetylation relaxes the chromatin structure by reducing the interaction between the positively charged histones and the negatively charged DNA, as acetylation removes the positive charge on histones. Thus, it allows transcription factors to gain access to the promoter region and initiate transcription activation (22). On the contrary, histone deacetylation represses transcription. The general acetylation state of histones is based on the balance between HATs and HDACs (19). Histone methylation can activate or inhibit gene contingent on the site of methylation and regulate the number of methyl groups that are added (2, 53). For example, methylation of histone H3 lysine 4 (H3K4) induces gene expression, whereas that of H3K9 and H3K27 induces gene repression (3, 4, 19). It is important to recognize that DNA methylation and histone modification are linked (24). Accordingly, when MBD proteins bind methylated cytosines, they recruit HDACs, resulting in heterochromatin conformation that inhibits the transcription machinery (1, 3, 54).
The role of histone methyltransferase, enhancer of zeste homolog 2 (EZH2), was recently studied in SSc FBs and endothelial cells (55). EZH2 catalyzes the trimethylation of histone H3 lysine 27 (H3K27me3) to repress transcription. This enzyme has a role in T cell differentiation, endothelial cell angiogenesis (56), and myofibroblast transformation. The levels of expression of EZH2 and H3K27me3 are elevated in SSc FBs when compared with control cells. Inhibition of EZH2 by DZNep decreases fibrosis both in vitro and in vivo. DZNep decreases the expression of EZH2, HEK27me3, COL1A1, TGFB, FRA2, and LRRC16A in a dose-dependent manner. Similarly, DZNep decreases the expression of DNMT1, DNMT3A, and MECP2, resulting in reduced DNA methylation. In addition, DZNep and GSK126 (another EZH2 inhibitor) prevent bleomycin-induced skin fibrosis. Exposure of SSc FBs to GSK126 results in decreased matrix gel contraction, indicating decreased myofibroblast contractility. The effect of overexpression of EZH2 in normal FBs was analyzed using a wound closure model. The results showed that the overexpression of EZH2 resulted in increased wound closure, supporting a role of EZH2 in myofibroblast contraction. Migration of FBs was negatively affected when EZH2 was overexpressed in LRRC16A (a gene encoding cell membrane cytoskeleton-associated protein) knockdown FBs, indicating a significant role of LRRC16A in EZH2-mediated FB migration.
Similarly, the collagen suppressor gene FLI1 was found to have more deacetylated H3 and H4 and more methylated regions in its promoter region as compared with controls. The addition of a HDAC inhibitor (trichostatin A, TSA) and a DNA methyltransferase inhibitor normalized the expression of type I collagen in SSc FBs (24). Moreover, TSA can reduce TGF-β-induced FB activation by decreasing the nuclear translocation of SMAD3/SMAD4 and DNA binding of SMAD transcription factors (57). As TSA is a broad deacetylase inhibitor, its clinical use is limited by its safety profile; therefore, more specific HDAC inhibitors are required for clinical use. Specifically inhibiting HDAC7 using small interfering RNA resulted in decreased TGF-β-induced accumulation of type I and type III collagen (58). Another HDAC inhibitor is suberoylanilide hydroxamic acid (SAHA), which was found to prevent TGF-β-induced collagen deposition and FB activation (59).
Another important profibrotic factor is the HAT p300 that is regulated by SIRT1 (60). P300 modifies transcription factors affecting the regulatory region of the collagen gene. Levels of SIRT1 are significantly decreased in SSc dermal FBs compared with controls. A SIRT1 activator resulted in decreased response of FBs to TGF-β stimulation and reduced collagen production (61). However, another study revealed opposite effects of the SIRT1 activator on FB response (62); more studies are required to clarify the effects of SIRT1.
The overexpression of EZH2 in SSc endothelial cells affected cell adhesion and migration (63–65). The knockdown of EZH2 in SSc endothelial cells significantly increased angiogenesis, which is similar to the effect of the addition of DZNep to cell cultures. The treatment with DZNep upregulated the expression of notch ligands JAG1, JAG2, DLL4, notch receptor NOTCH2, and notch target gene HES1, whereas it downregulated the expression of notch signaling inhibitors NOTCH1, NOTCH3, NUMB, and FBXW7. These results suggest that EZH2 activates certain genes and inhibits others. When compared with normal endothelial cells, SSc endothelial cells showed increased levels of JAG2 and NUMB, and decreased levels of DLL4, HES1 and HEY2. Moreover, it was found that EZH2 inhibited SSc endothelial cells tube formation by repressing the notch ligand DLL4 through increased binding of EZH2 and H3K23me3 at the promoter region of DLL4 (55). The effect on tube formation was reversed when endothelial cells were treated with an EZH2 inhibitor.
Histone modifications such as increased H4 acetylation and decreased H3K methylation were associated with activating B cell genes that are responsible for the production of antibodies (66).
