Abstract
Background
Early detection appears to be the most effective approach to improve the overall survival of patients with hepatocellular carcinoma (HCC). We evaluated the potential performance of plasma SEPT9 methylation (mSEPT9) as a noninvasive biomarker for the diagnosis of patients with HCC.
Methods
A total of 373 subjects were included, and the group consisted of 104 HCC patients, 95 with an at-risk disease, and 174 healthy controls (HC). The methylation of mSEPT9 was determined using methylation-specific fluorescence quantitative PCR. The diagnostic performance of plasma mSEPT9 for HCC was assessed in a single-blind manner.
Results
The receiver operating characteristic (ROC) curve showed that plasma mSEPT9 can be used to detect and discriminate HCC with an area under the ROC curve (AUROC) of 0.961, a sensitivity of 82.7%, and specificity of 96.0% from HC. These results showed that plasma mSEPT9 had better diagnostic performance than serum alpha fetoprotein (AFP) (AUROC 0.881, sensitivity 57.7%, and specificity 98.3%). Similar results were noted in the detection of early-stage HCC. When combined with serum AFP, the sensitivity increased to 91.3% and 87.7% for the detection of HCC and early-stage HCC,respectively. Notably, the levels of plasma mSEPT9 dramatically decreased after surgery (P = 0.001).
Conclusions
Plasma SEPT9 methylation might serve as a useful and noninvasive biomarker for the diagnosis of HCC and can be used to evaluate the therapeutic efficacy of HCC treatment.
1. Introduction
Liver cancer is currently the sixth most common malignant disease and the fourth leading cause of cancer death worldwide with approximately 841,000 new cases and 782,000 deaths annually [1]. In China, the incidence and mortality of liver cancer ranks fourth and second, respectively, in newly diagnosed cancer cases and cancer deaths, which accounts for approximately half of the new cases and deaths worldwide [2]. Hepatocellular carcinoma (HCC) is the most common form of liver cancer, and it is closely linked to cirrhosis, chronic infection with hepatitis B virus (HBV) or hepatitis C virus (HCV), and metabolic and alcoholic liver disease [3]. Despite improvements in the diagnosis and treatment of this cancer, the prognosis and survival remain very poor, with a five-year survival rate of less than 30%. This rate is primarily due to it being diagnosed at a late stage and the missed opportunity to receive optimal therapy for HCC patients [4]. Early-stage detection and receipt of curative therapy appear to be the most effective approach to improve the overall survival of HCC patients [5].
Currently, the serum alpha fetoprotein (AFP) assay and ultrasonography (US) are the primary methods used to detect and diagnose HCC [6]. However, AFP has been questioned due to its low sensitivity (25–65%) at the commonly used cut-off value of 20 ng/mL [7]. In addition, US sensitivity for early-stage HCC appears similar to that of serum AFP and relies largely on the presence of cirrhosis, the size of the tumor, and equipment and examiner expertise. Therefore, serum AFP and US are not satisfactory methods for the early diagnosis of HCC, and new effective biomarkers are urgently needed.
It has been recently reported that HCC is characterized by multiple genetic and epigenetic changes [8]. In contrast to genetic modifications, epigenetic modifications do not change the DNA sequence, but they change the DNA conformation and expression, which plays an important role in tumorigenesis and progression [9]. DNA hypermethylation is an epigenetic modification and an early cancer event that can be detected in plasma-derived cell-free DNA, making it an ideal and useful biomarker for early detection and staging of cancer [10]. Many studies have demonstrated that DNA methylation analysis of specific genes, such as p16, p15, RASSF1A, APC, and SOCS1, can be applied for the early detection and diagnosis of HCC patients [10, 11]. However, to date, no circulating epigenetic biomarker has been approved to diagnose HCC at the individual level.
The septin9 gene (SEPT9) methylation test is the first blood-based methylation test approved by the United State Food and Drug Administration (US FDA) as a colorectal cancer screening test [12]. Previous work has suggested that SEPT9 plays a key role in cell division and is a candidate tumor suppressor gene whose hypermethylation is associated with carcinogenesis [13, 14]. SEPT9 expression is frequently silenced via aberrant promoter hypermethylation in liver cancer, and it is a significant epidriver gene in liver carcinogenesis [15]. Recently, it has been reported that among patients with cirrhosis, the SEPT9 methylation (mSEPT9) test constitutes a promising circulating epigenetic biomarker for HCC diagnosis at the individual patient level [16]. Given that the underlying causes of liver cancer in Chinese populations are different from those in Europe, this study was designed to investigate whether SEPT9 methylation can be detected in the plasma of HCC patients in Chinese populations and subsequently be used as a noninvasive marker for HCC detection.
