Abstract
Aflatoxin B1 (AFB1) and ochratoxin A (OTA), which are toxic metabolites of ubiquitously occurring molds, show diverse toxicological effects such as hepatotoxicity, genotoxicity, and immunotoxicity in human and animals. Despite poultry show sensitivity to AFB1 and OTA, the mechanism of these mycotoxins in chickens has not been fully investigated. Here, we aimed to elucidate the molecular mechanism induced by AFB1 and/or OTA in chicken hepatic cells using transcriptomic analysis. Aflatoxin B1 and OTA induced cytotoxic effects in a dose-dependent manner at 48 h after exposure. Furthermore, correlation effect indicated an antagonism between the 2 toxins. The mRNA sequencing of AFB1-treated or OTA-treated chicken hepatocarcinoma and functional analysis revealed the pathways that were commonly regulated by both mycotoxins, especially PPAR signaling, focal adhesion, and MAPK signaling. Based on these findings, a possible hypothesis is that AFB1 and OTA have similar toxic mechanisms and compete for some steps in the chicken liver, and it is expected that the mycotoxins would have antagonistic effects. In addition, genes identified through transcriptome analysis provide candidates for further study of AFB1 and OTA toxicity and targets for efforts to improve the health of chickens exposed to mycotoxins.
Key words: aflatoxin, ochratoxin, hepatoxicity, chicken liver, transcriptomics
Introduction
Aflatoxins (AF) are a group of extremely toxic metabolites of certain molds, such as Aspergillus flavus and Aspergillus parasiticus, and can occur as natural contamination of foodstuffs and feed (Oznurlu et al., 2012). There are 4 types of AF named according to their fluorescence (blue or green) under UV light: aflatoxin B1 (AFB1), B2, G1, and G2. Aflatoxin B1 is the most toxic and is prevalent worldwide (Rawal et al., 2010). Since the discovery of AF, their negative effects on animal health have been researched in various areas of toxicology. Aflatoxin B1 is recognized as being hepatotoxic, carcinogenic, and mutagenic in human and animal (Hamid et al., 2013).
Ochratoxins (A, B, and C) are a group of toxic metabolites produced by a variety of molds including Aspergillus ochraceous and Penicillium verrucosum. Among these, ochratoxin A (OTA) is the most toxic and prevalent member and is ubiquitously found in many foodstuffs and feeds. Ochratoxin A has been classified as a possible carcinogen (group 2B) based on the large number of evidences of carcinogenicity from several animal researches by the International Agency for Research on Cancer (IARC, 1993). Numerous studies have shown that OTA exerts diverse toxicological effects, including hepatotoxicity (Qi et al., 2015), nephrotoxicity (Zeferino et al., 2016), genotoxicity (Zheng et al., 2013), and immunotoxicity (Verma et al., 2004).
A wide variation exists in species susceptibility to aflatoxicosis and ochratoxicosis. Poultry were known to be extremely sensitive to AFB1 and OTA. The presence of the mycotoxins in poultry diets induced reduction of body weight, egg production, and hatchability and increased susceptibility to disease mortality in chickens (Yarru et al., 2009; Sun et al., 2015; Zhang et al., 2016). According to a report by the Council for Agricultural Science and Technology, losses due to mycotoxicosis to the United States poultry industry exceeded $143 million annually (CAST, 1989).
The liver is the main target organ for AFB1 toxicity. Ochratoxin A accumulation, biotransformation, and detoxification also occurs in the liver, although the first target organ of ochratoxicosis is the kidney (Qi et al., 2015). A chicken hepatocellular carcinoma cell line, LMH, has been widely used to study the toxic mechanisms of mycotoxins (Liu et al., 2019; Markowiak et al., 2019). The cells retain a number of differentiated phenotypic traits of chicken hepatocytes and are of potential usefulness for study about toxicity (Kawaguchi et al., 1987).
Toxicogenomics provides the ability to determine the underlying molecular events that precede and accompany toxicity in a greater detail (Nuwaysir et al., 1999). Transcriptomics using microarray or mRNA sequencing technology has provided the opportunity to examine changes in the expression of a large number of genes simultaneously, thereby obtaining information on cellular mechanisms induced by toxic compounds (Merrick et al., 2013).
To the best of our knowledge, there are reports about transcriptomic analysis regarding aflatoxicosis in chicken liver, but hepatic gene expression study about ochratoxicosis has not been demonstrated (Yarru et al., 2009). Therefore, in the current study, we focused on the AFB1-induced or OTA-induced toxicity and their combination effects in chicken to gain further insights into the molecular events underlying the mycotoxin hepatic toxicity.
Materials and methods
Chemicals
Aflatoxin B1 (CAS:1162-65-8, molecular weight 312.27, >98% pure as reported by the supplier) and OTA (CAS:303-47-9, molecular weight 403.81, >98% pure as reported by the supplier) were obtained from Sigma-Aldrich. These mycotoxins were dissolved in dimethyl sulfoxide (DMSO).
Cell Culture
The chicken hepatic carcinoma cell line LMH (CRL-2117) was purchased from American Type Culture Collection (ATCC, Manassas, VA). Before the various mycotoxin treatments, cells were grown in Waymouth's MB 752/1 cell culture medium supplemented with 0.1% penicillin/streptomycin, and 10% fetal bovine serum in 0.1% gelatin-coated (Gelatin, from porcine skin, G1890, Sigma, MO) cell culture dish. All cell culture plates were maintained in a humidified incubator containing 5% CO2 at 37°C. Final concentrations of test mycotoxins were achieved by adding culture media, giving a final DMSO concentration of 0.1% (v/v).
MTT Assay
Cells were exposed to a serial concentration of AFB1 (0–3.0 μmol/L), OTA (0–20 μmol/L), or 0 to 3.0 μmol/L of each mycotoxin combination for 48 h. Then, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) solution (5 mg/mL) was then added to each well. After 4 h of incubation, the medium was removed, and 100 μL DMSO was added to dissolve the purple formazan. The optical density of each well was quantified by measuring absorbance at 540 nm. The viabilities of treated groups were expressed as a percentage of that of the control group, which was assumed to be 100%. The IC50 value of each mycotoxin was calculated using the GraphPad Prism, version 7, for Windows (La Jolla, CA).