Histone deacetylases regulate the proliferation and migration of endothelial cells. The expression of HDAC5, an antiangiogenic factor, is significantly increased in SSc MVECs, and it may play a significant role in SSc vasculopathy. Vascular damage in SSc is an early event that occurs before the onset of tissue fibrosis (67). The proposed mechanism for HDAC5 in inhibiting angiogenesis is that HDAC5 represses pro-angiogenic genes. The pro-angiogenic genes identified after HDAC5 was knockdown were FGF2, SLIT2, EPHB4, PVRL2 (cell adhesion molecule that improves angiogenic ability of MVECs), FSTL1 (plays a role in fibrosis and MVEC proliferation and tube formation), and CYR61 (a member of the CCN protein family that supports angiogenesis). Moreover, knockdown of HDAC5 increased the levels of bFGF, which is encoded by FGF2 and increased the expression of FSTL1. Although these observations are interesting, the results infer limited potential clinical utility of HDAC inhibitors as antifibrotic therapy because of their detrimental effects on MVECs that could potentially contribute to SSc vasculopathy. The ideal HDAC inhibitors as a potential therapeutic agent must have a specific target profile with no effects on multiple genes and multiple processes in different cell types.
Other histone modifications such as histone hypoacetylation in WIF1 (68), H3 and H4 hyperacetylation in NR4A1 (69), and H4 hyperacetylation in COL1A2 (60) were observed. Table 1 summarizes the biological consequences.
Non-coding RNA mechanisms
Non-coding RNAs (ncRNAs) are functional RNA molecules that are transcribed from DNA but not translated into proteins. These RNA molecules are biologically active and can affect gene expression, epigenetic modulation, and post-translational modification throughout the body (4, 70). The ncRNA molecules are divided into different groups based on the number of nucleotides: long ncRNAs (lncRNAs) that have more than 200 nucleotides and can be present in both the nucleus and the cytoplasm; medium-sized ncRNAs (<200 nucleotides) that include small nucleolar RNAs (snoRNAs) and promoter-associated small RNAs (PASRs); and small ncRNAs (<50 nucleotides) that include miRNAs and PIWI-interacting RNAs (piRNAs) (4).
MicroRNAs are a group of small non-coding RNAs, ranging from 18 to 22 nucleotides that are synthesized initially as a longer precursor, which is degraded to miRNAs (71). MiRNAs are involved in post-transcriptional silencing and regulation of gene expression (23, 72, 73) by binding to the complementary sequence in the 3′ prime region of the mRNA, resulting in translational repression or mRNA degradation (74–76). Therefore, the upregulation of miRNAs results in gene repression, whereas their downregulation results in gene activation. The expression of miRNAs is regulated through epigenetic mechanisms; for example, miRNAs can be silenced by DNA methylation. MiRNAs differ from siRNAs in that siRNAs target a single gene, whereas miRNAs can target multiple genes (77, 78). The involvement of miRNAs in tissue fibrosis was initially reported in cardiac fibrosis after myocardial infarction. It was found that the expression of the miR-29 family decreased in the cardiac cells adjacent to the infarct area (79). As miR-29 regulates fibrosis-related genes, its downregulation resulted in tissue fibrosis (9).
In 2010, the first study focusing on miRNA levels in SSc dermal FBs found that miR-29a was downregulated. The same finding was observed in bleomycin-induced skin fibrosis (80). Interestingly, downregulating miR-29a in normal dermal FBs increased the formation of collagen types I and III, and overexpressing it in SSc FBs decreased the expression of collagen. Furthermore, miR-29a plays an important role in liver (81) and kidney fibrosis (82). Profibrotic cytokines, such as TGF-β1 and IL-4, decrease the levels of miR-29a (9). Further analysis of miRNAs showed that in SSc skin, 9 miRNAs were upregulated and 15 miRNAs were downregulated. Of these, the expression of miR-206, miR-125b, and let-7g was confirmed by real-time polymerase chain reaction (PCR). As miR-125b functions as a regulator of multiple molecules involved in SSc pathology, including SMAD5, interleukin (IL)-1F10, IL-6R, and IL-13, its downregulation results in increased levels of these molecules. Moreover, the expression of multiple collagen-related miRNAs is decreased in SSc FBs and TGF-β-stimulated normal dermal FBs (83). Alpha 1 and α 2 type I collagen are regulated by miR-196a (84) and let-7a, the expression of both is reduced in SSc FBs, both in vivo and in vitro (9). Transfection by miR-196a and let-7a inhibitors resulted in increased expression of α 1 and α 2 type I collagens, whereas transfection with their mimics resulted in decreased expression. It is suggested that the activation of TGF-β in SSc dermal FBs results in miR-196a and let-7a downregulation, which in turn upregulates the expression of collagen. Interestingly, levels of miR-196a decreased in shafts of hairs obtained from patients with SSc (85).
Another miRNA with an important role in SSc is miR-150 that is underexpressed in SSc (86). MiR-150 is a regulator of intergrin-β3, a known inducer of TGF-β. Interestingly, the overexpression of miR-150 in SSc dermal FBs resulted in decreased integrin-β3, phosphorylated SMAD3, and type I collagen deposition. Knocking down miR-150 resulted in opposite changes.
