Abstract
The application of transcriptomics to the study of the reproductive tract in male turkeys can significantly increase our current knowledge regarding the specifics of bird reproduction. To characterize the complex transcriptomic changes that occur in the testis, epididymis, and ductus deferens, deep sequencing of male turkey RNA samples (n = 6) was performed, using Illumina RNA-Seq. The obtained sequence reads were mapped to the turkey genome, and relative expression values were calculated to analyze differentially expressed genes (DEGs). Statistical analysis revealed 1,682; 2,150; and 340 DEGs in testis/epididymis, testis/ductus deferens, and epididymis/ductus deferens comparisons, respectively. The expression of selected genes was validated using quantitative real-time reverse transcriptase-polymerase chain reaction. Bioinformatics analysis revealed several potential candidate genes involved in spermatogenesis, spermiogenesis and flagellum formation in the testis, and in post-testicular sperm maturation in the epididymis and ductus deferens. In the testis, genes were linked with the mitotic proliferation of spermatogonia and the meiotic division of spermatocytes. Histone ubiquitination and protamine phosphorylation were shown to be regulatory mechanisms for nuclear condensation during spermiogenesis. The characterization of testicular transcripts allowed a better understanding of acrosome formation and development and flagellum formation, including axoneme structures and functions. Spermatozoa motility during post-testicular maturation was linked to the development of flagellar actin filaments and biochemical processes, including Ca2+ influx and protein phosphorylation/dephosphorylation. Spermatozoa quality appeared to be controlled by apoptosis and antioxidant systems in the epididymis and ductus deferens. Finally, genes associated with reproductive system development and morphogenesis were identified. To the best of our knowledge, this is the first genome-wide functional investigation of genes associated with tissue-specific processes in turkey reproductive tract. A catalog of genes worthy of further studies to understand the avian reproductive physiology and regulation was provided.
Key words: transcriptomics, male reproductive tract, turkey
Introduction
The anatomy and physiology of the avian reproductive system are unique among vertebrates. The testes of birds are located within the body cavity; thus, birds are able to maintain efficient spermatogenesis at relatively high body temperatures (40–41°C; Beaupré et al., 1997; Aire, 2007). Spermatogenesis in birds occurs 4-fold faster and produces 4-fold the number of spermatozoa/g testis as that in mammals (Jones and Lin, 1993). Birds lack accessory sex glands (Lake, 1971), and the nonciliated cells of the ductuli efferentes, ductus epididymidis, and ductus deferens contribute to the production of seminal plasma through apocrine secretion (Hess et al., 1976). Spermatozoa develop their fertilization abilities in the testis (Asano and Tajima, 2017), and the epididymis was identified as the site where bird spermatozoa develop progressive motility (Nixon et al., 2014). The post-testicular sperm maturation mechanism that confers spermatozoa motility remains poorly understand in birds. The introduction of a new comprehensive analysis method, such as next-generation sequencing (NGS), may significantly increase current knowledge regarding the specifics of bird reproduction.
Transcriptomics is the study of the transcriptome, the complete set of RNA transcripts produced by the genome under specific circumstances or in specific cells, using high-throughput methods, including NGS and microarray platforms (Liang, 2013). Measuring the expression of an organism's genes, in different tissues, under different conditions, or at different time points, can provide information regarding how genes are regulated and reveal details of an organism's biology (Lowe et al., 2017). The introduction of transcriptomics into the study of mammalian reproductive tracts has facilitated significant progress in our understanding of the molecular mechanisms that underlie gonadal development, spermatogenesis, and spermatozoa maturation (Chalmel and Rolland, 2015; Kordonowy and MacManes, 2016; Lecluze et al., 2018; Tang et al., 2018; Long, 2020).
NGS has recently been used to research male bird reproductive processes and has focused primarily on chickens. Candidate microRNAs and long noncoding RNAs associated with sperm motility have been described for the chicken testis (Liu et al., 2017a; 2018). Some candidate microRNAs, including those involved in the regulation of the spermatogenesis process, spermatozoa function, and testicular metabolism, have been identified (Wu et al., 2017). The combs and testis of the homozygous rooster, the Rose-comb Silky chicken, were subjected to transcriptional profiling, in an attempt to reveal the causes of subfertility (Wang et al., 2017). In addition, transcriptome profiling by microarray analysis has been described for chicken sperm (Qi et al., 2020). However, few transcriptomic analyses have focused on specific components of the reproductive tract in birds, and the transcriptome of the turkey reproductive tract has not been sequenced, thus far. Therefore, in the present study, NGS was used, for the first time, to profile the testis, epididymis, and ductus deferens transcriptomes in turkeys. The aim of this study was to identify the key genes and pathways associated with specificity in the turkey reproductive tract.
Materials and methods
Birds, Housing, and Tissue Collection
Turkeys (British United Turkeys Big 6, Grelier, Saint-Laurent-de-la-Plaine, France) were maintained under standard husbandry conditions at the Turkey Testing Farm of the Department of Poultry Science (University of Warmia and Mazury in Olsztyn, Poland). Feed and water were provided ad libitum. Males were photostimulated at 26 wks of age (14 h light to 10 h darkness) and produced semen by 30 wks of age. Semen was collected at 1-wk intervals, by abdominal massage (Burrows and Quinns, 1937). The sperm concentration, viability, motility, and volume were 9.5 ± 1.5 × 109 sperm/mL, 97.9 ± 0.3%, 85.2 ± 9.6%, and 0.6 ± 0.05 mL, respectively. Male reproductive tract tissue samples were obtained from six, 38-wk-old turkeys that were killed in a local slaughterhouse. The tissues were immediately frozen in liquid nitrogen and designated for total RNA isolation. All experiments were performed in accordance with the International Guiding Principles for Biomedical Research Involving Animals, as proposed by the Society for the Study of Reproduction, and the Polish Animal Welfare Act. No ethics approval was required for these experiments.
RNA Isolation and the Evaluation of RNA Integrity
RNA isolation was performed, according to previously described methods (Słowińska et al., 2014). Briefly, total RNA was extracted from samples, using a Total RNA Mini kit (A&A Biotechnology, Gdynia, Poland), according to the manufacturer's instructions. The RNA concentration and quality were determined spectrophotometrically, using an ND-1000 spectrophotometer (NanoDrop Technologies LLC, Wilmington, DE). Genomic DNA was removed from RNA samples through DNase I digestion (Sigma Aldrich, St. Louis, MO). The RNA integrity was evaluated by microfluidic electrophoresis, using a 2100 Bioanalyzer, with an RNA 6000 Nano LabChip Kit (Agilent Technologies, Santa Clara, CA). Only samples with RNA integrity numbers (RINs: 28S/18S ratio) above 8.0 and rRNA ratios above 1.0 were used for NGS.
Library Preparation and Sequencing Procedures
For library preparation, poly-A–containing mRNA, using poly-T–attached magnetic beads was purified. Then, mRNA was fragmented, using divalent cations under increasing temperature conditions. RNA fragments were attached to the first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen, Waltham, MA). Second-strand cDNA synthesis was performed using DNA Polymerase I and RNase H. To prevent the ligation of fragments to each other, single “A” nucleotides were added to the 3′ ends. The synthesized sequences were purified and amplified through polymerase chain reaction (PCR), and the final cDNA library was established. To create strand-specific RNA sequencing libraries, the TruSeq Stranded mRNA Library Prep Kit (Illumina, San Diego, CA) was used, according to the manufacturer's protocol. The total RNA concentration was calculated using Quant-IT RiboGreen (Invitrogen, Waltham, MA). To assess the integrity of total RNA, samples were run on TapeStation RNA screentape (Agilent Technologies, Waldbronn, Germany). Indexed cDNA libraries were sequenced using the NovaSeq 6000 platform (Illumina, San Diego, CA). The paired-end, 2 × 100-bp sequences obtained during this study and all expression data have been submitted to the Gene Expression Omnibus database, under accession no. GSE142428 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142428).
Quality Control and Mapping Process
The quality of raw reads was investigated using the FastQC software, version 0.11.7 (Bioinformatics Group at the Babraham Institute, Cambridge, United Kingdom; www.bioinformatics.babraham.ac.uk). Reads with high levels of Illumina adaptors and low quality (Phred cutoff score ≤ 20; with 10-bp frameshifts) were removed from downstream analyses. All processed reads were trimmed to equal lengths (90 bp), using the Trimmomatic software, v. 0.38 (Bolger et al., 2014). Surviving paired-end reads were mapped to the reference turkey genome (Meleagris_gallopavo.UMD2.dna_sm.toplevel.fa), using ENSEMBL/GENCODE annotation (Meleagris_gallopavo.UMD2.91.gtf), by applying the mapper in the STAR software, v. 2.4 (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; https://github.com/alexdobin/STAR) (Dobin et al., 2013). The conversion from BAM format to SAM format was performed using the SAMtools software (Genome Research Ltd, Wellcome Trust Genome Campus, Hinxton, United Kingdom; https://github.com/samtools/samtools) (Li et al., 2009a). StringTie, v. 1.3.3 (https://ccb.jhu.edu/software/stringtie) (Pertea et al., 2015) was used to enrich annotations by merging the Ensembl GTF file with all BAM files.
Expression Profiling and Differentially Expressed Genes
Differential expression analysis was performed using 2 independent methods, the Ballgown method (Frazee et al., 2014) and Cufflinks software (Trapnell et al., 2012). An expression-count matrix was created by the prepDE.py script for use with the Ballgown method. Using enriched annotation (stringtie.gtf), Cufflinks software was used to calculate expression values (using Cuffquant), normalize data (using Cuffnorm), and perform differential analyses (using Cuffdiff). Full-length transcriptome sequences were extracted, using a gffread script (https://github.com/gpertea/gffread), and deposited in the Gene Expression Omnibus database, as GSE142428_transcriptome_Meleagris_gallopavo.fa.gz file. To obtain stringent results, transcripts with significant test values (adjusted P-value [padjusted] < 0.01 and logarithmic values of expression fold-changes [log2FC > 1.0]) that were confirmed using both methods were qualified as differentially expressed genes (DEGs). DEGs that were identified using both statistical methods were visualized in MA, volcano, and heatmap plots, using gplots (Warnes et al., 2009) and ggplot2 (Wickham, 2016) and R Bioconductor packages (R Core Team, 2017).
All memory-intensive processes were performed by the Regional IT Center of the University of Warmia and Mazury in Olsztyn, using a 60-core CPU and a 136 GB RAM server.
Functional Enrichment Analysis
The functional profiling of DEGs into 3 gene ontology categories, biological processes, cellular components, and molecular functions, was performed using G:Profiler (Raudvere et al., 2019; Meleagris gallopavo as the reference organism) with P < 0.05. For additional evaluation of DEGs related to epididymis and ductus deferens, PANTHER 14.1 software was used (Thomas et al., 2003; Mi et al., 2013; Gallus gallus as the organism). To establish possible interactions between identified genes and the network of predicted associations for genes, all identified DEGs were submitted to STRING (Search Tool for Retrieval of Interacting Genes; Szklarczyk et al., 2019), with a medium confidence score cutoff of 0.4.
Quantitative Real-Time Reverse Transcriptase PCR
Gene expression validation experiments were performed using the same RNA samples as those used for NGS. The endogenous mRNA expression levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), acrosin (ACR), catalase (CAT), DNA meiotic recombinase 1 (DMC1), dynein axonemal heavy chain 8 (DNAH8), Enah/Vasp-like (EVL), never in mitosis gene a-related kinase 2 (NEK2), phospholipase A2 group XIIB (PLA2G12b), protein phosphatase 1 regulatory inhibitor subunit 1B (PPP1R1B), protein kinase cAMP-dependent type II regulatory subunit alpha (PRKAR2A), receptor interacting serine/threonine kinase 1 (RIPK1), tubulin alpha 8 (TUBA8), and ubiquitin like with PHD and ring finger domains 1 (UHRF1) were measured using Custom TaqMan Gene Expression Assays (Applied Biosystems by Thermo Fisher Scientific). Primer-probe sets were designed based on sequences obtained from the National Center for Biotechnology Information, using the Primer3Plus online tool (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). Gene names and primer-probe sequences are presented in Supplementary Table 1. Reactions were performed using an ABI ViiA 7 sequence detection system (Applied Biosystems by Life Technologies), under the following conditions: 10 min at 95°C, 45 cycles of 15 s at 95°C, and 1 min at 60°C. All results were normalized against GAPDH contents and analyzed using the PCR Miner algorithm (Zhao and Fernald, 2005).
Statistical Analysis
For the analysis of TUBA8, ACR, UHRF1, PLA2G12b, and CAT in the male reproductive tract, a one-way analysis of variance (ANOVA), followed by Tukey's multiple comparison test, was performed. Other gene expression data (DMC1, NEK2, and DNAH8) were analyzed using the nonparametric Kruskal-Wallis test (a nonparametric ANOVA), followed by Dunn's multiple comparison test. Analyses were performed using the statistical software program GraphPad5 (GraphPad PRISM, v 5.0; GraphPad Software Inc., San Diego, CA). Data are expressed as the mean ± SD.
Results
Sequencing Results
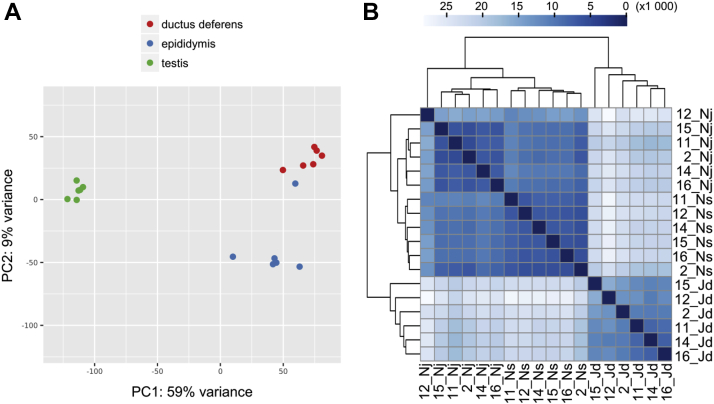
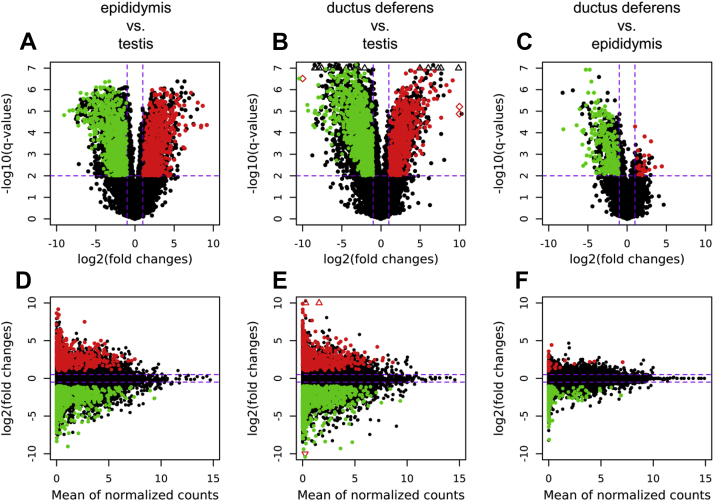
Sequencing the turkey reproductive tract transcriptome resulted in approximately 2,669 million reads (100 bp in length). Statistical data for the reads obtained from each RNA library are presented in Table 1. After removing short and low-quality reads (PHRED33 < 20), the surviving 2,236 million paired-end sequences (ranging from approximately 102 million to 176 million reads per sample) were mapped to the ENSEMBL turkey genome (Meleagris_gallopavo.UMD2.91). The percentage of unique reads that mapped to the turkey genome ranged from 96.63 to 98.87%. The overall numbers of transcript active regions engaged in the reproductive tract were 31,013; 35,232; and 56,037 for the ductus deferens, epididymis, and testis, respectively. Correlation coefficients between biological replicates ranged between 0.77-0.99, 0.63-0.99, and 0.83-0.99 for the testis, epididymis, and ductus deferens, respectively (P < 0.0001; Figure 1). The expression profiles of genes in each tissue differed significantly. Changes [padjusted < 0.01 and abs(log2FC) > 1.0] in transcript expression levels for each comparison (testis vs. epididymis; testis vs. ductus deferens; and epididymis vs. ductus deferens) are visualized in Figure 2, Figure 3, Figure 4.
Table 1.
