Abstract
Avian leukosis virus subgroup J (ALV-J) was first isolated from broiler chickens in China in 1999; subsequently, it was rapidly introduced into layer chickens and Chinese local chickens. Recently, the incidence of ALV-J in broiler and layer chickens has significantly decreased. However, it has caused substantial damage to Chinese local chickens, resulting in immense challenges to their production performance and breeding safety. To systematically analyze the molecular characteristics and the epidemic trend of ALV-J in Chinese local chickens, 260 clinical samples were collected for the period of 2013–2018; 18 ALV-J local chicken isolates were identified by antigen-capture enzyme-linked immunosorbent assay and subgroup A-, B-, and J-specific multiplex PCR. The whole genomic sequences of 18 isolates were amplified with PCR and submitted to GenBank. Approximately, 55.5% (10/18) of the 18 isolates demonstrated a relatively high homology (92.3–95.4%) with 20 ALV-J early-isolated local strains (genome sequences obtained from GenBank) in gp85 genes clustering in a separated branch. The 3ʹ untranslated region (3ʹ UTR) of the 18 isolates showed a 195–210 and 16–28 base pair deletion in the redundant transmembrane region and in direct repeat 1, respectively; 55.5% (10/18) of the 18 isolates retained the 147 residue E element. The U3 gene of 61.1% (11/18) of the 18 isolates shared high identity (94.6–97.3%) with ALV-J early-isolated local strains. These results implied that the gp85 and U3 of ALV-J local chicken isolates have rapidly evolved and formed a unique local chicken branch. In addition, it was determined that the gene deletion in the 3′UTR region currently serves as a unique molecular characteristic of ALV-J in China. Hence, the obtained results built on the existing ALV-J molecular epidemiological data and further elucidated the genetic evolution trend of ALV-J in Chinese local chickens.
Key words: subgroup J avian leukosis virus, Chinese local chicken, molecular characterization, molecular epidemiology, genetic evolution
Introduction
Avian leukosis is induced by avian leukosis viruses (ALV), which belong to a group in the genus Alpharetrovirus of the family Retroviridae. These viruses can cause malignant or benign tumorigenic diseases in poultry. Avian leukosis viruses can be differentiated into 11 subgroups, namely A–K, based on viral envelope interference, host range, and cross-reactivity with neutralizing antibodies (Payne et al., 1991; Weiss, 1992; Coffin et al., 1997; Payne and Nair, 2012; Zhao et al., 2018). Subgroups A, B, C, D, J, and K, which prevail in chickens, are considered exogenous ALV; the E subgroup is considered an endogenous ALV and is not pathogenic. Subgroups A and B are recognized as common pathogenic viruses that induce lymphoid leukosis and sarcoma, whereas subgroups C and D have only rarely been reported. However, avian leukosis virus subgroup J (ALV-J), a new exogenous ALV, is the most prevalent in chickens and has exhibited increased pathogenicity and faster transmission ability in recent years (Payne et al., 1992a; Jirong et al., 1998; Lai et al., 2011; Payne and Nair, 2012), primarily inducing myeloid leukosis in meat-type chickens (Venugopal et al., 2000). Since the first strain of ALV-J was isolated from meat-type breeder chickens in the UK in 1988, it has rapidly spread around the world, causing severe economic losses to the poultry industry (Payne et al., 1992b).
Since 2004, ALV-J outbreaks have occurred in chicken farms in several provinces in China, resulting in morbidity and mortality rates as high as 50% and huge economic losses to the Chinese layer and local chicken industry (Cheng et al., 2005; Sun and Cui, 2007; Gao et al., 2010; Zhang et al., 2011; Li et al., 2018). After the successful eradication programs, ALV-J has been optimally controlled, and its incidence in white meat-type chicken and layer chicken farms has decreased yearly in China. However, ALV-J infections in Chinese local chicken have become increasingly devastating in recent years (Meng et al., 2018; Zhang et al., 2018). Avian leukosis virus subgroup J causes an adverse decline in egg production and results in high mortality; additionally, it poses a grave threat to the safety of local chicken breeding sources in China.
Although there are a few prior studies on the isolation and sequencing of ALV-J strains from local chickens (Cheng et al., 2005; Sun and Cui, 2007; Wang et al., 2012b; Li et al., 2016; Zhang et al., 2018), no systematic epidemiological investigation and study of the molecular characteristics of ALV-J isolates from local chickens has been performed. Thus, in the present study, 260 samples were collected from local chickens in the 5 main local chicken breeding provinces of China (Jiangsu, Guangxi, Hunan, Shanghai, and Hebei) from 2013 to 2018. A total of 18 ALV-J strains were isolated, and their complete proviral genomes were sequenced. The molecular epidemiology and the evolutionary trend of the ALV-J isolates from local chickens were systematically analyzed.
Materials and methods
Clinical Samples
A total of 260 samples (including kidney, liver, spleen, or whole blood) were collected from 5 provinces in China (Jiangsu, Guangxi, Hunan, Shanghai, and Hebei) between 2013 and 2018. A tissue sample (the liver, spleen, or kidney collected randomly) represents a chicken, and a tube of whole blood represents a chicken. The local chickens showed clinical signs such as abnormal enlarged livers, spleens, and kidneys, slow growth, and decreased fertility. The tissue samples frozen at −20°C and whole blood stored at 4°C were transported from the field to the laboratory, and virus isolation was performed immediately.
Virus Isolation and Identification
Avian leukosis virus isolation and identification procedures in cell cultures were performed as previously described (Gao et al., 2012). First, tissue samples were homogenized in 1 mL sterile phosphate-buffered saline (8.0 g NaCl, 0.2 g KCl, 1.15 g Na H2PO4, and 0.2 g KH2PO4 in 1 L ddH2O) and centrifuged at 3,000 × g for 10 min. Whole-blood samples were centrifuged at 3,000 × g for 5 min. Second, the supernatant from a tissue sample or the plasma from whole blood was filtered through a 0.22-μm Millipore membrane, then added 0.5 mL of filtered supernatant to monolayer DF-1 cell cultures in 12-well plates, which are known to be susceptible only to exogenous ALV (Maas et al., 2006). Lastly, following virus adsorption, the supernatant was removed, and the DF-1 cells were maintained in Dulbecco's modified Eagle's medium (Thermo Scientific, Rockford, IL), supplemented with 1% (v/v) heat-inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO) and 1% (v/v) penicillin/streptomycin (Summus, Beijing, China) solution at 38.5°C under a humid atmosphere with 5% CO2 for 7 d. After 3 freeze-thaw cycles, the virus was serially passaged 3 times in DF-1 cells according to the virus isolation method described above.