2. Materials and Methods
2.1. Study Design and Patient Selection
In this study, patients with signs and symptoms of liver malignancy, including 104 hepatocellular carcinoma (HCC) patients, 95 with at-risk liver disease (cirrhosis, hepatitis, and focal nodular hyperplasia), and 174 healthy controls (HC) who had been deemed healthy after undergoing a physical examination from Beijing Ditan Hospital Capital Medical University, were enrolled from July 2018 to January 2019. All of the participants provided plasma samples, and the mSEPT levels were measured in a blinded manner. The final diagnostic result was based on at least two imaging methods (ultrasound, CT, and MRI) and a histopathological confirmation. Patients who had coexisting tumors, metastatic carcinoma, or an undefined pathological status were excluded. Plasma samples were also collected from 15 HCC patients prior to and 10 days after tumor resection to evaluate the postsurgical prognostic value of plasma mSEPT9. All of the study subjects, including 199 patients and 174 healthy controls, underwent simultaneous detection for AFP. The normal range for AFP in this study was determined to be 0–20 ng/mL, and values greater than 20 were considered abnormal. This study was approved by the Beijing Ditan Hospital Capital Medical University Ethics Committee, and all of the subjects received informed consent.
2.2. Sample Collection and Storage
Standard operating protocols and blood collections were performed as previously described [17]. A 10 mL venous blood sample was collected from each subject using K2EDTA anticoagulant tubes (BD Biosciences, USA). Plasma was isolated within four hours post blood collection using centrifugation at room temperature (1350 g, 12 min). Plasma samples were centrifuged again (1350 g, 12 min) to remove residual blood cells. The storage and transportation of samples was conducted following the user manual of the Diagnostic Kit for Septin9 Gene Methylation Assay (BioChain (Beijing) Science and Technology, Inc., Beijing, P.R. China). Sample quality was assessed as previously described [18].
2.3. Quantitative Methylation-Specific PCR (qMSP) for SEPT9 Assay
DNA extraction and bisulfite conversion were performed manually following the manufacturer's instructions for the Diagnostic Kit for Septin9 Gene Methylation Assay (BioChain (Beijing) Science and Technology, Inc., Beijing, P.R. China). Methylated target sequences in the bisulfite-treated DNA template were amplified using real-time PCR on an ABI7500 PCR device (Life Technologies). PCR was performed in a single 60 μL PCR with 25 μL of template DNA per well and run through the reaction system. This process consisted of an initial 30-minute activation at 94°C, followed by 45 cycles at 62°C for 5 seconds, 55.5°C for 35 seconds, and 93°C for 30 seconds, and then cooling at 40°C for 5 seconds. In the quantitative methylation-specific PCR, β-actin (ACTB) was used as an internal control to assess the quantity of the input DNA and the validity of the PCR amplification. The validity of each sample batch was determined on the basis of the methylated SEPT9 and ACTB cycle threshold (CT) values for the positive and negative controls. Positive and negative controls were provided in the kit and run in parallel with the samples each time. The detection threshold and the reaction factors were examined and optimized using clinical plasma samples.
2.4. Statistical Analysis
The statistical analysis was performed using the SPSS 20.0 software (IBM Corporation, Armonk, NY), and graphs were made using GraphPad Prism 5.0 for Windows. The differences in the CT value of the plasma SEPT9 methylation assay between different groups were analyzed using Student's t test. The association between the plasma mSEPT9 status of the HCC patients and their clinicopathological parameters was analyzed using Fisher's test. To assess the diagnostic performance of SEPT9 gene methylation either alone or in combination with AFP between HCC or early-stage HCC and HC or at-risk disease, the models were assessed using logistic regression. After this, the new variable predicted probability (P) was subjected to receiver operating characteristic curve (ROC) analysis. The ROC was generated to calculate the respective areas under the curves (AUCs) with a 95% confidence interval (CI), sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of the SEPT9 gene methylation either alone or in combination with AFP for HCC diagnosis at the determined cut-off value. Differences in the efficacy evaluation for HCC patients were evaluated using a paired t test. A two-tailed probability of P < 0.05 was considered to be statistically significant.
3. Results
3.1. Methylation Status of SEPT9 in Plasma DNA within Different Groups
The SEPT9 promoter methylation (mSEPT9) status in plasma from 104 patients with HCC, 95 patients with at-risk disease, and 174 healthy controls (HC) was assayed. The baseline characteristics of the participants are shown in Table 1. Since the baseline characteristics were not detected using the assay for most normal controls, the cycle threshold (CT) value was set at 45 (the maximal number of PCR cycles in the assay) for those not detected samples. The CT value of mSEPT9 in the HCC patients was significantly lower than in the HCs (HCC vs. HCs, P < 0.0001) and in the at-risk disease patients (HCC vs. at-risk, P < 0.0001), suggesting elevated levels of mSEPT9 in the plasma of HCC patients (Figure 1 and Table S1). We also analyzed the same data with the Wilcoxon-Mann-Whitney test and got the same conclusion (Figure S1).
Table 1.