IL-17A is known to have antifibrogenic effects; it stimulates the overexpression of miR-129-5p, which in turn downregulates the production of α 1 type I collagen. IL-17 receptor is downregulated in SSc FBs in association with decreased expression of miR-129-5p and overproduction of α 1 type I collagen (87).
Circulating levels of miRNAs are proposed as sensitive biomarkers of disease activity, as changes in miRNAs appear earlier than those in proteins. However, miRNAs are unstable when present extracellularly; they are rapidly degraded by RNases despite multiple proposed conditions to preserve their extracellular stability. Nonetheless, levels of few miRNAs are lower in SSc than in controls, and those of others are the same. However, the rank order is different, indicating different expression patterns. An example of different rank order is the levels of miR-7g, miR-21, miR-29b, miR-125, miR-145, and miR-206 between SSc and controls (71, 80, 88, 89).
MiR-196a was measured by PCR in cultured skin FBs and sera from patients with SSc and controls. Levels of miR-196a were significantly lower in SSc FBs compared with control FBs. However, there were no significant differences in the levels in the serum. Intriguingly, the lower serum miR-196a levels in SSc were associated with higher modified Rodnan skin score (mRSS), indicating more extensive skin fibrosis. Similarly, it was found that lower serum let-7a levels were associated with higher mRSS scores. Furthermore, it was found that patients with lower let-7a levels had a lower frequency of anticentromere antibodies, proposing a potential role for let-7a in regulating the immune system (9). Lower miR-30b serum levels were also associated with higher mRSS scores. Of note, miR-30b is a negative regulator of platelet-derived growth factor beta (PDGFB), and its lower levels may increase the levels of PDGFB (90).
Levels of other miRNAs vary depending on the disease phenotype; for example, miR-7 was found to be overexpressed in SSc FBs both in vivo and in vitro (91), whereas it was found to be downregulated in lcSSc FBs in vivo (92).
The use of bortezomib, a proteasome inhibitor that downregulates miR-21 (profibrotic mRNA that is upregulated in SSc dermal FBs), blocked TGF-β-induced fibrosis in an SSc animal model (93). Moreover, the topical application of miR-155 antagonist decreased the production of collagen in a mouse model (94).
The potential use of miRNAs in SSc therapy is emerging but is still in the experimental stage. A therapeutic role for let-7a in bleomycin-induced skin fibrosis has been shown. The intraperitoneal injection of let-7a combined with atelocollagen, for the protection of let-7a from in vivo degradation by RNases, resulted in the overexpression of let-7a in the skin with a concomitant decrease in collagen production (83).
Other examples of overexpressed miRNAs in SSc include miR-142 (95), miR-92a (96), and miR-483-5p (97). An underexpressed miRNA is miR-152 (98). Table 1 summarizes their biological consequences.
The future of miRNAs as a therapeutic option for SSc is promising. However, this approach needs to overcome several obstacles. The most troubling is the potential for miRNAs to alter the function of several genes that may result in undesirable outcomes. Moreover, appropriate dosing and the method of delivery are other obstacles in this emerging field.
Conclusion
In this review, we provided evidence for a key role of epigenetic regulation in the pathogenesis of SSc involving disparities in DNA methylation, anomalies in the histone code, and altered expression of miRNAs in different tissues and cell types (Figure 1). Although it is likely that environmental cues trigger epigenetic regulatory mechanisms, this needs to be confirmed in detail, possibly in a longitudinal cohort study starting with epigenetic profiling of individuals at risk of developing SSc and repeating the epigenetic profile for those who develop the disease. This would provide a better understanding of how environmental stimuli interact and trigger the epigenetic regulatory mechanisms. Furthermore, this approach will provide us a better understanding of whether these epigenetic variations among individuals are a cause or a result of the disease process. In addition, we should develop an experimental model of SSc that we can use for further analysis to obtain an epigenetic map for each cell type involved in the disease process, including endothelial cells, T cells, and FBs. A huge collaborative effort, similar to genome-wide association studies, is required to reveal the epigenetic map. With the ever-expanding discoveries of epigenetic targets, understanding the epigenetic basis of SSc is important for finding potential therapeutics. It is possible that in the near future, epigenetic research may lead to the development of epigenomic tools that can both uncover the risk and offer effective therapeutic options.
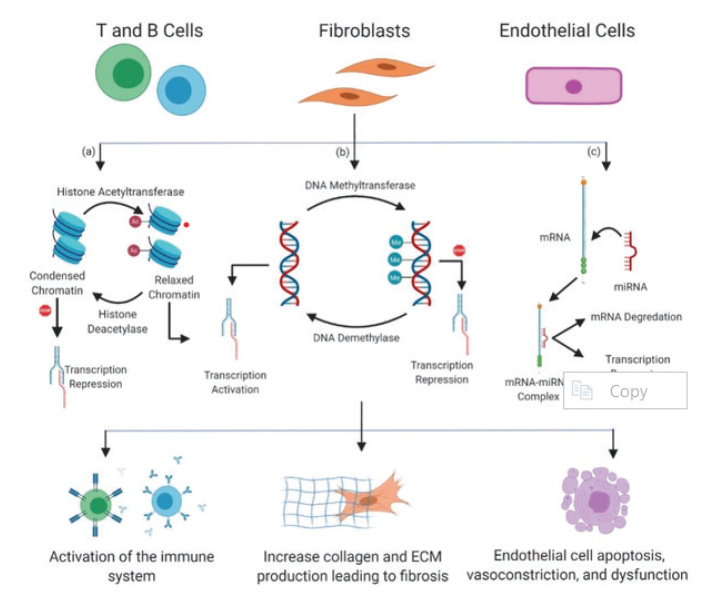
Figure 1.