The supernatant of infected DF-1 cells was harvested and tested for the presence of the ALV group-specific antigen (p27) by antigen-capture enzyme-linked immunosorbent assay (ELISA) using an ALV antigen test kit (Yun et al., 2013). Proviral DNA was directly extracted from positive cultured DF-1 cells using a Tissue DNA Extract Kit (Axygen Scientific, Inc., Union City, CA) according to the manufacturer's instructions. To further identify the subgroup of isolates, the genomic DNA was detected using subgroup A-, B-, and J-specific multiplex PCR with specific primers (Table 1) (Gao et al., 2014). The PCR was performed in a 25-μL system containing 2 μL DNA, 12.5 μL Premix Taq DNA polymerase (TaKaRa, Dalian, China), 2 μL PF (10 pM), 0.8 μL AR(10 pM), 1 μL BR, 1 μL CR (10 pM), and 5.7 μL double-distilled water (ddH2O). The PCR conditions were as follows: initial denaturation at 95°C for 5 min; 30 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 1 min; and a final elongation step at 72°C for 7 min. The PCR products were evaluated by 1.0% agarose gel electrophoresis.
Table 1.
Primers used in the present study.
| Primer | Sequence (5′-3′) | Size (bp) | Amplification target |
|---|---|---|---|
| PF | CGGAGAAGACACCCTTGCT | ||
| AR | GCATTGCCACAGCGGTACTG | 715 | ALV-A |
| BR | GTAGACACCAGCCGGACTATC | 515 | ALV-B |
| JR | CGAACCAAAGGTAACACACG | 422 | ALV-J |
| JAF | TGTAGTGTTATGCAATACTCTTATGTAACG | 2,758 | JA |
| JAR | TTGAGCGGAATAGCCAGATGTAG | ||
| JBF | AGGGAGTATCCTGGGAAGAG | 2,818 | JB |
| JBR | ACAACGGAAATAATAACCACGC | ||
| JCF | GGAGAAGACACCCTTGCTGCC | 2,460 | JC |
| JCR | TGAAGCCATCCGCTTCATGC |
PCR Amplification, Cloning, and Sequencing
To obtain the genome sequences of the ALV isolates, 3 primer pairs (JAF/JAR, JBF/JBR, and JCF/JCR) were designed for amplification of the whole genomic sequences of the ALV isolates (Table 1) based on ALV-J prototype strain HPRS-103 (GenBank No. Z46390). The PCR was performed in a 50-μL system containing 2 μL DNA, 25 μL PrimeSTAR Max premix DNA high-fidelity polymerase (TaKaRa), 2 μL upstream primer (JAF, JBF, or JCF) (10 pM), 2 μL downstream primer (JAR, JBR, or JCR) (10 pM), and 19 μL of ddH2O. The PCR conditions were as follows: initial denaturation at 98°C for 5 min; 35 cycles of 98°C for 15 s, 52°C for 15 s, and 72°C for 50 s; and a final elongation step of 72°C for 10 min. The PCR products were excised from a 1.0% agarose gel and purified using the AxyPrep DNA gel extraction kit (Axygen), cloned into the pMD18-T vector (TaKaRa) and then transformed into DH5α cells. Three different clones of each fragment were confirmed by sequencing (Kumei, Changchun, China).
Sequence Analysis
The genomic nucleotide sequences of 3 gene fragments (termed JA, JB, and JC) were edited and spliced using the SeqMan software of the DNASTAR package. Phylogenetic analysis of the ALV-J sequences was performed using the neighbor-joining method with 1,000 bootstrap replicates using the MEGA, version 6.0, program. The sequences obtained in the present study have been submitted to GenBank. The ALV reference strains used in the present study included ALV-J broiler isolates (prototype strain HPRS-103, American meat-type chickens, and broiler chickens in China) and ALV-J layer isolates. To systematically analyze the molecular characterization and genetic evolution trends of Chinese local chicken ALV-J, 20 ALV-J early-isolated local strains in China (genome sequences obtained from GenBank) were also selected as the reference strains (Meng et al., 2018).
Data Availability
The complete proviral genome sequences of all 18 isolates were deposited at GenBank with accession numbers MN735292 to MN735309.
Results
Isolation and Identification of ALV-J From Chinese Local Chickens
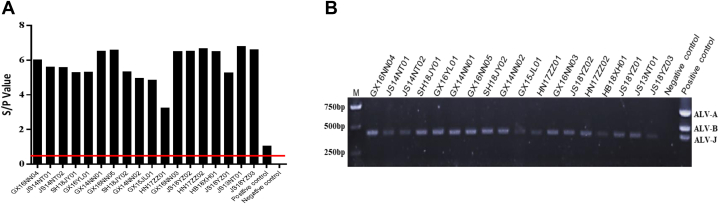
All clinical samples (including samples from kidney, liver, spleen, or whole blood) from local chickens were inoculated into DF-1 cells; after 7 d, the culture supernatants of the infected DF-1 cells were subjected to ALV p27 antigen ELISA (Figure 1A). The results showed that 18 culture supernatants were ALV positive. To further identify the subgroup, proviral DNA was extracted from the 18 positive culture supernatants and amplified by subgroup A-, B-, and J-specific multiplex PCR (Gao et al., 2014). Only the primers specific to the J subgroup (PF and JR) amplified a specific 422-base pair (bp) sequence; no specific fragments were observed for primers specific to subgroups A (PF and AR) and B (PF and BR) (Figure 1B). These results indicated that the 18 isolates belonged to the ALV-J subgroup and were designated JS13NT01, JS14NT01, JS14NT02, GX14NN01, GX14NN02, GX15JL01, GX16NN03, GX16NN04, GX16NN05, GX16YL01, HN17ZZ01, HN17ZZ02, HB18XH01, SH18JY01, SH18JY02, JS18YZ01, JS18YZ02, and JS18YZ03, respectively.
Figure 1.
Isolation and identification of ALV-J local chicken isolates. (A) The supernatants of DF-1 cells were assessed using ALV p27 antigen ELISA (42). Following OD650 measurement, the S/P ratios were calculated and used to express the S/P ratio per sample. S/P values greater than or equal to 0.2 were considered positive. The red line indicates the cut-off value for ELISA. (B) Identification of 18 isolates by subgroup A-, B-, and J-specific multiplex PCRs. DF-1 cells infected with Rous-associated virus type 1 (subgroup A), Rous-associated virus type 2 (subgroup B), and HPRS-103 (subgroup J) were used as the positive control for PCR with primer pairs PF/AR, PF/BR, and PF/JR, respectively. Uninfected DF-1 cells served as the negative control. Abbreviation: ALV-J, Avian leukosis virus subgroup J.
Sequence Analysis of the Proviral Genomes of the 18 Isolates
The complete proviral genomes of the 18 isolates from Chinese local chickens were amplified and sequenced to systematically analyze their molecular characteristics. The sequences of the 18 isolates have been submitted to GenBank (the accession numbers are listed in Table 2). The full-length proviral genome sequences of the 18 isolates were 7,450 to 7,635 bp in length. All genome sequences displayed a typical replication-competent type C retrovirus genetic organization (5′LTR-leader-gag-pol-env-3′LTR), lacking viral oncogenes (Ruddell, 1995). The complete genome sequences of the 18 isolates shared 93.6 to 98.9% identity with each other. The env genes of the 18 isolates were 1,512 to 1,518 bp in length, and their nucleotide sequences showed a maximum divergence of 10.7%, with nucleotide sequence identities ranging from 89.3 to 99.8%. The gag and pol nucleotide sequences of the 18 isolates also shared high sequence identity (94.3–100% and 95.8–100%, respectively).
