Abstract
This study investigated the factors that caused the differences in egg production during the development of ovarian follicles in Leizhou black ducks. Leizhou black ducks population was divided into 2 groups as high-yield group (HG) and low-yield group (LG). The number of eggs (NE), age at first egg (AFE), weight at first egg, and egg weight (EW) of both groups were recorded, and differences were analyzed using the t test. The logistic model was used to simulate the egg production curves to analyze the production rules. The ovarian follicles of both duck groups were collected to count the number of different grades sized follicles, weigh the ovaries, and observe follicular sections to analyze the developmental differences. Ovarian transcriptomic sequencing was performed to investigate differentially expressed genes and signal pathways in both duck groups. The results revealed a significant difference (P < 0.01) in the NE laid, AFE, and EW between both groups. Comparatively, HG had significantly more (P < 0.01) large yellow follicles (LYF) than LG. The density of medullary layer cells of the follicle section was greater in HG than LG ducks. Transcriptome sequencing revealed a total of 1,027 differentially expressed genes between the HG and LG ducks of which 495 genes were upregulated, and 532 genes were downregulated. Fifty genes were related to reproduction and reproductive processes. Kyoto Encyclopedia of Genes and Genomes–enriched signaling pathways revealed 274 signal pathways enriched in these differentially expressed genes of which the steroid biosynthesis pathway was significantly enriched. Analysis (Q < 0.05) showed that HSD3β → gonadotropin-releasing hormone (GnRH) and estrogen receptor (ESR) → LHβ/ERK1/2 were enriched in the steroid biosynthesis signal pathway. Follicle-stimulating hormone signal pathway mediated by HSD3β → GnRH and ESR → LHβ/ERK1/2 may be involved in ovarian follicle development to regulate LYF reserve process and affect its ovulation cycle, which in turn influence the egg production of Leizhou black ducks.
Key words: laying traits, Leizhou black duck, ovarian follicles, transcriptome sequencing
Introduction
The ovary, the female reproductive organ, has in recent times gained much attention in research as it produces and releases eggs. It also functions as an endocrine gland for female hormones production and discharge. Therefore, duck breeders in recent times have aimed at the ovary to explore and examine egg production and differentially expressed genes (DEG) that regulate egg production (Zhu et al., 2017; Asiamah Amponsah et al., 2019). The functional unit of the ovary is the ovarian follicles which contain oocytes that may be ovulated, fertilized to form an embryo that has been used as a study material to enhance reproduction in poultry (Wu et al., 2016a; Han et al., 2016; Gan et al., 2017; Zhu et al., 2017; Cui et al., 2019; Shen et al., 2019). The development process of poultry ovarian follicles undergoes a series of complex processes which include recruitment, selection, dominance, growth, and maturation before ovulation (Regan et al., 2018a). Studies have shown that the process from poultry follicle collection to ovulation is in a strict hierarchical order, and the follicular reserve at each stage may affect the ovulation cycle and ultimately affect egg production (Johnson, 2015; Regan et al., 2018b; Ghanem and Johnson, 2018). Ovarian follicular development and ovulation are regulated by hypothalamic (gonadotropin-releasing hormone, GnRH) and pituitary (follicle-stimulating hormone, FSH and luteinizing hormone, LH) hormones (Shimizu, 2016).
The development of genomic and transcriptomic studies of the ovaries has aided to expound mechanisms underlying reproductive physiology and to identify DEG in the ovary. Several studies have been done to identify specific ovarian genes in livestock such as cattle, sheep, goats, yaks, and pigs (Lan et al., 2014, 2016; Chen et al., 2015; Zhang et al., 2015; Zhao et al., 2015; Miao et al., 2016) and poultry (Luan et al., 2014; Ding et al., 2015; Tao et al., 2017; Zhu et al., 2017). In Jinding ducks ovary, 5 of the DEG, melanocortin 5 receptor, apolipoprotein D, ORAI calcium release-activated calcium modulator 1, dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4, and G protein–regulated inducer of neurite outgrowth 2 were confirmed to significantly play roles in reproductive activities and egg production (Tao et al., 2017). Again, in Shan Ma ducks, DEG identified in the ovary were involved in the GnRH signaling, insulin signaling, and transforming growth factor-beta (TGF-β) signaling, which collectively is important for physiologic and reproductive activities (Zhu et al., 2017).
Therefore, this study analyzed the follicular development of Leizhou black ducks and ovarian transcriptome sequencing to explore the regulatory and molecular mechanism affecting egg production traits in Leizhou black ducks and provide a theoretical basis for molecular breeding of Leizhou black ducks.
Materials and methods
Test Materials
The Leizhou black ducks used in this experiment were obtained from Zhanjiang Hengcheng Breeding Cooperative and given the same feeding conditions and nutritional levels. One hundred thirty female ducks of the same generation, size, and health status were randomly selected in a single cage. Using the week as the unit, the number of eggs (NE) produced was counted from 16 wk to 43 wk to group the ducks in high and low production groups. The average number of eggs of each Leizhou black duck is 122.2. Those which produced eggs >150 were defined as high-yield group (HG), whereas those with <90 eggs were defined as low-yield group (LG). The number of eggs (NE), age at first egg (AFE), weight at first egg (WFE), and egg weight (EW) of both groups were recorded. The ovarian tissues of 4 each of HG and LG at 43 wk were collected, weighed, sectioned, and follicular differences were analyzed. Three HG and 2 LG ovarian medulla layers were collected to construct gene pools for transcriptome sequencing analysis.
All the animals were maintained and studied following the National Institute of Health guidelines for care and use of laboratory animals, and all protocols were approved in advance by the Animal Care Committee of Guangdong Ocean University of China (No. NXY20160172).
Test Method
Analysis of Laying Rules
The NE, AFE, WFE, and EW were recorded for HG and LG ducks, and the differences were analyzed by the t test. The logistic model was used to simulate egg production curves of HG and LG to analyze the egg production characteristics of the 2 groups with the formula: y = A2 + (A1-A2)/(1+ (x/x0) ˆ p) (in the equation, the egg production period 16–43 wk is corresponding to 1–28 wk).
Analysis of Ovarian Follicles
The ovarian parenchyma masses were separated from the follicles and compared between the 2 duck groups, and the ovarian cell development was compared using a microscope. According to the follicle diameter, the follicles of different grades were divided, the developmental differences of both duck follicles were analyzed, and the egg production characteristics of Leizhou black duck were inferred based on the egg production curve characteristics. Follicle grade classification criteria were as follows: small white follicle (SWF): <4 mm, large white follicle (LWF): 4.49 ± 0.43 mm, small yellow follicle (SYF): 5.81 ± 0.37 mm, large yellow follicle (LYF): 6.94 ± 0.29 mm, F6: 10.63 ± 0.88 mm, F5: 14.11 ± 0.61 mm, F4: 17.33 ± 1.13 mm, F3: 24.31 ± 1.24 mm, F2: 30.70 ± 1.42 mm, and F1: 34.34 ± 0.44 mm. The ovarian follicles were rinsed in normal saline, quickly placed in 10% formaldehyde fixative, and stored at room temperature for 48 h. After fixation, washing, dehydration, clearing, wax dipping and embedding, sectioning, spreading, sticking, and sealing after drying and staining were performed, ovarian follicle sections were observed with an optical microscope.
Transcriptome Sequencing
Sequencing Methods and Analysis Methods
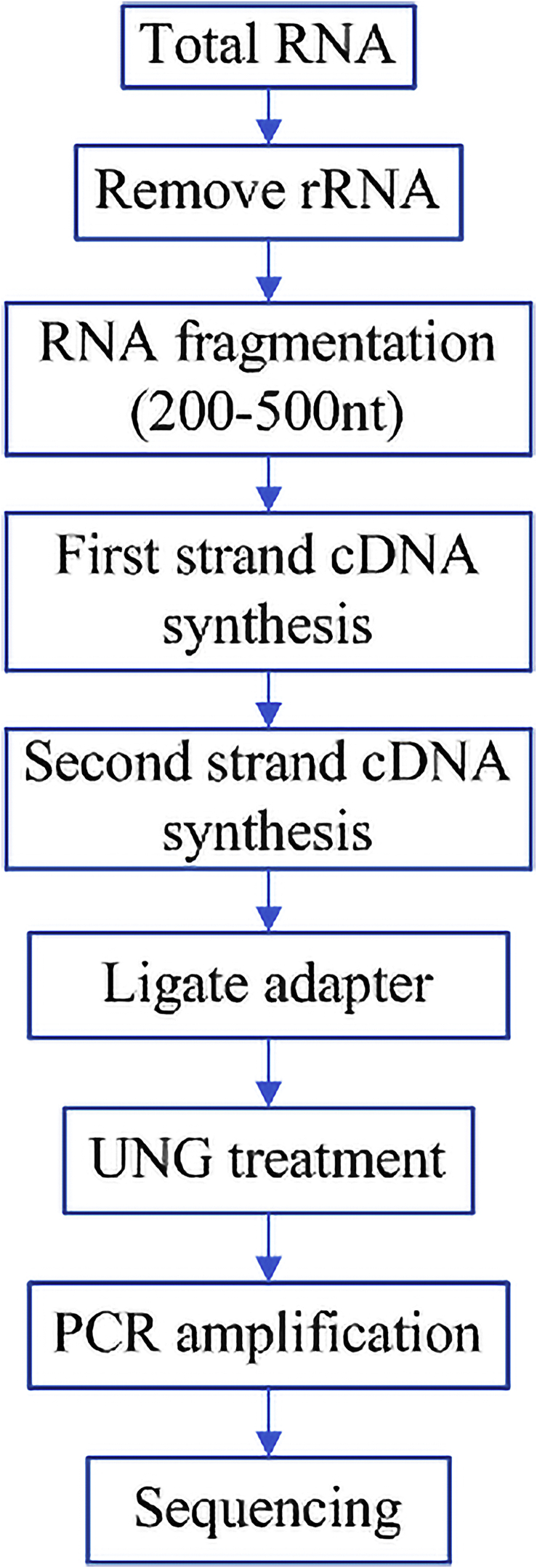
The medullary layer of the ovarian tissues of both HG and LG ducks were collected and stored in liquid nitrogen and dry ice and transported to Guangzhou Gidio Gene Denovo for transcriptome sequencing. The sequencing platform used Illumina HiSeq2500, and the Fold Change of the difference analysis was P < 0.05. The experimental process and database construction method are shown in Figures 1 and 2, and the subsequent analysis was performed according to (Li et al., 2017).