An overview of the effect of epigenetics on immune cells (T and B cells), fibroblasts, and endothelial cells that contribute to the pathogenesis of SSc. Histone acetylation switches chromatin configuration from condensed to relaxed, permitting the transcription machinery to access the DNA to initiate transcription. This process is catalyzed by histone acetyltransferases and reversed by histone deacetylases. DNA methylation results in transcription repression, as the addition of methyl groups to DNA prevents transcription factors from accessing the DNA. This process is catalyzed by DNA methyltransferases and reversed by DNA demethylases. Inhibition of gene expression by miRNAs through translational repression and degradation of mRNA. MiRNAs can upregulate profibrotic molecules or downregulate antifibrotic molecules. The results of epigenetic alterations in SSc are the activation of the immune system leading to autoimmunity; increased collagen and ECM production resulting in tissue/organ fibrosis; and endothelial cell injury and vascular dysfunction. The figure was created using BioRender.com.
Main Points.
Systemic sclerosis is the result of complex interaction between genetic susceptibility and environmental epigenetic factors.
There is evidence for epigenetic changes in key pathways in the pathogenesis of the disease.
Oxidation injury and hypoxia might be the trigger for epigenetic alterations in SSc.
Footnotes
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - B.K., A.R.; Design - B.K., A.R.; Supervision - B.K., N.A.; Data Collection and/or Processing - A.R.; Analysis and/or Interpretation - B.K., N.A., A.R.; Literature Search - B.K., N.A., A.R.; Writing Manuscript - B.K., N.A., A.R.; Critical Review - B.K., N.A.
Conflict of Interest: The authors have no conflict of interest to declare.
Financial Disclosure: The authors declared that this study has received no financial support.
References
- 1.Altorok N, Almeshal N, Wang Y, Kahaleh B. Epigenetics, the holy grail in the pathogenesis of systemic sclerosis. Rheumatology (Oxford) 2015;54:1759–70. doi: 10.1093/rheumatology/keu155. [DOI] [PubMed] [Google Scholar]
- 2.Altorok N, Kahaleh B. Epigenetics and systemic sclerosis. Semin Immunopathol. 2015;37:453–62. doi: 10.1007/s00281-015-0504-6. [DOI] [PubMed] [Google Scholar]
- 3.Aslani S, Sobhani S, Gharibdoost F, Jamshidi A, Mahmoudi M. Epigenetics and pathogenesis of systemic sclerosis; the ins and outs. Hum Immunol. 2018;79:178–87. doi: 10.1016/j.humimm.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Tsou PS, Sawalha AH. Unfolding the pathogenesis of scleroderma through genomics and epigenomics. J Autoimmun. 2017;83:73–94. doi: 10.1016/j.jaut.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walczyk M, Paradowska-Gorycka A, Olesinska M. Epigenetics: The future direction in systemic sclerosis. Scand J Immunol. 2017;86:427–35. doi: 10.1111/sji.12595. [DOI] [PubMed] [Google Scholar]
- 6.Murdaca G, Contatore M, Gulli R, Mandich P, Puppo F. Genetic factors and systemic sclerosis. Autoimmun Rev. 2016;15:427–32. doi: 10.1016/j.autrev.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Pattanaik D, Brown M, Postlethwaite BC, Postlethwaite AE. Pathogenesis of systemic sclerosis. Front Immunol. 2015;6:272. doi: 10.3389/fimmu.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun X, Xiao Y, Zeng Z, Shi Y, Tang B, Long H, et al. All-trans retinoic acid induces CD4+CD25+FOXP3+ regulatory t cells by increasing FOXP3 demethylation in systemic sclerosis CD4+ t cells. J Immunol Res. 2018;2018 doi: 10.1155/2018/8658156. 8658156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Makino T, Jinnin M. Genetic and epigenetic abnormalities in systemic sclerosis. J Dermatol. 2016;43:10–8. doi: 10.1111/1346-8138.13221. [DOI] [PubMed] [Google Scholar]
- 10.Zuo X, Zhang L, Luo H, Li Y, Zhu H. Systematic approach to understanding the pathogenesis of systemic sclerosis. Clin Genet. 2017;92:365–71. doi: 10.1111/cge.12946. [DOI] [PubMed] [Google Scholar]
- 11.Rubio-Rivas M, Moreno R, Corbella X. Occupational and environmental scleroderma. Systematic review and meta-analysis. Clin Rheumatol. 2017;36:569–82. doi: 10.1007/s10067-016-3533-1. [DOI] [PubMed] [Google Scholar]
- 12.Ciechomska M, Zarecki P, Merdas M, Swierkot J, Morgiel E, Wiland P, et al. The role of microRNA-5196 in the pathogenesis of systemic sclerosis. Eur J Clin Invest. 2017;47:555–64. doi: 10.1111/eci.12776. [DOI] [PubMed] [Google Scholar]
- 13.Lafyatis R. Editorial: Epigenetics in systemic sclerosis. Arthritis Rheumatol. 2016;68:2841–4. doi: 10.1002/art.39830. [DOI] [PubMed] [Google Scholar]