Table 2.
ALV-J strains used in the present study.
| No.1 | Strains | Origin | Yr | Accession no. | Host2 |
|---|---|---|---|---|---|
| 1 | JS13NT01 | Jiangsu | 2013 | MN735301 | Local |
| 2 | JS14NT01 | Jiangsu | 2014 | MN735308 | Local |
| 3 | JS14NT02 | Jiangsu | 2014 | MN735302 | Local |
| 4 | GX14NN01 | Guangxi | 2014 | MN735292 | Local |
| 5 | GX14NN02 | Guangxi | 2014 | MN735293 | Local |
| 6 | GX15JL01 | Guangxi | 2015 | MN735294 | Local |
| 7 | GX16NN03 | Guangxi | 2016 | MN735295 | Local |
| 8 | GX16NN04 | Guangxi | 2016 | MN735296 | Local |
| 9 | GX16NN05 | Guangxi | 2016 | MN735309 | Local |
| 10 | GX16YL01 | Guangxi | 2016 | MN735297 | Local |
| 11 | HN17ZZ01 | Hunan | 2017 | MN735299 | Local |
| 12 | HN17ZZ02 | Hunan | 2017 | MN735300 | Local |
| 13 | HB18XH01 | Hebei | 2018 | MN735298 | Local |
| 14 | SH18JY01 | Shanghai | 2018 | MN735306 | Local |
| 15 | SH18JY02 | Shanghai | 2018 | MN735307 | Local |
| 16 | JS18YZ01 | Jiangsu | 2018 | MN735303 | Local |
| 17 | JS18YZ02 | Jiangsu | 2018 | MN735304 | Local |
| 18 | JS18YZ03 | Jiangsu | 2018 | MN735305 | Local |
| 19 | HPRS-103 | UK | 1989 | Z46390 | Broiler |
| 20 | ADOL-Hc1 | USA | 1993 | AF097731 | Broiler |
| 21 | UD5 | USA | 2000 | AF307952 | Broiler |
| 22 | YZ9902 | Jiangsu | 1999 | HM235670 | Broiler |
| 23 | SD9901 | Shangdong | 1999 | AY897220 | Broiler |
| 24 | SD0002 | Shangdong | 2000 | AY897224 | Broiler |
| 25 | NX0101 | NingXia | 2001 | AY897227 | Broiler |
| 26 | BJ0303 | Beijing | 2003 | AY897232 | Broiler |
| 27 | JL08CH3-1 | Jilin | 2008 | HQ634809 | Layer |
| 28 | LN09SY31 | Liaoning | 2008 | HQ634803 | Layer |
| 29 | GL09DP02 | Shangdong | 2009 | JN378887 | Layer |
| 30 | SD09DP04 | Shangdong | 2009 | HQ634808 | Layer |
| 31 | HLJ09SH01 | Heilongjiang | 2009 | HQ634806 | Layer |
| 32 | JS09GY3 | Jilin | 2009 | GU982308 | Layer |
| 33 | JS09GY6 | Jilin | 2009 | GU982310 | Layer |
| 34 | HLJ10SH04 | Heilongjiang | 2010 | HQ634814 | Layer |
| 35 | JL10HW01 | Jilin | 2010 | HQ634800 | Layer |
| 36 | JL093-1 | Jilin | 2012 | JN624878 | Layer |
| 37 | GD0510 A | Shangdong | 2005 | EF103132 | Local |
| 38 | GD0512 | Shangdong | 2005 | EF103133 | Local |
| 39 | SCAU-0901 | Guangdong | 2009 | FJ619190 | Local |
| 40 | XG-09 | Guangdong | 2009 | HM775332 | Local |
| 41 | WN100401 | Shangdong | 2010 | HQ271447 | Local |
| 42 | WN100402 | Anhui | 2010 | HQ271448 | Local |
| 43 | WN100403 | Anhui | 2010 | HQ333257 | Local |
| 44 | SD12HN01 | Guangdong | 2012 | KM873187 | Local |
| 45 | SD12HN02 | Guangdong | 2012 | KM873188 | Local |
| 46 | WSC512 | Guangdong | 2012 | KJ631320 | Local |
| 47 | GX13QZ14 | Guangxi | 2013 | KT598477 | Local |
| 48 | GX14HG01 | Guangxi | 2014 | KU997685 | Local |
| 49 | GDKP1202 | Guangdong | 2012 | JX453210 | Local |
| 50 | GD1406-H | Guangdong | 2014 | KU500033 | Local |
| 51 | GX14YL03 | Guangxi | 2014 | KT598470 | Local |
| 52 | GX15PP03 | Guangxi | 2015 | KU848762 | Local |
| 53 | GX16ZS01 | Guangxi | 2016 | KY983549 | Local |
| 54 | GX16ZS748 | Guangxi | 2016 | KY983552 | Local |
| 55 | GX16ZSE10 | Guangxi | 2016 | KY983553 | Local |
| 56 | GX16ZS352 | Guangxi | 2016 | KY983550 | Local |
Abbreviation: ALV-J, Avian leukosis virus subgroup J.
Isolates 1 to 18 are ALV-J local chicken isolates identified in the present study. Isolates 19 to 26 are ALV-J broiler reference strains. Isolates 27 to 36 are ALV-J layer reference strains. Isolates 37 to 56 are ALV-J early-isolated local strains. The genome sequences of ALV reference strains are available in GenBank.
ALV-J isolate hosts: Broiler, meat-type chicken; Layer: Layer chicken; Local: Local chicken.
