Abstract
Gut microbiota play an important role in animal health. For livestock, an understanding of the effect of husbandry interventions on gut microbiota helps develop methods that increase sustainable productivity, animal welfare, and food safety. Poultry microbiota of the mid-gut and hind-gut can only be investigated postmortem; however, samples from the terminal cloaca may be collected from live animals. This study tests whether cloacal microbiota reflect cecal microbiota in European broiler poultry by evaluating total and paired cecal and cloacal microbiomes from 47 animals. 16S amplicon libraries were constructed and sequenced with a MiSeq 250 bp PE read metric. The composition of cloacal and cecal microbiomes were significantly affected by the age and location of animals, but the effect was very small. Bacilli were relatively more abundant in ceca and Clostridia in cloaca. There was an overlap of 99.5% for the abundances and 59% for the types of taxa between cloacal and cecal communities, but the small fraction of rare nonshared taxa were sufficient to produce a signal for differentiation between cecal and cloacal communities. There was a significant positive correlation between specific taxa abundances in cloacal and cecal communities (Rho = 0.66, P = 2 × 10−16). Paired analyses revealed that cloacal communities were more closely related to cecal communities from the same individual than expected by chance. This study is in line with the only other study to evaluate the relationship between cecal and cloacal microbiomes in broiler poultry, but it extends previous findings by analyzing paired cecal–cloacal samples from the same birds and reveals that abundant bacterial taxa in ceca may be reasonably inferred by sampling cloaca. Together, the findings from Europe and Australasia demonstrate that sampling cloaca shows promise as a method to estimate cecal microbiota, and especially abundant taxa, from live broiler poultry in a manner which reduces cost and increases welfare for husbandry and research purposes.
Key words: 16S rRNA amplicon sequencing, poultry microbiome, cloacal communities, cecal communities, 3Rs
Introduction
Poultry is a significant source of human dietary protein, and 1 estimate for the worldwide broiler chicken population is approximately 23 billion birds (Bennett et al., 2018). Elucidating methods that increase livestock health is both desirable from animal welfare and food safety perspectives and generally aligns with increased food-supply sustainability. Poultry health status is usually evaluated by investigating performance, welfare, intestinal morphology, and histological features (Biasato et al., 2016), but there are increasing reports of the role that gut microbiota play in health and physiology. Some bacterial species are associated with enteritis in poultry and are thus considered signs of poor health status (Yegani and Korver, 2008). These species typically include Clostridium perfringens, Escherichia coli, Pasteurella multocida, Erysipelothrix rhusiopathiae, Mycobacterium avium, and several serotypes of Salmonella (Porter, 1998); in addition, human food-borne disease agents are found in poultry gut microbiota (Clavijo and Flórez, 2018).
Microbiota have traditionally been studied using culture-dependent methods (Shapiro and Sarles, 1949), and several studies have shown the majority of species in the poultry gut are not yet culturable (Shang et al., 2018), but the exact percentage of culturable bacteria from the avian gastrointestinal tract (GIT) is unknown (Grond et al., 2018). Next-generation culture independent DNA sequencing is now common place and has been applied to avian intestinal microbiota in several studies, mainly to investigate the effect of feed ingredients on microbial communities, including essential oils (Amerah et al., 2011), pomegranate (Saeed et al., 2018), and probiotic bacteria such as Lactobacillus (Nakphaichit et al., 2011; Baldwin et al., 2018). Other studies have evaluated microbiome differences that correlate with disease in poultry (Tong et al., 2018) and the influence of photoperiod in broiler roosters (Wang et al., 2018).