Calculation of Combination Index Values
The interaction between AFB1 and OTA was analyzed with the help of the Chou-Talalay method (Chou and Talalay, 1984). The LMH cells were cultured in 96-well plates and co-incubated for 48 h with various concentrations of binary combinations of AFB1 and OTA in a ratio of 1 to 12. The correlation between the mycotoxin combinations was carried out using CompuSyn, version 1.0. The combination index (CI) values near 1 (0.90 < CI < 1.10) indicate an additive effect in the mycotoxins binary combination, whereas CI < 0.9 or CI > 1.1 indicates synergism or antagonism, respectively (Wang et al., 2014).
mRNA Isolation and High-Throughput mRNA Sequencing (RNA Sequencing)
The cells were cultured with medium containing 1.0 μmol/L of AFB1, 15 μmol/L of OTA, or 0.1% DMSO (positive control) for 48 h (n = 3, each treatments). Total RNA was extracted by Trizol reagent (Ambion, Austin, TX) according to the manufacturer's instructions. RNA quality of each sample was analyzed before library preparation using the Agilent 2100 bioanalyzer RNA kit (Agilent Technologies, Santa Clara, CA). The library was constructed following the manufacturer's protocol with reagents supplied by TruSeq Stranded mRNA kit (Illumina, San Diego, CA). The quality of libraries was evaluated using the Agilent 2100 bioanalyzer DNA kit (Agilent Technology) and CFX96 Real Time System (Bio-Rad, Hercules, CA). The libraries were sequenced on the BGI-MGISEQ2000 sequencer (Illumina) paired-end sequencing with read length of 99 bp.
The FastQC tool (ver.0.11.5) was used to examine raw read quality, and Skewer (ver.0.2.2) was used to reduce the adapter sequence. Next, we mapped the sequencing reads to chicken genome (galGal4 from UCSC genome), and gene annotation was done using STAR (ver.2.6).
Gene expression level was estimated using Fragment Per Kilo bases per Million reads method. Differentially expressed genes (DEG) were detected based on Cuffquant, Cuffnorm, and Cuffdiff of Cufflinks package, version 2.2.1, with |log2(fold-change)| ≥ 2 and P < 0.05. To gain insights into the DEG functions, the Database for Annotation, Visualization and Integrated Discovery, version 6.8, was used to attain Gene Ontology (GO) results and Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology-Based Annotation System, version 3.0, was carried out to establish the KEGG pathways associated with the up/downregulated genes.
Real-Time Quantitative PCR
mRNAs were used for RNA sequencing (n = 3), and additional samples (n = 5) were used for RT-qPCR to confirm the gene expression using iTaq Universal SYBR Green One-Step Kit (Bio-Rad) and ABI 7500 Real-Time PCR System (Applied Biosystems, Foster city, CA). The primer sequence and annealing temperature condition are shown in Table 1. The thermal cycling program was as follows: 95°C for 5 min, followed by 40 cycles of 95°C for 30 s, 60°C or 62°C for 30 s, and 72°C for 30 s. Following the amplification, melting curve analysis was conducted by heating from 65°C to 95°C with increments of 0.5°C for 5 s to determine the specificity of the PCR reactions. The beta-actin gene was used as internal control for normalization of gene expression and relative gene expression was evaluated using the 2−ΔΔCt method (Zhang et al., 2010).
Table 1.
Details of primers used for RT-qPCR.
| Linked pathway | Gene symbol (Accession no.) | Primer sequences (5' → 3′) | PCR product (bp) | Ta (°C) | References | |
|---|---|---|---|---|---|---|
| Lipid metabolism | FABP2 (NM_001007923) | F R |
TGGCATTTAACGGTACTTGGA TCAGATTATCGTGGGCTCCT |
111 | 60 | (Larkina et al., 2011) |
| CYP7A1 (NM_001001753.1) | F R |
CATTCTGTTGCCAGGTGATGTT GCTCTCTCTGTTTCCCGCTTT |
106 | 62 | (Wu et al., 2012) | |
| CYP27A1 (XM_422056.6) | F R |
AGGACTTTCGTCTGGCTCT CTCCGCATCGGGTATTT |
184 | 60 | (Hu et al., 2017) | |
| Drug metabolism | CYP1A4 (NM_205147) | F R |
TAAGGACGTCAATGCTCGTTTC CGTCCCGAATGTGCTCCTTAT |
88 | 62 | (Hickey et al., 2009) |
| GSTT1 (NM_205365.1) | F R |
CGGAGACTTCACCCTAGCAG GAGCATTAGCCCGGATGTTA |
155 | 60 | This study | |
| Reference gene | beta-actin (L08165.1) | F R |
ATGAAGCCCAGAGCAAAAGA GGGGTGTTGAAGGTCTCAAA |
223 | 60 | This study |
Abbreviations: F, forward; R, reverse; Ta, annealing temperature.
Statistical Analysis
Data are presented by descriptive analysis as mean ± SD of samples. Statistical analysis for RT-qPCR was performed by using GraphPad prism (ver. 7). The comparisons were performed using one-way ANOVA followed by Dunnett's test, and P-value below 0.05 was accepted as the level of significance.
Results and discussion
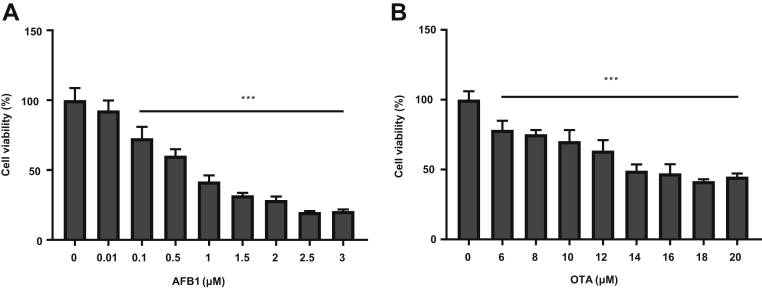
This is the first study, to the best of our knowledge, that analyzed the transcriptome response to AFB1 and OTA in chicken hepatocarcinoma cell line (LMH cells). The cytotoxic effects of AFB1 at doses of 0.01 to 3.0 μmol/L on LMH cell line were measured by the MTT assay. After 48 h of exposure, AFB1 induced a marked decrease in viable cells in a dose-dependent response (Figure 1A). Similarly, exposure to OTA at doses of 6 to 20 μmol/L for 48 h resulted in a concentration-dependent viability decrease (Figure 1B). In cultured LMH cells, the AFB1 concentration of 1.04 μmol/L showed 50% inhibition of cell viability (IC50); however, OTA required concentration of 12.58 μmol/L to obtain similar cytotoxicity. We exposed LMH cells to 1.0 μmol/L of AFB1 and 15 μmol/L of OTA for subsequent experiments.
Figure 1.