Baseline characteristics of the participants.
| HCC (%) | At-risk (%) | HC (%) | Statistics χ2 | P value | |
|---|---|---|---|---|---|
| N | 104 | 95 | 174 | ||
| Gender | |||||
| Male | 83 (79.8) | 56 (58.9) | 100 (57.5) | 15.562 | <0.001 |
| Female | 21 (20.2) | 39 (41.1) | 74 (42.5) | ||
| Age | |||||
| 0-50 | 33 (31.7) | 36 (37.9) | 83 (47.7) | 7.307 | 0.026 |
| >50 | 71 (68.3) | 59 (62.1) | 91 (52.3) | ||
| Cirrhosis | |||||
| Yes | 80 (76.9) | 63 (66.3) | NA | 2.762 | 0.097 |
| No | 24 (23.1) | 32 (33.7) | NA | ||
| Etiology | |||||
| HBV | 89 (85.6) | 59 (62.1) | NA | 23.353 | <0.001 |
| HCV | 10 (9.6) | 7 (7.4) | NA | ||
| Alcoholic | 2 (1.9) | 8 (8.4) | NA | ||
| Others | 3 (2.9) | 21 (22.1) | NA | ||
| AFP | |||||
| 0-20 ng/mL | 44 (42.3) | 82 (86.3) | 171 (98.3) | 129.220 | <0.001 |
| >20 ng/mL | 60 (57.7) | 13 (13.7) | 3 (1.7) | ||
| BCLC stage | |||||
| A | 46 (44.2) | ||||
| B | 27 (26.0) | ||||
| C/D | 23 (22.1) | ||||
| Unknown | 8 (7.7) |
HCC indicates hepatocellular carcinoma. HC indicates healthy control. At-risk indicates noncancerous liver diseases, including cirrhosis and hepatitis. HBV indicates hepatitis B virus, HCV indicates hepatitis C virus, AFP indicates alpha fetoprotein, and BCLC stage indicates Barcelona Clinic Liver Cancer.
Figure 1.
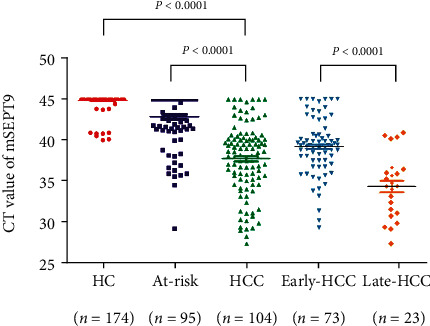
The plasma SEPT9 methylation (mSEPT9) level in cell-free DNA of plasma assayed using methylation-specific fluorescence quantitative PCR. Quantitative PCR showed a significantly decreased median CT value of SEPT9 methylation assay in hepatocellular carcinoma (HCC, n = 104) when compared to the healthy control (HC, n = 174) and at-risk disease (n = 95). The at-risk disease category indicates noncancerous liver diseases, including cirrhosis and hepatitis. CT indicates cycle threshold. Early-HCC represents patients with early-stage (stages A and B) HCC. Late-HCC represents patients with late-stage (stages C and D) HCC. The full line shows the mean CT of each group.
3.2. Diagnostic Value of Plasma mSEPT9 in Patients with HCC and Early-Stage HCC
To achieve the observed test performance, ROC curve analysis was used to evaluate the area under the curve (AUC), sensitivity, and specificity and to determine the best cut-off CT value of mSEPT9 for HCC diagnosis. Compared with the HC, AUC was estimated to be 0.961 (95% CI 0.933–0.989), and mSEPT9 had a sensitivity of 82.7% (86/104) and a specificity of 96.0% (167/174) at the CT cut-off value of 41. In addition, the positive predictive value (PPV), negative predictive value (NPV), and accuracy were 92.5%, 90.3%, and 91.0%, respectively (Figure 2(a) and Table 2). Similar results were obtained from a comparison between patients with HCC and those with at-risk diseases. AUC was estimated to be 0.843 (95% CI 0.787–0.899), and mSEPT9 had a sensitivity of 82.7% (86/104) and a specificity of 83.2% (79/95) at the CT cut-off value of 41. In addition, the PPV, NPV, and accuracy were 84.3%, 81.4%, and 82.9%, respectively (Figure 2(c) and Table 2).
Figure 2.