Sequence Analysis of the ALV-J gp85 Gene of the 18 Isolates
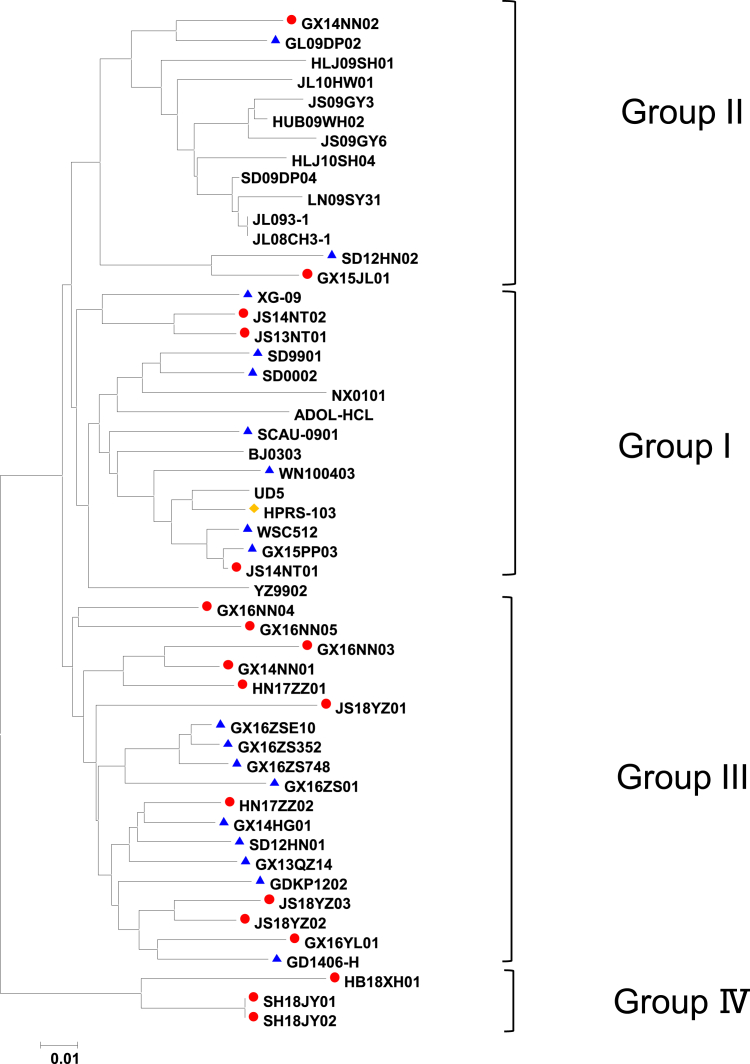
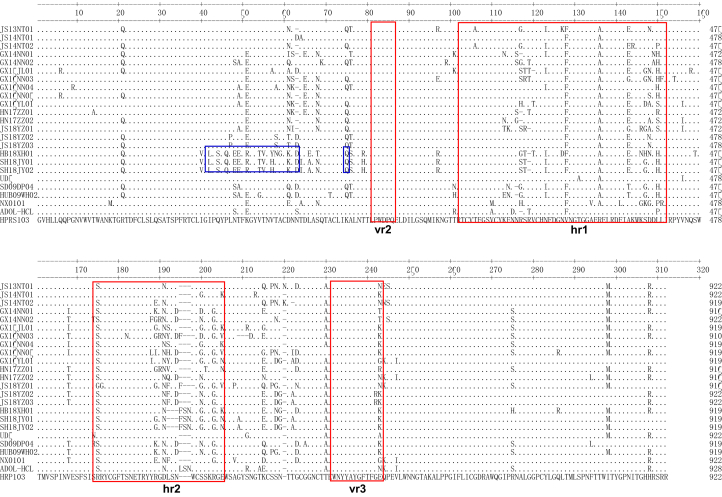
The gp85 is encoded by the env gene. The gp85 sequences of the 18 isolates were 912 to 924 bp in length, with 1 open reading frame encoding a protein of 304 to 308 amino acids, respectively. To further understand the evolutionary origins of ALV-J in local chickens, the gp85 genes of the 18 isolates were compared with sequences from reference strains (Table 2). The phylogenetic analysis of the gp85 genes indicated that 3 isolates (3/18; JS13NT01, JS14NT01, and JS14NT02) were more closely related to ALV-J broiler isolates and showed high sequence identity (95.1–97.9%) with the ALV-J prototype strain HPRS-103 (Bai, 1995), designated as group I. Two isolates (2/18; GX14NN02 and GX15JL01) shared high sequence identity (92.5–94.4%) with ALV-J layer isolates designated as group II. Ten isolates (10/18; GX14NN01, GX16NN04, HN17ZZ01, GX16NN03, JS18YZ01, HN17ZZ02, GX16NN05, GX16YL01, JS18YZ02, and JS18YZ03) shared relatively high sequence identity (92.3–95.4%) with ALV-J early-isolated local strains, designated as group III. The sequence identity of 3 isolates (3/18; SH18JY02, SH18JY01, and HB18XH01) was <90% for all reference strains; these isolates were distributed in a separate branch from groups I, II, and III; they were designated as a new branch, namely group IV (Figure 2). In addition, 12 unique amino acid substitutions (41V, 43L, 45S, 47Q, 49E, 50E, 52R, 55T, 56V, 58Y/H, 61K, and 76S) near the vr2 regions of SH18JY02, SH18JY01, and HB18XH01 in group IV were found (Figure 3). Taken together, these data indicated that the gp85 of most ALV-J local chicken isolates form a unique branch, belonging to group III.
Figure 2.
Phylogenetic analysis of the gp85 gene of ALV-J local chicken isolates. The tree was constructed using the neighbor-joining method with MEGA6.0 software. Bootstrap values were calculated with 1,000 replicates of the alignment. The 4 groups are marked. The red circles represent the 18 ALV-J local chicken isolates in the present study. The blue triangles represent ALV-J early-isolated local strains. The yellow diamond represents ALV-J prototype strain HPRS-103. Abbreviation: ALV-J, Avian leukosis virus subgroup J.
Figure 3.
Sequence Analysis of the gp85 protein of ALV-J local chicken isolates. The variable domains (vr2, hrl, hr2, and vr3) are indicated in red boxes and marked. Twelve unique amino acid substitutions (SH18JY02, SH18JY01, and HB18XH01) in group IV are indicated in blue boxes. Abbreviation: ALV-J, Avian leukosis virus subgroup J.
Sequence Analysis of the 3′ Untranslated Region of the 18 ALV-J Isolates
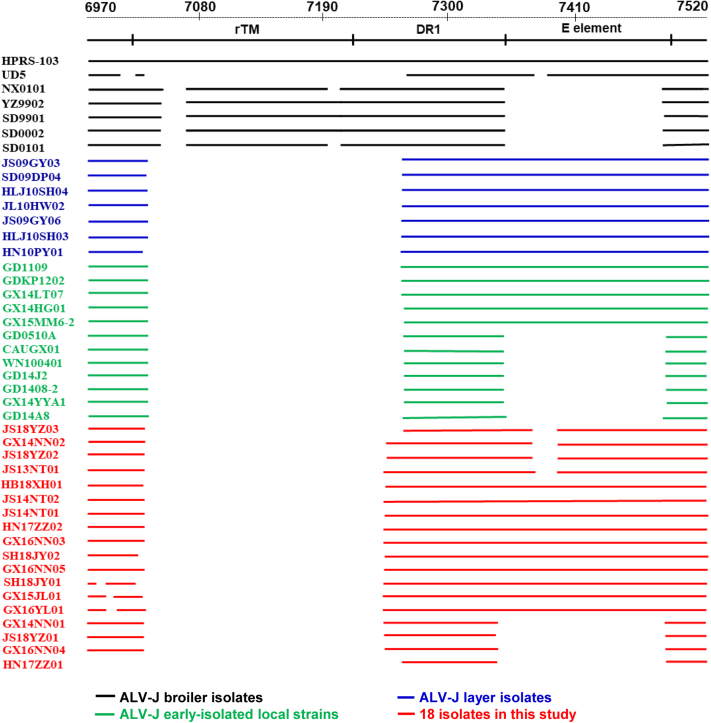
The 3′ untranslated region (UTR) nucleotide sequences of the 18 isolates had full lengths of 125–275 bp. The redundant transmembrane region (rTM), DR-1, and the E element are located in the 3′UTR (Tsichlis et al.,1982). A comparison of the 3′UTR nucleotide sequences of all 18 isolates with the corresponding sequences in the ALV-J broiler and layer chicken isolates showed that a section of 195–210 bp was deleted from the 3′ end of the rTM region, whereas a section of 16–28 bp was deleted from 5′ end of the direct repeat 1 (DR-1) region in all 18 isolates and ALV-J early-isolated local strains (Figure 4). These deletions were similar to mutations in the 3′UTRs of Chinese ALV-J layer isolates.