Effects of mycotoxins on chicken hepatocyte viability. Cell viability was determined using the MTT assay after 48 h after exposed to different doses of mycotoxins (A; AFB1, B; OTA). Data are expressed as the mean ± SD; ∗∗∗P < 0.001 compared with mock control (0.1% DMSO). Abbreviations: AFB1, Aflatoxin B1; DMSO, dimethyl sulfoxide; OTA, ochratoxin A.
In previous studies, cell viability was decreased by AFB1 or OTA treatment in liver and kidney cells in a time-dependent and dose-dependent manner (Renzulli et al., 2004; Costa et al., 2007; Stec et al., 2007; Ramyaa and Padma, 2013; Wang et al., 2014). The differences in mycotoxin-induced cytotoxicity might be correlated with experimental model (human, rat, pig, or chicken) and target organ (liver, kidney, or lymphocytes). Based on toxin concentrations, AFB1 was approximately 1.4, 3, and 9 times more potent as an inhibitor at different time points, respectively, than OTA in liver cells (Renzulli et al., 2004). The liver being the target organ of toxicity for AFB1 could be the reason for the observed different sensitivities to AFB1 or OTA-induced cytotoxicity. Consistent with these findings, in the current study, the dose–response curves of AFB1 and OTA in LMH cell line showed a dose-dependent toxicity, and the potency of AFB1 in inhibiting the cell viability was found to be over 10 times higher than that of OTA (Figure 1).
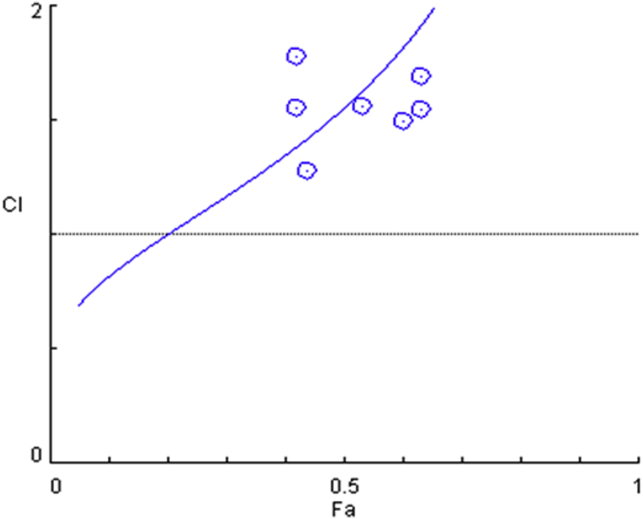
To calculate the combined effects of AFB1 and OTA, the CI values were analyzed on the basis of equipotent IC50 values obtained from individual exposure experiments, so that the contribution to the effect of AFB1 and OTA would be equal (Chou, 2006; Sobral et al., 2018). The interactive effect of AFB1 and OTA can be described as an antagonistic effect with high levels of cytotoxicity (Figure 2 and Table 2). These findings suggest that the co-occurrence of AFB1 and OTA in food commodities and diet may decrease their cytotoxic potential than the presence of either mycotoxin alone. In contrast, previous reports related to AFB1 and OTA mixtures have indicated an additive effect of this combination for cytotoxicity (Golli-Bennour et al., 2010; Corcuera et al., 2011), suggesting that AFB1 and OTA do not share toxic pathways in their mode of action, or they share only some steps. However, Corcuera et al. (2011) reported that the combination treatments showed a significant decrease in DNA damage compared with that after AFB1 single treatment, suggesting that AFB1 and OTA compete for the same CYP enzymes that represent a bioactivation route for AFB1 (Corcuera et al., 2011). These results indicate that AFB1 and OTA may share some common mechanisms of action, such as the induction of DNA adducts in chicken liver.
Figure 2.
Mycotoxin combination plots based on Chou and Talalay combination index theorem after 48 h of exposure of LMH cells. Combination index plot for combination of AFB1 and OTA. CI < 1, CI = 1, and CI > 1 indicate synergistic, additive, and antagonistic effect, respectively. Abbreviations: AFB1, Aflatoxin B1; CI, combination index; Fa, fraction affected; OTA, ochratoxin A.
Table 2.
CI values and combined effect of AFB1 and OTA.
| Concentration | CI | Description |
|---|---|---|
| IC10 | 0.81846 | Moderate synergism |
| IC20 | 1.00137 | Nearly additive |
| IC30 | 1.16940 | Slight antagonism |
| IC40 | 1.34784 | Moderate antagonism |
| IC50 | 1.55553 | Common antagonism |
| IC60 | 1.81909 | Common antagonism |
| IC70 | 2.19117 | Common antagonism |
| IC80 | 2.81075 | Common antagonism |
| IC90 | 4.26901 | Strong antagonism |
IC10 to IC90 are the doses required to inhibit proliferation 10 to 90%, respectively. LMH cells were exposed for 48 h to AFB1 + OTA at a ratio of 1:12.
Abbreviations: AFB1, aflatoxin B1; CI, combination index; OTA, ochratoxin A.
Transcriptomic analysis was performed to demonstrate the mechanisms of 2 mycotoxins and compare the regulation of gene expression during AFB1-induced and OTA-induced cytotoxicity in chicken hepatocarcinoma cells. An Illumina MGI sequencing platform was utilized. The average of 93,100,867 reads of a paired-end 99 bp, with Q30 values 90 to 92% were collected for libraries of control and each mycotoxin treatment groups (Supplementary Table 1). After filtering of adapters and trimming of ambiguous and low-quality reads, an average of 93,100,800 high-quality and clean reads were obtained, accounting for 99.9% of total raw reads, respectively. Approximately 80% of the quality-trimmed reads mapped to the annotated chicken gene set (Supplementary Table 2).