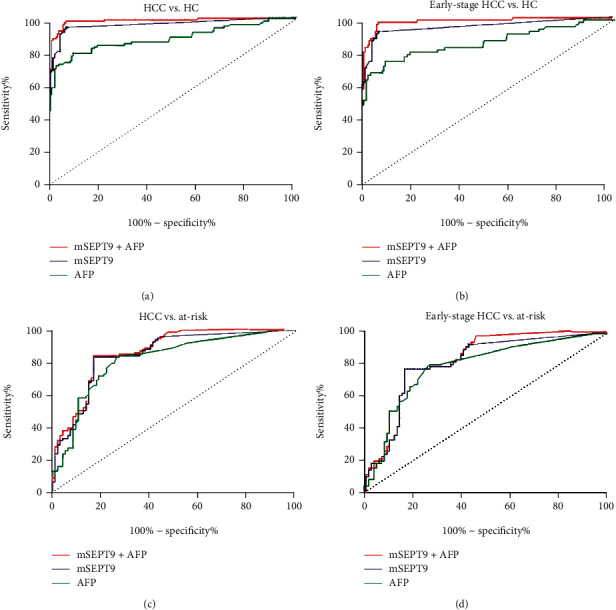
The receiver operating characteristic (ROC) curve of plasma SEPT9 methylation (mSEPT9) and serum alpha fetoprotein (AFP) in the diagnosis of HCC and early-stage HCC from the healthy control group and the at-risk disease group. The ROC curves of plasma mSEPT9 with and without the inclusion of AFP and AFP alone for HCC patients versus HC (a), early-stage HCC patients versus HC (b), HCC patients versus at-risk disease (c), and early-stage HCC patients versus at-risk disease (d). HCC indicates hepatocellular carcinoma. HC indicates healthy control. At-risk indicates noncancerous liver diseases including cirrhosis and hepatitis.
Table 2.
Performances of plasma SEPT9 methylation (mSEPT9) and serum AFP in the diagnosis of hepatocellular carcinoma (HCC) or early-stage HCC.
| Cut-off value | AUC | Sensitivity (%) | Specificity (%) | PPV (%) |
NPV (%) |
Accuracy (%) | ||
|---|---|---|---|---|---|---|---|---|
| Value | 95% CI | |||||||
| HCC vs. HC | ||||||||
| mSEPT9 | CT < 41 | 0.961 | 0.933-0.989 | 82.7 | 96.0 | 92.5 | 90.3 | 91.0 |
| AFP | > 20 ng/mL | 0.881 | 0.833-0.928 | 57.7 | 98.3 | 95.2 | 79.5 | 83.1 |
| SEPT9+AFP | 0.986 | 0.974-0.999 | 91.3 | 94.8 | 91.3 | 94.8 | 93.5 | |
| Early-stage HCC vs. HC | ||||||||
| mSEPT9 | CT < 41 | 0.946 | 0.907-0.985 | 76.7 | 96.0 | 88.9 | 90.8 | 90.3 |
| AFP | >20 ng/mL | 0.852 | 0.789-0.914 | 52.1 | 98.3 | 92.7 | 83.0 | 84.6 |
| SEPT9+AFP | 0.980 | 0.962-0.999 | 87.7 | 94.8 | 87.7 | 94.8 | 92.7 | |
| HCC vs. at-risk | ||||||||
| mSEPT9 | CT < 41 | 0.843 | 0.787-0.899 | 82.7 | 83.2 | 84.3 | 81.4 | 82.9 |
| AFP | >20 ng/mL | 0.812 | 0.751-0.873 | 57.7 | 86.3 | 82.2 | 65.1 | 71.4 |
| SEPT9+AFP | 0.863 | 0.812-0.914 | 91.3 | 76.8 | 81.2 | 89.0 | 84.4 | |
| Early-stage HCC vs. at-risk | ||||||||
| mSEPT9 | CT < 41 | 0.799 | 0.73-0.868 | 76.7 | 83.2 | 77.8 | 82.3 | 80.4 |
| AFP | >20 ng/mL | 0.779 | 0.707-0.852 | 52.1 | 86.3 | 74.5 | 70.1 | 71.4 |
| SEPT9+AFP | 0.817 | 0.753-0.881 | 87.7 | 76.8 | 74.4 | 89.0 | 81.5 | |
HCC indicates hepatocellular carcinoma. HC indicates healthy control. Early-stage HCC represents patients with early-stage hepatocellular carcinoma. At-risk indicates noncancerous liver diseases, including cirrhosis and hepatitis. AFP indicates alpha fetoprotein. CT indicates cycle threshold. AUC represents area under curve. CI indicates confidence interval. PPV indicates positive predictive value. NPV means negative predictive value.
The CT value of plasma mSEPT9 in patients with late-stage HCC (stages C/D) was significantly decreased than those in patients with early-stage HCC (stages A/B) (P < 0.001, Figure 1), which indicated that plasma mSEPT9 positively correlated with HCC malignancy. The ROC curve showed that plasma mSEPT9 demonstrated a remarkable performance in the diagnosis of patients with early-stage HCC as compared to the HC participants (AUC 0.946, 95% CI 0.907–0.985, sensitivity 76.7%, and specificity 96.0%), and at-risk disease (AUC 0.799, 95% CI 0.730–0.868, sensitivity 76.7%, and specificity 83.2%) (Figures 2(b) and 2(d) and Table 2). All of these results demonstrate that plasma mSEPT9 can be used as an excellent biomarker for the diagnosis of HCC and detection of early-stage HCC in clinical settings.