Summary of sequencing, quality control, and mapping processes.
| Tissue samples of turkey reproductive tract | 11_Jd | 11_Nj | 11_Ns | 12_Jd | 12_Nj | 12_Ns | 14_Jd | 14_Nj | 14_Ns | 15_Jd | 15_Nj | 15_Ns | 16_Jd | 16_Nj | 16_Ns | 2_Jd | 2_Nj | 2_Ns |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Row reads | 127.9 | 176.9 | 167.7 | 151.1 | 126.4 | 139.6 | 165.8 | 138.1 | 154.3 | 143.1 | 160.1 | 161.2 | 150.3 | 136.5 | 150.1 | 142.9 | 174.2 | 102.6 |
| Trimmed reads | 110.7 | 151.8 | 137.6 | 128.1 | 96.0 | 115.4 | 140.6 | 113.6 | 127.5 | 122.5 | 136.7 | 136.5 | 125.1 | 114.8 | 124.9 | 115.9 | 151.9 | 86.2 |
| Percent of trimmed | 86.5 | 85.8 | 82.0 | 84.8 | 76.0 | 82.7 | 84.8 | 82.3 | 82.6 | 85.6 | 85.4 | 84.7 | 83.2 | 84.1 | 83.2 | 81.1 | 87.2 | 84.1 |
| Mapped | 78.0 | 112.8 | 89.4 | 94.9 | 64.8 | 78.9 | 100.0 | 78.8 | 89.9 | 83.1 | 103.3 | 99.9 | 88.9 | 81.2 | 88.2 | 80.7 | 117.3 | 62.3 |
| Uniquely mapped | 76.2 | 111.5 | 88.2 | 93.6 | 62.6 | 77.8 | 97.6 | 77.7 | 88.3 | 81.8 | 101.7 | 98.4 | 86.9 | 80.0 | 87.0 | 79.1 | 115.8 | 61.3 |
| Percent of uniquely mapped | 97.7 | 98.9 | 98.6 | 98.6 | 96.6 | 98.5 | 97.6 | 98.6 | 98.2 | 98.4 | 98.5 | 98.4 | 97.7 | 98.6 | 98.6 | 98.0 | 98.7 | 98.4 |
The names of samples are similar to names in the GEO and SRA databases, where Jd, Nj, and Ns refer to the testis, epididymis, and ductus deferens, respectively. All values for read numbers are expressed in millions.
Figure 1.
(A) Graphical representation of the first (PC1) and second (PC2) principal components (PC) affecting gene expression patterns in turkey reproductive tract tissues. Red, blue, and green dots represent the ductus deferens, epididymis, and testis samples, respectively. (B) Poisson distance matrices illustrating the RNA-seq library integrity within tissue groups. The color scale represents the distances between biological replicates. The names of samples are similar to names in the GEO and SRA databases, where Jd, Nj, and Ns refer to the testis, epididymis, and ductus deferens, respectively.
Figure 2.
(A–C) Volcano plots presenting differentially expressed genes [DEGs; padjusted < 0.01 and abs(log2 fold change > 1.0)], as confirmed by 2 separate methods (Ballgown and Cufflinks). Red, green, and black dots represent upregulated, downregulated, and nonsignificant genes, respectively. (D–F) MA plot depicted DEGs on a logarithmic scale, showing the fragments per kilobase of transcript per million mapped reads (FPKM) value.
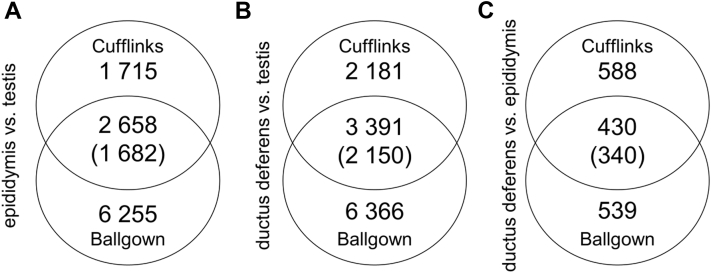
Figure 3.
Venn diagram showing the number of differentially expressed genes (DEGs) identified using both the Cufflinks and Ballgown methods. Overlapping circles represent transcripts that are common between the specific tests. The results in the brackets are considered to be significant genes, with known annotations in the ENSEMBL database, based on values of padjusted < 0.01 and an absolute value of logarithmic fold change >1.0. (A) Epididymis vs. testis; (B) ductus deferens vs. testis; and (C) ductus deferens vs. epididymis.
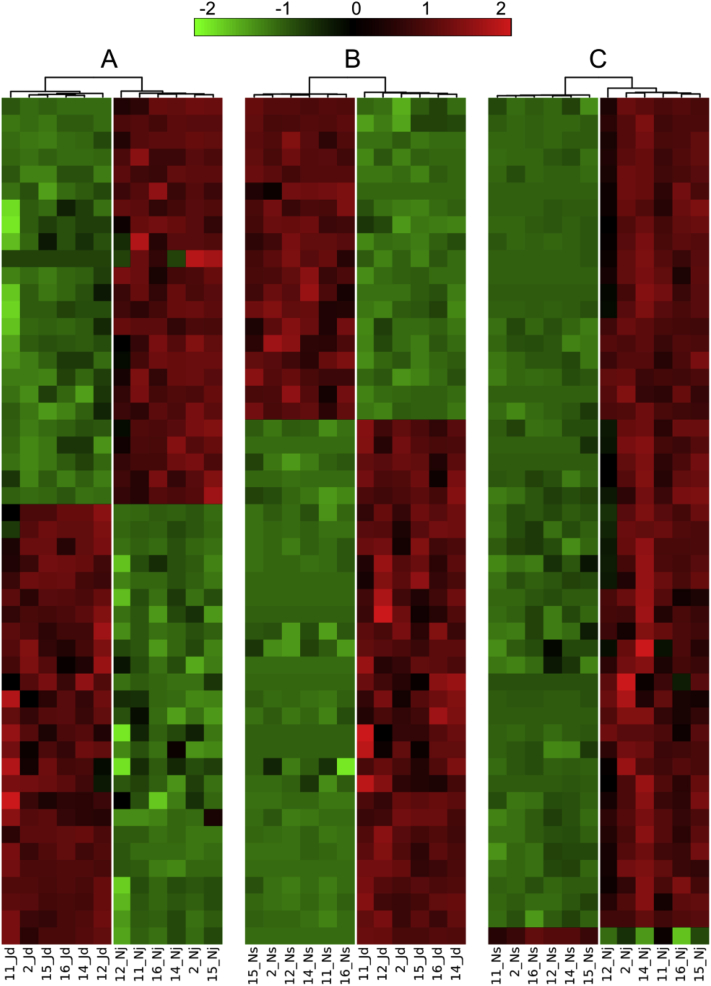
Figure 4.
The heatmaps of scaled expression data (Z-score) for the top 50 differentially expressed genes (DEGs), with the smallest SDs in expression profiles within the tissue groups. (A) Epididymis vs. testis; (B) ductus deferens vs. testis; (C) ductus deferens vs. epididymis. Complete expression data are presented in Supplementary Tables 3–5. The names of samples are similar to names in the GEO and SRA databases, where Jd, Nj, and Ns refer to the testis, epididymis, and ductus deferens, respectively.
DEGs in the Turkey Reproductive Tract
DEG analyses between the testis and epididymis, testis and ductus deferens, and epididymis and ductus deferens are presented in Supplementary Tables 2–4, respectively. Using the criteria of padjusted < 0.01 and abs(log2FC) > 1.5, a total of 1,682 DEGs were identified between the testis and epididymis (954 DEGs were upregulated in the testis, and 728 DEGs were upregulated in the epididymis). The log2FC values for DEGs ranged from +9.44, for cytochrome P450 family 17 subfamily A member 1 (CYP17A1), which was upregulated in the testis, to -9.51, for solute carrier family 6 member 14 (SLC6A14), which was upregulated in the epididymis (Supplementary Table 2). A total of 434 DEGs were nonannotated (Supplementary Table 2).
A total of 2,150 DEGs were identified between the testis and ductus deferens (1,327 DEGs were upregulated in the testis, and 823 DEGs were upregulated in the ductus deferens). The log2FC values for DEGs ranged from +11, for CYP17A1, which was upregulated in the testis, to –10.6, for fatty acid-binding protein 7 (FABP7), which was upregulated in the ductus deferens (Supplementary Table 3). A total of 542 DEGs were nonannotated (Supplementary Table 3).
A total of 340 DEGs were identified between the epididymis and ductus deferens (305 DEGs were upregulated in the epididymis, and 35 DEGs were upregulated in the ductus deferens). The log2FC values for DEGs ranged from +9.8, for DNAH5, which was upregulated in the epididymis, to -4.7, for keratin 19 (KRT19), which was upregulated in the ductus deferens (Supplementary Table 4). A total of 105 DEGs were nonannotated (Supplementary Table 4).
Functional Classification of DEGs
Pathway enrichment analyses showed significant overlap among several categories that were enriched in the testis compared with the epididymis and ductus deferens (Table 2). Biological processes connected with cell cycle processes, including mitosis and meiosis, and reproduction were found to be enriched in the testis compared with both the epididymis and ductus deferens. However, processes connected with cell movement were found to differentiate the testis from the ductus deferens. Upregulated DEGs identified in the testis were primarily associated with nuclear and chromosomal cellular component categories. In addition to biological processes, DEGs upregulated in the testis compared with the ductus deferens were associated with microtubules, cilia, and sperm flagella. The molecular functions enriched in the testis were associated with binding and catalytic activity (Table 2).
Table 2.
Gene ontology analysis of the differentially expressed genes (DEGs) identified in the testis compared with those in the epididymis and ductus deferens.
|
P value |
No DEGs |
Gene onthology |
P value |
No DEGs |
|---|---|---|---|---|
| Testis vs. epididymis | Testis vs. ductus deferens | |||
| Biological process | ||||
| Cell cycle | ||||
| 1.690E-13 | 96 | Cell cycle | 1.764E-11 | 112 |
| 3.135E-13 | 82 | Cell cycle process | 1.562E-10 | 93 |
| 1.726E-08 | 72 | Chromosome organization | 5.773E-07 | 84 |
| 2.750E-09 | 62 | DNA metabolic process | 9.569E-07 | 69 |
| 6.606E-14 | 43 | Chromosome segregation | 1.414E-10 | 45 |
| 4.458E-14 | 47 | Nuclear division | 5.553E-08 | 45 |
| 8.737E-10 | 34 | Nuclear chromosome segregation | 1.226E-05 | 33 |
| 9.497E-03 | 32 | DNA repair | 3.965E-03 | 40 |
| 3.673E-03 | 29 | Cell cycle phase transition | 1.064E-02 | 34 |
| 3.344E-05 | 25 | DNA replication | 3.999E-08 | 34 |
| 4.722E-05 | 23 | Sister chromatid segregation | 9.632E-03 | 23 |
| 4.585E-03 | 16 | DNA-dependent DNA replication | 3.157E-03 | 19 |
| 8.777E-07 | 13 | Homologous chromosome segregation | ||
| 4.123E-02 | 11 | Chromosome separation | ||
| 1.009E-03 | 8 | DNA replication initiation | ||
| Meiosis | ||||
| 1.608E-05 | 13 | Chromosome organization involved in meiotic cell cycle | 3.059E-02 | 11 |
| 6.448E-05 | 11 | Male meiotic nuclear division | 1.276E-02 | 10 |
| 4.985E-07 | 16 | Meiosis I | ||
| 7.445E-07 | 16 | Meiosis I cell cycle process | ||
| 1.662E-12 | 29 | Meiotic cell cycle | 8.020E-07 | 26 |
| 5.564E-11 | 26 | Meiotic cell cycle process | 1.145E-05 | 23 |
| 1.588E-06 | 15 | Meiotic chromosome segregation | 3.031E-02 | 12 |
| 2.091E-11 | 25 | Meiotic nuclear division | 5.162E-06 | 22 |
| Mitosis | ||||
| 7.940E-08 | 57 | Mitotic cell cycle | 8.212E-08 | 69 |
| 9.456E-03 | 26 | Mitotic cell cycle phase transition | ||
| 1.875E-06 | 48 | Mitotic cell cycle process | 5.440E-06 | 57 |
| 4.073E-05 | 27 | Mitotic nuclear division | 1.531E-02 | 27 |
| 1.200E-04 | 20 | Mitotic sister chromatid segregation | ||
| Reproduction | ||||
| 5.123E-09 | 34 | Cellular process involved in reproduction in multicellular organism | 1.935E-04 | 32 |
| 3.552E-03 | 38 | Developmental process involved in reproduction | ||
| 2.571E-10 | 45 | Gamete generation | 6.086E-07 | 47 |
| 4.132E-04 | 22 | Germ cell development | ||
| 5.576E-09 | 36 | Male gamete generation | 1.275E-07 | 40 |
| 2.764E-10 | 50 | Multicellular organism reproduction | 1.636E-06 | 52 |
| 1.820E-10 | 50 | Multicellular organismal reproductive process | 1.114E-06 | 52 |
| 1.069E-08 | 51 | Multiorganism reproductive process | 7.508E-06 | 55 |
| 4.636E-07 | 32 | Spermatogenesis | 1.644E-05 | 35 |
| 1.714E-09 | 70 | Reproduction | 2.358E-10 | 87 |
| 1.562E-09 | 70 | Reproductive process | 2.107E-10 | 87 |
| 6.031E-09 | 48 | Sexual reproduction | 6.886E-06 | 51 |
| Cell movement | ||||
| Axonemal dynein complex assembly | 3.776E-05 | 10 | ||
| Axoneme assembly | 5.973E-05 | 13 | ||
| Cilium assembly | 1.831E-09 | 42 | ||
| Cilium movement | 1.554E-06 | 14 | ||
| Cilium organization | 9.106E-11 | 45 | ||
| Epithelial cilium movement | 4.190E-02 | 6 | ||
| Flagellated sperm motility | 7.720E-07 | 17 | ||
| Inner dynein arm assembly | 1.844E-03 | 7 | ||
| Microtubule bundle formation | 3.974E-05 | 16 | ||
| 6.823E-03 | 36 | Microtubule cytoskeleton organization | 3.954E-07 | 54 |
| Microtubule-based movement | 3.812E-03 | 28 | ||
| 4.303E-02 | 44 | Microtubule-based process | 1.890E-09 | 74 |
| Sperm motility | 1.223E-07 | 18 | ||
| Other | ||||
| 3.605E-02 | 78 | Cellular response to stress | ||
| 2.691E-02 | 44 | Protein modification by small protein conjugation | ||
| 2.713E-03 | 53 | Protein modification by small protein conjugation or removal | ||
| 9.280E-03 | 43 | Protein ubiquitination | ||
| Cellular component | ||||
| Cell part | ||||
| 7.530E-08 | 503 | Cell | 5.626E-13 | 674 |
| 5.872E-08 | 500 | Cell part | 1.748E-13 | 671 |
| 4.979E-02 | 3 | Central element | ||
| 1.190E-02 | 35 | Centrosome | 1.280E-04 | 48 |
| 1.231E-02 | 75 | Cytoskeletal part | 1.813E-08 | 115 |
| 3.181E-02 | 87 | Cytoskeleton | 1.599E-06 | 128 |
| Nucleus | ||||
| 2.065E-05 | 39 | Nuclear chromosome | 6.220E-07 | 50 |
| 5.719E-04 | 34 | Nuclear chromosome part | 2.581E-05 | 44 |
| 7.360E-07 | 174 | Nuclear lumen | 1.072E-06 | 219 |
| 1.470E-10 | 202 | Nuclear part | 2.306E-11 | 257 |
| Nuclear ubiquitin ligase complex | 2.946E-03 | 10 | ||
| 6.971E-05 | 137 | Nucleoplasm | 1.660E-06 | 180 |
| 3.601E-11 | 274 | Nucleus | 2.304E-10 | 345 |
| Chromosome | ||||
| 3.987E-06 | 57 | Chromosomal part | 1.956E-08 | 75 |
| 9.117E-05 | 26 | Chromosomal region | 5.403E-05 | 31 |
| 1.363E-06 | 62 | Chromosome | 8.355E-09 | 81 |
| 1.553E-02 | 16 | Chromosome. Centromeric region | 1.121E-02 | 19 |
| 1.506E-08 | 23 | Condensed chromosome | 1.658E-05 | 22 |
| Condensed chromosome kinetochore | 2.874E-02 | 7 | ||
| 1.585E-02 | 5 | Condensed chromosome outer kinetochore | ||
| 2.605E-06 | 15 | Condensed nuclear chromosome | 5.989E-04 | 14 |
| Centriole | 2.667E-02 | 14 | ||
| Kinetochore | 2.477E-02 | 15 | ||
| 2.749E-02 | 22 | Spindle | 5.091E-03 | 28 |
| Microtubule/cilium | ||||
| 9 + 2 Motile cilium | 2.322E-05 | 15 | ||
| Axoneme | 5.710E-03 | 13 | ||
| Ciliary part | 2.657E-04 | 37 | ||
| Ciliary plasm | 5.710E-03 | 13 | ||
| Cilium | 3.269E-07 | 50 | ||
| Cytoplasm | 5.600E-03 | 425 | ||
| 3.301E-02 | 22 | Microtubule | 2.308E-03 | 29 |
| 6.755E-06 | 69 | Microtubule cytoskeleton | 1.072E-11 | 100 |
| 2.763E-04 | 46 | Microtubule organizing center | 5.210E-07 | 63 |
| Microtubule organizing center part | 1.342E-03 | 20 | ||
| Motile cilium | 1.848E-07 | 22 | ||
| Sperm | ||||
| Sperm flagellum | 5.648E-05 | 14 | ||
| Sperm part | 1.202E-04 | 18 | ||
| Molecular function | ||||
| Binding | ||||
| 1.472E-03 | 91 | Adenyl nucleotide binding | 3.998E-04 | 118 |
| 1.267E-03 | 91 | Adenyl ribonucleotide binding | 3.326E-04 | 118 |
| 6.819E-04 | 90 | ATP binding | 2.280E-04 | 116 |
| Chromatin binding | 3.349E-02 | 32 | ||
| 3.486E-02 | 4 | DNA replication origin binding | ||
| 9.735E-04 | 97 | Drug binding | 1.939E-03 | 122 |
| 2.279E-02 | 210 | Heterocyclic compound binding | 3.194E-02 | 274 |
| 3.857E-02 | 110 | Nucleoside phosphate binding | ||
| 3.857E-02 | 110 | Nucleotide binding | ||
| 2.126E-02 | 212 | Organic cyclic compound binding | 3.689E-02 | 276 |
| 3.246E-02 | 101 | Purine nucleotide binding | 1.953E-02 | 131 |
| 1.411E-02 | 100 | Purine ribonucleoside triphosphate binding | 9.773E-03 | 129 |
| 2.655E-02 | 101 | Purine ribonucleotide binding | 1.535E-02 | 131 |
| 3.637E-02 | 101 | Ribonucleotide binding | 1.449E-02 | 132 |
| 4.831E-02 | 23 | Tubulin binding | 4.722E-02 | 28 |
| Activity | ||||
| 3.020E-02 | 246 | Catalytic activity | 1.903E-02 | 325 |
| 9.752E-04 | 19 | Catalytic activity. Acting on DNA | 1.142E-04 | 24 |
| 2.636E-02 | 10 | Exonuclease activity | ||
Compared with the testis, DEGs upregulated in the epididymis and ductus deferens were primarily annotated in the following categories: 1) development, differentiation, and cell proliferation; 2) morphogenesis; 3) apoptosis; 4) cell communication and response to stimulus; 5) cell motility; 6) adhesion; and 7) oxidative stress (Table 3). The cellular component categories for DEGs upregulated in the epididymis and ductus deferens compared with the testis were primarily associated with the cell membrane and cell functions, including cell junctions and cell movement. The molecular functions enriched in the epididymis compared with the testis were associated with calcium ion binding. In contrast, protein and enzyme binding functions were enriched in the ductus deferens compared with the testis (Table 3).