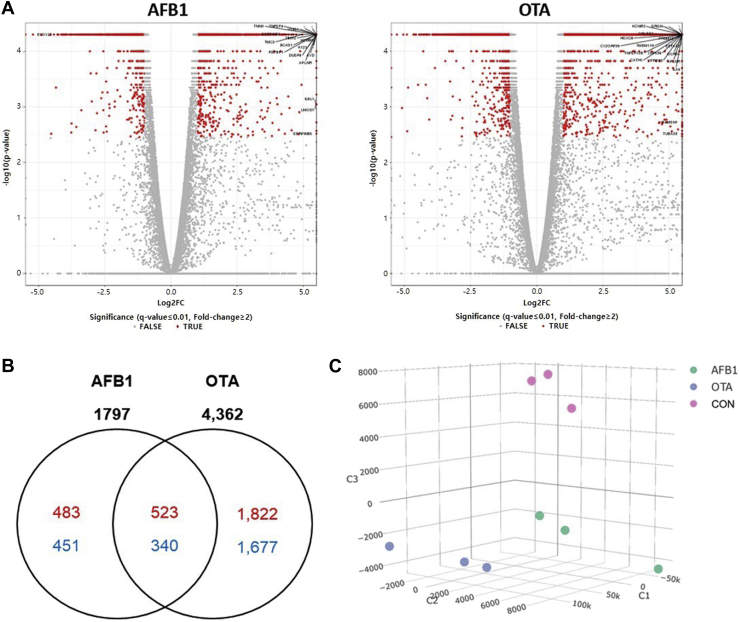
Differential expression analysis was performed to determine the gene expression changes in response to the mycotoxin treatments. The DEG caused by individual treatments of the mycotoxins are shown as Volcano plots (Figure 3A). Compared with the control treatment, a total of 1,797 and 4,362 genes were differentially expressed after AFB1 and OTA treatments, respectively (|log2(FC)| ≥ 1, FDR < 0.01). In AFB1-treated groups, 1,006 upregulated and 791 downregulated genes were found (|log2(FC)| ≥ 1, FDR < 0.01), whereas OTA treatment resulted in 2,345 upregulated and 2,017 downregulated transcripts (Figure 3B). Interestingly, the number of significant DEG in response to AFB1 and OTA treatment in the present study were much higher than those previously reported in liver cell lines from human (53 DEG to OTA; Arbillaga et al. 2008, 1,125 DEG to AFB1; Jennen et al. 2010, and 2,131 DEG to AFB1; Josse et al. 2012) or rats (785 DEG to AFB1; Yang et al. 2014). The differences between these reports could be attributed to the development of sequencing technology and analysis methods and different experimental conditions, which include the dosage of mycotoxins, exposure time, and animal species (Zeferino et al., 2016). Zeferino et al. (2016) had analyzed RNA sequencing data in chicken using mRNA reference for Homo sapiens because the functional annotation of chicken genome was not as complete as those for humans.
Figure 3.
Gene expression profiling of LMH cells after AFB1 or OTA treatments. (A) Visualization of differentially expressed genes (DEG) in LMH cells exposed to AFB1 or OTA. (B) Venn diagrams for DEG in each mycotoxin treatments compared to control. Totals for the AFB1 and OTA groups of each experiments are shown above each ellipse. (red, upregulated; down, downregulated). (C) Principal component analysis (PCA) plot of the DEG. The 9 samples shown that the mycotoxins treatments and untreated cells were well-clustered and distinguishable from each other. Control group vs. 1 μmol/L AFB1-treatment group, control groups vs. 15 μmol/L OTA (n = 3, respectively). Abbreviations: AFB1, aflatoxin B1; OTA, ochratoxin A.
The DEG was used in principal component analysis to visualize AFB1-induced and OTA-induced toxic effects on gene expression before and after exposure to the mycotoxins (Figure 3C). The principal component analysis revealed that the mycotoxins treatments (0.1 μmol/L of AFB1 or 15 μmol/L of OTA), or untreated cells were well-clustered and distinguishable from each other.
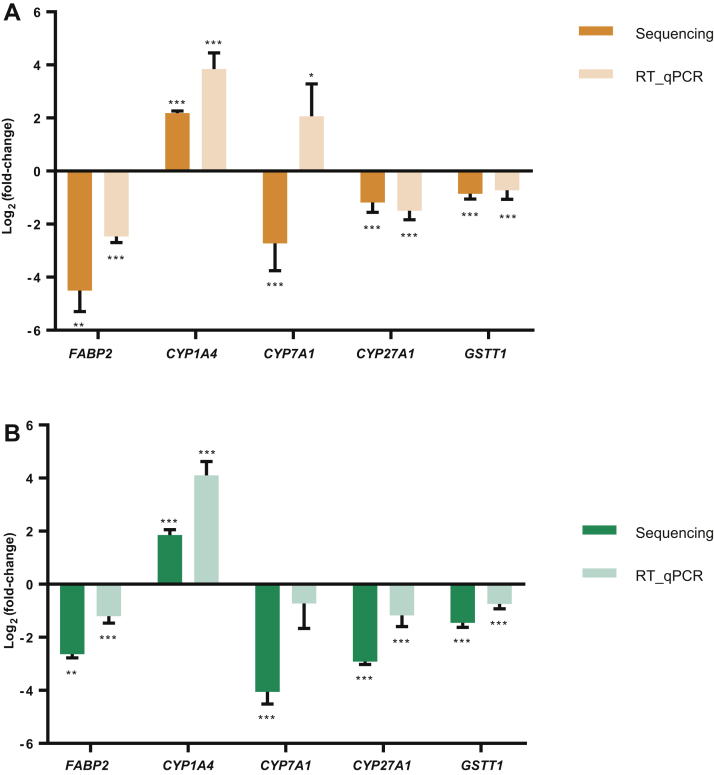
The mRNA expression levels of 5 DEG related to bile acid or xenobiotic metabolism was measured by RT-qPCR for validation of RNA seq data done using RT-qPCR. The CYP1A4 and GSTT1 genes were involved with drug metabolism in chickens (Rawal et al., 2010; Hafner et al., 2011), and downregulation of FABP2, CYP7A1, and CYP27A1 genes modulate the synthesis of bile acids (Chiang, 2009). The expression of CYP7A1 gene was shown different pattern with mRNA sequencing in AFB1 treatment and not significant in OTA treatment. However, the results showed that mRNA expressions of FABP2, CYP27A1, CYP1A4, and GSTT1 were significantly decreased by each mycotoxin treatment (Figure 4), in accordance with the RNA-seq results. These results demonstrate the validity, accuracy, and statistical power of sequencing data despite the relatively small cohort being considered.
Figure 4.
Validation of the gene expression profile by real-time PCR. The x-axis represents the gene name, the y-axis represents the log2 ratio (mycotoxin treatments/control), and the different colors represent data from mRNA sequencing or RT-qPCR as indicated in the key. (A, 1 μmol/L AFB1; B, 15 μmol/L OTA) Asterisk means significant difference with control group (∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001). Abbreviations: AFB1, aflatoxin B1; OTA, ochratoxin A.