3.3. The Diagnostic Value of Combining Serum AFP Concentration and Plasma mSEPT9 in Patients with HCC and Early-Stage HCC
Serum AFP is the most accepted and used biomarker for HCC detection and diagnosis. In this study, the differential diagnosis performances of plasma mSEPT9, AFP, and their combination were evaluated. It was shown that the use of serum AFP (AUC 0.881, 95% CI 0.833–0.928, sensitivity 57.7%, and specificity 98.3%), plasma mSEPT9 (AUC 0.961, 95% CI 0.933–0.989, sensitivity 82.7%, and specificity 96.0%), or their combination (AUC 0.986, 95% CI 0.974–0.999, sensitivity 91.3%, and specificity 94.8%) significantly improved the diagnostic ability of HCC in healthy individuals (Figure 2(a) and Table 2). The diagnosis between HCC and those with at-risk diseases also showed that plasma mSEPT9 and the combination were more sensitive than AFP alone (Figure 2(c) and Table 2).
Similar results were also obtained using a comparison between patients with early-stage, HC controls, and those with at-risk diseases. At a cut-off of 20 ng/mL, AFP had an AUC (0.852, 95% CI 0.789–0.914) sensitivity of 52.1% and specificity of 98.3% in distinguishing early-stage HCC from HC. The combination plasma mSEPT9 with serum AFP improved AUC (0.980, 95% CI 0.962–0.999) and sensitivity (87.7%) and lessened the decreased diagnostic specificity (94.8%) (Figure 2(b) and Table 2). The diagnosis between early-stage HCC and those at-risk for the disease also showed that the plasma mSEPT9 and the combination were more sensitive than AFP alone (Figure 2(d) and Table 2).
The positive detection rate of plasma mSEPT9 and serum AFP for controls and each HCC stage are presented as a histogram in Figure 3. There was a trend that plasma mSEPT9 assay detected more late-stage HCC cases than early-stage HCC cases (Figure 3). Combining the assay of plasma mSEPT9 test with serum AFP enhanced the test sensitivity in HCC detection. These results suggested that plasma mSEPT9 assay exhibited high sensitivity for HCC at each stage in opportunistic screening. When considering AFP status, it was found that the positive rates of mSEPT9 diagnosis in the AFP-negative and AFP-positive HCC patients were comparable (79.5% vs. 85.0%, P = 0.468) (Figure 4), which indicated the diagnosis of mSEPT9 in HCC cases irrespective of the AFP status. These results demonstrated that plasma mSEPT9 can complement the diagnosis of AFP-negative HCC and AFP-limited HCC patients with a remarkable discriminating performance.
Figure 3.
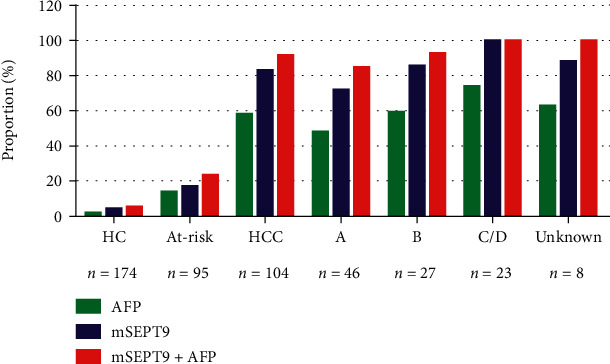
Positive detection rate of plasma SEPT9 methylation (mSEPT9) and serum AFP for controls and at all stages of HCC. HC indicates healthy control. At-risk indicates noncancerous liver diseases including cirrhosis and hepatitis. HCC indicates hepatocellular carcinoma (a–d), and unknown indicates stages of HCC according to the Barcelona Clinic Liver Cancer (BCLC) stage system. AFP means alpha fetoprotein.
Figure 4.
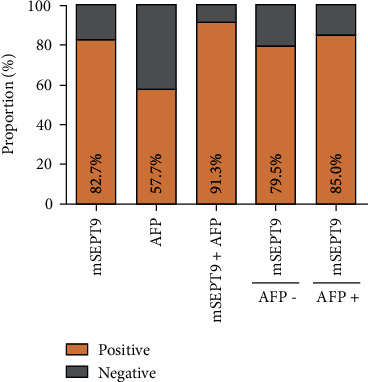
The positive rate for mSEPT9, serum AFP, or the combination by AFP status in HCC. The positive rates of mSEPT9 diagnosis in AFP-negative and AFP-positive HCC patients were comparable (79.5% vs. 85.0%, P = 0.468), which indicated the diagnosis of mSEPT9 in HCC irrespective of AFP status. HCC indicates hepatocellular carcinoma. AFP means alpha fetoprotein.
3.4. Correlation of Plasma mSEPT9 with the Clinicopathological Characteristics of Patients with HCC
The association of plasma mSEPT9 and the clinicopathological characteristics of the patients with HCC are shown in Table S2. Plasma mSEPT9 was not corrected for gender, serum AFP level, or HBeAg status. Plasma mSEPT9 was more frequently observed in older patients (>50) and with late-stage HCC than in those who were younger (P = 0.002) and with early-stage disease (P = 0.03).