Figure 1.
Processes of sequencing.
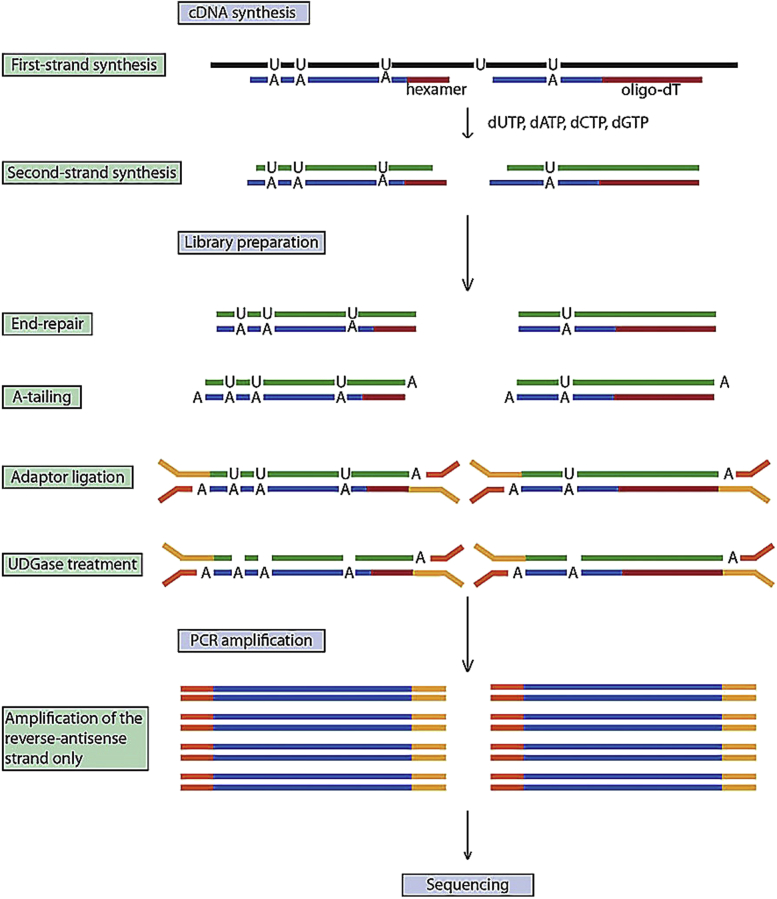
Figure 2.
Database construction principle of sequencing.
Gene Ontology Classification and Kyoto Encyclopedia of Genes and Genomes Pathway Analysis of the Differentially Expressed Genes
For gene ontology (GO) database, Blast2GO was used to determine and compare the functions of the DEG (Conesa et al., 2005; Götz et al., 2008). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were assigned to classify unigenes using the online KEGG Automatic Annotation Server (http://www.genome.jp/kegg/kaas/) (Kanehisa et al., 2008). In all tests, P-values were calculated using the Benjamini-corrected modified Fisher's exact test, and significant differences were set at P-values < 0.05.
Real-Time Quantitative Polymerase Chain Reaction Analysis
To validate the transcriptome sequencing results, 8 DEG were selected at random and detected by real-time quantitative PCR. NCBI-Primer Blast tool was employed to design the quantitative primers according to the gene accession number. The primer sequences are presented in Table 1. Quantitative β-actin was used as the internal reference gene. The reaction system was 10 μL ChamQTM SYBR qPCR Master Mix (1 × ), 0.4 μL PCR Forward Primer (0.4 μM), 0.4 μL PCR Reverse Primer (0.4 μM), 0.4 μL ROX Reference Dye (1 × ), 2.0 μL cDNA, 6.8 μL ddH2O, total volume 20 μL. Reaction program: 94°C, 35 s; (94°C, 20 s; 57°C, 35 s; lighting; 72°C, 25 s) 40 cycles; 95°C, 15 s; 60°C, 1 min; 95°C, 15 s.
Table 1.
Nucleotide sequences of primers used for qRT-PCR amplification.
| Gene | Sequence (5′-3′) | Product length (bp) | Annealing temperature (°C) | Accession ID |
|---|---|---|---|---|
| DIAPH2 | F:AACAGCTACGAACACAGGCT | 207 | 59.7 | XM_027465243.1 |
| R:TAAGCGGCAGCGAAGGTAG | ||||
| KMT2E | F:AGTGAAACGTGCAGGGTCAT | 170 | 59.9 | XM_027463760.1 |
| R:CAGCCGCTCATCCAAAAAGG | ||||
| KSR1 | F:ACTGAGTCAGGTGGGGAGTT | 217 | 59.5 | XP_015736661.1 |
| R:AGGTCCATGACTGGAGGAGA | ||||
| ESR2 | F:CAGTGCTACCTGTGACCAGA | 168 | 60.0 | XM_021274553.2 |
| R:TGCAGCCTTCACATGACCAG | ||||
| ASCF2 | F:TATGGTGGACTCCAAGCTGC | 215 | 59.5 | XP_012951750.1 |
| R:TTTCGCCAATGATGTTGCCA | ||||
| A2M | F:ACAGTTAGCCTGGAAGAGCG | 176 | 59.5 | XM_027449874.1 |
| R:TCCTGAATAGCAATCAAGGGGT | ||||
| BMPR1B | F:TTAGAGGGCTCGGACTTCCA | 128 | 59.9 | XM_013104107.3 |
| R:CGATTTTTCAGCGGAGGCAG | ||||
| ACVR1 | F:GTCAGGCTTGCTGGCCC | 162 | 60.3 | XM_005027629.4 |
| R:AACGATCTCCGTTCCCACAG | ||||
| β-actin | F:CGCAAATGCTTCTAAACC | 167 | 55.9 | NM_001310421.1 |
| R:AGACTGCTGCTGATACCTT |
Western Blot Analysis
To further screen the candidate genes related to reproduction, DEG related to reproduction, estrogen receptor 2 (ESR2) and bone morphogenetic protein receptor beta (BMPR1B) were selected to perform Western blot tests. The primary antibodies were from mouse ESR2 and BMPR1B, and the secondary antibodies were from mouse IgG. For specific methods of blotting, refer to (Riaz et al., 2019).
Results
Analysis of Laying Rules
The egg production performance of 13 LG and 17 HG ducks showed that the average NE, AFE, WFE, and EW were respectively 79, 137.6154, 1330.1, and 56.38462 for LG and 154.9231, 103, 1308.354, and 47.43846 for HG. The NE, AFE, and EW of HG ducks were significantly different (P < 0.01) from those of LG ducks (Table 2).
Table 2.
Egg production performance between HG and LG.
| Ducks | NE |
AFE |
WFE |
EW |
||||
|---|---|---|---|---|---|---|---|---|
| LG | HG | LG | HG | LG | HG | LG | HG | |
| 1 | 62 | 150 | 151 | 106 | 1275.4 | 1406.8 | 55 | 55.6 |
| 2 | 71 | 150 | 132 | 105 | 1444.8 | 1236.6 | 43.4 | 46.4 |
| 3 | 71 | 152 | 125 | 105 | 1275.4 | 1259.2 | 57.7 | 38.5 |
| 4 | 72 | 153 | 123 | 105 | 1244.2 | 1506.2 | 46 | 48.1 |
| 5 | 72 | 153 | 143 | 101 | 1407.6 | 1398.2 | 61.5 | 63.5 |
| 6 | 77 | 154 | 116 | 101 | 1241.8 | 1157.6 | 63.6 | 37 |
| 7 | 83 | 154 | 146 | 100 | 1316.4 | 1558.4 | 59.3 | 47.4 |
| 8 | 84 | 154 | 143 | 98 | 1242.8 | 1201.2 | 53.5 | 30.5 |
| 9 | 85 | 155 | 146 | 102 | 1500.2 | 1191.4 | 62.8 | 49 |
| 10 | 86 | 156 | 144 | 106 | 1217.2 | 1071.8 | 52.4 | 52.8 |
| 11 | 87 | 157 | 138 | 106 | 1495.4 | 1230.2 | 59.9 | 49.9 |
| 12 | 88 | 162 | 144 | 103 | 1207.9 | 1421.2 | 53.2 | 46.2 |
| 13 | 89 | 164 | 138 | 101 | 1422.2 | 1369.8 | 64.7 | 51.8 |
| 14 | 164 | 103 | 1352.2 | 45.6 | ||||
| 15 | 164 | 104 | 1008.8 | 44.9 | ||||
| 16 | 166 | 103 | 1339.6 | 61.6 | ||||
| 17 | 168 | 97 | 1370.2 | 63.2 | ||||
| Average | 79 | 154.9231 | 137.6154 | 103 | 1330.1 | 1308.354 | 56.38462 | 47.43846 |
| P-value | 1.13839E-22∗∗ | 1.65474E-13∗∗ | 0.525363398 | 0.017840088∗∗ | ||||
∗∗ indicate difference is significant (P < 0.01), P-value for LG vs. HG.