Figure 4.
Diagram of the deletions in the 3′UTR of ALV-J Local Chicken, Broiler, and Layer Isolates. The top 2 lines represent the base numbers and elements in the genomic proviral DNA of HPRS-103. The deletions are indicated by empty spaces between the lines. Abbreviations: 3′UTR, 3′ untranslated region; ALV-J, Avian leukosis virus subgroup J.
Ten isolates (10/18; GX16YL01, GX16NN05, GX16NN03, GX15JL01and HN17ZZ02, JS14NT02, JS14NT01, SH18JY01, SH18JY02, and HB18XH01) had a complete E element, 147 bp in length. Four isolates (4/18; JS18YZ02, JS18YZ03, GX14NN02, and JS13NT01) contained a 16–24 bp deletion in the E element, whereas the other 4 isolates (4/18; GX16NN04, GX14NN01, JS18YZ01, and HN17ZZ01) showed a 127-bp deletion in the E element (Figure 4).
Sequence Analysis of the 3′Long Terminal Repeat of the 18 Isolates
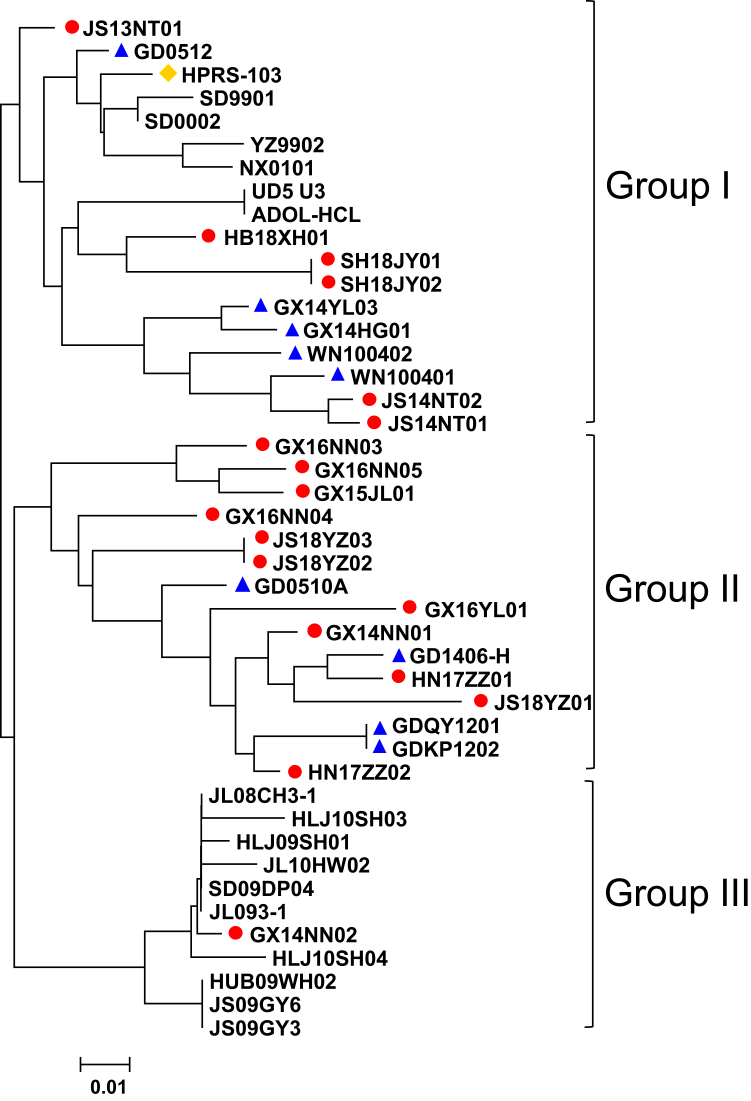
The 3′ long terminal repeat of the 18 isolates was composed of U3, R, and U5; it comprised a length of 313–325 bp. The U5 and R regions were conserved compared with those of all reference strains, sharing 95.2–96.8% sequence identity. The U3 regions of the 18 isolates were 214–226 bp in length. Phylogenetic analysis of the U3 sequences of the 18 isolates showed that 6 isolates (6/18; JS13NT01, JS14NT01, JS14NT02, HB18XH01, SH18JY02, and SH18JY01), and the ALV-J broiler isolates belonged to a branch designated as group I (Figure 5). Eleven isolates (11/18; GX16NN03, GX16NN05, GX15JL01, HN17ZZ02, JS18YZ01, JS18YZ02, JS18YZ03, GX14NN01, GX16NN04, HN17ZZ01, and GX16YL01) exhibited the greatest sequence identity (94.6–97.3%) with ALV-J early-isolated local strains and belonged to group II (Figure 5). One isolate (1/18; GX14NN02) was closely related to ALV-J layer isolates, with a high identity of 96.8 to 99.6% and belonged to group III (Figure 5). These data confirmed that the U3 gene of most ALV-J local chicken isolates formed a unique branch.
Figure 5.
Phylogenetic analysis of the U3 region of ALV-J local chicken isolates. The 3 groups are marked. The red circles represent the 18 isolates in this study. The blue triangles represent ALV-J early-isolated local strains. The yellow diamond represents the ALV-J prototype strain HPRS-103. Abbreviation: ALV-J, Avian leukosis virus subgroup J.
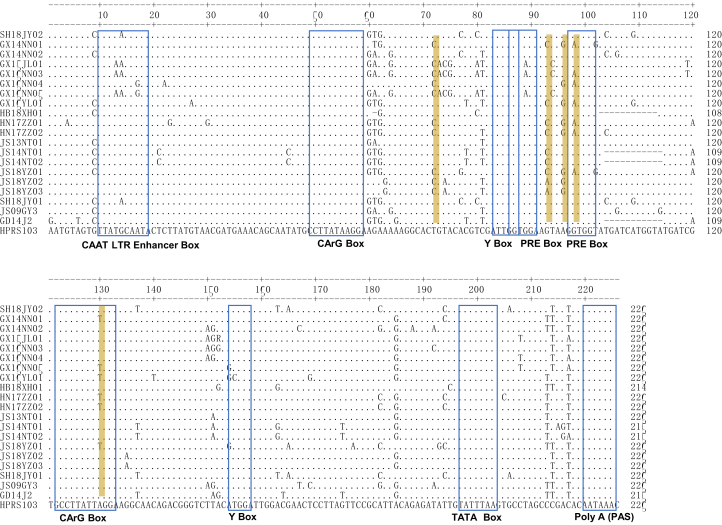
The U3 region contains many transcriptional regulatory elements, which are also prone to nucleotide substitutions and deletions (Ruddell, 1995). Our results showed that all transcriptional regulation elements were relatively conserved in the 18 isolates, including 2 CArG boxes, a TATA box, a C/EBP, 2 Y boxes, and 2 PRE motifs. However, compared with the U3 region of ALV-J broiler and layer isolates, 5 unique nucleotide substitutions (T72 C, G93 A/C, A96 G, G98 A, and A130 T) were observed in ALV-J local chicken isolates belonging to group II (Figure 6).