Table 3.
Gene ontology analysis of differentially expressed genes (DEGs) identified in the epididymis and ductus deferens compared with those in the testis.
|
P value |
No DEGs |
Gene onthology |
P value |
No DEGs |
|---|---|---|---|---|
| Epididymis | Ductus deferens | |||
| Biological process | ||||
| Development, differentiation, and cell proliferation | ||||
| 2.717E-03 | 148 | Anatomical structure development | 1.902E-05 | 183 |
| 2.208E-02 | 93 | Animal organ development | ||
| 2.095E-03 | 43 | Animal organ morphogenesis | ||
| 5.369E-03 | 34 | Blood vessel development | 8.487E-03 | 38 |
| Cell differentiation | 1.748E-05 | 133 | ||
| 1.068E-02 | 64 | Cell population proliferation | 1.393E-06 | 86 |
| Cellular developmental process | 1.082E-05 | 140 | ||
| 8.588E-03 | 45 | Circulatory system development | 1.188E-02 | 51 |
| 4.266E-03 | 154 | Developmental process | 1.327E-06 | 196 |
| 1.403E-04 | 32 | Epithelial cell differentiation | 3.656E-04 | 35 |
| 5.215E-06 | 52 | Epithelium development | 2.954E-05 | 57 |
| 1.845E-02 | 12 | Formation of primary germ layer | 7.590E-04 | 15 |
| 8.345E-04 | 24 | Gland development | ||
| Multicellular organism development | 1.165E-02 | 156 | ||
| 1.527E-02 | 34 | Vasculature development | 2.577E-02 | 38 |
| 4.915E-07 | 76 | Tissue development | 4.564E-06 | 84 |
| 8.180E-05 | 49 | Tube development | 5.650E-05 | 56 |
| Morphogenesis | ||||
| 9.471E-03 | 45 | Anatomical structure formation involved in morphogenesis | 9.246E-05 | 57 |
| 1.564E-04 | 90 | Anatomical structure morphogenesis | 2.557E-07 | 113 |
| 2.089E-02 | 26 | Angiogenesis | 3.628E-03 | 31 |
| 1.375E-02 | 30 | Blood vessel morphogenesis | 3.255E-02 | 33 |
| 1.259E-03 | 15 | Branching morphogenesis of an epithelial tube | 2.596E-03 | 16 |
| Cell morphogenesis | 2.583E-05 | 53 | ||
| Cell morphogenesis involved in differentiation | 2.465E-04 | 40 | ||
| Cellular component morphogenesis | 1.810E-05 | 57 | ||
| Epithelial tube morphogenesis | 2.765E-02 | 22 | ||
| 8.967E-04 | 17 | Morphogenesis of a branching epithelium | 2.636E-03 | 18 |
| 5.797E-04 | 18 | Morphogenesis of a branching structure | 8.485E-03 | 18 |
| 5.131E-03 | 27 | Morphogenesis of an epithelium | 2.075E-02 | 29 |
| 2.069E-03 | 23 | Wound healing | ||
| 1.459E-03 | 32 | Tissue morphogenesis | 5.434E-04 | 37 |
| 5.905E-04 | 41 | Tube morphogenesis | 8.916E-04 | 46 |
| Apoptosis | ||||
| 3.437E-02 | 58 | Apoptotic process | ||
| 4.722E-03 | 7 | Apoptotic process involved in development | ||
| 1.130E-02 | 6 | Apoptotic process involved in morphogenesis | ||
| 3.909E-02 | 59 | Programmed cell death | ||
| 1.290E-02 | 52 | Regulation of apoptotic process | ||
| 1.177E-02 | 56 | Regulation of cell death | ||
| 1.103E-02 | 53 | Regulation of programmed cell death | ||
| Cell communication and respond to stimulus | ||||
| 2.481E-02 | 17 | Cell chemotaxis | ||
| Cell communication | 2.103E-03 | 206 | ||
| 4.590E-07 | 100 | Cellular response to chemical stimulus | 6.126E-05 | 108 |
| 4.733E-03 | 73 | Cellular response to organic substance | 1.074E-02 | 83 |
| Cellular response to stimulus | 1.920E-02 | 227 | ||
| Regulation of cell communication | 1.123E-04 | 131 | ||
| 2.155E-03 | 120 | Regulation of response to stimulus | 1.337E-05 | 149 |
| Negative regulation of cell communication | 7.280E-04 | 62 | ||
| 8.888E-03 | 58 | Negative regulation of response to stimulus | 8.958E-05 | 73 |
| 4.748E-02 | 67 | Positive regulation of response to stimulus | ||
| 2.061E-03 | 223 | Response to stimulus | 5.852E-03 | 261 |
| Cell motility | ||||
| Actin cytoskeleton organization | 4.425E-06 | 46 | ||
| Actin filament organization | 1.872E-03 | 28 | ||
| 3.340E-02 | 35 | Actin filament-based process | 1.664E-05 | 48 |
| 4.059E-02 | 50 | Cell migration | 3.402E-04 | 64 |
| 1.585E-02 | 9 | Cell migration involved in sprouting angiogenesis | ||
| Cell motility | 1.884E-03 | 66 | ||
| Locomotion | 3.707E-03 | 72 | ||
| Movement of cell or subcellular component | 4.228E-02 | 75 | ||
| 1.242E-02 | 26 | Positive regulation of cell migration | 1.629E-02 | 29 |
| Negative regulation of locomotion | 2.372E-02 | 21 | ||
| Regulation of cell migration | 6.945E-05 | 48 | ||
| Regulation of cell motility | 1.450E-04 | 49 | ||
| Regulation of locomotion | 1.184E-04 | 52 | ||
| 9.743E-03 | 28 | Positive regulation of locomotion | 6.284E-03 | 32 |
| 1.330E-02 | 10 | Actin polymerization/depolymerization | ||
| 2.782E-02 | 26 | Positive regulation of cell motility | 1.432E-02 | 30 |
| Adhesion | ||||
| Cell-cell adhesion | 5.274E-03 | 37 | ||
| Cell-matrix adhesion | 2.133E-02 | 18 | ||
| 5.491E-04 | 24 | Cell-substrate adhesion | 4.543E-03 | 25 |
| Negative regulation of cell adhesion | 2.706E-04 | 20 | ||
| 1.125E-02 | 30 | Regulation of cell adhesion | 1.820E-05 | 40 |
| 3.549E-05 | 54 | Biological adhesion | 1.738E-07 | 67 |
| 2.798E-05 | 54 | Cell adhesion | 1.281E-07 | 67 |
| Oxidative stress | ||||
| 3.655E-02 | 16 | Reactive oxygen species metabolic process | ||
| 8.575E-04 | 49 | Response to oxygen-containing compound | 3.930E-02 | 49 |
| 2.880E-02 | 96 | Response to stress | ||
| 1.320E-02 | 9 | Positive regulation of oxygen species | ||
| Other | ||||
| 4.913E-03 | 31 | Transmembrane receptor protein tyrosine kinase signaling pathway | ||
| 2.278E-02 | 24 | Regulation of hormone levels | ||
| Cellular component | ||||
| Cell part | ||||
| 1.544E-08 | 27 | Apical part of cell | 4.019E-04 | 23 |
| 5.237E-05 | 32 | Cell surface | 2.662E-02 | 30 |
| 7.274E-05 | 66 | Extracellular region | ||
| 1.544E-04 | 51 | Extracellular region part | 5.767E-03 | 54 |
| 9.221E-06 | 43 | Extracellular space | 1.149E-02 | 41 |
| 4.052E-02 | 52 | Vesicle | ||
| Cytoplasm | 1.236E-02 | 308 | ||
| Endoplasmic reticulum | 3.518E-05 | 67 | ||
| Membrane | ||||
| 4.969E-07 | 21 | Apical plasma membrane | 2.515E-02 | 16 |
| 3.425E-02 | 14 | Cell projection membrane | ||
| Endomembrane system | 5.972E-03 | 127 | ||
| 7.276E-03 | 250 | Membrane | 1.644E-03 | 303 |
| 2.620E-02 | 206 | Membrane part | ||
| 5.671E-07 | 131 | Plasma membrane | 1.837E-07 | 155 |
| 1.604E-05 | 75 | Plasma membrane part | 1.255E-04 | 84 |
| 3.548E-04 | 37 | Plasma membrane region | 6.810E-03 | 39 |
| 2.656E-02 | 18 | Side of membrane | ||
| Junction | ||||
| 1.853E-03 | 19 | Adherens junction | 7.537E-07 | 27 |
| 4.196E-03 | 19 | Anchoring junction | 6.057E-07 | 28 |
| 2.609E-02 | 33 | Cell junction | 1.009E-05 | 47 |
| Cell-cell junction | 6.441E-04 | 28 | ||
| 1.103E-07 | 137 | Cell periphery | 3.261E-08 | 162 |
| 2.538E-02 | 13 | Cell-substrate adherens junction | 6.923E-06 | 20 |
| 4.005E-02 | 13 | Cell-substrate junction | 2.807E-06 | 21 |
| Cell movement | ||||
| 4.531E-02 | 24 | Actin cytoskeleton | 7.409E-06 | 36 |
| Actin filament bundle | 2.414E-03 | 10 | ||
| Actomyosin | 4.166E-03 | 10 | ||
| 2.921E-03 | 13 | Cluster of actin-based cell projections | 2.781E-02 | 13 |
| Contractile actin filament bundle | 9.785E-04 | 10 | ||
| 1.767E-02 | 13 | Contractile fiber | 1.052E-02 | 15 |
| 1.653E-02 | 12 | Contractile fiber part | 7.428E-03 | 14 |
| 3.204E-03 | 10 | I band | 3.510E-03 | 11 |
| 1.060E-02 | 13 | Myofibril | 5.845E-03 | 15 |
| Sarcolemma | 5.392E-03 | 10 | ||
| 8.075E-03 | 12 | Sarcomere | 3.208E-03 | 14 |
| Stress fiber | 9.785E-04 | 10 | ||
| 4.838E-03 | 9 | Z disc | 5.470E-04 | 11 |
| Molecular function | ||||
| Protein binding | 2.490E-06 | 238 | ||
| Enzyme binding | 1.890E-02 | 85 | ||
| Cytoskeletal protein binding | 3.340E-03 | 51 | ||
| Protein-containing complex binding | 7.662E-04 | 48 | ||
| 3.559E-04 | 40 | Calcium ion binding | ||
| Protein phosphorylation | 5.390E-03 | 26 | ||
| 4.321E-02 | 37 | Protein-containing complex binding | ||
| Actin binding | 9.651E-04 | 31 | ||
| Actin filament binding | 3.261E-02 | 15 | ||
| 9.534E-03 | 13 | Glycosaminoglycan binding | ||
| 2.321E-02 | 13 | Sulfur compound binding | ||
| 4.189E-03 | 11 | Heparin binding | ||
| Extracellular matrix binding | 2.588E-02 | 7 | ||
| S100 protein binding | 2.216E-03 | 6 | ||
| 1.518E-02 | 4 | Bicarbonate transmembrane transporter activity | ||
DEGs upregulated in the epididymis compared with the ductus deferens were primarily associated with biological processes, such as cell movement and metabolic processes (Table 4). The cellular component categories for DEGs upregulated in the epididymis compared with the ductus deferens were associated with the cilium and plasma membrane. Thirty-five DEGs enriched in the turkey ductus deferens were associated with membranes and integral components of membranes, according to the STRING analysis (false discovery rate = 0.0239; Supplementary Figure 1).
Table 4.