Abbreviations: AFE, age at first egg; EW, egg weight; HG, high-yield group; LG, low-yield group; NE, number of eggs; WFE, weight at first egg.
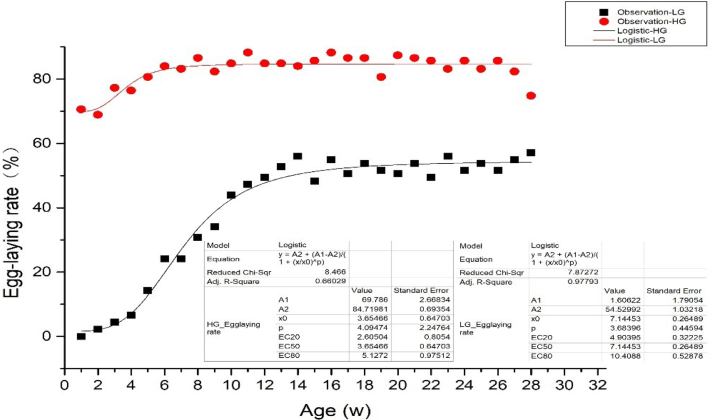
As shown in Figure 3, the HG and LG logistic model expressions are y = 69.8/(1+ (x/3.6) ˆ 4.1) (R2 = 0.66) and y = 1.6/(1+ (x/7.1) ˆ 3.7) (R2 = 0.98), respectively. The results showed that the growth period, stable period (high yield period), and inflection point of HG ducks were 16 to 20 wk, 21 to 42 wk, and 20 wk, respectively, with a slight downward trend at 43 wk. In LG ducks, the growth period, high yield period, and the inflection point were 17 to 28 wk, 29 to 43 wk, and 28 wk, respectively (Figure 3). Comparatively, the egg production rate of HG from 16 to 43 wk was higher (P < 0.01) with a shorter growth period than LG. The duration of the high production of HG (22 wk) was longer than that of LG (15 wk).
Figure 3.
The logistic model of the egg-laying curve between HG and LG. Abbreviations: HG, high-yield group; LG, low-yield group.
Differential Analysis of Ovarian Follicles
Comparison of Parenchymal Quality and Quantity of HG and LG Ovaries
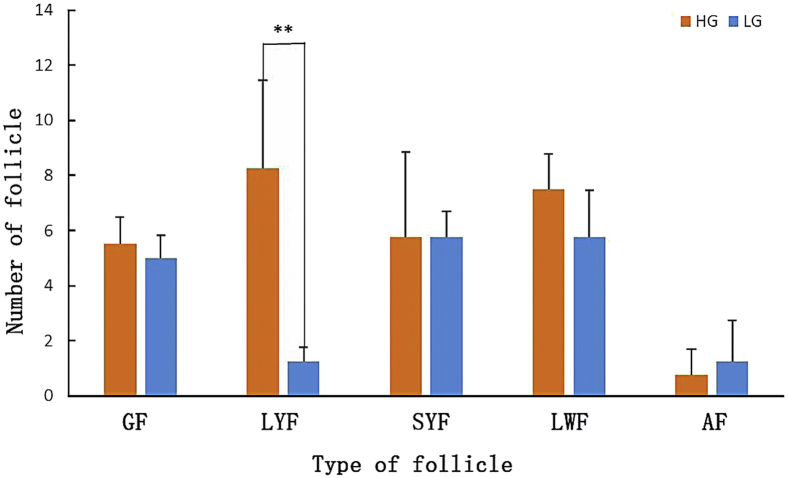
Comparing the ovarian parenchymal mass, the average mass of HG ovarian parenchyma was higher than that of LG (Figure 4), but the difference was not significant (P > 0.05). The difference in ovarian parenchymal mass between the 2 groups indicates that the stability of LG ovaries during development may be lower than that of HG. Based on the anatomy of HG and LG ovaries and the statistics of the number of follicles in different grades (Figure 5), the results showed that both HG and LG have grade follicles, LYF, SYF, LWF, and SWF, atretic follicles, and follicular atresia occurred in grade follicles (F5-F4). The number of LYF in HG was significantly higher (P < 0.01) than LG. The results of the anatomical map (Figure 6) showed that among the reserves of LYF, the selected LYF (see arrow in Figure 6) has the advantage of preferential development. Its size is slightly larger than LYF but smaller than F6, and its yolk color is significantly different from other LYF, and LYF selection also occurred before F1 ovulation. Combining the characteristics of the egg production curve, it is speculated that the reserve of LYF may be an important factor that causes the number of eggs of Leizhou black ducks to vary.
Figure 4.
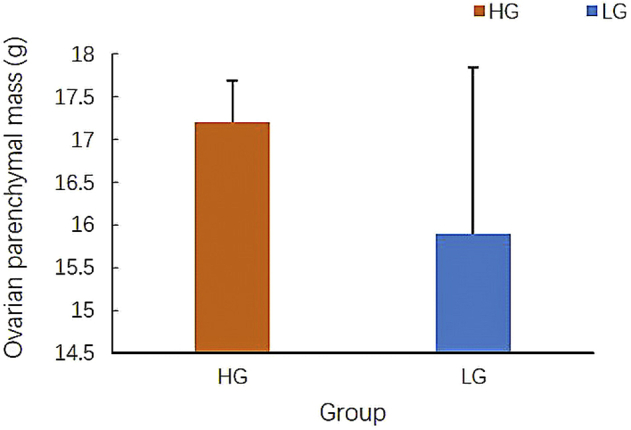
Ovarian parenchymal mass. Abbreviations: HG, high-yield group; LG, low-yield group.
Figure 5.
The number of different follicles between HG and LG. Note: ∗∗ indicates the difference is significant (P < 0.01). Abbreviations: AF, atresia follicles; GF, grade follicles; HG, high-yield group; LG, low-yield group; LWF, large white follicles; LYF, large yellow follicles; SYF, small yellow follicles.
Figure 6.
Ovarian follicle anatomy of HG and LG. The red arrow shows selected LYF from the LYF reserves. Abbreviations: F, follicles; H1-H4, high yield ducks 1-4; HG, high-yield group; L1-L4, low yield ducks 1-4; LG, low-yield group; LWF, large white follicles; LYF, large yellow follicles; SWF, small white follicles; SYF, small yellow follicles.
Comparison of Ovarian Follicular Sections between HG and LG
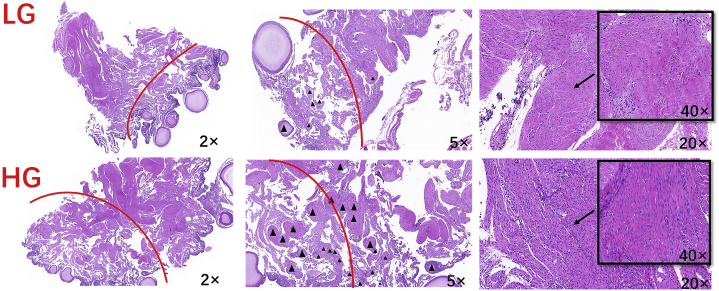
In this study, HG and LG ovary sections were observed (Figure 7). It can be seen that the density of medulla cells in the HG medullary layer unit area of the ovary cortex and medulla is greater than that of LG. The number of secondary follicles in HG was significantly higher than that in LG.
Figure 7.
Section map of HG and LG ovarian follicles. Note: Triangle shape denotes secondary follicles. The red line is an approximate junction, below it is the cortex region, and above is the medullar region. Abbreviations: HG, high-yield group; LG, low-yield group.
Ovarian Transcriptome Sequencing Analysis
Sequencing Data Statistics
A total of 5 ovarian tissue of HG and LG transcriptome libraries were constructed (Table 3). To ensure the quality of the data, quality control and filtering were performed on the data, and high-quality clean reads were obtained by removing connector-containing reads and low-quality reads. The Q20 and Q30 were both greater than 94%, and the GC content was greater than 44%. Finally, 50 Gb of Clean Data was obtained, each sample was about 10 Gb, and the comparison rate between each sample and the reference genome (Anas platyrhynchos) exceeded 81% (Table 3). The sequence saturation of the 5 samples was above 90%, and their random distributions were consistent with the normal distribution. The analysis of the above sequencing data shows that the quality of the data is reliable and can be analyzed in the next step.
Table 3.