- 14.O’Reilly S, Ciechomska M, Fullard N, Przyborski S, van Laar JM. IL-13 mediates collagen deposition via STAT6 and microRNA-135b: A role for epigenetics. Sci Rep. 2016;6:25066. doi: 10.1038/srep25066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schleithoff C, Voelter-Mahlknecht S, Dahmke IN, Mahlknecht U. On the epigenetics of vascular regulation and disease. Clin Epigenetics. 2012;4:7. doi: 10.1186/1868-7083-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szerlong HJ, Hansen JC. Nucleosome distribution and linker DNA: Connecting nuclear function to dynamic chromatin structure. Biochem Cell Biol. 2011;89:24–34. doi: 10.1139/O10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 18.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–32. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Sun YE. Epigenetic regulation of stem cell differentiation. Pediatr Res. 2006;59:21R–5R. doi: 10.1203/01.pdr.0000203565.76028.2a. [DOI] [PubMed] [Google Scholar]
- 20.Matouk CC, Marsden PA. Epigenetic regulation of vascular endothelial gene expression. Circ Res. 2008;102:873–87. doi: 10.1161/CIRCRESAHA.107.171025. [DOI] [PubMed] [Google Scholar]
- 21.Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. J Cell Physiol. 2007;213:384–90. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 22.Luo Y, Wang Y, Wang Q, Xiao R, Lu Q. Systemic sclerosis: Genetics and epigenetics. J Autoimmun. 2013;41:161–7. doi: 10.1016/j.jaut.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 23.Yan MS, Matouk CC, Marsden PA. Epigenetics of the vascular endothelium. J Appl Physiol (1985) 2010;109:916–26. doi: 10.1152/japplphysiol.00131.2010. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. 2006;54:2271–9. doi: 10.1002/art.21948. [DOI] [PubMed] [Google Scholar]
- 25.Qi Q, Guo Q, Tan G, Mao Y, Tang H, Zhou C, et al. Predictors of the scleroderma phenotype in fibroblasts from systemic sclerosis patients. J Eur Acad Dermatol Venereol. 2009;23:160–8. doi: 10.1111/j.1468-3083.2008.03016.x. [DOI] [PubMed] [Google Scholar]
- 26.Altorok N, Tsou PS, Coit P, Khanna D, Sawalha AH. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis. 2015;74:1612–20. doi: 10.1136/annrheumdis-2014-205303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fish JE, Marsden PA. Endothelial nitric oxide synthase: Insight into cell-specific gene regulation in the vascular endothelium. Cell Mol Life Sci. 2006;63:144–62. doi: 10.1007/s00018-005-5421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Kahaleh B. Epigenetic regulation in scleroderma: High-throughput DNA methylation profiling of Ssc fibroblasts and microvascular endothelial cells and the central role for Nos3 and Fli1 epigenetic repression in the emergence of Ssc cellular phenotype. American College of Rheumatology; Annual scientific meeting; 2007. [Google Scholar]
- 29.Wang Y, Kahaleh B. Epigenetic repression of bone morphogenetic protein receptor II expression in scleroderma. J Cell Mol Med. 2013;17:1291–9. doi: 10.1111/jcmm.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alqahtani A. Genome-wide DNA methylation pattern in systemic sclerosis microvascular endothelial cells: Identification of epigenetically affected key genes and pathways. 2017 ACR/ARHP Annual Meeting; September 18, 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuschiotti P, Larregina AT, Ho J, Feghali-Bostwick C, Medsger TA., Jr Interleukin-13-producing CD8+ T cells mediate dermal fibrosis in patients with systemic sclerosis. Arthritis Rheum. 2013;65:236–46. doi: 10.1002/art.37706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lei W, Luo Y, Lei W, Luo Y, Yan K, Zhao S, et al. Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol. 2009;38:369–74. doi: 10.1080/03009740902758875. [DOI] [PubMed] [Google Scholar]
- 33.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor beta in human disease. N Engl J Med. 2000;342:1350–8. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 34.Sawalha AH, Richardson B. MEK/ERK pathway inhibitors as a treatment for inflammatory arthritis might result in the development of lupus: Comment on the article by Thiel et al. Arthritis Rheum. 2008;58:1203–4. doi: 10.1002/art.23382. [DOI] [PubMed] [Google Scholar]
- 35.Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–78. doi: 10.1038/gene.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antiga E, Quaglino P, Bellandi S, Volpi W, Del Bianco E, Comessatti A, et al. Regulatory T cells in the skin lesions and blood of patients with systemic sclerosis and morphoea. Br J Dermatol. 2010;162:1056–63. doi: 10.1111/j.1365-2133.2010.09633.x. [DOI] [PubMed] [Google Scholar]