Gene ontology analysis of differentially expressed genes (DEGs) identified in the epididymis compared with those in the ductus deferens.
| Gene ontology | P value | No DEGs |
|---|---|---|
| Biological process | ||
| Cellular movement | ||
| Axoneme assembly | 1.010E-10 | 11 |
| Cilium assembly | 2.519E-10 | 20 |
| Axonemal dynein complex assembly | 4.819E-10 | 9 |
| Cilium organization | 6.731E-10 | 20 |
| Cilium movement | 4.508E-09 | 10 |
| Microtubule bundle formation | 1.425E-08 | 11 |
| Inner dynein arm assembly | 4.214E-06 | 6 |
| Outer dynein arm assembly | 8.743E-05 | 5 |
| Cilium-dependent cell motility | 2.345E-04 | 8 |
| Cilium or flagellum-dependent cell motility | 2.345E-04 | 8 |
| Motile cilium assembly | 8.542E-04 | 5 |
| Microtubule cytoskeleton organization | 5.323E-03 | 16 |
| Microtubule-based process | 1.256E-02 | 19 |
| Microtubule-based movement | 1.271E-02 | 11 |
| Reproduction | ||
| Sperm motility | 7.175E-05 | 8 |
| Flagellated sperm motility | 7.175E-05 | 8 |
| Reproductive process | 1.680E-02 | 19 |
| Reproduction | 1.734E-02 | 19 |
| Metabolic process | ||
| Oxoacid metabolic process | 2.888E-05 | 24 |
| Organic acid metabolic process | 4.011E-05 | 24 |
| Carboxylic acid metabolic process | 6.568E-05 | 23 |
| Dicarboxylic acid metabolic process | 1.126E-03 | 7 |
| Monocarboxylic acid metabolic process | 6.281E-03 | 14 |
| Alpha-amino acid metabolic process | 3.109E-02 | 9 |
| Small molecule metabolic process | 1.085E-05 | 35 |
| Cell organization | ||
| Cell projection organization | 2.987E-06 | 31 |
| Plasma membrane bounded cell projection organization | 1.004E-05 | 30 |
| Organelle assembly | 1.090E-05 | 23 |
| Cellular component assembly | 5.002E-04 | 42 |
| Cellular component biogenesis | 2.977E-03 | 42 |
| Cell projection assembly | 2.132E-08 | 22 |
| Plasma membrane bounded cell projection assembly | 1.267E-07 | 21 |
| Cell differentiation | ||
| Multiciliated epithelial cell differentiation | 3.930E-02 | 3 |
| Cellular component | ||
| Cilium | ||
| Cilium | 3.588E-10 | 23 |
| Ciliary part | 5.120E-06 | 16 |
| Motile cilium | 4.639E-05 | 9 |
| Ciliary plasm | 6.388E-04 | 7 |
| Axoneme | 6.388E-04 | 7 |
| Plasma membrane | ||
| Plasma membrane bounded cell projection | 7.367E-06 | 30 |
| Cell projection | 1.020E-05 | 30 |
| Plasma membrane bounded cell projection part | 8.776E-05 | 22 |
| Cell projection part | 8.776E-05 | 22 |
| Plasma membrane bounded cell projection cytoplasm | 1.923E-02 | 7 |
| Photoreceptor inner segment | 4.066E-02 | 4 |
Validation of Selected DEGs by qRT-PCR
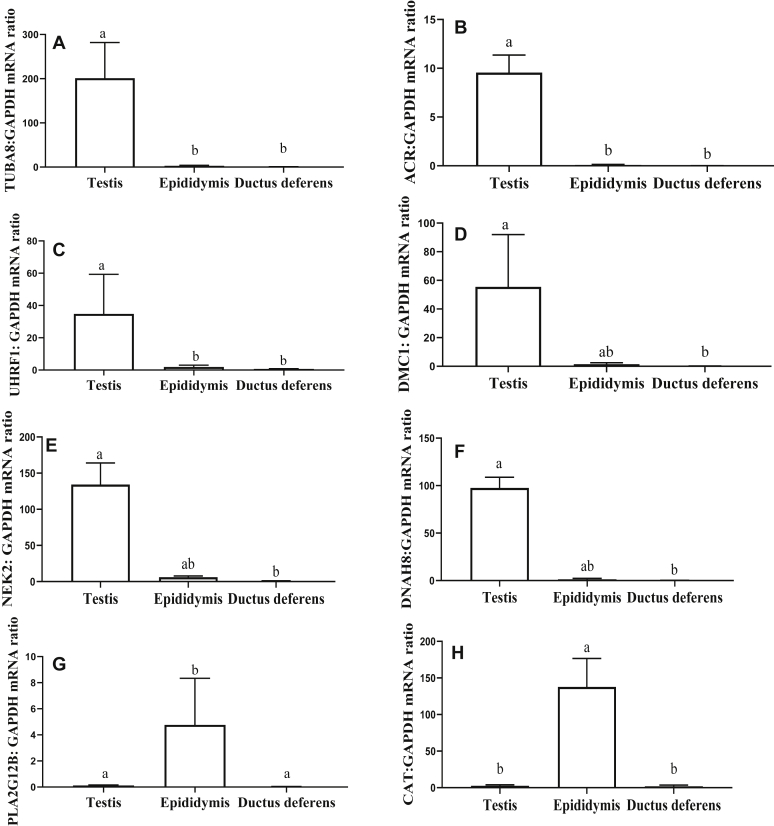
To validate the NGS results, 11 DEGs (6 upregulated and 5 downregulated DEGs in the testis) were selected for qRT-PCR analysis. The DEGs were chosen for the important physiological processes identified for testis and epididymis such as spermatogenesis (DMC1), nuclear condensation (NEK2), histone ubiquitination (UHRF1), acrosome development (ACR), testicular sperm motility (TUBA8 and DNAH8), calcium ion binding (PLA2G12b), and antioxidant protection (CAT). Among these 11 DEGs, the expression level differences for 8 genes were confirmed by qRT-PCR analysis (Figure 5). The observed differences in the mRNA expression levels of TUBA8, ACR, and UHRF1 confirmed that these genes were expressed at higher levels in the testis than in the epididymis and ductus deferens (Figures 5A–5C). Statistical analyses of mRNA expression level differences for 3 DEGs, DMC1, NEK2, and DNAH8, showed a lack of normality for the ductus deferens data. Analyses using a nonparametric one-way ANOVA showed higher expression levels in the testis than in the ductus deferens (Figures 5D–5F). However, it was not possible to determine differences in expression levels between the testis and epididymis (Figures 5D–5F). The mRNA expression level differences for PLA2G12b and CAT confirmed that these genes were expressed at higher levels in the epididymis than in the testis and ductus deferens (Figures 5G and 5H).
Figure 5.
Real-time validation of selected differentially expressed genes (DEGs), identified in the testis, epididymis, and ductus deferens by RNA-Seq. The validation was performed using the same RNA samples as were used for NGS. Data are expressed as the mean ± SD. (A) Expression of tubulin alpha 8 (TUBA8); (B) acrosin (ACR); (C) ubiquitin like with PHD and ring finger domains 1 (UHRF1); (D) DNA meiotic recombinase 1 (DMC1); (E) never in mitosis gene a-related kinase 2 (NEK2); (F) dynein axonemal heavy chain 8 (DNAH8); (G) phospholipase A2 group XIIB (PLA2G12 B); and (H) catalase (CAT). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
This study focused on identifying mRNA transcripts that reflect the specificity of particular components of the male reproductive system in turkeys, and this approach appeared to be successful. The gene expression analysis revealed different expression patterns for different components of the reproductive tract, and 1,682; 2,150; and 340 predicted DEGs were identified in testis/epididymis, testis/ductus deferens, and epididymis/ductus deferens comparisons, respectively. In the testis, the identified upregulated DEGs were primarily involved in the cell cycle, meiosis, mitosis, and reproductive processes, including gamete generation and development and spermatogenesis. In the epididymis and ductus deferens, the identified DEGs were involved in development, differentiation and cell proliferation, morphogenesis, apoptosis, cell communication, and response to stimulus. DEGs contributing to cell movement processes were identified in the testis, epididymis, and ductus deferens. These identified DEGs can potentially be used to characterize the specific functions of particular components of the reproductive system and to determine specificity during maturation processes for bird semen.
Spermatogenesis
Functional analyses performed on the transcripts from the testis revealed that the biological process identified by g:Profiler were specifically related to cell cycle processes, including mitotic and meiotic processes (Table 2), such as reduction division, which is an indication of spermatogenesis. The mitotic proliferation of spermatogonia and the meiotic division of primary spermatocytes into round spermatids are the primary events that occur during spermatogenesis in the avian testis (Asano and Tajima, 2017). Forty-two DEGs isolated from the turkey testis that were associated with spermatogenesis are likely to be involved in mitosis (18 DEGs) and meiosis (13 DEGs) (Supplementary Table 5). Among the DEGs identified in our study, DNA meiotic recombinase 1 (DMC1), DNA repair protein RAD51 homolog (RAD51), RAD21 cohesin complex component like 1, and synaptonemal complex protein 1 (SYCP1) are known to participate in bird meiosis (Zheng et al., 2009; Pigozzi, 2016). DMC1 and RAD51 have specifically been correlated with histone γ-H2AX phosphorylation during meiotic recombination (Pigozzi, 2016), RAD21 participates in the 3-dimensional arrangement of chromosomes during meiosis (Rutkowska and Badyaev, 2008), and SYCP1 is found in the meiotic axes in avian meiocytes and in chicken gonads at the moment of meiosis (Zheng et al., 2009). Our results indicated the presence of several transcripts that have not been previously associated with avian meiosis, which were primarily involved with meiotic chromosome segregation (Supplementary Table 5). The identification of these transcripts will be important for further experiments that aim to understand the detailed meiosis mechanisms in avian species.
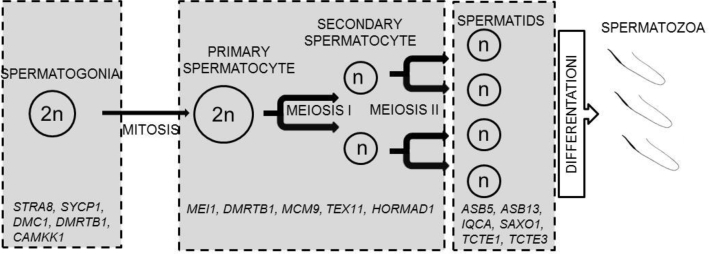
In our study, genes that were expressed during specific stages of spermatogenesis in turkeys were identified (Figure 6). Stimulated by retinoic acid 8, SYCP1 and DMC1 were identified in our study as DEGs that were expressed in differentiating spermatogonia, whereas meiotic double-stranded break formation protein 1 was expressed in spermatocytes (Zhu et al., 2016; Hermann et al., 2018). Moreover, in the testis, several genes expressed during spermatogonia that have been described in mammals were identified but have not yet been associated with turkey, including spermatogenesis genes such as DMRT like family B with proline-rich C-terminal 1 (DMRTB1) and calcium/calmodulin-dependent protein kinase 1 (CAMKK1), spermatocyte genes, such as DMRTB1, minichromosome maintenance 9 homologous recombination repair factor, testis expressed 11, HORMA domain containing 1, and spermatid genes, such as ankyrin repeat and SOCS box 5 and 13 (ASB5 and ASB13), IQ motif containing with AAA domain 1, stabilizer of axonemal microtubules 1, and t-complex-associated-testis-expressed (TCTE1 and TCTE3) (Zhu et al., 2016; Hermann et al., 2018). The results of our study indicated the expression of genes associated with specific cell cycle stages during turkey spermatogenesis, such as the mitotic proliferation of spermatogonia and the meiotic division of spermatocytes. These identified genes may represent potential genetic markers for differentiating cells during avian spermatogenesis.
Figure 6.
Genes expressed during specific stages of spermatogenesis in turkeys. Gene symbols: ankyrin repeat and SOCS box containing 13 (ASB13); ankyrin repeat and SOCS box containing 5 (ASB5); calcium/calmodulin dependent protein kinase kinase 1 (CAMKK1); DNA meiotic recombinase 1 (DMC1); DMRT like family B with proline rich C-terminal 1 (DMRTB1); HORMA domain containing 1 (HORMAD1); IQ motif containing with AAA domain 1 (IQCA); minichromosome maintenance 9 homologous recombination repair factor (MCM9); meiotic double-stranded break formation protein 1 (MEI1); stabilizer of axonemal microtubules 1 (SAXO1); stimulated by retinoic acid 8 (STRA8); synaptonemal complex protein 1 (SYCP1); t-complex-associated-testis-expressed 1 (TCTE1); t-complex-associated-testis-expressed 3 (TCTE3); testis expressed 11 (TEX11).
Nuclear Condensation
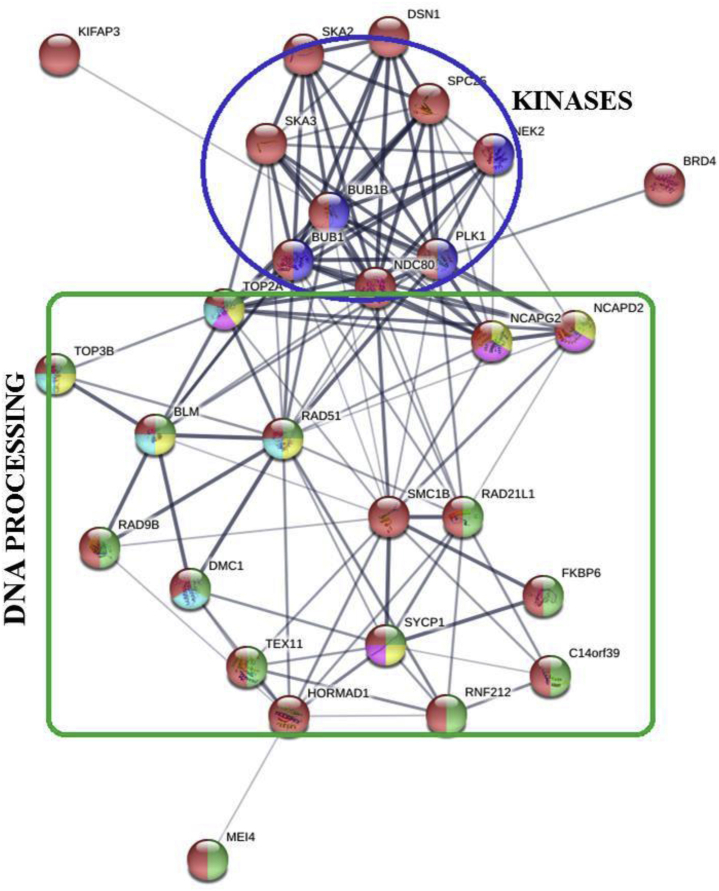
Nuclear condensation occurs during spermiogenesis and represents the final stage of spermatozoa formation. During this period, haploid, round spermatids undergo morphological differentiation to become spermatozoa (Asano and Tajima, 2017). In higher vertebrates, including birds, somatic histones are largely replaced during this stage by protamines, which are small, highly basic proteins that bind strongly to DNA, resulting in a high level of compaction (Chiva et al., 1987; Ausió et al., 2014). The results of our study indicate the expression of 28 DEGs associated with chromosome condensation in turkeys (Figure 7). Among these genes, a group of genes involved in DNA processing and genes encoding proteins containing kinase domains were distinguished (Figure 7). DNA conformational changes, DNA packing, metabolic processes, and catalytic activity can reflect changes to the DNA structure during the displacement of histones from nucleosomes by protamines and the formation of DNA-protamine toroids (Leuba and Brewer, 2009). The identification of kinase transcripts, such as BUB1 mitotic checkpoint serine/threonine kinases (BUB1 and BUB1B), NEK2, and polo-like kinase 1, likely reflects protamine phosphorylation, which has been described in mammals during spermatozoa development (Papoutsopoulou et al., 1999; Balhorn, 2007) suggesting that the phosphorylation of protamines may also occur during nuclear condensation in birds. This possibility was further supported by the presence of the phosphorylation site SRSRSR in bird protamines, which was recently described by Lüke et al. (2016).
Figure 7.
Protein-protein interaction networks for DEGs overexpressed in the testis are involved in chromatin condensation, according to an analysis by the STRING online database, version 11.0. The line thickness indicates the strength of the data (tight lines indicate high scores >0.9; thin lines indicate medium scores >0.4). Red: condensed chromosome. Dark blue: protein serine/threonine kinase activity. Green: DNA metabolic process. Yellow: DNA conformation change. Light blue: catalytic activity, acting on DNA. Pink: DNA packing. Gene symbols: Bloom syndrome RecQ like helicase (BLM); bromodomain containing 4 (BRD4); BUB1 mitotic checkpoint serine/threonine kinase (BUB1); BUB1 mitotic checkpoint serine/threonine kinase B (BUB1B); chromosome 14 open reading frame 39 (C14ofr39); DNA meiotic recombinase 1 (DMC1); DSN1 component of MIS12 kinetochore complex (DSN1); FK506 binding protein 6 (FKBP6); HORMA domain containing 1 (HORMAD1); kinesin associated protein 3 (KIFAP3); meiotic double-stranded break formation protein 4 (MEI4); non-SMC condensin I complex subunit D2 (NCAPD2); non-SMC condensin II complex subunit G2 (NCAPG2); NDC80 kinetochore complex component (NDC80); never in mitosis gene a-related kinase 2 (NEK2); polo like kinase 1 (PLK1); RAD21 cohesin complex component like 1 (RAD21L1); DNA repair protein RAD51 homolog (RAD51); RAD9 checkpoint clamp component B (RAD9B); ring finger protein 212 (RNF212); spindle and kinetochore associated complex subunit 2 (SKA2); spindle and kinetochore associated complex subunit 3 (SKA3); structural maintenance of chromosomes 1B (SMC1B); SPC25 component of NDC80 kinetochore complex (SPC25); synaptonemal complex protein 1 (SYCP1); testis expressed 11 (TEX11); DNA topoisomerase II alpha (TOP2A); DNA topoisomerase III beta (TOP3B).
Protein Ubiquitination
The ubiquitination of proteins in the testis occurs during many processes required for mature spermatozoa progression (Meistrich, 1989). The ubiquitin system appears to be involved in cell cycle regulation during the first 2 phases of spermatogenesis and regulates protein degradation during the cellular remodeling of haploid spermatids. Ubiquitination is a critical modification for the replacement of histones by protamines during spermiogenesis. Histones are among the key proteins that undergo proteolysis in early elongating spermatids, which permits chromatin condensation (Liu et al., 2005; Richburg et al., 2014). In our study, 43 DEGs that may be involved in protein ubiquitination in turkey testis were identified. These transcripts are associated with particular roles during the ubiquitin cascade (Supplementary Table 6), such as ubiquitin conjugation and ligation, which are performed by ubiquitin-conjugating enzymes and ubiquitin ligases (E3s), respectively (Zheng and Shabek, 2017). In mammals, E3 ligases have been implicated in the removal and degradation of histones and the condensation of sperm DNA (Richburg et al., 2014). Five of the identified DEGs in our study were directly assigned to histone ubiquitination, including tripartite motif-containing 37, lysine demethylase 1A, ring finger proteins (RNF2 and RNF168), and ubiquitin-like with PHD and ring finger domains 1 (UHRF1) (Supplementary Table 6). The identification of DEGs associated with testicular ubiquitination supports the finding reported by Agell et al. (1983), who demonstrated a high level of ubiquitination for histone H2As before the replacement of histones with protamines in rooster spermatids. When taken together, the findings of our study extended the knowledge of fowl spermatogenesis by identifying several transcripts associated with ubiquitin-dependent processes in the testis.