Sequencing data reads and comparison rate statistics.
| Sample | Total reads | Unmapped reads | Unique mapped reads | Multiple mapped reads | Mapping ratio (%) |
|---|---|---|---|---|---|
| L1 | 972,500,64 | 18087600 (18.60%) | 78531126 (80.75%) | 631338 (0.65%) | 81.40 |
| L2 | 972,547,76 | 17760970 (18.26%) | 78891260 (81.12%) | 602546 (0.62%) | 81.74 |
| H1 | 956,274,54 | 17418028 (18.21%) | 77627662 (81.18%) | 581764 (0.61%) | 81.79 |
| H2 | 912,164,14 | 16110032 (17.66%) | 74573840 (81.75%) | 532542 (0.58%) | 82.34 |
| H3 | 106,386,128 | 19502696 (18.33%) | 86149258 (80.98%) | 734174 (0.69%) | 81.67 |
Differential Gene Screening
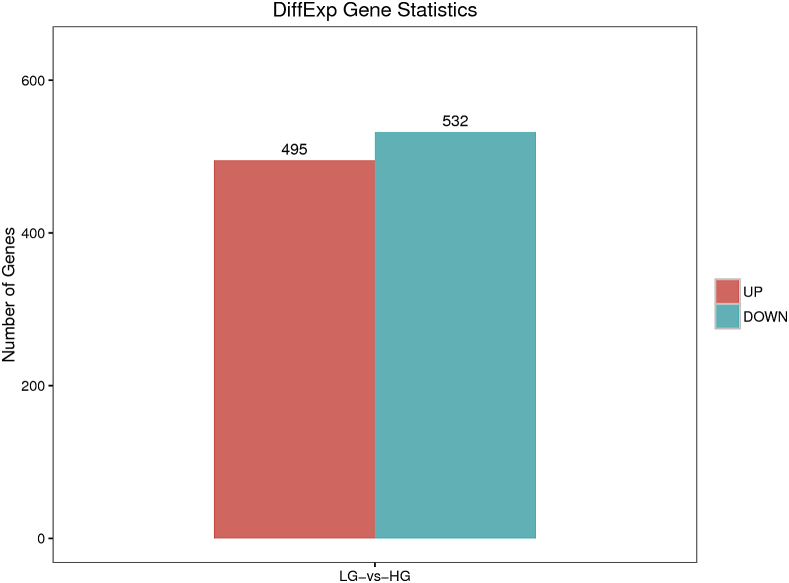
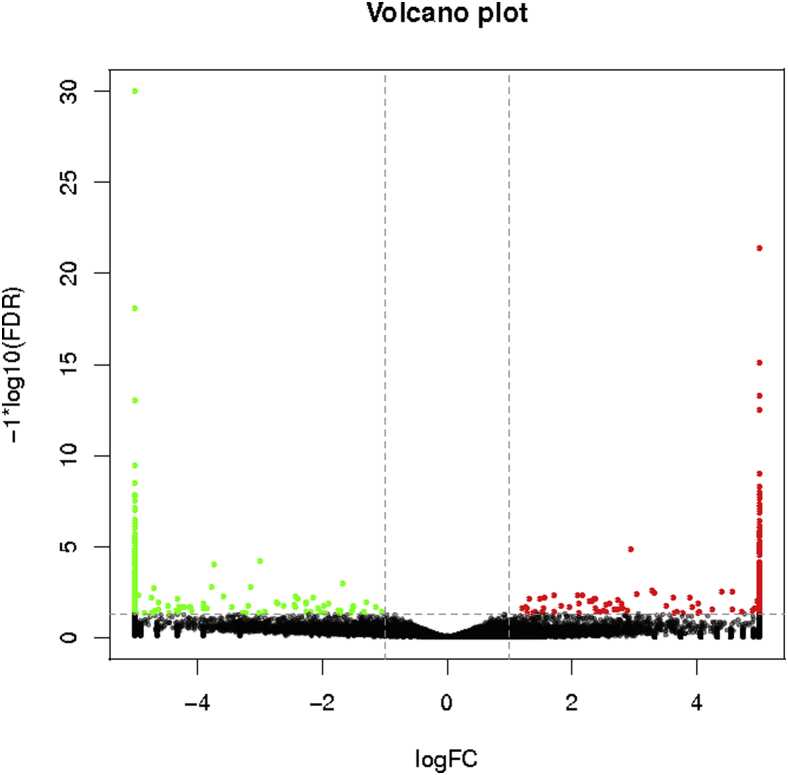
Through statistical analysis of the expression results of all mRNA transcripts sequenced from 5 ovarian tissue samples from the 2 duck groups (Figures 8 and 9), 39,904 transcripts were expressed, of which 31,937 (80.03%) were known transcripts and 9,710 new transcripts. In HG ducks, 30,236 (75.77%) transcripts were known with 9,143 new transcripts, and in LG ducks, 28,711 (71.95%) were known transcripts with 8,615 new transcripts. A total of 1,027 genes were differentially expressed between the HG and LG groups, out of which 532 genes were downregulated and 495 genes were upregulated (Figure 8). Among the 1,027 DEG, 155 were related to duck development, and 25 related to reproductive performance. The DEG related to reproduction included prolactin receptor (PRLR), ESR2, BMPR1B, RNA helicase Mov10l1, septin-4 isoform X4, activin receptor type-1 isoform X1, disks large homolog 1 isoform X15, nucleoside diphosphate kinase homolog 5 isoform X1, ovomucoid (LOC101798642), ovostatin (LOC101801176), ovalbumin-related protein Y-like (LOC101802317), and others (Table 4).
Figure 8.
Statistics of differentially expressed genes.
Figure 9.
Volcano map of differentially expressed gene.
Table 4.
Detailed information on 18 upregulated and downregulated differentially expressed genes.
| Gene ID | Gene name | Log2 fold change | P-value | Up/Down (LG/HG) | Gene description |
|---|---|---|---|---|---|
| XM_005020857.4 | MOV10L1 | 10.52160044 | 2.42024E-05 | Up | RNA helicase Mov10l1 |
| XM_005025066.4 | KITLG | 10.16323035 | 0.000658319 | Up | Kit ligand isoform X3 |
| XM_027446002.1 | SEPTIN4 | 9.076815597 | 0.028012697 | Up | Septin-4 isoform X4 |
| XM_027459894.1 | NEURL1 | 8.395177077 | 0.032304708 | Up | E3 ubiquitin-protein ligase NEURL1 isoform X2 |
| XM_027460138.1 | PLEKHA1 | 11.49018234 | 0.015183216 | Up | Pleckstrin homology domain-containing family A member 1 isoform X4 |
| XM_021274553.2 | ESR2 | 9.409390936 | 0.013866488 | Up | Estrogen receptor beta |
| XM_005027629.4 | ACVR1 | −8.471675214 | 0.022375246 | Down | Activin receptor type-1 isoform X1 |
| XM_013097218.3 | BBS4 | −7.936637939 | 0.044748637 | Down | Bardet-Biedl syndrome 4 protein isoform X1 |
| XM_013104107.3 | BMPR1B | −8.276124405 | 0.038425964 | Down | Bone morphogenetic protein receptor type-1B, partial |
| XM_021268752.2 | DLG1 | −8.696967526 | 0.036312672 | Down | Disks large homolog 1 isoform X15 |
| XM_027455561.1 | PLCB1 | −9.103287808 | 0.027094995 | Down | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 isoform X2 |
| XM_027468487.1 | NME5 | −9.33315535 | 0.047456583 | Down | Nucleoside diphosphate kinase homolog 5 isoform X1 |
| XM_027469909.1 | CIT | −4.584962501 | 0.013212023 | Down | Citron Rho-interacting kinase, partial |
| XM_027470906.1 | STRBP | −8.906890596 | 0.015474849 | Down | Spermatid perinuclear RNA-binding protein isoform X1 |
| NM_001310400.1 | PRLR | −10.53138146 | 1.14591E-07 | Down | Prolactin receptor |
| XM_005027912.4 | LOC101798642 | −5.045073075 | 2.97012E-06 | Down | Ovomucoid |
| XM_027449874.1 | LOC101801176 | −12.89462832 | 6.47574E-07 | Down | Ovostatin |
| XM_027451683.1 | LOC101802317 | −10.7368247 | 4.37836E-06 | Down | Ovalbumin-related protein Y-like |
Abbreviations: HG, high-yield group; LG, low-yield group.
Functional Enrichment Analysis of Differentially Expressed Genes
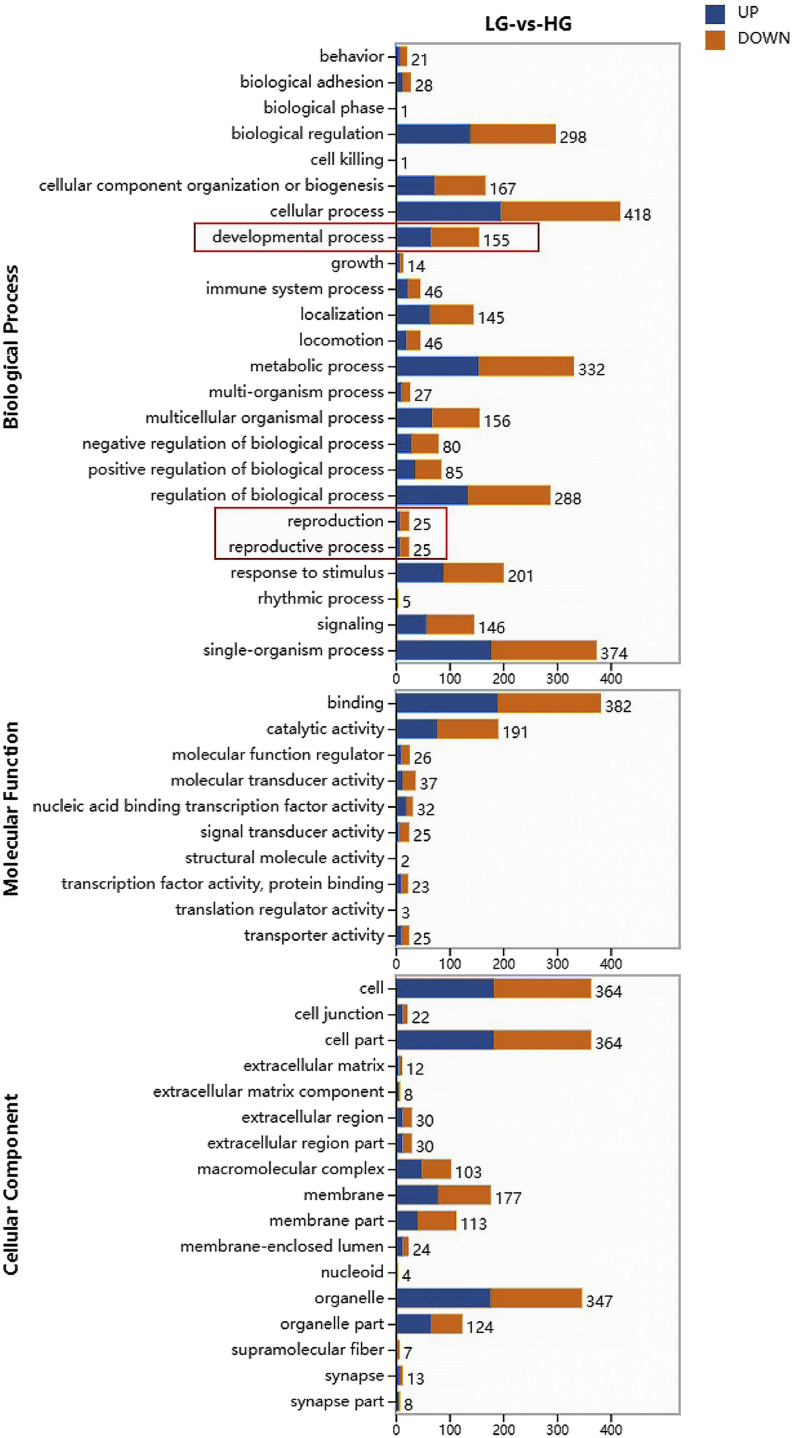
To further analyze the biological functions of 1,027 DEG, functional enrichment analysis of the DEG was performed using Blast2GO. The results exhibited that the DEG were involved in 51 biological functions, of which 24 (47%) are for biological processes, 17 (33%) for cellular components, and 10 (20%) for molecular functions (Figure 10). In biological processes, the DEG between the 2 groups accounted for the largest proportion of cellular processes. Cells and cell parts account for the largest proportion of cell functions. In molecular function, differential genes accounted for the highest proportion of binding. It can be seen from the red line in Figure 10 that in the biological processes related to duck breeding, 25 DEG are involved in reproduction, 25 DEG in the reproductive process, and 155 DEG in the developmental process.