3.5. Plasma mSEPT9 Can Monitor the Efficacy of Treatment in Patients with HCC
Dynamic changes in biomarkers that reflect a patient's condition can provide clinical guidance for doctors. In this study, the plasma mSEPT9 levels (CT values) in paired plasma samples from 15 cases undergoing surgery were assessed. The mean CT value of plasma mSEPT9 was 37.9 (range: 29.1–43.7) before surgery and 42.2 (range: 36.3–45.0) after 10 days (n = 15, P = 0.001) (Figure 5). Moreover, the CT plasma mSEPT9 values in all 13 patients with positive mSEPT9 (CT<41) before surgery decreased dramatically after 10 days. Therefore, dynamic changes in the plasma mSEPT9 levels of HCC patients can be used to monitor the efficacy of treatment.
Figure 5.
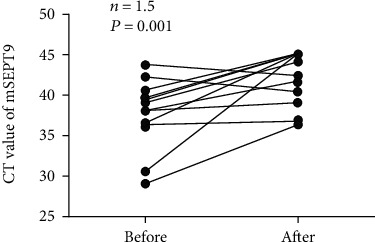
The mSEPT9 levels (CT values) for HCC patients before and 10 days after surgery. Before and After represent before surgery and after surgery, respectively. HCC indicates hepatocellular carcinoma. CT means cycle threshold.
4. Discussion
In the diagnosis of HCC, plasma mSEPT9 showed remarkable sensitivity and specificity for detection in HCC patients, even in the early stages from a healthy individual and those with at-risk diseases. Combined with serum AFP, plasma mSEPT9 showed significantly superior sensitivity than AFP alone in distinguishing HCC patients from both controls. Plasma mSEPT9 can be used to compliment the diagnosis of AFP-negative HCC and AFP-limited HCC patients with a remarkable discriminating performance. Moreover, dynamic changes in the plasma mSEPT9 levels in HCC patients can be used to monitor the efficacy of treatment. These findings showed that plasma mSEPT9 can be used as a noninvasive diagnosis biomarker for HCC patients.
DNA hypermethylation is an important event in carcinogenesis that can be detected in plasma-derived cell-free DNA, which makes it an ideal and useful biomarker for early detection and staging of cancer. Many studies have demonstrated that DNA methylation analysis of specific genes can be used for the early diagnosis of cancer, such as plasma mSEPT9 in colorectal cancer [19], bronchoalveolar lavage fluid SHOX2 and RASSF1A in lung cancer, [20], and urine GSTP1 in prostate cancer [21]. However, there is no FDA-approved circulating epigenetic biomarker that has been shown to be useful to diagnose HCC.
Septins are an evolutionarily conserved family of GTPases with diverse functions that include roles in cytokinesis, vesicle trafficking, apoptosis, and the maintenance of cell polarity [22]. SEPT9, a member of the septin family, acts as a tumor suppressor gene in carcinogenesis and is silenced or decreased by aberrant promoter methylation, which can be reactivated by treatment with azacytidine, providing evidence of epigenetic mechanisms of this gene during carcinogenesis [14, 23]. Several studies have suggested that SEPT9 acts a tumor suppressor gene and plays an important role in the regulation of cell division during its hypermethylation in liver carcinogenesis [15, 24]. It has been recently reported that plasma mSEPT9, as a novel circulating cell-free DNA-based epigenetic biomarker, can be used to diagnose HCC at the individual patient level with 80.8% sensitivity and 95.8% specificity [16]. In Europe, the etiology of cirrhosis and HCC is primarily hepatitis C and alcoholic cirrhosis, while in China, the focus is primarily on hepatitis B. In this study, the diagnostic performance of plasma mSEPT9 in a Chinese population [25] was studied. It was found that the diagnostic performance of plasma mSEPT9 was in accordance with a previous study conducted in Europe. Plasma mSEPT9 exhibited a high diagnostic performance for HCC, and thus, it could be considered a promising biomarker for diagnosing HCC among healthy individuals and patients with at-risk diseases.
HCC is the most common type of primary liver cancer that is usually diagnosed at advanced stages due to a lack of effective diagnostic biomarkers. Serum AFP is the most widely used biomarker for the diagnosis of HCC. However, the serum AFP level lacks adequate sensitivity and specificity for early diagnosis of HCC, with nearly one-third of early-stage HCC patients testing negative for AFP [7]. Therefore, AFP is not a satisfactory biomarker for early diagnosis and prognostic evaluation. In this study, the differential diagnosis performances of plasma mSEPT9, AFP, and their combination were evaluated. It was shown that, in comparison with serum AFP, plasma mSEPT9 or the combination significantly improved the diagnostic ability between HCC and healthy individuals and between HCC and those with at-risk diseases. Plasma mSEPT9 complemented the diagnosis of AFP-negative HCC and AFP-limited HCC patients with a remarkable discriminating performance. In addition, dynamic changes of plasma mSEPT9 in HCC patients can be used to monitor treatment efficacy. Therefore, it is conceivable that plasma mSEPT9 is a promising noninvasive diagnostic biomarker for HCC patients.