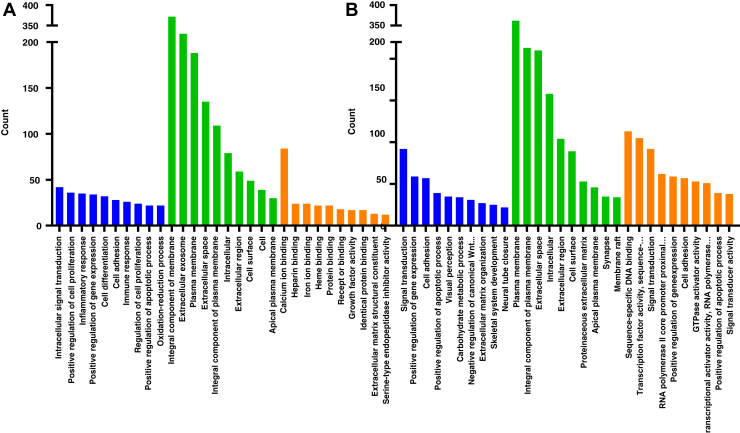
Next, we investigated the potential biological functions of the DEG. A total of 2,185 and 4,904 DEG (|log2(FC)| > 1, P < 0.05) were used to analyze the functional annotation using Database for Annotation, Visualization and Integrated Discovery and KEGG Orthology-Based Annotation System. The GO term and pathways with at least 2 genes modulated, and a P < 0.05 were considered statistically significant. The top 10 groups in the 3 domains are shown in Figure 5.
Figure 5.
Gene ontology (GO) term enrichment analysis. Gene ontology categories included molecular function, cellular component, and biological process. GO categories for each function were sorted by decreasing order of gene counts, based on the GO enrichment. (A; AFB1, B; OTA, Blue; biological process, green; cellular component, orange; molecular function). Abbreviations: AFB1, aflatoxin B1; OTA, ochratoxin A.
Within the “biological process” category, the most abundant terms commonly included “positive regulation of gene expression”, “cell adhesion”, “cell mutation” and “positive regulation of apoptotic process” in AFB1 and OTA treatments. Interestingly, we observed a high percentage of DEG assigned to “cell component” domain including “integral component of plasma membrane”, “plasma membrane”, “extracellular space”, and “extracellular region”. The pathways identified in this investigation were shown similar aspect with previously reported study about OTA toxicity in rat liver (Marin-Kuan et al., 2006). Those results suggest that the toxic mechanisms regarding AFB1 or OTA were resembled with metabolism of mammalian liver.
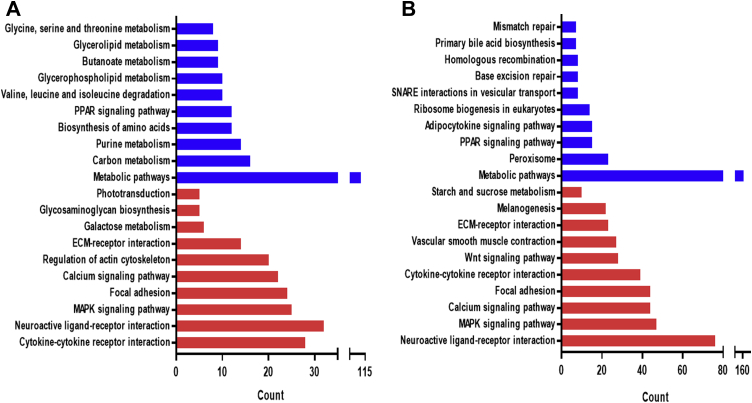
The DEG was mapped to reference pathways in KEGG and assigned to 38 (10 up, 28 down) and 21 (11 up, 10 down) pathways for AFB1 and OTA treatment, respectively, including many metabolic and signaling pathways that are not directly related to xenobiotic metabolism. The top 10 terms in the 3 main categories are shown in Figure 6. A similar tendency of the functional annotation results was observed between AFB1 and OTA treatments. By comparing the mRNA expression between AFB1 and OTA treatments, 3 and 5 commonly enriched pathways were identified to contain upregulated and downregulated DEG, respectively (Figure 6).
Figure 6.
KEGG pathway enrichment analysis of upregulated /downregulated differentially expressed genes in LMH cells upon exposure to mycotoxins. The transcripts in the top 10 pathways are summarized. The x-axis indicates functional pathways, and the y-axis indicates the number of genes in the same pathway. Red bars show the pathways involved with the downregulated genes and blue bars are pathways associated with the upregulated genes (A, 1 μmol/L AFB1; B, 15 μmol/L OTA). Abbreviations: AFB1, aflatoxin B1; KEGG, Kyoto Encyclopedia of Genes and Genomes; OTA, ochratoxin A.
Furthermore, the 8 pathways, among which “Metabolic pathways”, “PPAR signaling pathway”, “Primary bile acid biosynthesis”, “ECM-receptor interaction”, “Calcium signaling pathway”, “MAPK signaling pathway”, “Focal adhesion”, and “Neuroactive ligand-receptor interaction” were commonly enriched in both mycotoxin treatments. Some of the signaling pathways are well known for their roles in lipid and xenobiotic metabolisms. These results indicate that same pathways are triggered after AFB1-induced and OTA-induced cytotoxicity in chicken hepatocarcinoma cells.
PPAR are nuclear receptors that are known to play a role in lipid metabolism and homeostasis (Pineda Torra et al., 2003). PPAR signaling is also involved in anti-inflammation (Wang et al., 2017) and xenobiotic metabolism (Pawlak et al., 2015). An aberration of PPAR leads to deregulation of genes in metabolic pathway, thereby contributing to etiology of metabolic syndrome (Qi et al., 2015). The avian PPAR show high homology with mouse, rat, and human PPAR (Ringseis et al., 2012), and similar expression pattern of PPARα is observed in tissues of chickens and mammals (Shen et al., 2008). PPARα gene expression was linearly decreased by AFB1 concentration (0.11–0.21 mg/kg) in duckling liver (Chen et al., 2014). PPAR is also reported to influence bile acid formation and bile composition. Farnesoid X receptor is a bile acid-activated nuclear receptor that mediates the effects of bile acids on gene expression and plays a major role in bile acid and possibly also in lipid metabolism. In this study, the genes regulating bile acid synthesis, CYP7A1 and CYP27A1, were downregulated by AFB1 and OTA treatments in LMH. The lipophilic xenobiotics were recognized as a “toxic bile acid”, leading to the suppression of bile acid biosynthesis, promoted by modulation of CYP7A1 and CYP27A1 gene expression (Handschin et al., 2002; Chiang, 2004).