- 37.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10:490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 38.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: Mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang YY, Wang Q, Sun XH, Liu RZ, Shu Y, Kanekura T, et al. DNA hypermethylation of the forkhead box protein 3 (FOXP3) promoter in CD4+ T cells of patients with systemic sclerosis. Br J Dermatol. 2014;171:39–47. doi: 10.1111/bjd.12913. [DOI] [PubMed] [Google Scholar]
- 40.Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, et al. DNA methylation controls Foxp3 gene expression. Eur J Immunol. 2008;38:1654–63. doi: 10.1002/eji.200838105. [DOI] [PubMed] [Google Scholar]
- 41.Lian X, Xiao R, Hu X, Kanekura T, Jiang H, Li Y, et al. DNA demethylation of CD40l in CD4+ T cells from women with systemic sclerosis: a possible explanation for female susceptibility. Arthritis Rheum. 2012;64:2338–45. doi: 10.1002/art.34376. [DOI] [PubMed] [Google Scholar]
- 42.Valentini G, Romano MF, Naclerio C, Bisogni R, Lamberti A, Turco MC, et al. Increased expression of CD40 ligand in activated CD4+ T lymphocytes of systemic sclerosis patients. J Autoimmun. 2000;15:61–6. doi: 10.1006/jaut.2000.0387. [DOI] [PubMed] [Google Scholar]
- 43.Jiang H, Xiao R, Lian X, Kanekura T, Luo Y, Yin Y, et al. Demethylation of TNFSF7 contributes to CD70 overexpression in CD4+ T cells from patients with systemic sclerosis. Clin Immunol. 2012;143:39–44. doi: 10.1016/j.clim.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Shu Y, Xiao Y, Wang Q, Kanekura T, Li Y, et al. Hypomethylation and overexpression of ITGAL (CD11a) in CD4(+) T cells in systemic sclerosis. Clin Epigenetics. 2014;6:25. doi: 10.1186/1868-7083-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu B, Gharaee-Kermani M, Wu Z, Phan SH. Essential role of MeCP2 in the regulation of myofibroblast differentiation during pulmonary fibrosis. Am J Pathol. 2011;178:1500–8. doi: 10.1016/j.ajpath.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altorok N, Tsou PS, Coit P, Khanna D, Sawalha AH. Genome-wide DNA methylation analysis in dermal fibroblasts from patients with diffuse and limited systemic sclerosis reveals common and subset-specific DNA methylation aberrancies. Ann Rheum Dis. 2015;74:1612–20. doi: 10.1136/annrheumdis-2014-205303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans IC, Barnes JL, Garner IM, Pearce DR, Maher TM, Shiwen X, et al. Epigenetic regulation of cyclooxygenase-2 by methylation of c8orf4 in pulmonary fibrosis. Clin Sci (Lond) 2016;130:575–86. doi: 10.1042/CS20150697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noda S, Asano Y, Nishimura S, Taniguchi T, Fujiu K, Manabe I, et al. Simultaneous downregulation of KLF5 and Fli1 is a key feature underlying systemic sclerosis. Nat Commun. 2014;5:5797. doi: 10.1038/ncomms6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dees C, Schlottmann I, Funke R, Distler A, Palumbo-Zerr K, Zerr P, et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis. 2014;73:1232–9. doi: 10.1136/annrheumdis-2012-203194. [DOI] [PubMed] [Google Scholar]
- 50.Almanzar G, Klein M, Schmalzing M, Hilligardt D, El Hajj N, Kneitz H, et al. Disease manifestation and inflammatory activity as modulators of Th17/Treg balance and RORC/FoxP3 methylation in systemic sclerosis. Int Arch Allergy Immunol. 2016;171:141–54. doi: 10.1159/000450949. [DOI] [PubMed] [Google Scholar]
- 51.Waterborg JH. Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem Cell Biol. 2002;80:363–78. doi: 10.1139/o02-080. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Li C, Sun G. Histone acetylation and its modifiers in the pathogenesis of diabetic nephropathy. J Diabetes Res. 2016;2016:11. doi: 10.1155/2016/4065382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kramer M, Dees C, Huang J, Schlottmann I, Palumbo-Zerr K, Zerr P, et al. Inhibition of H3K27 histone trimethylation activates fibroblasts and induces fibrosis. Ann Rheum Dis. 2013;72:614–20. doi: 10.1136/annrheumdis-2012-201615. [DOI] [PubMed] [Google Scholar]
- 54.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–50. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 55.Tsou PS, Campbell P, Amin MA, Coit P, Miller S, Fox DA, et al. Inhibition of EZH2 prevents fibrosis and restores normal angiogenesis in scleroderma. Proc Natl Acad Sci U S A. 2019;116:3695–702. doi: 10.1073/pnas.1813006116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pei-Suen Tsou PC, Khanna Dinesh, Sawalha Amr H. EZH2 modulates angiogenesis and fibrosis in scleroderma. Arthritis Rheumatol. 2016;68:1091–2. [Google Scholar]