Acrosome
Sperm acrosome formation represents an important stage of spermiogenesis. Acrosome biogenesis in birds has been classified into 4 different phases: the Golgi phase, the cap phase, the acrosome phase, and the maturation phase (Asano and Tajima, 2017). Although the functional analysis performed for transcripts identified in the testis did not associate any identified DEGs with acrosomogenesis, specific DEGs could be connected with each of the 4 stages of acrosome formation in turkeys, based on available literature (Supplementary Table 7), including 3 in the Golgi phase, 2 in the cap phase, and 2 in the acrosome and maturation phase, respectively (Supplementary Table 7). In our study, transcripts that encoded acrosomal matrix proteins in the turkey testis were identified. Many well-characterized acrosomal matrix proteins were associated with transcripts identified in the testis in mammals, including acrosin (ACR), zona pellucida binding protein (ZPBP and ZPBP2), acrosin-binding protein (ACRBP), casein kinase II α (CSNK2A2), and proprotein convertase subtilisin 4/Kexin type (PCSK4) (Supplementary Table 7). Three of these, ACR, ZPBP, and ZPBP2, were previously described in the bird acrosome (Lin et al., 2007; Słowińska et al., 2010). In mammals, ACR is thought to play essential roles during fertilization, including during the recognition, binding, and penetration of the ovum zona pellucida (Klemm et al., 1991). The presence of the proacrosin/acrosin system has previously been confirmed and described in detail for turkey spermatozoa (Słowińska et al., 2010). In mammals, the ZPBP1 and ZPBP2 proteins participate in secondary binding between acrosome-reacted sperm and the egg-specific extracellular matrix (Clark, 2010). The coexistence of both the ZPBP1 and ZPBP2 genes has also previously been reported in birds (Lin et al., 2007). ACRBP, which has not previously been identified in bird spermatozoa, is a binding protein for both the precursor and intermediate forms of ACR (Baba et al. 1989; 1994). The mature form of ACRBP appears to play roles during the conversion of proacrosin into intermediate forms and the packaging of acrosomal matrix proteins during spermatogenesis (Baba et al., 1994; Foster, 2013). Finally, CSNK2A2 was identified, which, in mammals, participates in the formation of the acrosome during vesicle trafficking, and PCSK4, which is involved in the processing/activation of acrosomal proteins, acrosomal exocytosis, and zona pellucida binding (Chen et al., 2016; Foster and Gerton, 2016). Our study revealed the presence of several important transcripts associated with acrosome biogenesis, structure, and function. The mammalian and bird acrosomes are well known to differ significantly (Aire, 2014). In mammals, the acrosomal vesicle is a hollow, round structure containing an electron-dense granule, and the acrosomal granule is surrounded by an electron-lucent matrix. In birds, the acrosome vesicle is a membrane-bound vesicle, filled with homogenous, moderately electron-dense material (Aire, 2014). Our study identified a number of transcripts that may be useful for future detailed studies examining the specific mechanisms necessary for acrosome formation in birds.
Sperm Motility
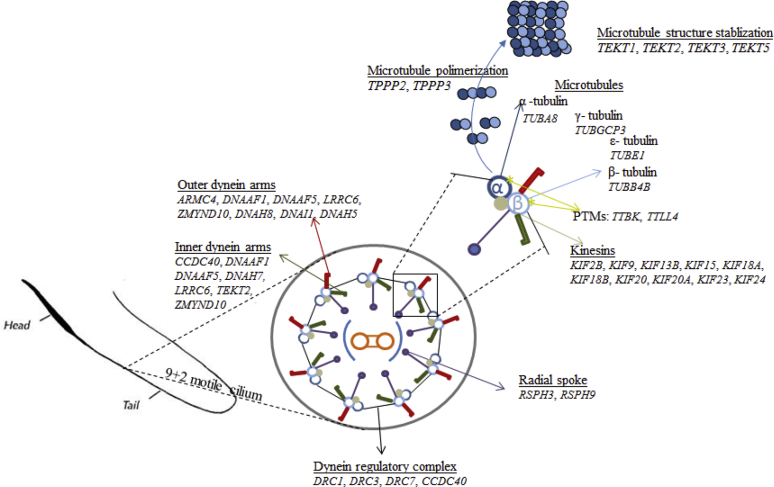
The DEGs upregulated in the turkey testis were found to be associated with several flagellar structures, including microtubules, outer and inner dynein arms, radial spokes, the dynein regulatory complex (DRC), and tektin, a microtubule-stabilizing protein (Figure 8). These results support the formation of flagella from the proximal and distal centrioles, which is typical during spermatogenesis in birds (Asano and Tajima, 2017). Some of the DEGs identified in our study were previously described for the mammalian axoneme (Figure 8; Pereira et al., 2017). In our study, DEGs that participate in the formation of the 9 + 2 motile cilium (Table 2) were identified, which likely reflects the structure of turkey spermatozoa axonemes, which are composed of nine outer doublet microtubules and 2 central singlet microtubules, referred to as the 9 + 2 structure (Figure 8; Thurston and Hess, 1987; Jamieson, 2007).
Figure 8.
The DEGs overexpressed in the testis are associated with the axoneme components of turkey spermatozoa flagella. Gene symbols: armadillo repeat containing 4 (ARMC4); coiled-coil domain containing 40 (CCDC40), dynein axonemal assembly factor 1, 5 (DNAAF1, DNAAF5); dynein axonemal heavy chain 5, 7, 8 (DNAH5, DNAH7, DNAH8); dynein axonemal intermediate chain 1 (DNAI1); dynein regulatory complex subunit 1, 3, 7 (DRC1, DRC3, DRC7); kinesin family member 9, 2B,13 B, 15, 18A, 18B, 20, 20A, 23, 24, (KIF9, KIF2B, KIF13 B, KIF15, KIF18 A, KIF18 B, KIF20, KIF20 A, KIF23, KIF24), leucine rich repeat containing 6 (LRRC6); radial spoke head 3 homolog, 9 homolog (RSPH3, RSPH9); tektin 1, 2, 3, 5 (TEKT1, TEKT2, TEKT3, TEKT5); tubulin polymerization promoting protein family member 2, 3 (TPPP2, TPPP3); tau tubulin kinase 2 (TTBK); tubulin tyrosine ligase like 4 (TTLL4), tubulin alpha 8 (TUBA8); tubulin beta 4B class IVb (TUBB4B); tubulin epsilon 1 (TUBE1), tubulin gamma complex associated protein 3 (TUBGCP3), zinc finger MYND-type containing 10 (ZMYND10).
Microtubules, which are major components of the eukaryotic cytoskeleton, are formed by the heterodimeric α- and β-tubulin proteins (Kierszenbaum, 2002), and both tubulin alpha 8 (TUBA8) and tubulin beta 4B class IVb were identified in our study (Figure 8). Both α- and β-tubulins, together with tubulin polymerization promoting protein (TPPP2 and TPPP3, Figure 8), are involved in microtubule polymerization (Orosz and Ovádi, 2008). Tektin transcripts (TEKT1, TEKT2, TEKT3, and TEKT5), which encode proteins that stabilize the microtubule structure, were also identified in our study (Amos, 2008). In addition to the α- and β-tubulins, other tubulins, such as γ- and ε-tubulins, which may be involved in axoneme formation (Kierszenbaum, 2002; Inaba, 2003), were identified in our study. The α- and β-tubulins undergo several types of posttranslational modifications, such as acetylation, palmitoylation, tyrosine phosphorylation, and polyglutamylation (Kierszenbaum, 2002; Inaba, 2003). These posttranslational modifications play roles in microtubule stability, the interactions between microtubules and associated proteins, and participate in axonemal motility (Gagnon et al., 1996; Huitorel et al., 1999). The identification of transcripts that encode proteins responsible for tubulin phosphorylation and polyglutamylation, such as tau tubulin kinase-2 and tubulin tyrosine ligase like 4, respectively, further supported the occurrence of posttranslational tubulin modifications in the turkey testis.
Microtubules serve as tracks for 2 classes of motor proteins, namely kinesins and dyneins (Cooper, 2000; Berg et al., 2002). In our study, 10 DEGs associated with kinesin microtubule motor activity were identified (Figure 8). This study is the first to report the presence of a kinesin system in the bird testis. Moreover, 8 and 7 DEGs identified in our study specifically encoded outer and inner dynein arms, respectively (Figure 8). Dyneins are motor proteins that convert the chemical energy released by ATP hydrolysis into the mechanical energy of movement (Pereira et al., 2017). The outer and inner dynein arms are composed of heavy, intermediate, and light chains. Six genes that encoded heavy chain genes (DNAH1, DNAH3, DNAH7, DNAH8, and DNAH17) and 1 that encoded an intermediate chain gene (DNAI1) were identified in our study (Figure 8). Similar to the study by Kollmar et al. (2016), dynein 6 or 9 were not identified, which are likely absent in birds. Dynein ATPase activity in bird spermatozoa has previously been reported in chickens, where dynein-ATPase activity was observed to be involved in reversible temperature-dependent immobilization (Ashizawa et al., 2013). Dynein activity is controlled by the combinatorial actions of several regulatory proteins, including the DRC and radial spokes (Inaba, 2003). Four genes in our study were identified as components of the DRC (DRC1, DRC3, DRC7, and coiled-coil domain containing 40), and 2 genes were identified as radial spokes (RSPH3 and RSPH9) (Figure 8). Understanding the dynein regulatory genes associated with sperm motility mechanisms in birds may contribute to our understanding of reproductive disorders because, in mammals, mutations in DRC and radial spoke genes lead to anomalies in the central pair complex and the inner dynein arm and result in primary ciliary dyskinesia (Antony et al., 2013; Onoufriadis et al., 2014).
Post-testicular Development of Sperm Motility
In birds, testicular spermatozoa are characterized by low motility, suggesting the importance of post-testicular sperm maturation to improve spermatozoa motility (Asano and Tajima, 2017). The epididymis was identified as the location where bird spermatozoa develop progressive motility (Nixon et al., 2014). In our study, DEGs associated with flagellated sperm motility in the epididymis were identified, which appear to primarily be involved in inner and outer dynein arm assembly (Table 4, Supplementary Table 8). In addition to the identification of common DEGs that participate in dynein arm formation in both the testis and epididymis, the transcript that codes for DNAH5 had the highest expression level in the epididymis (Supplementary Table 8). The protein encoded by DNAH5 appears to be very important for male fertility in mammals (Pereira et al., 2015). Therefore, the analysis of testicular and epididymal DEGs suggested that flagellar structures are primarily formed in the testis in turkeys (Figure 8), and processes associated with the dynein arm continue in the epididymis, where the final stages of sperm motility development occur. Presumably, the dynein arm assembly process is completed within the epididymis because the levels of transcripts associated with this process are low in the ductus deferens.
Calcium Ion-Binding Protein
In our study, in addition to DEGs that were common between both the testis and epididymis, transcripts that were enriched in the epididymis, including those that encode calcium-binding proteins and participate in reproductive processes, were also identified (Table 3, Supplementary Table 8). Calcium ions play a key role in the stimulation of fowl sperm motility, in vivo (Nguyen et al., 2016; Asano and Tajima, 2017). A regulatory role for Ca2+ has been documented for axonemal functions in fowl semen (Ashizawa et al., 1994; Deviche et al., 2011), and intracellular free Ca2+ is essential for the maintenance of chicken sperm motility. In our study, epididymal DEGs that encoded calcium/calmodulin-dependent protein kinase (CAMKK1 and CAMK1D), phosphatidylinositol kinase (PIP5K1B), phospholipase A2 (PLA2G12b), and myosin light chain kinase (MYLK and MYLK2), which have previously been demonstrated to be involved in calcium-regulated flagellar motility in fowl sperm, were also identified (Froman and Feltmann, 2005; Ashizawa et al., 2009; Nguyen, 2019). Moreover, our study revealed, for the first time, 40 DEGs that were upregulated in the epididymis associated with calcium ion binding, according to g:Profiler (Table 3), suggesting the importance of Ca2+-binding proteins during the regulation of fowl sperm motility in the epididymis and providing a more complete picture of the Ca2+ regulation process than was previously known.
Lipid Metabolism
Epididymal DEGs that were associated with reproductive processes could be also linked with lipid metabolism (Supplementary Table 8). Lipid changes during epididymal maturation have been well described for mammalian spermatozoa, resulting in the development of membrane fluidity, stability, and fusion capacity (Amann et al., 1993; Fouchecourt et al., 2000; Jervis and Robaire, 2001). Phospholipids and cholesterol components in mammalian spermatozoa are primarily affected during sperm maturation (Jones, 1998; Whitfield et al., 2015; de Souza et al., 2017). The DEGs identified in our study associated with lipid metabolism in turkeys are potentially involved in fatty acid binding and cholesterol homeostasis (hepatocyte nuclear factor 4 alpha [HNF4A]), the retinoic acid biosynthetic process (retinol dehydrogenase 10), the prostaglandin metabolic process (hematopoietic prostaglandin D synthase), and the phospholipid biosynthetic process (hexosaminidase subunit beta). Phospholipids and free cholesterol are present in turkey spermatozoa and are lost during the liquid storage of turkey spermatozoa (Zaniboni and Cerolini, 2009). Moreover, disturbances in lipid metabolism have previously been associated with yellow semen syndrome (YSS) in turkeys, during which numerous lipid vacuoles can be found in the ductuli efferentes epithelia of the epididymis (Hess et al., 1982; Thurston and Korn, 1997; Słowińska et al., 2018; 2019). Although detailed studies regarding the maturation mechanisms for bird spermatozoa in the epididymis are lacking, the results of our study strongly suggest a possible role for lipids during turkey sperm maturation. Further studies should focus on detailed characterizations of lipid changes during epididymal maturation and the identification of lipid transporters in bird sperm.
Actin
Functional analyses of the transcripts identified in the epididymis and ductus deferens revealed the presence of several transcripts encoding actin-binding proteins, proteins involved in actin filament-based processing and organization, and actin polymerization/depolymerization (Table 3, Supplementary Table 8). Actin is a well-known cytoskeletal protein, and its localization along the entire length of the flagellum and in the post acrosomal region of the head indicates its crucial involvement during several processes, such as the development of progressive motility in epididymal spermatozoa, the hyperactivation of motility during capacitation, and acrosomal exocytosis, in mammals (Clarke et al., 1982; Flaherty et al., 1986; Fouquet and Kann, 1992; de las Heras et al., 1997; Breitbart and Finkelstein, 2018). Actin polymerization/depolymerization occurs during spermatozoa capacitation in mammals, and the depolarization of F-actin in the tail allows the development of hyperactivated motility (Breitbart and Finkelstein, 2018). In our study, 10 DEGs associated with actin polymerization/depolymerization were identified in the turkey epididymis (actin-related protein 2/3 complex subunit 5 [ARPC5], capping protein regulator and myosin 1 linker 1, EVL, gelsolin [GSN], IQ motif containing GTPase-activating protein 2, microtubule-associated monooxygenase, calponin and LIM domain containing 2, slit guidance ligand 2, spectrin beta nonerythrocytic 1, twinfilin actin-binding protein 1, and WD repeat domain 1). Moreover, transcripts encoding proteins that control F-actin formation (Rho GTPase-activating proteins [ARHGAP17 and ARHGAP24] and ARPC5) were identified (Ducummon and Berger, 2006; Breitbart and Finkelstein, 2018). Actin filament elongation during sperm capacitation is controlled by the activation of phospholipase D and calcium/calmodulin-dependent protein kinase and the inactivation of gelsolin and cofilin (Jungnickel et al., 2006; Etkovitz et al., 2007). In our study, similar transcripts (phospholipase D family member 5, CaMKK1, CaMKK1D, GSN, and cofilin 2) were identified. In summary, our results indicated that several transcripts encoding actin and related proteins that participate in the actin polymerization/depolymerization processes are likely also involved in the development of progressive motility in both the epididymis and ductus deferens of birds.