Focal adhesion is the structural link between the extracellular matrix (ECM) and actin cytoskeleton and plays essential roles in signal transduction pathways causing various biological processes, including cell survival, motility, proliferation, differentiation, and regulation of gene expression. Previous study suggested that the cell adhesion proteins function to sense and respond to extracellular environment through the mediation of bi-directional (inside-out and outside-in) transmembrane signaling (Wu, 2007). Therefore, changes in the expression of genes involved in cell structure may negatively affect cell function. Fang et al. (2013) performed transcriptomic analysis during different transitions of bladder cancer and reported that ECM-receptor interaction and focal adhesion were important pathways involved in cancer progression. The focal adhesion and ECM-receptor interaction pathways were prominently annotated with commonly expressed genes responding to various concentrations of AFB1 treatment (0.03, 0.1, 0.2 μmol/L) in rat hepatic epithelial cells (WB-F344) (Yang et al., 2014). Yarru et al. (2009) hypothesized that changes in gene expression associated with cell skeletal structure observed in the liver of AFB1-supplemented chickens. In this study, the genes involved in focal adhesion pathway were upregulated in both AFB1-treated and OTA-treated LMH cells compared with that in the control. These alterations were likely linked to the response of cell adhesion proteins to xenobiotics, including transmembrane signaling and regulation of gene expression.
The MAPK signaling pathway is known to play important roles in cell survival, antiapoptotic activity, and cancer development (Zhao et al., 2017). Dai et al. (2014) performed miRNA profiling and suggested that MAPK signaling pathway is critical in nephrotoxicity by OTA treatment in rat. In addition, MAPK signaling pathway was also enriched in rat liver (Qi et al., 2015) and kidney (Ali et al., 2011) after OTA administration. The liver and kidney consist of a variety of cell types. Some genes may be specifically expressed or deregulated in hepatic or renal cells. However, from the perspective of mRNA expression, the trend in changes in the gene expression related to MAPK signaling pathway induced by AFB1 and OTA were consistent with the renal results obtained in previous studies. MAPK activity also associated with the disruption of tight junction of cell, and this process could interact with pathways related to lipid metabolism to influence the deposition of fat (Cui et al., 2012).
In conclusion, although AFB1 and OTA showed high cytotoxicity in LMH cells, the mycotoxins were weakly cytotoxic when in combination than alone, thereby showing an antagonistic effect. To demonstrate the toxic mechanisms of AFB1 and OTA and compare their mode of action, mRNA sequencing and bioinformatics analysis were performed in AFB1 and OTA-treated LMH cells. The GO term analysis indicated that the treatments regulate gene expression associated with inflammatory response, cell adhesion, and apoptotic process. In KEGG pathway analysis, “metabolic pathway”, “PPAR signaling pathway”, “MAPK signaling pathway”, and “focal adhesion” were commonly significantly affected by the AFB1 and OTA treatments. A possible hypothesis for these findings may be that AFB1 and OTA have similar toxic mechanisms and compete for some steps in chicken liver, and it is expected that they would have antagonistic effects. Further investigation using the in vivo or comparable study which can represent the molecular mechanism of mycotoxins in chicken liver is required for the substantiation of AFB1-induced or OTA-induced carcinogenesis and their interaction in the poultry.
Acknowledgments
This study was supported by National Research Foundation of Korea (Project number: 2017R1A2B2012125).
Conflict of Interest Statement: The authors did not provide any conflict of interest statement.
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.05.058.
Supplementary data
References
- Ali R., Mittelstaedt R.A., Shaddock J.G., Ding W., Bhalli J.A., Khan Q.M., Heflich R.H. Comparative analysis of micronuclei and DNA damage induced by Ochratoxin A in two mammalian cell lines. Mutat. Res. 2011;723:58–64. doi: 10.1016/j.mrgentox.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Arbillaga L., Vettorazzi A., Gil A.G., van Delft J.H., García-Jalón J.A., López de Cerain A. Gene expression changes induced by ochratoxin A in renal and hepatic tissues of male F344 rat after oral repeated administration. Toxicol. Appl. Pharmacol. 2008;230:197–207. doi: 10.1016/j.taap.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Chen X., Horn N., Cotter P.F., Applegate T.J. Growth, serum biochemistry, complement activity, and liver gene expression responses of Pekin ducklings to graded levels of cultured aflatoxin B1. Poult. Sci. 2014;93:2028–2036. doi: 10.3382/ps.2014-03904. [DOI] [PubMed] [Google Scholar]
- Chiang J.Y.L. Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004;40:539–551. doi: 10.1016/j.jhep.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Chiang J.Y. Hepatocyte nuclear factor 4α regulation of bile acid and drug metabolism. Expert Opin. Drug Metab. Toxicol. 2009;5:137–147. doi: 10.1517/17425250802707342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.-C. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- Chou T.C., Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Corcuera L.A., Arbillaga L., Vettorazzi A., Azqueta A., López de Cerain A. Ochratoxin A reduces aflatoxin B1 induced DNA damage detected by the comet assay in Hep G2 cells. Food Chem. Toxicol. 2011;49:2883–2889. doi: 10.1016/j.fct.2011.07.029. [DOI] [PubMed] [Google Scholar]
- Costa S., Utan A., Speroni E., Cervellati R., Piva G., Prandini A., Guerra M.C. Carnosic acid from rosemary extracts: a potential chemoprotective agent against aflatoxin B1. An in vitro study. J. Appl. Toxicol. 2007;27:152–159. doi: 10.1002/jat.1186. [DOI] [PubMed] [Google Scholar]