Figure 10.
GO analysis of differentially expressed genes. Abbreviations: GO, gene ontology; HG, high-yield group; LG, low-yield group.
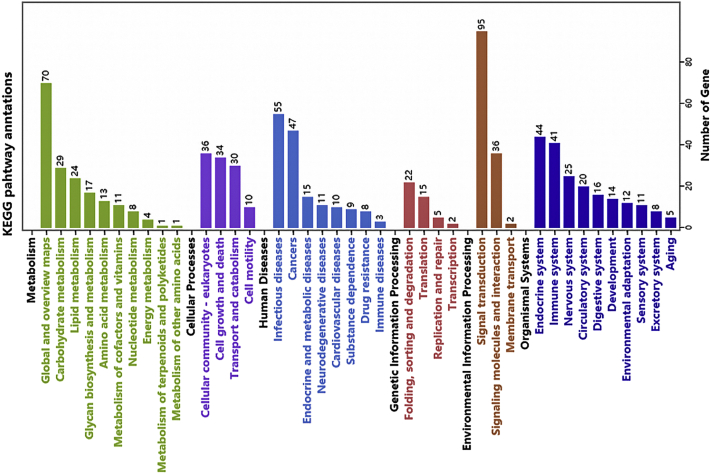
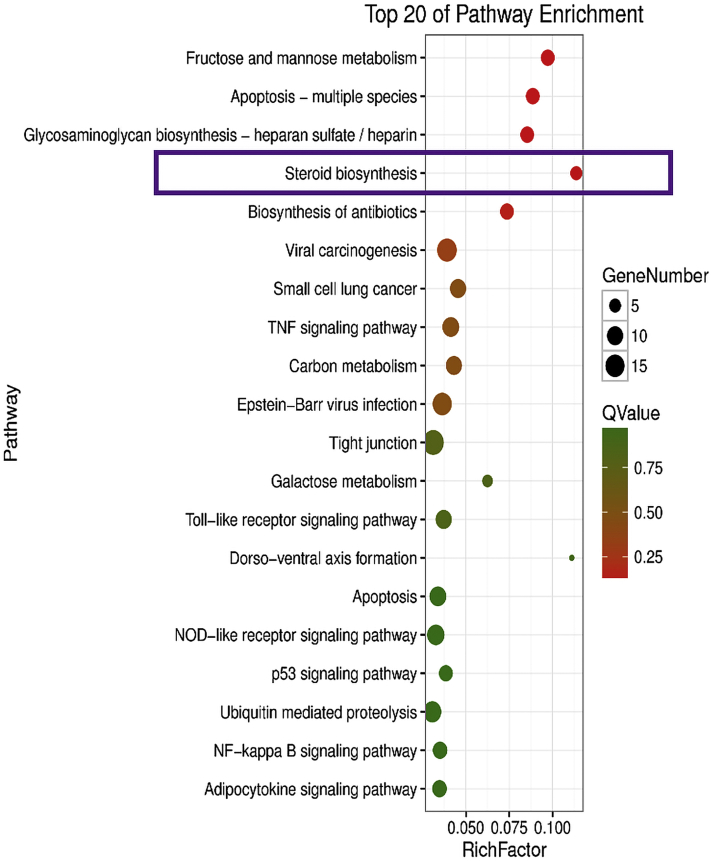
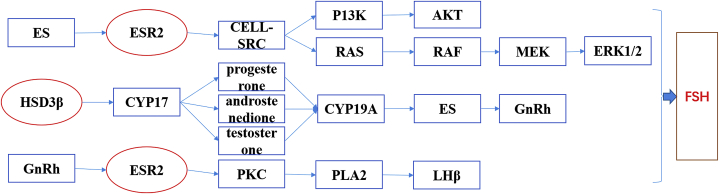
For further annotation of the main biochemical metabolic pathways and signaling pathways of these DEG, KEGG was employed to analyze the pathway of the DEG. As shown in Figure 11, 274 signal pathways were enriched in these DEG. The pathways involved in reproductive performance included estrogen signaling pathway, ovarian steroidogenesis, and GnRH signaling pathway. To obtain more valuable reference pathways, differential signal pathways were enriched again with Q < 0.05 as the significant condition, and 11 signal pathways were enriched. The DEG signal pathways between groups are shown in Figure 12, according to the top 20 significant differences. The differential gene enrichment analysis was performed with Q < 0.05 as the significant condition, and a steroid biosynthesis pathway (HSD3β → GnRH, ESR → LHβ/ERK1/2) directly related to reproduction was enriched. HSD3β → GnRH, ESR → LHβ/ERK1/2 jointly mediated the key signaling pathway, FSH that may affect the egg production differences of Leizhou black duck (Figure 13). These DEG may be involved in ovarian follicular development and ovulation during hormone regulation.
Figure 11.
KEGG annotation statistics of differential expressed gene. Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Figure 12.
KEGG enriched significant bubble map between HG and LG. Abbreviations: HG, high-yield group; KEGG, Kyoto Encyclopedia of Genes and Genomes; LG, low-yield group.
Figure 13.
Signaling pathway of FSH. Abbreviation: FSH, follicle-stimulating hormone.
Differential Gene Screening and Verification
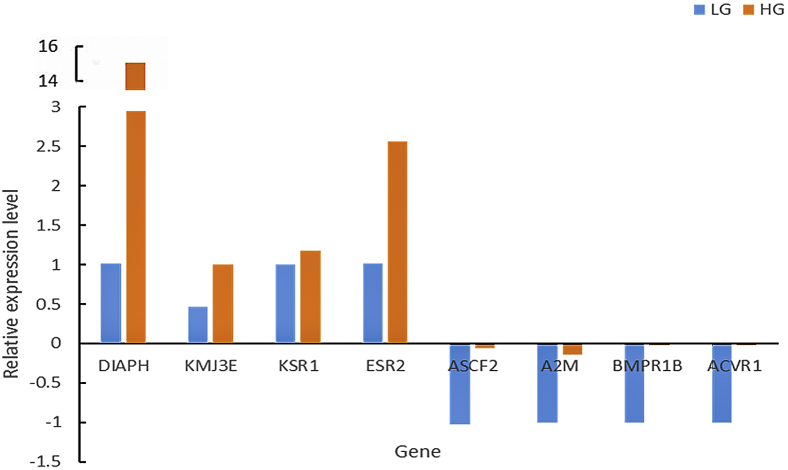
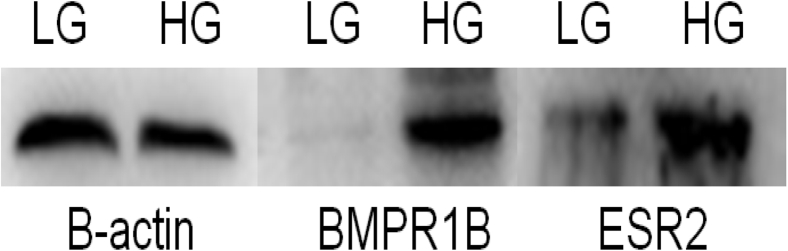
The results from qPCR of the 8 randomly selected DEG (Figure 14) from transcriptomic sequence data revealed that the expression trend of the 8 genes was consistent with the transcriptome sequencing data, indicating that the transcriptome sequencing data had high accuracy. The Western blot results showed that both ESR2 and BMPR1B proteins were expressed in the ovarian tissue of Leizhou black duck (Figure 15). The expression of ESR2 protein was significantly higher in HG than LG, whereas the expression of BMPR1B proteins was higher in LG than HG, which were consistent with the mRNA expression trend (Figure 14).