Phosphorylation/dephosphorylation
Phosphorylation plays critical roles in the regulation of many physiological processes. Fowl sperm flagellar motility appears to be controlled by various protein phosphorylation-dephosphorylation systems (Asano and Tajima, 2017). DEGs encoding several serine/threonine kinases were identified, which are responsible for protein phosphorylation and are involved in the regulation of sperm motility in fowl, including isoforms of protein kinase A (PRKAR2A, PRKAG3, and PRKACB; Nguyen et al., 2014), protein kinase C (PRKCA, PRKCQ, and PACSIN2; Ashizawa et al., 1994), and mitogen-activated protein kinase (MAPK1 and MAPK6 (Ashizawa et al., 1995; 1997). Moreover, several other kinases that may participate in protein phosphorylation in the turkey epididymis/ductus deferens were identified in our study, such as alpha kinase 1; cyclin-dependent kinase (CAMK1D and CDK18); eukaryotic translation initiation factor 2 alpha kinase 2; Golgi-associated secretory pathway pseudokinase (FAM20 A, FAM20 C); fyn-related Src family tyrosine kinase; MYLK2; MYLK; NEK7; pyruvate dehydrogenase kinase 4; protein kinase domain containing, cytoplasmic; protein kinase, cGMP-dependent, type I; ribosomal protein S6 kinase A3; serine/threonine/tyrosine kinase 1; and tribbles pseudokinase 2. The specific roles played by these kinases remain unknown at present. Further studies should focus on the characterization of these kinases and their target proteins in relation to particular reproductive processes in the turkey, particularly in the epididymis and ductus deferens.
In addition to phosphorylation, 5 DEGs encoding protein phosphatases (PPs) in the turkey epididymis/ductus deferens (type 1: PPP1R12 A and PPP1R1B; type 2: PPP2R3; and serine/threonine-protein phosphatase: PPP3CA) were identified, which are known to affect sperm motility through protein dephosphorylation (Nguyen, 2019). The activation of type 1 PPs, which are present in the fowl sperm axoneme and/or as accessory cytoskeletal components, leads to the inhibition of sperm motility (Labas et al., 2015). In mammals, PP1s are involved in the inhibition of sperm motility through interactions with A-kinase anchoring proteins (AKAPs) and calcium/calmodulin-dependent protein kinase (CAMKII) (Han et al., 2007), which were also identified in our study (AKAP13 and CAMK1D, respectively). The results of our study indicated a role for phosphorylation in the acquisition of sperm motility and a role for dephosphorylation in the inhibition of sperm motility.
Apoptosis
In our study, a large number of apoptosis-related DEGs were identified, for the first time, in the turkey reproductive tract (Supplementary Table 9). Identified DEGs were found to be directly associated with apoptosis and apoptosis-related pathways such as the transforming growth factor (TGF)-beta signaling pathway, the MAPK signaling pathway, signaling by interleukins, and the p53 signaling pathway. MAPK and p53 signaling pathways are well-known regulators of apoptosis (Wada and Penninger, 2004; Pietsch et al., 2008; Wan et al., 2016). In mammals, apoptosis contributes to germ cell degeneration and seasonal testicular regression (Shaha et al., 2010; Aitken et al. 2011), which has also been observed in seasonal bird species when testosterone levels fall and testicular size decreases (Thurston and Korn, 2000; Santiago-Moreno et al., 2016). The results of our study strongly suggested that, in contrast with mammals, in which apoptosis occurs in the testis (Aitken et al., 2011), the epididymis and ductus deferens may be the primary sites for apoptosis in the turkey reproductive tract. Therefore, in mammals, apoptosis may be the primary regulatory mechanism for spermatogenesis (Liu et al., 2017b), whereas, in birds, apoptosis may primarily be involved in the regulation of post-testicular sperm maturation quality.
Response to Reactive Oxygen Species
High concentrations of polyunsaturated fatty acids in bird spermatozoa increase membrane sensitivity to lipid peroxidation in the presence of reactive oxygen species (Fujihara and Howarth, 1978; Surai et al., 1998; Cerolini et al., 2006). Spermatozoa can be protected from lipid peroxidation by the antioxidant system, which relies on the enzymatic activities of CAT, superoxide dismutase (SOD), and peroxiredoxin (PRDx) and has previously been described for bird semen (Słowińska et al., 2011; Partyka et al., 2012). In our study, transcripts that encoded all of these enzymes (Supplementary Table 10) were linked with post-testicular development in the epididymis and ductus deferens (CAT, SOD, and PRDx; Supplementary Table 8). In addition to these well-known antioxidant enzymes, other DEGs linked to defense again oxidative stress, such as sestrin 1, selenoprotein T, MAPK1, and amyloid-beta precursor protein, were identified (Supplementary Table 10). Further studies are necessary to confirm whether the antioxidant proteins identified in our study are also present in ejaculated semen and should aim to establish the relationships between newly identified antioxidant-related proteins and CAT, SOD, and PRDx.
Sperm Membrane Maturation
Thirty-five DEGs enriched in the turkey ductus deferens were linked by STRING analysis to membranes and integral components of membranes (Supplementary Figure 1). These results are in agreement with the studies reported by Morris et al. (1987) and Esponda (1991), indicating that, in birds, proteins secreted by the epithelial cells of the Wolffian duct (vas deferens) are involved in changes associated with the maturation of the sperm membrane. Moreover, 5 DEGs enriched in the ductus deferens were linked with motility (Supplementary Table 8) and were classified as being associated with solute:cation symporter activity and the response to estrogen. In mammals, the solute carrier SLC22A14 plays a pivotal role in the normal flagellar structure, motility, and fertility of spermatozoa (Maruyama et al., 2016). In our study, 2 additional solute carrier DEGs were identified (SLC15A1 and SLC34A2) that could be involved in sperm motility by transporting several substances, such as ions, amino acids, sugars, and metabolites, that are important for sperm motility. The role played by estrogen in sperm motility has been widely reported for mammals (Hess and Cooke, 2018). The balance between testosterone and estradiol was found to be critical for normal sperm maturation within the epididymis and for the proper functioning of the ductus deferens (Shetty et al., 1997; Hess et al., 2000). In our study, 2 identified DEGs (KRT19 and SLC34A2) were involved in the response to estrogens in the ductus deferens, suggesting a positive effect for estrogens on the final spermatozoa maturation process in the ductus deferens of birds. In mammals, elevated estradiol levels have been correlated with male infertility (Leavy et al., 2017). Similarly, in turkeys, excess estrogen has been correlated with the pathological condition YSS (Pardyak et al., 2018). Further studies should focus on determining whether DEGs related to estrogen response are associated with the YSS pathology in turkeys.
Reproductive System Development
In our study, DEGs that encode proteins associated with reproductive system development and morphogenesis in turkeys were identified. The testis was characterized by the presence of several DEGs involved in male sexual differentiation, testis growth, and development, including doublesex and mab-3–related transcription factor 2, nuclear receptor subfamily 5 group A member 1, and anti-Mullerian hormone (Kuroiwa et al., 2002). In addition, hundreds of DEGs identified in the epididymis and ductus deferens were associated with cell development, differentiation, and morphogenesis, primarily in relation to blood vessels and the epithelium.
Blood Vessel Development
DEGs involved in angiogenesis and blood vessel development in the turkey epididymis and ductus deferens have been assigned by PANTHER to key mammalian signaling pathways associated with angiogenesis, such as the vascular endothelial growth factor (VEGF), TGF-β, Toll receptor, and WNT signaling pathways (Supplementary Table 11). So far, only the VEGF signaling pathway has been found to induce angiogenesis in the male reproductive tract (Hyder and Stancel, 2000), and together with TGF-β, it is a known proangiogenic factor that regulates vasculogenesis, which represents the first stage of vascular system development (Guerrero and McCarty, 2016). The Toll receptor and WNT signaling pathways are involved in the promotion of angiogenesis, primarily through the stimulation of cell migration, cell permeability, and remodeling (Xu et al., 2013; Olsen et al., 2017).
To our knowledge, none of these pathways have previously been associated with angiogenesis and have not been described in the male reproductive tracts of birds. The only available information for birds concerns the female reproductive system and embryos, in which the VEGF and WNT signaling pathways have been associated with follicle selection and development in the avian ovary (Johnson, 2014), and the TGF-β-signaling pathways have found to be active in primordial germ cells in early chicken embryos (Whyte et al., 2015). Our study identified, for the first time, a set of genes involved in angiogenesis in the male reproductive tract of turkeys. These genes were assigned to known angiogenesis signaling pathways that have previously been reported in mammals. Our results suggest the importance of avian angiogenesis and blood vessel development in the epididymis and ductus deferens, indicating the need for the extensive remodeling of these organs. These findings should be verified by future experiments focused on angiogenesis in particular segments of the turkey reproductive tract during a reproductive season.
Epithelial Cell Development
Identified DEGs associated with epithelial cell development in the turkey epididymis and ductus deferens were specifically related to reproductive structure development and developmental processes during reproduction (Table 3). DEGs involved in developmental processes previously described the mammalian reproductive tracts of both males were identified, including amphiregulin (AREG), cytochrome P450 family 7 subfamily B member 1 (CYP7B1), hepatocyte nuclear factor 1-alpha (HNF1), HNF4A, lymphoid enhancer-binding factor 1 (LEF1), LDL receptor-related protein 2, MAPK1, polycystin 2 (PKD2), ROS proto-oncogene 1, receptor tyrosine kinase (ROS1), and serpin family E member 2 (SERPINE2) (Hudson et al., 2001; Van Praet et al., 2003; Li et al., 2009b; Lu et al., 2011; Wu et al., 2011; Nie and Arend, 2014; Breton et al., 2016; Browne et al., 2016; Vermillion et al., 2018), and females, including activin A receptor type 1, annexin A1, CYP7B1, and LEF1 (Ignotz et al., 2007; Mullen and Behringer, 2014; Haraguchi et al., 2017; Sharum et al., 2017). In the male reproductive tract, identified transcripts encoded proteins directly involved in male system development (PKD2; Nie and Arend, 2014), tissue repair (AREG; Vermillion et al., 2018), epithelial cell differentiation (ROS1, CYP7B1, HNF1, and HNF4; Hudson et al., 2001; Breton et al., 2016; Browne et al., 2016), and junction integrity (MAPK1; Dobin et al., 2013). SERPINE2 encodes a serine protease inhibitor considered to be an intrinsic sperm surface protein that binds to epididymal sperm in mammals and modifies the sperm surface (Lu et al., 2011). The results of our study strongly suggest that extensive processes associated with development, differentiation, and remodeling occur in the epithelium of the epididymis and ductus deferens in the turkey reproductive tract. The epithelial cells of the epididymis and ductus deferens are known to be actively secretory, and in birds, the secretory products produced by the epithelial cells of the epididymis and the Wolffian duct (vas deferens) lead to numerous functional changes during post-testicular sperm maturation, including the acquisition of motility and the increased capacity to fertilize (Morris et al., 1987; Esponda, 1991; Nixon et al., 2014).
Conclusions
In conclusion, the transcriptomic analysis of the turkey reproductive tract identified many candidate genes that may potentially be involved in spermatogenesis, spermiogenesis and flagellar formation in the testis, and in post-testicular sperm maturation in the epididymis and ductus deferens. In the testis, specific genes were linked with the mitotic proliferation of spermatogonia and the meiotic division of spermatocytes, which are the primary events that occur during bird spermatogenesis. The ubiquitination of histones and the phosphorylation of protamines were shown to be regulatory mechanisms for nuclear condensation during spermiogenesis. The identified testicular transcripts suggested the importance of acrosome formation and development of flagellar formation, including the axoneme structure (microtubule polymerization and stabilization) and function (regulation and the provision of energy for movement). The achievement of spermatozoa motility during post-testicular maturation appears to be associated with the development of flagellar actin filaments. Ca2+ influx and protein phosphorylation/dephosphorylation processes appear to be the primary regulatory mechanisms for sperm motility during post-testicular maturation. The quality of spermatozoa appears to be controlled by the apoptosis and antioxidant systems of the epididymis and ductus deferens. Finally, genes involved in reproductive system development and morphogenesis were also described for the turkey reproductive tract.
Acknowledgments
This work was supported by the National Science Centre, Poland (2016/21/B/NZ9/03583).
Conflict of interest: The authors declare no conflicts to disclose.
Footnotes
Supplementary data associated with this article can be found in the online version at http://doi.org/10.1016/j.psj.2020.07.031.