The plasma mSEPT9 assay was shown to be an excellent test for HCC detection in a previous study [16]. The application of three PCR reactions and the specific probe for mSEPT9 maximized the opportunity to detect a trace amount of methylated SEPT9 DNA. However, in this study, kits were made using BioChain to conduct the single-tube PCR with double the volume. This reduced the number of tests and the chance of contamination and lowered the costs of the assay. This method will benefit patients in future screenings. This simplified mSEPT9 assay reduced the complexity of the test process from three PCR reactions to one, which could potentially benefit many patients without reducing the detection performance. Hence, this is a simpler, cheaper, more efficient, and convenient method for HCC screening.
Although the plasma mSEPT9 displayed a good performance for HCC detection among healthy individuals and patients with at-risk diseases, it also exhibited a cross-specificity with colorectal cancer. Nevertheless, patients with at-risk diseases, such as those with cirrhosis, are more likely to develop HCC than any other type of cancer [26]. In addition, the positive rate of plasma mSEPT9 (4.1%) in the screening population and the prevalence of colorectal cancer was low, which resulted in a dramatically reduced positive predictive value (PPV). Thus, a positive plasma mSEPT9 test in a patient with an at-risk disease in the liver should initially evoke a high level of suspicion for HCC instead of colorectal cancer. Multicenter, large-scale cohort studies of patients would be helpful for evaluating the diagnostic accuracy of the plasma mSEPT9 test in screening settings.
5. Conclusions
This study demonstrated that plasma mSEPT9 can be a promising noninvasive and circulating epigenetic biomarker for HCC diagnosis at the individual level. In addition, the combination of plasma mSEPT9 and AFP could enhance the sensitivity of AFP alone in the diagnosis of HCC.
Acknowledgments
This work was supported by the Youth Talent Development Fund of Beijing Ditan Hospital, Capital Medical University (201601). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. We thank LetPub (http://www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Abbreviations
- SEPT9:
Septin9
- HCC:
Hepatocellular carcinoma
- HC:
Healthy control
- At-risk:
Noncancerous liver diseases, including cirrhosis and hepatitis
- HBV:
Hepatitis B virus
- HCV:
Hepatitis C virus
- AFP:
Alpha fetoprotein
- BCLC:
Barcelona Clinic Liver Cancer
- CT:
Cycle threshold.
Contributor Information
Xiaoliang Han, Email: sean.han@biochainbj.com.
Li Jiang, Email: movdush@139.com.
Baohua Li, Email: libaohua8@163.com.
Zhongtao Zhang, Email: zhangzht0527@163.com.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Ethical Approval
This study was approved by the Beijing Ditan Hospital Capital Medical University Ethics Committee, and all of the subjects received informed consent.
Conflicts of Interest
The authors declare that they have no competing interests.
Supplementary Materials
Figure S1: the plasma SEPT9 methylation (mSEPT9) level was analyzed by using the Wilcoxon-Mann-Whitney test. According to Donald's study (1), when standard deviations (SD) of two samples are unequal, a t test may have better type-1 error control than nonparametric alternatives, such as the Wilcoxon-Mann-Whitney test. In this case, the standard deviations of these groups are not equal, so we show the result of t test in text. Table S1: methylation-specific fluorescence quantitative PCR analysis of plasma SEPT9 (mSEPT9). Table S2: correlation between SEPT9 promoter methylation and the clinicopathological parameters in patients with HCC.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Sun K., Zheng R., et al. Cancer incidence and mortality in China, 2014. Chinese Journal of Cancer Research. 2018;30(1):1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong M. C. S., Huang J. L. W., George J., et al. The changing epidemiology of liver diseases in the Asia-Pacific region. Nature Reviews. Gastroenterology & Hepatology. 2019;16(1):57–73. doi: 10.1038/s41575-018-0055-0. [DOI] [PubMed] [Google Scholar]
- 4.Allemani C., Matsuda T., Di Carlo V., et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. The Lancet. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singal A. G., Pillai A., Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Medicine. 2014;11(4, article e1001624) doi: 10.1371/journal.pmed.1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omata M., Cheng A. L., Kokudo N., et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatology International. 2017;11(4):317–370. doi: 10.1007/s12072-017-9799-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzartzeva K., Obi J., Rich N. E., et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154(6):1706–1718.e1. doi: 10.1053/j.gastro.2018.01.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taniguchi K., Yamada T., Sasaki Y., Kato K. Genetic and epigenetic characteristics of human multiple hepatocellular carcinoma. BMC Cancer. 2010;10(1):p. 530. doi: 10.1186/1471-2407-10-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baylin S. B. DNA methylation and gene silencing in cancer. Nature Clinical Practice. Oncology. 2005;2(Supplement 1):S4–11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y. J., Wu H. C., Shen J., et al. Predicting hepatocellular carcinoma by detection of aberrant promoter methylation in serum DNA. Clinical Cancer Research. 2007;13(8):2378–2384. doi: 10.1158/1078-0432.CCR-06-1900. [DOI] [PubMed] [Google Scholar]