Figure 14.
Fluorescence quantification to verify the expression of differentially expressed genes in transcriptome. Abbreviations: HG, high-yield group; LG, low-yield group.
Figure 15.
The results of Western blot between ESR and BMPR1B. Abbreviations: BMPR1B, bone morphogenetic protein receptor beta; ESR, estrogen receptor; HG, high-yield group; LG, low-yield group.
Discussion
Analysis of Egg-Laying Traits and Ovarian Follicles of HG and LG Ducks
Egg production is an important indicator of measuring the economic traits of poultry. Studies have shown that egg production in poultry is directly influenced by ovarian follicular development. Based on previous studies, mature Leizhou black ducks are similar to chickens in the number of follicles, but the start of production is earlier than chickens, and the number of eggs produced by different individuals is very different (Zou et al., 2019). In this study, a comparison of the egg production traits of LG and HG showed that AFE, EW, and NE were significantly different between the 2 duck groups. The egg production curve results showed that the start period of HG is significantly earlier than that of LG. The difference in egg production between the 2 groups is mainly reflected in the egg production rate of HG at 16 to 43 wk was significantly higher than that of LH (P < 0.01). High-yield group had a significantly shorter growth period than LG (P < 0.05). A study in different breeds of geese with significantly different egg production showed that the histological characteristics of high and low egg-laying geese were mainly because of the number of follicles and the granule cell thickness of LYF, which were closely related to LH and FSH. That is, the greater the number of follicles and the thicker the LYF granular cells, the higher the egg production (Yang et al., 2019). Transcriptome analysis revealed that the differences in the development of ovaries and follicles may be closely related to the ovarian steroid synthesis pathway, including StAR, CYP17, HSD3B, etc. causing differences in egg-laying (Ren et al., 2019).
The reserve of HG in LYF follicles was higher than that of LG (P < 0.01). Observation of ovarian follicle sections revealed that the density of medulla cells per unit area of the HG medullary layer in the ovarian cortex and medulla was greater than LG and that the number of secondary follicles in HG was significantly higher than LG. Egg-laying traits are related to the onset period, which is regulated by ovarian follicular development, that is, the speed of ovarian maturity determines its onset period. After the ovary matures in poultry, the ovulation mechanism is based on a strict hierarchical order (F6-F1). After F1 ovulation, a follicle is selected from LYF to replace F6. The yolk material in the selected LYF follicle is quickly deposited for grade follicular development (Diaz et al., 2011). As an important reserve follicle that develops into grade follicles, LYF development and recruitment may affect the ovulation cycle of poultry (Chen and Jiang Xun Ping, 2006). Studies have also shown that the degradation of poultry follicles mainly occurs in the primitive follicular stage. After maturation of the ovary, the recruited SWF, LWF, SYF, and LYF have a high probability of entering the grade follicular stage (Braw-Tal, 2002; Johnson, 2015; Ghanem and Johnson, 2018). Therefore, the egg production within 43 wk of Leizhou black duck may be related to the maturation speed of ovarian follicles, and the difference in egg production rate between HG and LG may be directly related to the number of LYF follicle reserves. Also, at 43 wk, the egg production rate of HG showed a downward trend, and LG remained stable. This phenomenon may have a certain relationship with the 50 wk Leizhou black duck during molting. The molting period of HG appears earlier, that is, the continuous high yield of HG may have a certain effect on the development of the body (Hanguang, 2016). Therefore, it is undeniable that without a specific time limit, the number of eggs produced by LG may not be lower than that of HG, and its specific mechanism remains to be further studied.
Analysis of Differentially Expressed Genes Between HG and LG
In recent years, transcriptomic sequencing technology has been widely used in research in different fields, providing a convenient way to further explore the molecular mechanism between HG and LG. Ovarian transcriptomic studies in ducks help to reveal DEG related to egg production to elevate the breeding of higher egg-laying ducks to meet higher human demands and improve economic efficiency (Colonello-Frattini and Hartfelder, 2009; Zeng et al., 2015; Zhu et al., 2017). In this study, a total of 1,027 DEG were found between HG and LG ducks with 532 downregulated genes and 495 upregulated genes. To validate the accuracy of the transcriptomic sequencing data, 8 of the DEG were randomly selected for quantitative fluorescence verification from the original tissue which proved that the 8 genes were consistent with the transcriptome sequencing data.
Western blot analysis showed that both ESR2 and BMPR1B proteins were expressed in the ovarian tissues with significant expressions in HG and LG respectively. Genes that are highly expressed in reproductive cells or tissues may also indicate that the genes are activated as shown in previous studies (Gan et al., 2015; Zhang et al., 2015; Tao et al., 2017; Zhu et al., 2017). Thus, in our study, we identified 25 DEG related to reproduction in the ovaries of HG and LG ducks which included PRLR, ESR 2, BMPR1B, and others which may be involved in ovarian follicle development and may influence egg production. Prolactin and its receptor in poultry regulate follicle development and egg production and induce brooding behavior (Reddy et al., 2002; Wang et al., 2011; Yadav et al., 2018; Asiamah Amponsah et al., 2019). Other studies revealed that polymorphism in the PRL gene indicated PRL might be a candidate gene that influences egg-laying traits and reproduction in ducks (Wang et al., 2011; Chuekwon and Boonlum, 2017; Bai et al., 2019). Estrogen receptor 2 has been studied to play vital roles in the ovary during egg-laying stages and may be closely related to laying performance in ducks and may regulate key genes in granulosa cells essential for follicle maturation and ovulation (Wu et al., 2014; Khristi et al., 2018). Bone morphogenetic protein receptor beta is a member of the TGF-β superfamily which is firstly expressed in primordial follicles on the oocyte and granulosa cells of primary follicles throughout folliculogenesis (Regan et al., 2018a; Abdurahman et al., 2019; Zhang et al., 2019).
GO and KEGG Analysis of the DEG
To further explore the biological functions of the DEG, GO annotation and KEGG analysis were performed. From the GO analysis, the results exhibited that the DEG were assigned to 17 GO categories under cellular components of which “cell” and “cell part” accounted for the largest proportion followed by “organelle”, “membrane”, “organelle part”, and “membrane part”. This result indicates that genes involved in physiological activities of Leizhou Black ducks ovary are not only present in the cells but also on the membranes, which conforms to studies on the ovarian transcriptomes of Shan Ma ducks, Jinding ducks and Yaks (Lan et al., 2014; Tao et al., 2017; Zhu et al., 2017) suggesting that the cell membrane plays an essential role in reproduction and physiologic activities of the ovary. Gene ontology analysis results revealed that the DEG also involved in 10 categories in the molecular function of which the largest proportion was “binding” followed by “catalytic activity”. Previous studies have shown that a high percentage of proteins expressed in oocytes were binding proteins which suggest that binding proteins play significant roles in ovarian physiologic and reproductive activities (Mamo et al., 2011; Regassa et al., 2011; Zhu et al., 2017). Gene ontology analysis results exhibited that the DEG were also assigned to 24 categories under biological processes of which “cellular process” had the largest proportion followed by “single organism process”, “metabolic process”, and “biological regulation”. Also, there were 155 genes related to the developmental process and 25 genes each associated with reproduction and reproductive process. This implies that the development and reproductive related genes play different roles in HG and LG Leizhou black duck egg production which is consistent with studies in Shan Ma and Jinding ducks (Tao et al., 2017; Zhu et al., 2017). The GO analysis results were similar to previously reported studies in the ovaries of Shan Ma and Jinding ducks (Tao et al., 2017; Zhu et al., 2017).
To further comprehend and better annotated the DEG to ascertain the main metabolic and signaling transduction pathways the genes are involved, KEGG pathway enrichment analysis was performed. The outcome presented 274 signal pathways were enriched in these DEG of which the steroid biosynthesis pathway was the most enriched. The steroid biosynthesis pathway is known as an important target for endocrine-disrupting chemicals that impair reproduction (Sanderson, 2006). Ovarian steroid hormones such as estrogens (ESR) play an important signaling role in the female reproductive system and ensure normal follicular development by regulating the dynamic balance of follicular growth and development hormones (Findlay et al., 2009; Vrtačnik et al., 2014; Wu et al., 2014; Khristi et al., 2018; Prasasya and Mayo, 2019). In this study, ESR 2 was significantly higher in HG than LG ducks which may contribute to higher egg-laying performance in HG ducks. Follicle stimulating hormone signaling pathways mediated by HSD3β → GnRH, ESR2 → LHβ/ERK1/2 may affect the egg production of Leizhou black ducks.
ERK is a member of the MAPK family whose pathway is involved in regulating cell growth, development, and division. The MAPK signaling pathway is an important pathway for intercellular signaling and can participate in cell proliferation, differentiation, and follicle maturation (Mendoza et al., 2011; McCain, 2013). Studies have shown that FSH can regulate the Ca2+ concentration on follicle granulosa cells through FSHR and up-regulate the downstream protein Ras (Durán-Pastén and Fiordelisio, 2013). Ras activates the MAPK signal and enhances its cascaded ERK1/2 pathway (Mendoza et al., 2011; McCain, 2013). It promotes granulation by increasing the transcriptional activity of granule cells and the synthesis of cyclinD2 (Hatzirodos et al., 2014, 2015). MAPK-mediated FSHR signaling is an important pathway for follicular selection, development, and maturation. Therefore, in this study, the FSH signaling pathway mediated by ESR2 → LHβ/ERK1/2 may play an essential role in the egg-laying traits of Leizhou black ducks, and it is of great significance for the molecular regulatory network of egg-laying traits in poultry.