Supplementary data
References
- Agell N., Chiva M., Mezquita C. Changes in nuclear content of protein conjugate histone H2A-ubiquitin during rooster spermatogenesis. FEBS Lett. 1983;155:209–212. doi: 10.1016/0014-5793(82)80604-2. [DOI] [PubMed] [Google Scholar]
- Aire T.A. Spermatogenesis and testicular cycles. In: Jamieson B.G.M., editor. Reproductive Biology and Phylogeny of Birds: Phylogeny, Morphology, Hormones, Fertilization. vol. 6A. Science Publishers; Enfield, NH: 2007. pp. 279–347. [Google Scholar]
- Aire T.A. Taylor & Francis Online; 2014. Spermiogenesis in Birds. Spermatogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken R.J., Findlay J.K., Hutt K.J., Kerr J.B. Apoptosis in the germ line. Reproduction. 2011;141:139–150. doi: 10.1530/REP-10-0232. [DOI] [PubMed] [Google Scholar]
- Amann R.P., Hammerstedt R.H., Veeramachaneni D.N. The epididymis and sperm maturation: a perspective. Reprod. Fertil. Dev. 1993;5:361–381. doi: 10.1071/rd9930361. [DOI] [PubMed] [Google Scholar]
- Amos L.A. The tektin family of microtubule-stabilizing proteins. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony D., Becker-Heck A., Zariwala M.A., Schmidts M., Onoufriadis A., Forouhan M., Wilson R., Taylor-Cox T., Dewar A., Jackson C., Goggin P., Loges N.T., Olbrich H., Jaspers M., Jorissen M., Leigh M.W., Wolf W.E., Daniels M.L., Noone P.G., Ferkol T.W., Sagel S.D., Rosenfeld M., Rutman A., Dixit A., O'Callaghan C., Lucas J.S., Hogg C., Scambler P.J., Emes R.D., Uk10k, Chung E.M., Shoemark A., Knowles M.R., Omran H., Mitchison H.M. Mutations in CCDC39 and CCDC40 are the major cause of primary ciliary dyskinesia with axonemal disorganization and absent inner dynein arms. Hum. Mutat. 2013;34:462–472. doi: 10.1002/humu.22261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano A., Tajima A. Development and preservation of avian sperm. In: Sasanami T., editor. Avian Reproduction, Advances in Experimental Medicine and Biology. Springer Nature Singapore Pte Ltd; 2017. pp. 59–73. [DOI] [PubMed] [Google Scholar]
- Ashizawa K., Wishart G.J., Nishinakama K., Sakamoto T., Tsuzuki Y. Regulatory mechanisms of fowl sperm motility: possible role of endogenous myosin light chain kinase-like protein. J. Reprod. Fertil. 1995;104:141–148. doi: 10.1530/jrf.0.1040141. [DOI] [PubMed] [Google Scholar]
- Ashizawa K., Hashimoto K., Higashio M., Tsuzuki Y. The addition of mitogen-activated protein kinase and p34cdc2 kinase substrate peptides inhibits the flagellar motility of demembranated fowl spermatozoa. Biochem. Biophys. Res. Commun. 1997;240:116–121. doi: 10.1006/bbrc.1997.7626. [DOI] [PubMed] [Google Scholar]
- Ashizawa K., Oyama N., Katayama S., Narumi K., Tatemoto H., Tsuzuki Y. Regulation of fowl sperm motility: evidence for the indirect, but not direct, involvement of dynein-ATPase activity on the reversible temperature-dependent immobilization. Theriogenology. 2013;79:558–565. doi: 10.1016/j.theriogenology.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Ashizawa K., Katayama S., Kobayashi T., Tsuzuki Y. Possible role of protein kinase C in regulation of flagellar motility and intracellular free Ca2+ concentration of fowl spermatozoa. J. Reprod. Fertil. 1994;101:511–517. doi: 10.1530/jrf.0.1010511. [DOI] [PubMed] [Google Scholar]
- Ashizawa K., Omura Y., Katayama S., Tatemoto H., Narumi K., Tsuzuki Y. Intracellular signal transduction pathways in the regulation of fowl sperm motility: evidence for the involvement of phosphatidylinositol 3-kinase (PI3-K) cascade. Mol. Reprod. Dev. 2009;76:603–610. doi: 10.1002/mrd.20995. [DOI] [PubMed] [Google Scholar]
- Ausió J., González-Romero R., L C. Woodcock. Comparative structure of vertebrate sperm chromatin. J. Struct. Biol. 2014;188:142–155. doi: 10.1016/j.jsb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- Baba T., Michikawa Y., Kashiwabara S., Arai Y. Proacrosin activation in the presence of a 32-kDa protein from boar spermatozoa. Biochem. Biophys. Res. Commun. 1989;160:1026–1032. doi: 10.1016/s0006-291x(89)80105-6. [DOI] [PubMed] [Google Scholar]
- Baba T., Niida Y., Michikawa Y., Kashiwabara S., Kodaira K., Takenaka M., Kohno N., Gerton G.L., Arai Y. An acrosomal protein, sp32, in mammalian sperm is a binding protein specific for two proacrosins and an acrosin intermediate. J. Biol. Chem. 1994;269:10133–10140. [PubMed] [Google Scholar]
- Balhorn R. The protamine family of sperm nuclear proteins. Genome Biol. 2007;8:227. doi: 10.1186/gb-2007-8-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaupré C.E., Tressler C.J., Beaupré S.J., Morgan J.L., Bottje W.G., Kirby J.D. Determination of testis temperature rhythms and effects of constant light on testicular function in the domestic fowl (Gallus domesticus) Biol. Reprod. 1997;56:1570–1575. doi: 10.1095/biolreprod56.6.1570. [DOI] [PubMed] [Google Scholar]
- Berg J.M., Tymoczko J.L., Stryer L. W H Freeman and Co.; New York, NY: 2002. Biochemistry. [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart H., Finkelstein M. Actin cytoskeleton and sperm function. Biochem. Biophys. Res. Commun. 2018;506:372–377. doi: 10.1016/j.bbrc.2017.11.001. [DOI] [PubMed] [Google Scholar]
- Breton S., Ruan Y.C., Park Y.J., Kim B. Regulation of epithelial function, differentiation, and remodeling in the epididymis. Asian J. Androl. 2016;18:3–9. doi: 10.4103/1008-682X.165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne J.A., Yang R., Eggener S.E., Leir S.H., Harris A. HNF1 regulates critical processes in the human epididymis epithelium. Mol. Cell. Endocrinol. 2016;425:94–102. doi: 10.1016/j.mce.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows W.H., Quinns J.P. The collection of spermatozoa from the domestic fowl and Turkey. Poult. Sci. 1937;16:19–24. [Google Scholar]
- Cerolini S., Zaniboni L., Maldjian A., Gliozzi T. Effect of docosahexaenoic acid and alpha-tocopherol enrichment in chicken sperm on semen quality, sperm lipid composition and susceptibility to peroxidation. Theriogenology. 2006;66:877–886. doi: 10.1016/j.theriogenology.2006.02.022. [DOI] [PubMed] [Google Scholar]
- Chalmel F., Rolland A.D. Linking transcriptomics and proteomics in spermatogenesis. Reproduction. 2015;150:R149–R157. doi: 10.1530/REP-15-0073. [DOI] [PubMed] [Google Scholar]
- Chen S.R., Batool A., Wang Y.Q., Hao X.X., Chang C.S., Cheng C.Y., Liu Y.X. The control of male fertility by spermatid-specific factors: searching for contraceptive targets from spermatozoon's head to tail. Cell Death Dis. 2016;7:e2472. doi: 10.1038/cddis.2016.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiva M., Kasinsky H.F., Subirana J.A. Characterization of protamines from four avian species. FEBS Lett. 1987;215:237–240. doi: 10.1016/0014-5793(87)80153-9. [DOI] [PubMed] [Google Scholar]
- Clark G.F. The mammalian zona pellucida: a matrix that mediates both gamete binding and immune recognition? Syst. Biol. Reprod. Med. 2010;56:349–364. doi: 10.3109/19396360903524812. [DOI] [PubMed] [Google Scholar]
- Clarke G.N., Clarke F.M., Wilson S. Actin in human spermatozoa. Biol. Reprod. 1982;26:319–327. doi: 10.1095/biolreprod26.2.319. [DOI] [PubMed] [Google Scholar]
- Cooper G.M. Sinauer Associates; Sunderland (MA): 2000. The Cell: A Molecular Approach. [Google Scholar]
- de las Heras M.A., Valcarcel A., Pérez L.J., Moses D.F. Actin localization in ram spermatozoa: effect of freezing/thawing, capacitation and calcium ionophore-induced acrosomal exocytosis. Tissue Cell. 1997;29:47–53. doi: 10.1016/s0040-8166(97)80071-7. [DOI] [PubMed] [Google Scholar]
- de Souza A.P.B., Schorr-Lenz A.M., Lucca F., Bustamante-Filho I.C. The epididymis and its role on sperm quality and male fertility. Anim. Reprod. 2017;14:1234–1244. [Google Scholar]
- Deviche P., Hurley L.L., Fokidis H.B. Avian testicular structure, function and regulation. In: Norris D.O., Lopez K.H., editors. Hormones and Reproduction of Vertebrates. vol. 4. Academic Press; Cambridge, MA: 2011. pp. 27–70. [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducummon C.C., Berger T. Localization of the Rho GTPases and some Rho effector proteins in the sperm of several mammalian species. Zygote. 2006;14:249–257. doi: 10.1017/S0967199406003790. [DOI] [PubMed] [Google Scholar]
- Esponda P. Spermatozoon maturation in vertebrates with internal fertilization. Microsc. Electron Biol. Celular. 1991;15:1–23. [PubMed] [Google Scholar]
- Etkovitz N., Rubinstein S., Daniel L., Breitbart H. Role of PI3-kinase and PI4-kinase in actin polymerization during bovine sperm capacitation. Biol. Reprod. 2007;77:263–273. doi: 10.1095/biolreprod.106.056705. [DOI] [PubMed] [Google Scholar]
- Flaherty S.P., Winfrey V.P., Olson G.E. Localization of actin in mammalian spermatozoa: a comparison of eight species. Anat. Rec. 1986;216:504–515. doi: 10.1002/ar.1092160407. [DOI] [PubMed] [Google Scholar]
- Foster J.A. Baby brother acrosin-binding protein (ACRBP) says “look at me now!”. Biol. Reprod. 2013;88:106. doi: 10.1095/biolreprod.113.109413. [DOI] [PubMed] [Google Scholar]
- Foster J.A., Gerton G.L. The acrosomal matrix. Adv. Anat. Embryol. Cell Biol. 2016;220:15–33. doi: 10.1007/978-3-319-30567-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchecourt S., Metayer S., Locatelli A., Dacheux F., Dacheux J.L. Stallion epididymal fluid proteome: qualitative and quantitative characterization; secretion and dynamic changes of major proteins. Biol. Reprod. 2000;62:1790–1803. doi: 10.1095/biolreprod62.6.1790. [DOI] [PubMed] [Google Scholar]
- Fouquet J.P., Kann M.L. Species-specific localization of actin in mammalian spermatozoa: fact or artifact? Microsc. Res. Tech. 1992;20:251–258. doi: 10.1002/jemt.1070200304. [DOI] [PubMed] [Google Scholar]
- Frazee A.C., Pertea G., Jaffe A.E., Langmead B., Salzberg S.L., Leek J.T. Flexible analysis of transcriptome assemblies with Ballgown. BioRxiv. 2014:003665. [Google Scholar]
- Froman D.P., Feltmann A.J. Fowl (Gallus domesticus) sperm motility depends upon mitochondrial calcium cycling driven by extracellular sodium. Biol. Reprod. 2005;72:97–101. doi: 10.1095/biolreprod.104.033209. [DOI] [PubMed] [Google Scholar]
- Fujihara N., Jr Howarth B. Lipid peroxidation in fowl spermatozoa. Poult. Sci. 1978;57:1766–1768. doi: 10.3382/ps.0571766. [DOI] [PubMed] [Google Scholar]
- Gagnon C., White D., Cosson J., Huitorel P., Eddé B., Desbruyères E., Paturle-Lafanechère L., Multigner L., Job D., Cibert C. The polyglutamylated lateral chain of alpha-tubulin plays a key role in flagellar motility. J. Cell Sci. 1996;109:1545–1553. doi: 10.1242/jcs.109.6.1545. [DOI] [PubMed] [Google Scholar]
- Guerrero P.A., McCarty J.H. TGF-β activation and aignaling in angiogenesis. In: Simionescu D., Simionescu A., editors. Physiologic and Pathologic Angiogenesis: Signaling Mechanisms and Targeted Therapy. InTech; Croatia: 2016. pp. 3–22. [Google Scholar]
- Han Y., Haines C.J., Feng H.L. Role(s) of the serine/threonine protein phosphatase 1 on mammalian sperm motility. Arch. Androl. 2007;53:169–177. doi: 10.1080/01485010701314032. [DOI] [PubMed] [Google Scholar]
- Haraguchi R., Kitazawa R., Murashima A., Yamada G., Kitazawa S. Developmental contribution of Wnt-signal-responsive cells to mouse reproductive tract formation. Acta Histochem. Cytochem. 2017;50:127–133. doi: 10.1267/ahc.17017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann B.P., Cheng K., Singh A., Roa-De La Cruz L., Mutoji K.N., Chen I.C., Gildersleeve H., Lehle J.D., Mayo M., Westernströer B., Law N.C., Oatley M.J., Velte E.K., Niedenberger B.A., Fritze D., Silber S., Geyer C.B., Oatley J.M., McCarrey J.R. The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep. 2018;25:1650–1667. doi: 10.1016/j.celrep.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess R.A., Cooke P.S. Estrogen in the male: a historical perspective. Biol. Reprod. 2018;99:27–44. doi: 10.1093/biolre/ioy043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess R.A., Bunick D., Lubahn D.B., Zhou Q., Bouma J. Morphologic changes in efferent ductules and epididymis in estrogen receptor-alpha knockout mice. J. Androl. 2000;21:107–121. [PubMed] [Google Scholar]
- Hess R.A., Thurston R.J., Biellier H.V. Morphology of the epididymal region and ductus deferens of the Turkey (Meleagris gallopavo) J. Anat. 1976;122:241–252. [PMC free article] [PubMed] [Google Scholar]
- Hess R.A., Thurston R.J., Biellier H.V. Morphology of the epididymal region of turkeys producing abnormal yellow semen. Poult. Sci. 1982;61:531–539. doi: 10.3382/ps.0610531. [DOI] [PubMed] [Google Scholar]
- Hudson C.E., Schulte B.A., Sutter T.R., Norris J.S. Steroid hormones modulate expression of cytochrome P450 enzymes in male hamster reproductive tract and leiomyosarcomas. Carcinogenesis. 2001;22:763–770. doi: 10.1093/carcin/22.5.763. [DOI] [PubMed] [Google Scholar]
- Huitorel P., Audebert S., White D., Cossone J., Gagnon C. Role of tubulin epitopes in the regulation of flagellar motility. In: Gagnon C., editor. The Male Gamete: From Basic Science to Clinical Applications. Cache River Press; Illinois: 1999. pp. 475–491. [Google Scholar]
- Hyder S.M., Stancel G.M. Regulation of VEGF in the reproductive tract by sex-steroid hormones. Histol. Histopathol. 2000;15:325–334. doi: 10.14670/HH-15.325. [DOI] [PubMed] [Google Scholar]
- Ignotz G.G., Cho M.Y., Suarez S.S. Annexins are candidate oviductal receptors for bovine sperm surface proteins and thus may serve to hold bovine sperm in the oviductal reservoir. Biol. Reprod. 2007;77:906–913. doi: 10.1095/biolreprod.107.062505. [DOI] [PubMed] [Google Scholar]
- Inaba K. Molecular architecture of the sperm flagella: molecules for motility and signaling. Zoolog. Sci. 2003;20:1043–1056. doi: 10.2108/zsj.20.1043. [DOI] [PubMed] [Google Scholar]
- Jamieson B.G.M. Avian spermatozoa: structure and phylogeny. In: Jamiesson B.G.M., editor. Reproductive Biology and Phylogeny of Birds. Science Publishers; Enfield, NH: 2007. pp. 349–511. [Google Scholar]
- Jervis K.M., Robaire B. Dynamic changes in gene expression along the rat epididymis. Biol. Reprod. 2001;65:696–703. doi: 10.1095/biolreprod65.3.696. [DOI] [PubMed] [Google Scholar]
- Johnson A.L. The avian ovary and follicle development: some comparative and practical insights. Turk J. Vet. Anim. Sci. 2014;38:660–669. [Google Scholar]
- Jones R. Plasma membrane structure and remodelling during sperm maturation in the epididymis. J. Reprod. Fertil. Suppl. 1998;53:73–84. [PubMed] [Google Scholar]
- Jones R.C., Lin M. Spermatogenesis in birds. J. Reprod. Fertil. Suppl. 1993;15:233–264. [PubMed] [Google Scholar]
- Jungnickel M.K., Sutton K.A., Wang Y., Florman H.M. Phosphoinositide-dependent pathways in mouse sperm are regulated by egg ZP3 and drive the acrosome reaction. Dev. Biol. 2006;304:116–126. doi: 10.1016/j.ydbio.2006.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum A.L. Sperm axoneme: a tale of tubulin posttranslation diversity. Mol. Reprod. Dev. 2002;62:1–3. doi: 10.1002/mrd.10139. [DOI] [PubMed] [Google Scholar]
- Klemm U., Müller-Esterl W., Engel W. Acrosin, the peculiar sperm-specific serine protease. Hum. Genet. 1991;87:635–641. doi: 10.1007/BF00201716. [DOI] [PubMed] [Google Scholar]
- Kollmar M. Fine-tuning motile cilia and flagella: evolution of the dynein motor proteins from plants to humans at high resolution. Mol. Biol. Evol. 2016;33:3249–3267. doi: 10.1093/molbev/msw213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordonowy L.L., MacManes M.D. Characterization of a male reproductive transcriptome for Peromyscus eremicus (Cactus mouse) PeerJ. 2016;4:e2617. doi: 10.7717/peerj.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa A., Yokomine T., Sasaki H., Tsudzuki M., Tanaka K., Namikawa T., Matsuda Y. Biallelic expression of Z-linked genes in male chickens. Cytogenet. Genome Res. 2002;99:310–314. doi: 10.1159/000071609. [DOI] [PubMed] [Google Scholar]
- Labas V., Grasseau I., Cahier K., Gargaros A., Harichaux G., Teixeira-Gomes A.P., Alves S., Bourin M., Gérard N., Blesbois E. Qualitative and quantitative peptidomic and proteomic approaches to phenotyping chicken semen. J. Proteomics. 2015;112:313–335. doi: 10.1016/j.jprot.2014.07.024. [DOI] [PubMed] [Google Scholar]
- Lake P.E. The male reproduction. In: Bell D.J., Freeman B.M., editors. Physilogy and Biochemistry of the Domestic Fowl. Academic Press; New York, NY: 1971. pp. 1411–1447. [Google Scholar]
- Leavy M., Trottmann M., Liedl B., Reese S., Stief C., Freitag B., Baugh J., Spagnoli G., Kölle S. Effects of elevated β-estradiol levels on the functional morphology of the testis - new insights. Sci. Rep. 2017;7:39931. doi: 10.1038/srep39931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecluze E., Jégou B., Rolland A.D., Chalmel F. New transcriptomic tools to understand testis development and functions. Mol. Cell. Endocrinol. 2018;468:47–59. doi: 10.1016/j.mce.2018.02.019. [DOI] [PubMed] [Google Scholar]