Figure 6.
Analysis of the nucleotide sequences of the U3 region of ALV-J local chicken isolates. The letters indicate nucleotide substitutions; the dots indicate identical nucleotides; the dashes indicate gaps produced in the alignment. The putative transcription regulatory elements are indicated by blue boxes. Yellow shading indicates unique nucleotide substitutions in the ALV-J local chicken isolates. Abbreviation: ALV-J, Avian leukosis virus subgroup J.
Discussion
Avian leukosis virus subgroup J was first isolated from meat chickens in China (Du et al., 2000) and has caused serious loss to the poultry industry, especially in terms of Chinese local chickens (Li et al., 2016; Zhang et al., 2018). There are several varieties of local chickens in China, which are widely distributed; these account for one-third of the domestic chicken industry (Qin, 2016). Thus, the high pathogenicity and diversity of ALV-J pose a great threat to the performance and breeding safety of local chickens in China (Meng et al., 2018; Zhang et al., 2018). To understand the molecular characteristics of ALV-J in local chicken, the full-length genome sequence of 18 isolates from local chickens in the main local chicken breeding provinces in China during the duration 2013–2018 were collected, sequenced, and analyzed. Our results show that the gp85 and U3 genes of most ALV-J local chicken isolates formed a separate branch. The 195–210 bp deletion in the rTM and the 16–28 bp deletion in the DR-1 of the 3′UTR were found in all 18 isolates. To the best of our knowledge, this is the first systematic study of the molecular characterization of ALV-J isolates from Chinese local chickens.
The gp85 protein forms globular structures on the surface of the virus and is closely associated with the process of viral binding and the determination of the host-specificity of each viral subgroup (Chai and Bates, 2006; Guan et al., 2017). The gp85 is the most variable of the envelope glycoproteins and exhibits high diversity in the ALV-J genome (Venugopal et al., 1998; Gao et al., 2012; Wang et al., 2017). In the present study, phylogenetic analysis demonstrated that the gp85 genes of the 18 isolates were distributed among various genetic backgrounds and belonged to 4 different branches, indicating the complexity and diversity of variation in local chickens. Based on the phylogenetic and sequence analyses of gp85 from the ALV-J local chicken isolates, it was hypothesized that the evolution of ALV-J in local chickens can be divided into 3 stages. First, it was determined that the gp85 genes of ALV-J isolates from local chickens before 2014 were closely related to the corresponding genes of ALV-J broiler isolates, belonging to the first branch. Most studies indicated that certain local chicken companies practiced crossbreeding with white meat-type breeders in the past (Cui et al., 2003; Shen et al., 2014), which facilitated horizontal transmission of ALV-J. Thus, it was hypothesized that these isolates may originate from early white meat-type breeders. Second, with the introduction of ALV-J into local chickens from 2015 to 2018, the diversity of Chinese local chickens and the differences in their growth performance provided a good environment for ALV-J variation (Meng et al., 2018). Thus, ALV-J local chicken strains continually evolved and formed a separate branch. Third, from 2018, new variations have formed in 3 isolates (SH18JY02, SH18JY01, and HB18XH01) discussed in the present study, with <90% sequence identity with all reference strains and belonging to a new branch. These findings will provide an enhanced understanding of the molecular epidemiology and evolution trend of ALV-J in Chinese local chickens.
The gp85 contains 5 sequence variability regions, namely hr1, hr2, vr1, vr2, and vr3, based on the ALV-A to ALV-E gp85 sequences (Dorner, 1985; Bova et al., 1988). However, the gp85 gene of ALV-J shares only 40% sequence identity with the corresponding gene in the other subgroups (Bai, 1995). In the present study, it was found that a few mutations in ALV-J gp85 occurred before the vr1 region and between the hr2 and vr3 regions; in contrast, the vr2 and vr3 regions of ALV-J gp85 were relatively conserved (Figure 3). The gp85 gene-variation characteristics of ALV-J are completely different from ALV-A to ALV-E. In addition, our previous studies have shown that most ALV-J gp85 sequence variations occurred primarily near the hr1 and hr2 regions (Gao et al., 2012). Therefore, it was concluded that the division of gp85 gene-variation regions in the other subgroups (ALV-A to ALV-E) may not be conducive to ALV-J and that the host range region and variant regions of the ALV-J gp85 gene should be redefined.
The 3′UTR contains effective regulatory sequences that influence the expression of the chromosome and viral genes. It is a vital region that controls viral replication and pathogenesis (Zavala et al., 2007). A 205-bp deletion in the rTM and DR-1 region of the 3′UTR was first observed in ALV-J layer chicken isolates in China in 2012 (Gao et al., 2012); a 205-bp deletion in the rTM and DR-1 region was subsequently observed in ALV-J broiler isolates (Ma et al., 2018). In the present study, all the 18 isolates displayed a 195–210 bp deletion in the rTM and a 16–28 bp deletion in the DR-1 region. Based on the above analysis, it was hypothesized that these deletions in the rTM and DR-1 regions of the 3′UTR may constitute unique molecular characteristics of ALV-J strains in China. In addition, our previous studies have demonstrated that ALV-J strains containing a 205 bp deletion in the 3′UTR showed higher pathogenicity and oncogenicity in SPF chickens (Wang et al., 2012a). Thus, the deletions in the rTM and DR-1 regions may be the result of natural evolution of ALV-J, which was influenced by genetic selection of the breeding stock and immune pressures (Zavala et al., 2007; Gao et al., 2012).
The U3 region contains the promoter and enhancer, which are closely associated with viral replication, transcription, and translation (Zachow and Conklin, 1992). In addition, the U3 region is prone to variation in ALV-J strains. However, few studies have systematically analyzed the ALV-J U3 gene in local chicken. Our results indicated that the U3 sequences of 61.1% (11/18) of the 18 isolates shared high identity (94.6–97.3%) with ALV-J early-isolated local strains, forming their own unique genetic branch. Sequence analysis showed 5 unique nucleotide mutations in the U3 region in most ALV-J local chicken isolates. One of 5 nucleotide mutations occurred in the second CArG box, an element that is essential for cell type-specific expression of oncogenes (Cui et al., 2014). Thus, future studies should investigate the possible effects of this mutation on cell type-specific expression of oncogenes.