Gonadotropin-releasing hormone is an important signal factor connecting the hypothalamus–pituitary–ovarian axis to secrete gonadotropins (FSH and LH) through neural action which play vital roles in reproduction including sex hormone biosynthesis (Casañ et al., 2000; Lee et al., 2008; Clarke et al., 2009; Stamatiades and Kaiser, 2018). Under the action of gonadotropins, GnRH can promote ovarian development and maturation by up-regulating the expression of IGF1 and TGF-β (Wu et al., 2016b). Also, gonadotropins can promote the release of FSH from the corpus luteum, activity of follicular granule cells, and accelerate the selection and matured follicles (Clarke et al., 2009; Xu et al., 2012). Therefore, the HSD3β → GnRH signaling pathway mediated by the ovarian steroid production pathway enriched in this study may be an important factor affecting the egg production difference of Leizhou black ducks.
Conclusion
In summary, the variation in egg production between Leizhou black ducks HG and LG is closely related to the NE, AFE, EW, and the number of LYF. It was found through transcriptome sequencing that steroid biosynthesis (HSD3β → GnRH, ESR → LHβ/ERK1/2), which is directly related to reproduction, was enriched, and FSH signal pathway mediated by HSD3β → GnRH, ESR → LHβ/ERK1/2 is the key signaling pathways that may affect the egg production differences of Leizhou black ducks. Therefore, these differential genes may be involved in hormonal regulation of ovarian follicular development and ovulation, thereby affecting the egg production of Leizhou black ducks.
Acknowledgments
This work was sponsored by Guangdong Science and Technology Plan Project (2017A020208066), provincial-level major scientific research project in Guangdong Province (2017KZDXM043), the excellent dissertation cultivation project of Guangdong Ocean University (201610), the Postgraduate demonstration course construction project of Guangdong Ocean University (21411), National Natural Science Foundation of China (No. 30972085), and The Key Platform Project of Innovative Strong School Engineering by the Department of Education of Guangdong Province (2018302).
Conflict of Interest Statement: No conflict of interest exists in the submission of this manuscript, and it is approved by all authors for publication. All authors declare that the work described was original research that has not been published previously, and not under consideration for publication elsewhere, in whole or in part.
Contributor Information
Zhihui Zhao, Email: zhzhao@jlu.edu.cn.
Ying Su, Email: dwkxsy@163.com.
References
- Abdurahman A., Du X., Yao Y., Sulaiman Y., Aniwashi J., Li Q. Smad4 feedback enhances BMPR1B transcription in ovine granulosa cells. Int. J. Mol. Sci. 2019;20:1–14. doi: 10.3390/ijms20112732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asiamah Amponsah C., Zou K., li Lu L., Zhang S.W., Xue Y., Su Y., Zhao Z. Genetic effects of polymorphisms of candidate genes associated with ovary development and egg production traits in ducks. Anim. Reprod. Sci. 2019;211:1–9. doi: 10.1016/j.anireprosci.2019.106219. [DOI] [PubMed] [Google Scholar]
- Bai D.-P., Hu Y.-Q., Li Y.-B., Huang Z.-B., Li A. Polymorphisms analysis of the prolactin gene with egg production traits in two Chinese domestic ducks. Br. Poult. Sci. 2019;60:125–129. doi: 10.1080/00071668.2019.1567909. [DOI] [PubMed] [Google Scholar]
- Braw-Tal R. The initiation of follicle growth: the oocyte or the somatic cells? Mol. Cell. Endocrinol. 2002;187:11–18. doi: 10.1016/s0303-7207(01)00699-2. [DOI] [PubMed] [Google Scholar]
- Casañ E.M., Raga F., Bonilla-Musoles F., Polan M.L. Human oviductal gonadotropin-releasing hormone: possible implications in fertilization, early embryonic development, and implantation 1. J. Clin. Endocrinol. Metab. 2000;85:1377–1381. doi: 10.1210/jcem.85.4.6503. [DOI] [PubMed] [Google Scholar]
- Chen X.P., Jiang Xun Ping D.J.T. Apoptosis of goose granulosa cells and its relationship between reproductive hormones. Acta Vet. Zootech. Sin. 2006;37:982–986. [Google Scholar]
- Chen H.Y., Shen H., Jia B., Zhang Y.S., Wang X.H., Zeng X.C. Differential gene expression in ovaries of Qira black sheep and Hetian sheep using RNA-seq technique. PLoS One. 2015;10:e0120170. doi: 10.1371/journal.pone.0120170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuekwon K., Boonlum S. Association of prolactin gene with egg production in Khaki Campbell ducks. Walailak J. Sci. Tech. 2017;14:849–853. [Google Scholar]
- Clarke I.J., Smith J.T., Caraty A., Goodman R.L., Lehman M.N. Kisspeptin and seasonality in sheep. Peptides. 2009;30:154–163. doi: 10.1016/j.peptides.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonello-Frattini N.A., Hartfelder K. Differential gene expression profiling in mucus glands of honey bee (Apis mellifera) drones during sexual maturation. Apidologie. 2009;40:481–495. [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. Blast2GO: a universal tool for annotation, visualization, and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cui C., Han S., Yin H., Luo B., Shen X., Yang F., Liu Z., Zhu Q., Li D., Wang Y. FOXO3 is expressed in ovarian tissues and acts as an apoptosis initiator in granulosa cells of chickens. Biomed. Res. Int. 2019;2019:1–9. doi: 10.1155/2019/6902906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F.J., Anthony K., Halfhill A.N. Early avian follicular development is characterized by changes in transcripts involved in steroidogenesis, paracrine signaling, and transcription. Mol. Reprod. Dev. 2011;78:212–223. doi: 10.1002/mrd.21288. [DOI] [PubMed] [Google Scholar]
- Ding N., Han Q., Zhao X.Z., Li Q., Li J., Zhang H.F., Gao G.L., Luo Y., Xie Y.H., Su J., Wang Q.G. Differential gene expression in pre-laying and laying period ovaries of Sichuan white geese (Anser cygnoides) Genet. Mol. Res. 2015;14:6773–6785. doi: 10.4238/2015.June.18.20. [DOI] [PubMed] [Google Scholar]
- Durán-Pastén M.L., Fiordelisio T. GnRH-Induced Ca2+ Signaling patterns and gonadotropin secretion in pituitary gonadotrophs. Functional adaptations to both ordinary and extraordinary physiological demands. Front. Endocrinol. (Lausanne) 2013;4:127. doi: 10.3389/fendo.2013.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay J., Kerr J., Britt K., Liew S., Simpson E., Rosairo D., Drummond A. Ovarian physiology: follicle development, oocyte, and hormone relationships. Anim. Reprod. 2009;6:16–19. [Google Scholar]
- Gan X., Chen D., Deng Y., Yuan J., Kang B., Qiu J., Sun W., Han C., Hu J., Li L., Wang J. Establishment of an in vitro culture model of theca cells from hierarchical follicles in ducks. Biosci. Rep. 2017;37:1–10. doi: 10.1042/BSR20160491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L., Xie L., Zuo F., Xiang Z., He N. Transcriptomic analysis of Rongchang pig brains and livers. Gene. 2015;560:96–106. doi: 10.1016/j.gene.2015.01.051. [DOI] [PubMed] [Google Scholar]
- Ghanem K., Johnson A.L. Follicle dynamics and granulosa cell differentiation in the Turkey hen ovary. Poult. Sci. 2018;97:3755–3761. doi: 10.3382/ps/pey224. [DOI] [PubMed] [Google Scholar]
- Götz S., García-Gómez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talón M., Dopazo J., Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Zhao D., Liu Y., Liu Q., Huang X., Yang J., An F., Li Y. Quantitative proteomic analysis of duck ovarian follicles infected with duck Tembusu virus by label-free LC-MS. Front. Microbiol. 2016;7:1–11. doi: 10.3389/fmicb.2016.00463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanguang H. Guangdong Ocean University; Guangdong, CN: 2016. Study on the features and candidate genes of egg production in the base breeding group of Leizhou black duck. Master’s thesis. [Google Scholar]
- Hatzirodos N., Hummitzsch K., Irving-Rodgers H.F., Rodgers R.J. Transcriptome profiling of the theca interna in transition from small to large antral ovarian follicles. PLoS One. 2014;9:e97489. doi: 10.1371/journal.pone.0097489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzirodos N., Hummitzsch K., Irving-Rodgers H.F., Rodgers R.J. Transcriptome comparisons identify new cell markers for theca interna and granulosa cells from small and large antral ovarian follicles. PLoS One. 2015;10:1–13. doi: 10.1371/journal.pone.0119800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.L. Ovarian follicle selection and granulosa cell differentiation. Poult. Sci. 2015;94:781–785. doi: 10.3382/ps/peu008. [DOI] [PubMed] [Google Scholar]
- Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khristi V., Chakravarthi V.P., Singh P., Ghosh S., Pramanik A., Ratri A., Borosha S., Roby K.F., Wolfe M.W., Rumi M.A.K. ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell. Endocrinol. 2018;474:214–226. doi: 10.1016/j.mce.2018.03.012. [DOI] [PubMed] [Google Scholar]