- Leuba S.H., Brewer L.R. Single molecule studies of chromatin structure and dynamics. In: Knight A.E., editor. Single Molecule Biology. Elsevier; Amsterdam, The Netherlands: 2009. pp. 143–171. [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M.W., Mruk D.D., Cheng C.Y. Mitogen-activated protein kinases in male reproductive function. Trends Mol. Med. 2009;15:159–168. doi: 10.1016/j.molmed.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H.-K. Transcriptomics. In: Liang H.-K., editor. Bioinformatics for Biomedical Science and Clinical Applications. Woodhead Publishing Limited; Sawston, Cambridge, UK: 2013. pp. 49–80. [Google Scholar]
- Lin Y.N., Roy A., Yan W., Burns K.H., Matzuk M.M. Loss of zona pellucida binding proteins in the acrosomal matrix disrupts acrosome biogenesis and sperm morphogenesis. Mol. Cell. Biol. 2007;27:6794–6805. doi: 10.1128/MCB.01029-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sun Y., Li Y., Bai H., Xue F., Xu S., Xu H., Shi L., Yang N., Chen J. Analyses of long non-coding RNA and mRNA profiling using RNA sequencing in chicken testis with extreme sperm motility. Sci. Rep. 2017;7:9055. doi: 10.1038/s41598-017-08738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wang L., Chen H., Huang Y., Yang P., Ahmed N., Wang T., Liu Y., Chen Q. Molecular and cellular mechanisms of apoptosis during dissociated spermatogenesis. Front Physiol. 2017;8:188. doi: 10.3389/fphys.2017.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sun Y., Li Y., Bai H., Xu S., Xu H., Ni A., Yang N., Chen J. Identification and differential expression of microRNAs in the testis of chicken with high and low sperm motility. Theriogenology. 2018;122:94–101. doi: 10.1016/j.theriogenology.2018.09.010. [DOI] [PubMed] [Google Scholar]
- Liu Z., Oughtred R., Wing S.S. Characterization of E3Histone, a novel testis ubiquitin protein ligase which ubiquitinates histones. Mol. Cell. Biol. 2005;25:2819–2831. doi: 10.1128/MCB.25.7.2819-2831.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A. The 'omics' revolution: use of genomic, transcriptomic, proteomic and metabolomic tools to predict male reproductive traits that impact fertility in livestock and poultry. Anim. Reprod. Sci. 2020;19:106354. doi: 10.1016/j.anireprosci.2020.106354. [DOI] [PubMed] [Google Scholar]
- Lowe R., Shirley N., Bleackley M., Dolan S., Shafee T. Transcriptomics technologies. Plos Comput. Biol. 2017;13:e1005457. doi: 10.1371/journal.pcbi.1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.H., Lee R.K., Hwu Y.M., Chu S.L., Chen Y.J., Chang W.C., Lin S.P., Li S.H. SERPINE2, a serine protease inhibitor extensively expressed in adult male mouse reproductive tissues, may serve as a murine sperm decapacitation factor. Biol. Reprod. 2011;84:514–525. doi: 10.1095/biolreprod.110.085100. [DOI] [PubMed] [Google Scholar]
- Lüke L., Tourmente M., Dopazo H., Serra F., Roldan E.R. Selective constraints on protamine 2 in primates and rodents. BMC Evol. Biol. 2016;16:21. doi: 10.1186/s12862-016-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama S.Y., Ito M., Ikami Y., Okitsu Y., Ito C., Toshimori K., Fujii W., Yogo K. A critical role of solute carrier 22a14 in sperm motility and male fertility in mice. Sci. Rep. 2016;6:36468. doi: 10.1038/srep36468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meistrich M.L. Histones and basic nuclear proteins transitions in mammalian spermatogenesis. In: Hnilica L.S., Stein G.S., Stein J.L., editors. Histones and Other Basic Nuclear Proteins. CRC Press; Boca Raton, FL: 1989. pp. 165–196. [Google Scholar]
- Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S.A., Jr Howarth B., Crim J.W., Rodriguez de Cordoba A., Esponda P., Bedford J.M. Specificity of sperm-binding Wolffian duct proteins in the rooster and their persistence on spermatozoa in the female host glands. J. Exp. Zool. 1987;242:189–198. doi: 10.1002/jez.1402420210. [DOI] [PubMed] [Google Scholar]
- Mullen R.D., Behringer R.R. Molecular genetics of Müllerian duct formation, regression and differentiation. Sex Dev. 2014;8:281–296. doi: 10.1159/000364935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.M.D. Main signaling pathways involved in the control of fowl sperm motility. Poult. Sci. 2019;98:1528–1538. doi: 10.3382/ps/pey465. [DOI] [PubMed] [Google Scholar]
- Nguyen T.M., Duittoz A., Praud C., Combarnous Y., Blesbois E. Calcium channels in chicken sperm regulate motility and the acrosome reaction. FEBS J. 2016;283:1902–1920. doi: 10.1111/febs.13710. [DOI] [PubMed] [Google Scholar]
- Nguyen T.M., Alves S., Grasseau I., Métayer-Coustard S., Praud C., Froment P., Blesbois E. Central role of 5'-AMP-activated protein kinase in chicken sperm functions. Biol. Reprod. 2014;91:121. doi: 10.1095/biolreprod.114.121855. [DOI] [PubMed] [Google Scholar]
- Nie X., Arend L.J. Novel roles of Pkd2 in male reproductive system development. Differentiation. 2014;87:161–171. doi: 10.1016/j.diff.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon B., Ewen K.A., Krivanek K.M., Clulow J., Kidd G., Ecroyd H., Jones R.C. Post-testicular sperm maturation and identification of an epididymal protein in the Japanese quail (Coturnix coturnix japonica) Reproduction. 2014;147:265–277. doi: 10.1530/REP-13-0566. [DOI] [PubMed] [Google Scholar]
- Olsen J.J., Pohl S.Ö., Deshmukh A., Visweswaran M., Ward N.C., Arfuso F., Agostino M., Dharmarajan A. The role of Wnt Signalling in angiogenesis. Clin. Biochem. Rev. 2017;38:131–142. [PMC free article] [PubMed] [Google Scholar]
- Onoufriadis A., Shoemark A., Schmidts M., Patel M., Jimenez G., Liu H., Thomas B., Dixon M., Hirst R.A., Rutman A., Burgoyne T., Williams C., Scully J., Bolard F., Lafitte J.J., Beales P.L., Hogg C., Yang P., Chung E.M., Emes R.D., O'Callaghan C., UK10K, Bouvagnet P., Mitchison H.M. Targeted NGS gene panel identifies mutations in RSPH1 causing primary ciliary dyskinesia and a common mechanism for ciliary central pair agenesis due to radial spoke defects. Hum. Mol. Genet. 2014;23:3362–3374. doi: 10.1093/hmg/ddu046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orosz F., Ovádi J. TPPP orthologs are ciliary proteins. FEBS Lett. 2008;582:3757–3764. doi: 10.1016/j.febslet.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Papoutsopoulou S., Nikolakaki E., Chalepakis G., Kruft V., Chevaillier P., Giannakouros T. SR protein-specific kinase 1 is highly expressed in testis and phosphorylates protamine 1. Nucleic Acids Res. 1999;27:2972–2980. doi: 10.1093/nar/27.14.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardyak L., Kaminska A., Brzoskwinia M., Hejmej A., Kotula-Balak M., Jankowski J., Ciereszko A., Bilinska B. Differences in aromatase expression and steroid hormone concentrations in the reproductive tissues of male domestic turkeys (Meleagris gallopavo) with white and yellow. Br. Poult. Sci. 2018;59:591–603. doi: 10.1080/00071668.2018.1483576. [DOI] [PubMed] [Google Scholar]
- Partyka A., Łukaszewicz E., Niżański W. Lipid peroxidation and antioxidant enzymes activity in avian semen. Anim. Reprod. Sci. 2012;134:184–190. doi: 10.1016/j.anireprosci.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Pereira R., Oliveira J., Ferraz L., Barros A., Santos R., Sousa M. Mutation analysis in patients with total sperm immotility. J. Assist. Reprod. Genet. 2015;32:893–902. doi: 10.1007/s10815-015-0474-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R., Sá R., Barros A., Sousa M. Major regulatory mechanisms involved in sperm motility. Asian J. Androl. 2017;19:5–14. doi: 10.4103/1008-682X.167716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M., Pertea G.M., Antonescu C.M., Chang T.-C., T Mendell J., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietsch E.C., Sykes S.M., McMahon S.B., Murphy M.E. The p53 family and programmed cell death. Oncogene. 2008;27:6507–6521. doi: 10.1038/onc.2008.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigozzi M.I. The chromosomes of birds during meiosis. Cytogenet. Genome Res. 2016;150:128–138. doi: 10.1159/000453541. [DOI] [PubMed] [Google Scholar]
- Qi X.L., Xing K., Huang Z., Chen Y., Wang L., Zhang L.C., Sheng X.H., Wang X.G., Ni H.M., Guo Y. Comparative transcriptome analysis digs out genes related to antifreeze between fresh and frozen-thawed rooster sperm. Poult. Sci. 2020;99:2841–2851. doi: 10.1016/j.psj.2020.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: a language and environment for statistical computing. 2017. http://www.R-project.org/ Accessed April 2020.
- Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richburg J.H., Myers J.L., Bratton S.B. The role of E3 ligases in the ubiquitin-dependent regulation of spermatogenesis. Semin. Cell Dev. Biol. 2014;30:27–35. doi: 10.1016/j.semcdb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowska J., Badyaev A.V. Review. Meiotic drive and sex determination: molecular and cytological mechanisms of sex ratio adjustment in birds. Philos. Trans. R. Soc. Lond. B, Biol. Sci. 2008;363:1675–1686. doi: 10.1098/rstb.2007.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago-Moreno J., Esteso M.C., Villaverde-Morcillo S., Toledano-Déaz A., Castaño C., Velázquez R., López-Sebastián A., L Goya A., Martínez J.G. Recent advances in bird sperm morphometric analysis and its role in male gamete characterization and reproduction technologies. Asian J. Androl. 2016;18:882–888. doi: 10.4103/1008-682X.188660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaha C., Tripathi R., Mishra D.P. Male germ cell apoptosis: regulation and biology. Philos. Trans. R. So.C Lond. B, Biol. Sci. 2010;365:1501–1515. doi: 10.1098/rstb.2009.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharum I.B., Granados-Aparici S., Warrander F.C., Tournant F.P., Fenwick M.A. Serine threonine kinase receptor associated protein regulates early follicle development in the mouse ovary. Reproduction. 2017;153:221–231. doi: 10.1530/REP-16-0612. [DOI] [PubMed] [Google Scholar]
- Shetty G., Krishnamurthy H., Krishnamurthy H.N., Bhatnagar S., N R., Moudgal Effect of estrogen deprivation on the reproductive physiology of male and female primates. J. Steroid Biochem. Mol. Biol. 1997;61:157–166. [PubMed] [Google Scholar]
- Słowińska M., Hejmej A., Bukowska J., Liszewska E., Bilińska B., Hliwa P., Kozłowski K., Jankowski J., Ciereszko A. Expression and secretion of albumin in male Turkey (Meleagris gallopavo) reproductive tract in relation to yellow semen syndrome. Poult. Sci. 2019;98:1872–1882. doi: 10.3382/ps/pey490. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Jankowski J., Dietrich G.J., Karol H., Liszewska E., Glogowski J., Kozlowski K., Sartowska K., Ciereszko A. Effect of organic and inorganic forms of selenium in diets on Turkey semen quality. Poult. Sci. 2011;90:181–190. doi: 10.3382/ps.2010-00956. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Liszewska E., Nynca J., Bukowska J., Hejmej A., Bilińska B., Szubstarski J., Kozłowski K., Jankowski J., Ciereszko A. Isolation and characterization of an ovoinhibitor, a multidomain Kazal-like inhibitor from Turkey (Meleagris gallopavo) seminal plasma. Biol. Reprod. 2014;91:108. doi: 10.1095/biolreprod.114.118836. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Olczak M., Liszewska E., Watorek W., Ciereszko A. Isolation, characterization and cDNA sequencing of acrosin from Turkey spermatozoa. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 2010;157:127–136. doi: 10.1016/j.cbpb.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Słowińska M., Sallem H., Clench M.R., Ciereszko A. Metabolomic analysis of white and yellow seminal plasma in turkeys (Meleagris gallopavo) Poult. Sci. 2018;97:1059–1065. doi: 10.3382/ps/pex366. [DOI] [PubMed] [Google Scholar]
- Surai P.F., Blesbois E., Grasseau I., Chalah T., Brillard J.P., Wishart G.J., Cerolini S., Sparks N.H. Fatty acid composition, glutathione peroxidase and superoxide dismutase activity and total antioxidant activity of avian semen. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 1998;120:527–533. doi: 10.1016/s0305-0491(98)10039-1. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L., Mering C. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D., Huang Z., He X., Wu H., Peng D., Zhang L., Zhang X. Altered miRNA profile in testis of post-cryptorchidopexy patients with non-obstructive azoospermia. Reprod. Biol. Endocrinol. 2018;16:78. doi: 10.1186/s12958-018-0393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston R.J., Korn N. Semen quality in the domestic Turkey: the yellow semen syndrome. Avian Poult. Biol. Rev. 1997;8:109–121. [Google Scholar]
- Thurston R.J., Korn N. Spermiogenesis in commercial poultry species: anatomy and control. Poult. Sci. 2000;79:1650–1668. doi: 10.1093/ps/79.11.1650. [DOI] [PubMed] [Google Scholar]
- Thurston R.J., Hess R.A. Ultrastructure of spermatozoa from domesticated birds: comparative study of Turkey, chicken and Guinea fowl. Scanning Microsc. 1987;1:1829–1838. [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Praet O., Argraves W.S., Morales C.R. Co-expression and interaction of cubilin and megalin in the adult male rat reproductive system. Mol. Reprod. Dev. 2003;64:129–135. doi: 10.1002/mrd.10245. [DOI] [PubMed] [Google Scholar]
- Vermillion M.S., Ursin R.L., Kuok D.I.T., Vom Steeg L.G., Wohlgemuth N., Hall O.J., Fink A.L., Sasse E., Nelson A., Ndeh R., McGrath-Morrow S., Mitzner W., Chan M.C.W., Pekosz A., Klein S.L. Production of amphiregulin and recovery from influenza is greater in males than females. Biol. Sex Differ. 2018;9:24. doi: 10.1186/s13293-018-0184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T., Penninger J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene. 2004;23:2838–2849. doi: 10.1038/sj.onc.1207556. [DOI] [PubMed] [Google Scholar]
- Wan C., Xiang J., Li Y., Guo D. Differential gene expression patterns in chicken cardiomyocytes during hydrogen peroxide-induced apoptosis. PLoS ONE. 2016;11:e0147950. doi: 10.1371/journal.pone.0147950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J., Feng C., Zhao Y., Hu X., Li N. Transcriptome analysis of comb and testis from Rose-comb Silky chicken (R1/R1) and Beijing Fatty wild type chicken (r/r) Poult. Sci. 2017;96:1866–1873. doi: 10.3382/ps/pew447. [DOI] [PubMed] [Google Scholar]
- Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Machler M., Magnusson A., Moeller S. gplots: various R programming tools for plotting data. R. Package Version. 2009;2:1. [Google Scholar]
- Whitfield M., Pollet-Villard X., Levy R., Drevet J.R., Saez F. Posttesticular sperm maturation, infertility, and hypercholesterolemia. Asian J. Androl. 2015;17:742–748. doi: 10.4103/1008-682X.155536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte J., Glover J.D., Woodcock M., Brzeszczynska J., Taylor L., Sherman A., Kaiser P., McGrew M.J. FGF, insulin, and SMAD signaling cooperate for avian primordial germ cell self-renewal. Stem Cell Rep. 2015;5:1171–1182. doi: 10.1016/j.stemcr.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. Springer-Verlag; New York, NY: 2016. Ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- Wu S.R., Guo W., Li Y.L., Ren X.C., Lei X.Y., Li X.Y., Yao J.H., Yang X.J. miRNA and piRNA expression profiles of breeder cock testes detected by next-generation sequencing. Reprod. Domest. Anim. 2017;52:203–213. doi: 10.1111/rda.12880. [DOI] [PubMed] [Google Scholar]
- Wu X., Daniels G., Shapiro E., Xu K., Huang H., Li Y., Logan S., Greco M.A., Peng Y., Monaco M.E., Melamed J., Lepor H., Grishina I., Lee P. LEF1 identifies androgen-independent epithelium in the developing prostate. Mol. Endocrinol. 2011;25:1018–1026. doi: 10.1210/me.2010-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Zhou Y., Lin H., Hu H., Wang Y., Xu G. Toll-like receptor 2 in promoting angiogenesis after acute ischemic injury. Int. J. Mol. Med. 2013;31:555–560. doi: 10.3892/ijmm.2013.1240. [DOI] [PubMed] [Google Scholar]
- Zaniboni L., Cerolini S. Liquid storage of Turkey semen: changes in quality parameters, lipid composition and susceptibility to induced in vitro peroxidation in control, n-3 fatty acids and alpha-tocopherol rich spermatozoa. Anim. Reprod. Sci. 2009;112:51–65. doi: 10.1016/j.anireprosci.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Zhao S., Fernald R.D. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 2005;2005:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Shabek N. Ubiquitin ligases: structure, function, and regulation. Annu. Rev. Biochem. 2017;86:129–157. doi: 10.1146/annurev-biochem-060815-014922. [DOI] [PubMed] [Google Scholar]
- Zheng Y.H., Rengaraj D., Choi J.W., Park K.J., Lee S.I., Han J.Y. Expression pattern of meiosis associated SYCP family members during germline development in chickens. Reproduction. 2009;138:483–492. doi: 10.1530/REP-09-0163. [DOI] [PubMed] [Google Scholar]
- Zhu Z., Li C., Yang S., Tian R., Wang J., Yuan Q., Dong H., He Z., Wang S., Li Z. Dynamics of the transcriptome during human spermatogenesis: predicting the potential key genes regulating male gametes generation. Sci. Rep. 2016;6:19069. doi: 10.1038/srep19069. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.