The ALV-J host range has gradually expanded, and its pathogenicity has become more complex in China (Xu et al., 2004). Currently, ALV-J causes relatively serious damage in Chinese local chickens (Cheng et al., 2005; Zhang et al., 2018). The present study systematically analyzed the whole genome sequence of the 18 isolates from local chickens from 5 provinces in China from 2013 to 2018. The gp85 gene and U3 of ALV-J local chicken isolates showed obvious variation and differed from the corresponding entities in the ALV-J broiler isolates and ALV-J layer isolates, gradually forming a distinct branch. In addition, it was determined that the gene deletions in the rTM and DR-1 regions of the 3′UTR have proved to be a unique molecular characteristic of ALV-J in China. These findings will further expand on the epidemiological data of ALV-J in Chinese local chickens and contribute to a better understanding of the pathogenic mechanism of ALV-J in Chinese local chickens.
Acknowledgments
The present study was supported by the National Natural Science Foundation of China (31872482, 31761133002, and 31972671), the National Key Research and Development Program of China (No. 2016YFE0203200), and the China Agriculture Research System (CARS-41-G15).
Conflict of Interest Statement: The authors declare that they have no conflict of interest.
Contributor Information
Yanming Sun, Email: sym@shzu.edu.cn.
Yulong Gao, Email: gaoyulong@caas.cn.
References
- Bai J. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J. Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bova C.A., Olsen J.C., Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J. Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai N., Bates P. Na+/H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc. Nati. Acad. Sci. U. S. A. 2006;103:5531–5536. doi: 10.1073/pnas.0509785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.Q., Zhang L., Liu S.D., Zhang L.J., Cui Z.Z. Emerging of avian leukosis virus subgroup J in a flock of Chinese local breed. Acta Microbiol. Sinica. 2005;45:584–587. (In Chinese) [PubMed] [Google Scholar]
- Coffin J.M., Hughes S.H., Varmus H.E. Cold Spring Harbor Lab. Press; Plainview, NY: 1997. Retroviruses. [PubMed] [Google Scholar]
- Cui Z.Z., Du Y., Zhang Z., Silva R.F. Comparison of Chinese field strains of avian leukosis subgroup J viruses with prototype strain HPRS-103 and United States strains. Avian Dis. 2003;47:1321–1330. doi: 10.1637/6085. [DOI] [PubMed] [Google Scholar]
- Cui N., Su S., Chen Z.M., Zhao X.M., Cui Z.Z. Genomic sequence analysis and biological characteristics of a rescued clone of avian leukosis virus strain JS11C1, isolated from indigenous chickens. J. Gen. Virol. 2014;95:2512–2522. doi: 10.1099/vir.0.067264-0. [DOI] [PubMed] [Google Scholar]
- Dorner A.J. Molecular basis of host range variation in avian retroviruses. J. Virol. 1985;53:32–39. doi: 10.1128/jvi.53.1.32-39.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Cui Z.Z., Qin A., Silva R.F., Lee L.F. Isolation and partial sequence comparison of avian leukosis virus subgroup. J. Chin. J. Virol. 2000;16:341–346. (In Chinese) [Google Scholar]
- Gao Y.L., Qin L.T., Pan W., Wang Y.Q., Qi X.L., Gao H.L., Wang X.M. Avian leukosis virus subgroup J in layer chickens, China. Emerg. Infect. Dis. 2010;16:1637–1638. doi: 10.3201/eid1610.100780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.L., Yun B.L., Qin L.T., Pan W., Qu Y., Liu Z.S., Wang Y.Q., Qi X.L., Gao H.L., Wang X.M. Molecular epidemiology of avian leukosis virus subgroup J in layer flocks in China. J. Clin. Microbiol. 2012;50:953–960. doi: 10.1128/JCM.06179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Yun B.L., Wang Q., Jiang L.L., Zhu H.B., Gao Y.N., Qin L.T., Wang Y.Q., Qi X.L., Gao H.L., Wang X.M., Gao Y.L. Development and application of a multiplex PCR method for rapid differential detection of subgroup A, B, and J avian leukosis viruses. J. Clin. Microbiol. 2014;52:37–44. doi: 10.1128/JCM.02200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X.L., Zhang Y., Yu M.M., Ren C.Q., Gao Y.N., Yun B.L., Liu Y.Z., Wang Y.Q., Qi X.L., Liu C.J., Cui H.Y., Zhang Y.P., Gao L., Li K., Pan Q., Zhang B.S., Wang X.M., Gao Y.L. Residues 28 to 39 of the extracellular loop 1 of chicken Na+/H+ exchanger type I mediate cell binding and entry of subgroup J avian leukosis virus. J. Virol. 2017;92 doi: 10.1128/JVI.01627-17. e01627-01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirong B., Payne L.N., Skinner M.A. Sequence analysis of an infectious proviral clone of HPRS-103 shows that it represents a new avian retrovirus envelope subgroup (J) Avian Pathol. 1998;27:S92–S93. [Google Scholar]
- Lai H.Z., Zhang H.N., Ning Z.Y., Chen R.A., Zhang W.Y., Qing A.J., Xin C.A., Yu K.Z., Cao W.S., Liao M. Isolation and characterization of emerging subgroup J avian leukosis virus associated with hemangioma in egg-type chickens. Vet. Microbiol. 2011;151:275–283. doi: 10.1016/j.vetmic.2011.03.037. [DOI] [PubMed] [Google Scholar]
- Li X.J., Lin W.C., Chang S., Zhao P., Zhang X.H., Liu Y., Chen W.G., Li B.H., Shu D.M., Zhang H.M., Chen F., Xie Q.M. Isolation, identification and evolution analysis of a novel subgroup of avian leukosis virus isolated from a local Chinese yellow broiler in south China. Arch. Virol. 2016;161:2717–2725. doi: 10.1007/s00705-016-2965-x. [DOI] [PubMed] [Google Scholar]
- Li J.L., Meng F.F., Zhang W.H., Wang Y.X., Chang S., Zhao P., Cui Z.Z. Characterization of avian leukosis virus subgroup J isolated between 1999 and 2013 in China. Poult. Sci. 2018;10:3532–3539. doi: 10.3382/ps/pey241. [DOI] [PubMed] [Google Scholar]
- Ma M.G., Yu M.M., Xu C.T., Huang Q.H., Meng Z.J., Wang S.Y., Xing L.X., Chang F.F., Li Y., He X.J., Liu C.J., Qi X.L., Wang Y.Q., Sun Y.M., Wang X.M., Gao Y.L. Full-length genome sequence analysis of subgroup J avian leukosis viruses isolated from imported white feather broiler breeders. Chin J. Prevent Vet. Med. 2018;41:750–754. (In Chinese) [Google Scholar]