- 11.Um T.-H., Kim H., Oh B.-K., et al. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. Journal of Hepatology. 2011;54(5):939–947. doi: 10.1016/j.jhep.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Potter N. T., Hurban P., White M. N., et al. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clinical Chemistry. 2014;60(9):1183–1191. doi: 10.1373/clinchem.2013.221044. [DOI] [PubMed] [Google Scholar]
- 13.Robertson C., Church S. W., Nagar H. A., Price J., Hall P. A., Russell S. E. H. Properties of SEPT9 isoforms and the requirement for GTP binding. The Journal of Pathology. 2004;203(1):519–527. doi: 10.1002/path.1551. [DOI] [PubMed] [Google Scholar]
- 14.Burrows J. F., Chanduloy S., McIlhatton M. A., et al. Altered expression of the septin gene, SEPT9, in ovarian neoplasia. The Journal of Pathology. 2003;201(4):581–588. doi: 10.1002/path.1484. [DOI] [PubMed] [Google Scholar]
- 15.Villanueva A., Portela A., Sayols S., et al. DNA methylation-based prognosis and epidrivers in hepatocellular carcinoma. Hepatology. 2015;61(6):1945–1956. doi: 10.1002/hep.27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oussalah A., Rischer S., Bensenane M., et al. Plasma m _SEPT9_ : A Novel Circulating Cell-free DNA-Based Epigenetic Biomarker to Diagnose Hepatocellular Carcinoma. eBioMedicine. 2018;30:138–147. doi: 10.1016/j.ebiom.2018.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu D., Zhou G., Jin P., et al. Detection of Colorectal Cancer Using a Simplified _SEPT9_ Gene Methylation Assay Is a Reliable Method for Opportunistic Screening. The Journal of Molecular Diagnostics. 2016;18(4):535–545. doi: 10.1016/j.jmoldx.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Song L., Peng X., Li Y., et al. The SEPT9 gene methylation assay is capable of detecting colorectal adenoma in opportunistic screening. Epigenomics. 2017;9(5):599–610. doi: 10.2217/epi-2016-0146. [DOI] [PubMed] [Google Scholar]
- 19.Song L., Yu H., Jia J., Li Y. A systematic review of the performance of the SEPT9 gene methylation assay in colorectal cancer screening, monitoring, diagnosis and prognosis. Cancer Biomarkers. 2017;18(4):425–432. doi: 10.3233/CBM-160321. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C., Yu W., Wang L., et al. DNA methylation analysis of the SHOX2 and RASSF1A panel in bronchoalveolar lavage fluid for lung cancer diagnosis. Journal of Cancer. 2017;8(17):3585–3591. doi: 10.7150/jca.21368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woodson K., O’Reilly K. J., Hanson J. C., Nelson D., Walk E. L., Tangrea J. A. The usefulness of the detection of GSTP1 methylation in urine as a biomarker in the diagnosis of prostate cancer. The Journal of Urology. 2008;179(2):508–512. doi: 10.1016/j.juro.2007.09.073. [DOI] [PubMed] [Google Scholar]
- 22.Hall P. A., Russell S. E. H. The pathobiology of the septin gene family. The Journal of Pathology. 2004;204(4):489–505. doi: 10.1002/path.1654. [DOI] [PubMed] [Google Scholar]
- 23.Grützmann R., Molnar B., Pilarsky C., et al. Sensitive detection of colorectal cancer in peripheral blood by septin 9 DNA methylation assay. PLoS One. 2008;3(11, article e3759) doi: 10.1371/journal.pone.0003759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakehashi A., Ishii N., Shibata T., et al. Mitochondrial prohibitins and septin 9 are implicated in the onset of rat hepatocarcinogenesis. Toxicological Sciences. 2011;119(1):61–72. doi: 10.1093/toxsci/kfq307. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z., Jiang Y., Yuan H., et al. The trends in incidence of primary liver cancer caused by specific etiologies: results from the Global Burden of Disease Study 2016 and implications for liver cancer prevention. Journal of Hepatology. 2019;70(4):674–683. doi: 10.1016/j.jhep.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Sørensen H. T., Friis S., Olsen J. H., et al. Risk of liver and other types of cancer in patients with cirrhosis: a nationwide cohort study in Denmark. Hepatology. 1998;28(4):921–925. doi: 10.1002/hep.510280404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: the plasma SEPT9 methylation (mSEPT9) level was analyzed by using the Wilcoxon-Mann-Whitney test. According to Donald's study (1), when standard deviations (SD) of two samples are unequal, a t test may have better type-1 error control than nonparametric alternatives, such as the Wilcoxon-Mann-Whitney test. In this case, the standard deviations of these groups are not equal, so we show the result of t test in text. Table S1: methylation-specific fluorescence quantitative PCR analysis of plasma SEPT9 (mSEPT9). Table S2: correlation between SEPT9 promoter methylation and the clinicopathological parameters in patients with HCC.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