- Lan D., Xiong X., Huang C., Mipam T.D., Li J. Toward understanding the genetic basis of yak ovary reproduction: a characterization and comparative analyses of estrus ovary transcriptome in yak and cattle. PLoS One. 2016;11:e0152675. doi: 10.1371/journal.pone.0152675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan D.L., Xiong X.R., Wei Y.L., Xu T., Zhong J.C., Zhi X.D., Wang Y., Li J. RNA-Seq analysis of yak ovary: improving yak gene structure information and mining reproduction-related genes. Sci. China Life Sci. 2014;57:925–935. doi: 10.1007/s11427-014-4678-2. [DOI] [PubMed] [Google Scholar]
- Lee V.H.Y., Lee L.T.O., Chow B.K.C. Gonadotropin-releasing hormone: regulation of the GnRH gene. FEBS J. 2008;275:5458–5478. doi: 10.1111/j.1742-4658.2008.06676.x. [DOI] [PubMed] [Google Scholar]
- Li L., Miao Z., Xin Q., Zhu Z., Zhang L., Zhuang X., Zheng N. Transcriptome analysis of differential gene expression associated with testis tissue in mule duck and muscovy duck. Sci. Agric. Sin. 2017;50:3608–3619. [Google Scholar]
- Luan X., Liu D., Cao Z., Luo L., Liu M., Gao M., Zhang X. Transcriptome profiling identifies differentially expressed genes in Huoyan goose ovaries between the laying period and ceased period. PLoS One. 2014;9:e113211. doi: 10.1371/journal.pone.0113211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamo S., Carter F., Lonergan P., V Leal C.L., Al Naib A., McGettigan P., Mehta J.P., Evans A.C.O., Fair T. Sequential analysis of global gene expression profiles in immature and in vitro matured bovine oocytes: Potential molecular markers of oocyte maturation. BMC Genomics. 2011;12:151. doi: 10.1186/1471-2164-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCain J. The MAPK (ERK) Pathway: Investigational Combinations for the Treatment of BRAF- Mutated Metastatic Melanoma. P.T. 2013;38:96–108. [PMC free article] [PubMed] [Google Scholar]
- Mendoza M.C., Er E.E., Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem. Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao X., Luo Q., Qin X. Genome-wide transcriptome analysis in the ovaries of two goats identifies differentially expressed genes related to fecundity. Gene. 2016;582:69–76. doi: 10.1016/j.gene.2016.01.047. [DOI] [PubMed] [Google Scholar]
- Prasasya R.D., Mayo K.E. Regulation of follicle formation and development by ovarian signaling pathways. In: Leung P.C.K., Adashi E.Y., editors. The Ovary. 3rd ed. Academic Press; Cambridge, MA: 2019. pp. 23–49. [Google Scholar]
- Reddy I.J., David C.G., Sarma P.V., Singh K. The possible role of prolactin in laying performance and steroid hormone secretion in domestic hen (Gallus domesticus) Gen. Comp. Endocrinol. 2002;127:249–255. doi: 10.1016/s0016-6480(02)00034-5. [DOI] [PubMed] [Google Scholar]
- Regan S.L.P., Knight P.G., Yovich J.L., Leung Y., Arfuso F., Dharmarajan A. Involvement of bone morphogenetic proteins (BMP) in the regulation of ovarian function. Vitam. Horm. 2018;107:227–261. doi: 10.1016/bs.vh.2018.01.015. [DOI] [PubMed] [Google Scholar]
- Regan S.L.P., Knight P.G., Yovich J.L., Stanger J.D., Leung Y., Arfuso F., Almahbobi G., Dharmarajan A. The effect of ovarian reserve and receptor signaling on granulosa cell apoptosis during human follicle development. Mol. Cell. Endocrinol. 2018;470:219–227. doi: 10.1016/j.mce.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Regassa A., Rings F., Hoelker M., Cinar U., Tholen E., Looft C., Schellander K., Tesfaye D. Transcriptome dynamics and molecular cross-talk between bovine oocyte and its companion cumulus cells. BMC Genomics. 2011;12:57. doi: 10.1186/1471-2164-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Sun C., Chen L., Hu J., Huang X., Liu X., Lu L. Exploring differentially expressed key genes related to development of follicles by RNA-seq in Peking ducks (Anas Platyrhynchos) PLoS One. 2019;14:e0209061. doi: 10.1371/journal.pone.0209061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz H., Yousuf M.R., Liang A., Hua G.H., Yang L. Effect of melatonin on regulation of apoptosis and steroidogenesis in cultured buffalo granulosa cells. Anim. Sci. J. 2019;90:473–480. doi: 10.1111/asj.13152. [DOI] [PubMed] [Google Scholar]
- Sanderson J.T. The steroid hormone biosynthesis pathway as a target for endocrine-disrupting chemicals. Toxicol. Sci. 2006;94:3–21. doi: 10.1093/toxsci/kfl051. [DOI] [PubMed] [Google Scholar]
- Shen M., Li T., Zhang G., Wu P., Chen F., Lou Q., Chen L., Yin X., Zhang T., Wang J. Dynamic expression and functional analysis of circRNA in granulosa cells during follicular development in chicken. BMC Genomics. 2019;20:1–15. doi: 10.1186/s12864-019-5462-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T. Molecular and cellular mechanisms for the regulation of ovarian follicular function in cows. J. Reprod. Dev. 2016;62:323–329. doi: 10.1262/jrd.2016-044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatiades G.A., Kaiser U.B. Gonadotropin regulation by pulsatile GnRH: signaling and gene expression. Mol. Cell. Endocrinol. 2018;463:131–141. doi: 10.1016/j.mce.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z., Song W., Zhu C., Xu W., Liu H., Zhang S., Huifang L. Comparative transcriptomic analysis of high and low egg-producing duck ovaries. Poult. Sci. 2017;96:4378–4388. doi: 10.3382/ps/pex229. [DOI] [PubMed] [Google Scholar]
- Vrtačnik P., Ostanek B., Mencej-Bedrač S., Marc J. The many faces of estrogen signaling. Biochem. Med. 2014;24:329–342. doi: 10.11613/BM.2014.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liang Z., Yu W., Feng Y., Peng X., Gong Y., Li S. Polymorphism of the prolactin gene and its association with egg production traits in native Chinese ducks. South Afr. J. Anim. Sci. 2011;41:63–69. [Google Scholar]
- Wu S.J., Cheng Y.S., Liu H.L., Wang H.H., Huang H.L. Global transcriptional expression in ovarian follicles from Tsaiya ducks (Anas platyrhynchos) with a high-fertilization rate. Theriogenology. 2016;85:1439–1445.e1. doi: 10.1016/j.theriogenology.2016.01.005. [DOI] [PubMed] [Google Scholar]
- Wu X., Liu X.T., Lan J.J., Wan X.P., Yan M.J., Lian S.Y., Li A. Investigation of insulin-like growth factor-1 gene with egg-laying traits in the Muscovy duck (Cairina moschata) Can. J. Anim. Sci. 2016;214:203–214. [Google Scholar]
- Wu Y., Xiao H.W., Liang Z.H., Pan A.L., Shen J., Pi J.S., Pu Y.J., Du J.P., Chen Z.H. Differential expression profiling of estrogen receptor in the ovaries of two egg duck (Anas platyrhynchos) breeds. Czech J. Anim. Sci. 2014;59:238–243. [Google Scholar]
- Xu W., MeiJiao Y., LiPing L., SenYang L., XingYuan F., GuangYing W., Ang L. Genetic effects of gonadotropin-releasing hormone (GnRH) and growth hormone (GH) genes on the egg performance in muscovy duck (Cairina moschata) J. Agric. Biotechnol. 2012;20:289–295. [Google Scholar]
- Yadav S.K., Maurya S.K., Yadav A.K., Kumar A., Yadav K. Study of prolactin receptor gene (PRLR5) polymorphisms and its association with egg production in Kadaknath hens. Indian J. Anim. Res. 2018;52:1232–1235. [Google Scholar]
- Yang Y.Z., Yao Y., Cao Z.F., Gu T.T., Xu Q., Chen G.H. Histological characteristics of follicles and reproductive hormone secretion during ovarian follicle development in laying geese. Poult. Sci. 2019;98:6063–6070. doi: 10.3382/ps/pez278. [DOI] [PubMed] [Google Scholar]
- Zeng T., Zhang L., Li J., Wang D., Tian Y., Lu L. De novo assembly and characterization of Muscovy duck liver transcriptome and analysis of differentially regulated genes in response to heat stress. Cell Stress Chaperones. 2015;20:483–493. doi: 10.1007/s12192-015-0573-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Chang H.M., Zhu H., Liu R.Z., Leung P.C.K. BMP6 increases TGF-β1 production by up-regulating furin expression in human granulosa-lutein cells. Cell. Signal. 2019;55:109–118. doi: 10.1016/j.cellsig.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Zhang X., Huang L., Wu T., Feng Y., Ding Y., Ye P., Yin Z. Transcriptomic analysis of ovaries from pigs with high and low litter size. PLoS One. 2015;10:e0139514. doi: 10.1371/journal.pone.0139514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z.Q., Wang L.J., Sun X.W., Zhang J.J., Zhao Y.J., Na R.S., Zhang J.H. Transcriptome analysis of the Capra hircus ovary. PLoS One. 2015;10:e0121586. doi: 10.1371/journal.pone.0121586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.M., Miao Z.W., Chen H.P., Xin Q.W., Li L., Lin R.L., Lou Huang Q., Zheng N.Z. Ovarian transcriptomic analysis of Shan Ma ducks at peak and late stages of egg production. Asian-australasian J. Anim. Sci. 2017;30:1215–1224. doi: 10.5713/ajas.16.0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Kun, Cui Hongyan, Xue Yuan, Zhang Shaowei, Lu Lili, Zhao Zhihui S.Y. Correlation analysis of reproductive traits with FSHR and ESR1 in Leizhou black duck. Biotechnol. Bull. 2019;35:118–126. [Google Scholar]