- Maas R., Zoelen D.V., Oei H., Claassen I. Replacement of primary chicken embryonic fibroblasts (CEF) by the DF-1 cell line for detection of avian leucosis viruses. Biologicals. 2006;34:177–181. doi: 10.1016/j.biologicals.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Meng F.F., Li Q.C., Zhang Y.W., Zhang Z.H., Tian S.B., Cui Z.Z., Chang S., Zhao P. Characterization of subgroup J avian Leukosis virus isolated from Chinese indigenous chickens. Virol. J. 2018;15:33–40. doi: 10.1186/s12985-018-0947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne L.N., Howes K., Gillespie A.M., Smith L.M. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated. J. Gen. Virol. 1992;73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- Payne L.N., Gillespie A.M., Howes K. Myeloid leukaemogenicity and transmission of the HPRS-103 strain of avian leukosis virus. Leukemia. 1992;6:1167–1176. [PubMed] [Google Scholar]
- Payne L.N., Nair V. The long view: 40 years of avian leukosis research. Avian Pathol. 2012;41:11–19. doi: 10.1080/03079457.2011.646237. [DOI] [PubMed] [Google Scholar]
- Payne L.N., Brown S.R., Bumstead N., Howes K., Thouless M.E. A novel subgroup of exogenous avian leukosis virus in chickens. J. Gen. Virol. 1991;72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- Qin K.T. Current situation and development trend of high-quality local chicken in China. Guide to Chin. Poult. 2016;18:17–18. (In Chinese) [Google Scholar]
- Ruddell A. Transcription regulatory elements of the avian retroviral long terminal repeat. Virology. 1995;206:1–7. doi: 10.1016/s0042-6822(95)80013-1. [DOI] [PubMed] [Google Scholar]
- Shen Y.W., Cai L.M., Wang Y.M., Wei R.R., He M.L., Wang S.H., Wang G.H., Cheng Z.Q. Genetic mutations of avian leukosis virus subgroup J strains extended their host range. J. Gen. Virol. 2014;95:691–699. doi: 10.1099/vir.0.059915-0. [DOI] [PubMed] [Google Scholar]
- Sun S.H., Cui Z.Z. Epidemiological and pathological studies of subgroup J avian leukosis virus infections in Chinese local “yellow” chickens. Avian Pathol. 2007;36:221–226. doi: 10.1080/03079450701332345. [DOI] [PubMed] [Google Scholar]
- Tsichlis P.N., Donehower L., Hager G., Zeller N., Malavarca R., Astrin S., Skalka A.M. Sequence comparison in the crossover region of an oncogenic avian retrovirus recombinant and its nononcogenic parent: genetic regions that control growth rate and oncogenic potential. Mol. Cell. Biol. 1982;2:1331–1338. doi: 10.1128/mcb.2.11.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venugopal K., Howes K., Flannery D.M.J., Payne L.N. Isolation of acutely transforming subgroup J avian leukosis viruses that induce erythroblastosis and myelocytomatosis. Avian Pathol. 2000;29:497–503. doi: 10.1080/030794500750047252. [DOI] [PubMed] [Google Scholar]
- Venugopal K., Smith L.M., Howes K., Payne L.N. Antigenic variants of J subgroup avian leukosis virus: sequence analysis reveals multiple changes in the env gene. J. Gen. Virol. 1998;79:757–766. doi: 10.1099/0022-1317-79-4-757. [DOI] [PubMed] [Google Scholar]
- Wang Q., Gao Y.L., Wang Y.Q., Qin L.T., Qi X.L., Qu Y., Gao H.L., Wang X.M. A 205-nucleotide deletion in the 3' Untranslated region of avian leukosis virus subgroup J, currently emergent in China, contributes to its pathogenicity. J. Virol. 2012;86:12849–12860. doi: 10.1128/JVI.01113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Zhao P., Cui Z.Z. Identification of a new subtype of avian leukosis virus isolated from local chicken breeds in China. Chin. J. Virol. 2012;06:609–614. [PubMed] [Google Scholar]
- Wang P.K., Lin L.L., Li H.J., Yang Y.L., Huang T., Wei P. Diversity and evolution analysis of glycoprotein GP85 from avian leukosis virus subgroup J isolates from chickens of different genetic backgrounds during 1989-2016: Coexistence of five extremely different clusters. Arch. Virol. 2017;163:377–389. doi: 10.1007/s00705-017-3601-0. [DOI] [PubMed] [Google Scholar]
- Weiss R.A. Cellular receptors and viral glycoproteins involved in retrovirus entry. In: Levy J.A., editor. The Retroviruses. Plenum Press; New York, NY: 1992. pp. 1–108. [Google Scholar]
- Xu B.R., Dong W.X., Yu C.M., He Z.Q., Lv Y.L., Sun Y.Z., Feng X.Y., Li N., Lee L.F., Li M.X. Occurrence of avian leukosis virus subgroup J in commercial layer flocks in China. Avian Pathol. 2004;33:13–17. doi: 10.1080/03079450310001636237a. [DOI] [PubMed] [Google Scholar]
- Yun B.L., Li D.L., Zhu H.B., Liu W., Qin L.T., Liu Z.S., Wu G., Wang Y.Q., Qi X.L., Gao H.L., Wang X.M., Gao Y.L. Development of an antigen-capture ELISA for the detection of avian leukosis virus p27 antigen. J. Virol. Met. 2013;187:278–283. doi: 10.1016/j.jviromet.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Zachow K.R., Conklin K.F. CArG, CCAAT, and CCAAT-like protein binding sites in avian retrovirus long terminal repeat enhancers. J. Virol. 1992;66:1959–1970. doi: 10.1128/jvi.66.4.1959-1970.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavala G., Cheng S., Jackwood M.W. Molecular epidemiology of avian leukosis virus subgroup J and evolutionary history of its 3′ untranslated region. Avian Dis. 2007;51:942–953. doi: 10.1637/0005-2086(2007)51[942:MEOALV]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Guan X.L., Chen Z.W., Cao D.G., Kang Z.F., Shen Q.C., Lei Q.X., Li F.W., Li H.Q., Muhammad F.L., Wang Y.Q., Qi X.L., Wang X.M., Gao Y.L. The high conserved cellular receptors of avian leukosis virus subgroup J in Chinese local chickens contributes to its wide host range. Poul. Sci. 2018;97:4187–4192. doi: 10.3382/ps/pey331. [DOI] [PubMed] [Google Scholar]
- Zhang H.N., Lai H.Z., Qi Y., Zhang X.T., Ning Z.Y., Luo K.J., Xin C.A., Cao W.S., Liao M. An ALV-J isolate is responsible for spontaneous haemangiomas in layer chickens in China. Avian Pathol. 2011;40:261–267. doi: 10.1080/03079457.2011.560142. [DOI] [PubMed] [Google Scholar]
- Zhao Z.J., Rao M.Z., Liao M., Cao W.S. Phylogenetic analysis and pathogenicity assessment of the emerging recombinant subgroup K of avian leukosis virus in south China. Viruses. 2018;10:194–199. doi: 10.3390/v10040194. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete proviral genome sequences of all 18 isolates were deposited at GenBank with accession numbers MN735292 to MN735309.