- Council for Agricultural Science and Technology. Mycotoxins: economic and health risks. In: Nisi K.A., editor. Task Force Report No. 116. Council for Agricultural Science and Technology; Ames, IA: 1989. pp. 1–91. [Google Scholar]
- Cui H.-X., Liu R.-R., Zhao G.-P., Zheng M.-Q., Chen J.-L., Wen J. Identification of differentially expressed genes and pathways for intramuscular fat deposition in pectoralis major tissues of fast-and slow-growing chickens. BMC Genomics. 2012;13:213. doi: 10.1186/1471-2164-13-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Q., Zhao J., Qi X., Xu W., He X., Guo M., Dweep H., Cheng W.-H., Luo Y., Xia K., Gretz N., Huang K. MicroRNA profiling of rats with ochratoxin A nephrotoxicity. BMC Genomics. 2014;15:333. doi: 10.1186/1471-2164-15-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Z.-Q., Zang W.-D., Chen R., Ye B.-W., Wang X.-W., Yi S.-H., Chen W., He F., Ye G. Gene expression profile and enrichment pathways in different stages of bladder cancer. Genet. Mol. Res. 2013;12:1479–1489. doi: 10.4238/2013.May.6.1. [DOI] [PubMed] [Google Scholar]
- Golli-Bennour E.E., Kouidhi B., Bouslimi A., Abid-Essefi S., Hassen W., Bacha H. Cytotoxicity and genotoxicity induced by aflatoxin B1, ochratoxin A, and their combination in cultured Vero cells. J. Biochem. Mol. Toxicol. 2010;24:42–50. doi: 10.1002/jbt.20310. [DOI] [PubMed] [Google Scholar]
- Hafner M., Rezen T., Rozman D. Regulation of hepatic cytochromes p450 by lipids and cholesterol. Curr. Drug Metab. 2011;12:173–185. doi: 10.2174/138920011795016890. [DOI] [PubMed] [Google Scholar]
- Hamid A.S., Tesfamariam I.G., Zhang Y., Zhang Z.G. Aflatoxin B1-induced hepatocellular carcinoma in developing countries: geographical distribution, mechanism of action and prevention. Oncol. Lett. 2013;5:1087–1092. doi: 10.3892/ol.2013.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschin C., Podvinec M., Amherd R., Looser R., Ourlin J.-C., Meyer U.A. Cholesterol and bile acids regulate Xenosensor signaling in drug-mediated induction of cytochromes P450. J. Biol. Chem. 2002;277:29561–29567. doi: 10.1074/jbc.M202739200. [DOI] [PubMed] [Google Scholar]
- Hickey N.J., Crump D., Jones S.P., Kennedy S.W. Effects of 18 perfluoroalkyl compounds on mRNA expression in chicken embryo hepatocyte cultures. Toxicol. Sci. 2009;111:311–320. doi: 10.1093/toxsci/kfp160. [DOI] [PubMed] [Google Scholar]
- Hu Y., Sun Q., Zong Y., Liu J., Idriss A.A., Omer N.A., Zhao R. Prenatal betaine exposure alleviates corticosterone-induced inhibition of CYP27A1 expression in the liver of juvenile chickens associated with its promoter DNA methylation. Gen. Comp. Endocrinol. 2017;246:241–248. doi: 10.1016/j.ygcen.2016.12.014. [DOI] [PubMed] [Google Scholar]
- IARC . International Agency for Research on Cancer; Lyon, France: 1993. Ochratosin A. Monographs on the evaluation of carcinogenic risks to humans. Pages 489–521 in Some Naturally Occurring Substances: Food Items and Constituents, Heterocyclic Aromatic Amines and Mycotoxins. [Google Scholar]
- Jennen D.G.J., Magkoufopoulou C., Ketelslegers H.B., van Herwijnen M.H.M., Kleinjans J.C.S., van Delft J.H.M. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol. Sci. 2010;115:66–79. doi: 10.1093/toxsci/kfq026. [DOI] [PubMed] [Google Scholar]
- Josse R., Dumont J., Fautrel A., Robin M.-A., Guillouzo A. Identification of early target genes of aflatoxin B1 in human hepatocytes, inter-individual variability and comparison with other genotoxic compounds. Toxicol. Appl. Pharmacol. 2012;258:176–187. doi: 10.1016/j.taap.2011.10.019. [DOI] [PubMed] [Google Scholar]
- Kawaguchi T., Nomura K., Hirayama Y., Kitagawa T. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 1987;47:4460–4464. [PubMed] [Google Scholar]
- Larkina T.A., Sazanova A.L., Fomichev K.A., Barkova O.Y., Sazanov A.A., Malewski T., Jaszczak K. Expression profiling of candidate genes for abdominal fat mass in domestic chicken Gallus gallus. Russ. J. Genet. 2011;47:1012–1015. [PubMed] [Google Scholar]
- Liu Q., Wen J., Zhu J., Zhang T., Deng Y., Jiang J. Aromatic hydrocarbon receptor regulates chicken cytochrome P450 1A5 transcription: a novel insight into T-2 toxin-induced gene expression and cytotoxicity in LMH cells. Biochem. Pharmacol. 2019;168:319–329. doi: 10.1016/j.bcp.2019.07.023. [DOI] [PubMed] [Google Scholar]
- Markowiak P., Śliżewska K., Nowak A., Chlebicz A., Żbikowski A., Pawłowski K., Szeleszczuk P. Probiotic microorganisms detoxify ochratoxin A in both a chicken liver cell line and chickens. J. Sci. Food Agric. 2019;99:4309–4318. doi: 10.1002/jsfa.9664. [DOI] [PubMed] [Google Scholar]
- Marin-Kuan M., Nestler S., Verguet C., Bezencon C., Piguet D., Mansourian R., Holzwarth J., Grigorov M., Delatour T., Mantle P., Cavin C., Schilter B. A toxicogenomics approach to identify new plausible epigenetic mechanisms of ochratoxin a carcinogenicity in rat. Toxicol. Sci. 2006;89:120–134. doi: 10.1093/toxsci/kfj017. [DOI] [PubMed] [Google Scholar]
- Merrick B.A., Phadke D.P., Auerbach S.S., Mav D., Stiegelmeyer S.M., Shah R.R., Tice R.R. RNA-seq profiling reveals novel hepatic gene expression pattern in aflatoxin B1 treated rats (W Yan, Ed.) PLoS One. 2013;8:e61768. doi: 10.1371/journal.pone.0061768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir E.F., Bittner M., Trent J., Barrett J.C., Afshari C.A. Microarrays and toxicology: the advent of toxicogenomics. Mol. Carcinog. 1999;24:153–159. doi: 10.1002/(sici)1098-2744(199903)24:3<153::aid-mc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Oznurlu Y., Celik I., Sur E., Ozaydın T., Oğuz H., Altunbaş K. Determination of the effects of aflatoxin B1 given in ovo on the proximal tibial growth plate of broiler chickens: histological, histometric and immunohistochemical findings. Avian Pathol. 2012;41:469–477. doi: 10.1080/03079457.2012.712673. [DOI] [PubMed] [Google Scholar]