- 57.Huber LC, Distler JH, Moritz F, Hemmatazad H, Hauser T, Michel BA, et al. Trichostatin A prevents the accumulation of extracellular matrix in a mouse model of bleomycin-induced skin fibrosis. Arthritis Rheum. 2007;56:2755–64. doi: 10.1002/art.22759. [DOI] [PubMed] [Google Scholar]
- 58.Hemmatazad H, Rodrigues HM, Maurer B, Brentano F, Pileckyte M, Distler JH, et al. Histone deacetylase 7, a potential target for the antifibrotic treatment of systemic sclerosis. Arthritis Rheum. 2009;60:1519–29. doi: 10.1002/art.24494. [DOI] [PubMed] [Google Scholar]
- 59.Wang Z, Chen C, Finger SN, Kwajah S, Jung M, Schwarz H, et al. Suberoylanilide hydroxamic acid: A potential epigenetic therapeutic agent for lung fibrosis? Eur Respir J. 2009;34:145–55. doi: 10.1183/09031936.00084808. [DOI] [PubMed] [Google Scholar]
- 60.Ghosh AK, Bhattacharyya S, Lafyatis R, Farina G, Yu J, Thimmapaya B, et al. p300 is elevated in systemic sclerosis and its expression is positively regulated by TGF-beta: Epigenetic feed-forward amplification of fibrosis. J Invest Dermatol. 2013;133:1302–10. doi: 10.1038/jid.2012.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei J, Ghosh AK, Chu H, Fang F, Hinchcliff ME, Wang J, et al. The histone deacetylase sirtuin 1 is reduced in systemic sclerosis and abrogates fibrotic responses by targeting transforming growth factor beta signaling. Arthritis Rheum. 2015;67:1323–34. doi: 10.1002/art.39061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zerr P, Palumbo-Zerr K, Huang J, Tomcik M, Sumova B, Distler O, et al. Sirt1 regulates canonical TGF-beta signalling to control fibroblast activation and tissue fibrosis. Ann Rheum Dis. 2016;75:226–33. doi: 10.1136/annrheumdis-2014-205740. [DOI] [PubMed] [Google Scholar]
- 63.Maleszewska M, Vanchin B, Harmsen MC, Krenning G. The decrease in histone methyltransferase EZH2 in response to fluid shear stress alters endothelial gene expression and promotes quiescence. Angiogenesis. 2016;19:9–24. doi: 10.1007/s10456-015-9485-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dreger H, Ludwig A, Weller A, Stangl V, Baumann G, Meiners S, et al. Epigenetic regulation of cell adhesion and communication by enhancer of zeste homolog 2 in human endothelial cells. Hypertension. 2012;60:1176–83. doi: 10.1161/HYPERTENSIONAHA.112.191098. [DOI] [PubMed] [Google Scholar]
- 65.Mitic T, Caporali A, Floris I, Meloni M, Marchetti M, Urrutia R, et al. EZH2 modulates angiogenesis in vitro and in a mouse model of limb ischemia. Mol Ther. 2015;23:32–42. doi: 10.1038/mt.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang Y, Yang Y, Luo Y, Yin Y, Wang Q, Li Y, et al. Aberrant histone modification in peripheral blood B cells from patients with systemic sclerosis. Clin Immunol. 2013;149:46–54. doi: 10.1016/j.clim.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 67.Tsou PS, Wren JD, Amin MA, Schiopu E, Fox DA, Khanna D, et al. Histone deacetylase 5 is overexpressed in scleroderma endothelial cells and impairs angiogenesis via repression of proangiogenic factors. Arthritis Rheumatol. 2016;68:2975–85. doi: 10.1002/art.39828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Svegliati S, Marrone G, Pezone A, Spadoni T, Grieco A, Moroncini G, et al. Oxidative DNA damage induces the ATM-mediated transcriptional suppression of the Wnt inhibitor WIF-1 in systemic sclerosis and fibrosis. Sci Signal. 2014;7:ra84. doi: 10.1126/scisignal.2004592. [DOI] [PubMed] [Google Scholar]
- 69.Palumbo-Zerr K, Zerr P, Distler A, Fliehr J, Mancuso R, Huang J, et al. Orphan nuclear receptor NR4A1 regulates transforming growth factor-beta signaling and fibrosis. Nat Med. 2015;21:150–8. doi: 10.1038/nm.3777. [DOI] [PubMed] [Google Scholar]
- 70.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jinnin M. Various applications of microRNAs in skin diseases. J Dermatol Sci. 2014;74:3–8. doi: 10.1016/j.jdermsci.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 72.Goldberg AD, Allis CD, Bernstein E. Epigenetics: A landscape takes shape. Cell. 2007;128:635–8. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 73.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–76. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 74.Ceribelli A, Yao B, Dominguez-Gutierrez PR, Nahid MA, Satoh M, Chan EK. MicroRNAs in systemic rheumatic diseases. Arthritis Res Ther. 2011;13:229. doi: 10.1186/ar3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 76.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–21. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 77.Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–78. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 79.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, et al. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62:1733–43. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 81.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–18. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 82.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, et al. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2012;23:252–65. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Makino K, Jinnin M, Hirano A, Yamane K, Eto M, Kusano T, et al. The downregulation of microRNA let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. J Immunol. 2013;190:3905–15. doi: 10.4049/jimmunol.1200822. [DOI] [PubMed] [Google Scholar]
- 84.Honda N, Jinnin M, Kajihara I, Makino T, Makino K, Masuguchi S, et al. TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol. 2012;188:3323–31. doi: 10.4049/jimmunol.1100876. [DOI] [PubMed] [Google Scholar]
- 85.Wang Z, Jinnin M, Kudo H, Inoue K, Nakayama W, Honda N, et al. Detection of hair-microRNAs as the novel potent biomarker: Evaluation of the usefulness for the diagnosis of scleroderma. J Dermatol Sci. 2013;72:134–41. doi: 10.1016/j.jdermsci.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 86.Zhu H, Luo H, Zuo X. MicroRNAs: Their involvement in fibrosis pathogenesis and use as diagnostic biomarkers in scleroderma. Exp Mol Med. 2013;45:e41. doi: 10.1038/emm.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakashima T, Jinnin M, Yamane K, Honda N, Kajihara I, Makino T, et al. Impaired IL-17 signaling pathway contributes to the increased collagen expression in scleroderma fibroblasts. J Immunol. 2012;188:3573–83. doi: 10.4049/jimmunol.1100591. [DOI] [PubMed] [Google Scholar]
- 88.Li H, Yang R, Fan X, Gu T, Zhao Z, Chang D, et al. MicroRNA array analysis of microRNAs related to systemic scleroderma. Rheumatol Int. 2012;32:307–13. doi: 10.1007/s00296-011-2165-7. [DOI] [PubMed] [Google Scholar]
- 89.Zhu H, Li Y, Qu S, Luo H, Zhou Y, Wang Y, et al. MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol. 2012;32:514–22. doi: 10.1007/s10875-011-9647-y. [DOI] [PubMed] [Google Scholar]
- 90.Tanaka S, Suto A, Ikeda K, Sanayama Y, Nakagomi D, Iwamoto T, et al. Alteration of circulating miRNAs in SSc: miR-30b regulates the expression of PDGF receptor beta. Rheumatology (Oxford) 2013;52:1963–72. doi: 10.1093/rheumatology/ket254. [DOI] [PubMed] [Google Scholar]
- 91.Kajihara I, Jinnin M, Yamane K, Makino T, Honda N, Igata T, et al. Increased accumulation of extracellular thrombospondin-2 due to low degradation activity stimulates type I collagen expression in scleroderma fibroblasts. Am J Pathol. 2012;180:703–14. doi: 10.1016/j.ajpath.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 92.Etoh M, Jinnin M, Makino K, Yamane K, Nakayama W, Aoi J, et al. microRNA-7 down-regulation mediates excessive collagen expression in localized scleroderma. Arch Dermatol Res. 2013;305:9–15. doi: 10.1007/s00403-012-1287-4. [DOI] [PubMed] [Google Scholar]
- 93.Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai J, et al. MicroRNA-21 in scleroderma fibrosis and its function in TGF-beta-regulated fibrosis-related genes expression. J Clin Immunol. 2013;33:1100–9. doi: 10.1007/s10875-013-9910-5. [DOI] [PubMed] [Google Scholar]
- 94.Yan Q, Chen J, Li W, Bao C, Fu Q. Targeting miR-155 to treat experimental scleroderma. Sci Rep. 2016;6:20314. doi: 10.1038/srep20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Makino K, Jinnin M, Kajihara I, Honda N, Sakai K, Masuguchi S, et al. Circulating miR-142-3p levels in patients with systemic sclerosis. Clin Exp Dermatol. 2012;37:34–9. doi: 10.1111/j.1365-2230.2011.04158.x. [DOI] [PubMed] [Google Scholar]
- 96.Sing T, Jinnin M, Yamane K, Honda N, Makino K, Kajihara I, et al. microRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford) 2012;51:1550–6. doi: 10.1093/rheumatology/kes120. [DOI] [PubMed] [Google Scholar]
- 97.Chouri E, Servaas NH, Bekker CPJ, Affandi AJ, Cossu M, Hillen MR, et al. Serum microRNA screening and functional studies reveal miR-483-5p as a potential driver of fibrosis in systemic sclerosis. J Autoimmun. 2018;89:162–70. doi: 10.1016/j.jaut.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 98.Wang Y, Kahaly O, Kahaleh B. Down-regulated microRNA-152 induces aberrant DNA methylation in scleroderma endothelial cells by targeting DNA methyltransferase 1. Arthritis Rheum. 2010;62:1352. [Google Scholar]