- Pawlak M., Lefebvre P., Staels B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015;62:720–733. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- Pineda Torra I., Claudel T., Duval C., Kosykh V., Fruchart J.C., Staels B. Bile acids induce the expression of the human peroxisome proliferator-activated receptor α gene via activation of the farnesoid X receptor. Mol. Endocrinol. 2003;17:259–272. doi: 10.1210/me.2002-0120. [DOI] [PubMed] [Google Scholar]
- Qi X., Yang X., Chen S., He X., Dweep H., Guo M., Cheng W.-H., Xu W., Luo Y., Gretz N., Dai Q., Huang K. Ochratoxin A induced early hepatotoxicity: new mechanistic insights from microRNA, mRNA and proteomic profiling studies. Sci. Rep. 2015;4:5163. [Google Scholar]
- Ramyaa P., Padma V.V. Ochratoxin-induced toxicity, oxidative stress and apoptosis ameliorated by quercetin – modulation by Nrf2. Food Chem. Toxicol. 2013;62:205–216. doi: 10.1016/j.fct.2013.08.048. [DOI] [PubMed] [Google Scholar]
- Rawal S., Kim J.E., Coulombe R. Aflatoxin B1 in poultry: toxicology, metabolism and prevention. Res. Vet. Sci. 2010;89:325–331. doi: 10.1016/j.rvsc.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Renzulli C., Galvano F., Pierdomenico L., Speroni E., Guerra M.C. Effects of rosmarinic acid against aflatoxin B1 and ochratoxin-A-induced cell damage in a human hepatoma cell line (Hep G2) J. Appl. Toxicol. 2004;24:289–296. doi: 10.1002/jat.982. [DOI] [PubMed] [Google Scholar]
- Ringseis R., Wen G., Eder K. Regulation of genes involved in carnitine homeostasis by PPARa across different species (rat, mouse, pig, cattle, chicken, and human) PPAR Res. 2012;2012:1–11. doi: 10.1155/2012/868317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., Oesterling E., Stromberg A., Toborek M., MacDonald R., Hennig B. Zinc deficiency induces vascular pro-inflammatory parameters associated with NF-κB and PPAR signaling. J. Am. Coll. Nutr. 2008;27:577–587. doi: 10.1080/07315724.2008.10719741. [DOI] [PubMed] [Google Scholar]
- Sobral M.M.C., Faria M.A., Cunha S.C., Ferreira I.M.P.L.V.O. Toxicological interactions between mycotoxins from ubiquitous fungi: impact on hepatic and intestinal human epithelial cells. Chemosphere. 2018;202:538–548. doi: 10.1016/j.chemosphere.2018.03.122. [DOI] [PubMed] [Google Scholar]
- Stec J., Szczotka M., Ku J. Cytotoxicity of feed-borne mycotoxins to animal cell lines in vitro using the MTT assay. Bull. Vet. Inst. Pulawy. 2007;51:679–684. [Google Scholar]
- Sun L.H., Lei M.Y., Zhang N.Y., Gao X., Li C., Krumm C.S., Qi D.S. Individual and combined cytotoxic effects of aflatoxin B1, zearalenone, deoxynivalenol and fumonisin B1 on BRL 3A rat liver cells. Toxicon. 2015;95:6–12. doi: 10.1016/j.toxicon.2014.12.010. [DOI] [PubMed] [Google Scholar]
- Verma J., Johri T.S., Swain S., Ameena S. Effect of graded levels of aflatoxin, ochratoxin and their combinations on the performance and immune response of broilers. Br. Poult. Sci. 2004;45:512–518. doi: 10.1080/00071660412331286226. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang X., Sheng N., Zhou X., Cui R., Zhang H., Dai J. RNA-sequencing analysis reveals the hepatotoxic mechanism of perfluoroalkyl alternatives, HFPO2 and HFPO4, following exposure in mice: hepatotoxic mechanism of HFPO2 and HFPO4 in mice. J. Appl. Toxicol. 2017;37:436–444. doi: 10.1002/jat.3376. [DOI] [PubMed] [Google Scholar]
- Wang H.W., Wang J.Q., Zheng B.Q., Li S.L., Zhang Y.D., Li F.D., Zheng N. Cytotoxicity induced by ochratoxin A, zearalenone, and α-zearalenol: effects of individual and combined treatment. Food Chem. Toxicol. 2014;71:217–224. doi: 10.1016/j.fct.2014.05.032. [DOI] [PubMed] [Google Scholar]
- Wu C. A focal point in current cell biology and molecular medicine. Cell Adhes. 2007;1:7–12. doi: 10.4161/cam.1.1.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Fu W., Huang Y., Ni Y. Effects of Kisspeptin-10 on lipid metabolism in cultured chicken hepatocytes. Asian Austral. J. Anim. Sci. 2012;25:1229–1236. doi: 10.5713/ajas.2012.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ji J., Li G., Li J., Chen Z., Wang H. Transcriptomic analysis of aflatoxin B1-regulated genes in rat hepatic epithelial cells. Trans. Tianjin Univ. 2014;20:451–457. [Google Scholar]
- Yarru L.P., Settivari R.S., Antoniou E., Ledoux D.R., Rottinghaust G.E. Toxicological and gene expression analysis of the impact of aflatoxin B on hepatic function of male broiler chicks. Poult. Sci. 2009;88:360–371. doi: 10.3382/ps.2008-00258. [DOI] [PubMed] [Google Scholar]
- Zeferino C.P., Wells K.D., Moura A.S.A.M.T., Rottinghaus G.E., Ledoux D.R. Changes in renal gene expression associated with induced ochratoxicosis in chickens: activation and deactivation of transcripts after varying durations of exposure. Poult. Sci. 2016;96:pew419. doi: 10.3382/ps/pew419. [DOI] [PubMed] [Google Scholar]
- Zhang J.D., Ruschhaupt M., Biczok R. 2010. ddCt Method for qRT–PCR Data Analysis. Accessed Aug. 2020. http://bioconductor.jp/packages/2.14/bioc/vignettes/ddCt/inst/doc/rtPCR.pdf. [Google Scholar]
- Zhang N.Y., Qi M., Zhao L., Zhu M.K., Guo J., Liu J., Gu C.Q., Rajput S.A., Krumm C.S., Qi D.S., Sun L.H. Curcumin prevents aflatoxin B1 hepatoxicity by inhibition of cytochrome P450 isozymes in chick liver. Toxins. 2016;8:327. doi: 10.3390/toxins8110327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Qi X., Dai Q., He X., Dweep H., Guo M., Luo Y., Gretz N., Luo H., Huang K., Xu W. Toxicity study of ochratoxin A using HEK293 and HepG2 cell lines based on microRNA profiling. Hum. Exp. Toxicol. 2017;36:8–22. doi: 10.1177/0960327116632048. [DOI] [PubMed] [Google Scholar]
- Zheng J., Zhang Y., Xu W., Luo Y., Hao J., Shen X.L., Yang X., Li X., Huang K. Zinc protects HepG2 cells against the oxidative damage and DNA damage induced by ochratoxin A. Toxicol. Appl. Pharmacol. 2013;268:123–131. doi: 10.1016/j.taap.2013.01.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.