Ceca are usually targeted in microbiome studies as they play an important role in bird health and productivity (Stanley et al., 2015). Ceca are involved in water regulation, anaerobic carbohydrate digestion, and fermentation (including cellulose and starch; Clench and Mathias, 1995; Grant et al., 2018) which are strongly related to productivity (Díaz Carrasco et al., 2018). With up to 1011 cells per gram, ceca have the greatest bacterial biodiversity along the chicken GIT (Grant et al., 2018). Despite the importance of understanding microbial communities in this portion of the gut, cecal contents can only be accessed once an animal is slaughtered. One option is to sample fecal material to provide a window into animal GIT microbiota. There are only a few studies that have investigated the correlation between cecal and fecal microbiomes in a range of animal species; for example, there was no correlation between fecal and cecal microbial communities in mice (Pang et al., 2012) or pigs (Panasevich et al., 2018), and no correlation was reported between colon and cecal contents in urban Canadian geese (Drovetski et al., 2018). Ceca are close to the end of the GIT in birds, and thus, it is worth evaluating whether microbial communities from the adjacent gut terminal cloaca reflect cecal microbiomes in any way. Because cloaca may be sampled from live animals, this may potentially provide a method that avoids unnecessary animal sacrifice. In addition, the effect of diet or other interventions on the composition of animal microbiomes may best be answered by taking repeated samples from the same animal, which is clearly not possible if ceca are analyzed directly from euthanized animals. Cloacal sampling from live birds is easy to perform; moreover, sampling cloaca reduces any ambiguity and cross-contamination that are associated with sampling feces after excretion. While several studies have sampled cloaca to investigate gut microbiomes in different bird species (e.g., penguins; Barbosa et al., 2016; herring gulls; Merkeviciene et al., 2017; and wild mallards; Ganz et al., 2017), only a very few studies have focused on correlations between cloacal and cecal microbiota. Zhang et al. 2017 demonstrated that cloacal microbiomes in United States wild endangered Attwater's Prairie chicken partly reflect microbiome diversity of ileum, cecum, and large intestine, but only 8 birds were analyzed making the statistical power weak. We are aware of only 1 study comparing cloacal to cecal communities in broiler poultry. Stanley et al. (2015) evaluated 163 male Cobb 500 broilers in Australia and robustly showed significant overlaps with the types of species but that species abundances were less well correlated. Currently there are no data on the degree to which cloacal samples reflect cecal microbial communities in poultry systems outside of Australia, and here, we evaluate European systems with the Aviagen Ross 308 breed using next-generation culture independent DNA sequencing approaches to test whether cloacal samples may be used to evaluate poultry gut microbiomes and thus reduce experimental costs and unnecessary animal sacrifice.
Materials and methods
Sampling
Samples were collected from 47 healthy Ross 308 broiler chickens across 8 flocks, each from a separate farm, from 2 locations 50 km apart in Northern Ireland and managed under the same production system in December 2017. All flocks were fed from the same feed mill and had identical feed regimes: starter 300 g/bird (day 0–10); grower 900 g/bird (day 10–21); and finisher as required (day 21–clear). The animals' age ranged from 19 to 31 d and weighed 1284.5 ± 290.4 g (mean ± SD; Supplementary Table 1), meaning at the time of sampling 2 flocks were on grower and the rest on finisher feed. Cloacal samples were obtained with a sterile cotton swab inserted 10 to 12 mm in the cloacal opening and gently rotated. Cecal contents were collected by opening the birds immediately after euthanasia, cutting off 1 cecum, and manually squeezing the content over an exposed sterile swab. Swabs were immediately placed at −80°C and transported to the laboratory for DNA extraction.
DNA Extraction and Library Preparation
Total DNA was extracted using the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) following Hang et al., 2014. The swab tip was cut into a sterile tube with 450 μL of enzymatic lysis buffer containing 20 mmol/L Tris-HCl Ph 8.0 (Sigma Aldrich, St. Louis, MO), 2 mmol/L sodium EDTA (Sigma Aldrich), 1.2% Triton X-100 (Sigma Aldrich), and 20 mg/mL lysozyme, and samples were extracted and purified in spin columns following manufacturer's instructions. 16S rRNA (V4 region; 254 bp) amplicons and libraries were constructed by Earlham Institute (Norwich, UK) and were sequenced on an Illumina MiSeq instrument with a 250 PE technology. Sequence quality was evaluated with FastQC (Andrews, 2010). QIIME, version 2, was used to investigate the microbial communities (Bolyen et al., 2018). Paired end sequences were denoised by using dada2 (Callahan et al., 2016). Amplicon sequence variants (ASV) were clustered using vsearch with an identity of 0.97 (Rognes et al., 2016). Variance-stabilizing normalization (Muletz Wolz et al., 2018) was performed on the raw sequence counts in R (v1.0.153; R Core Team, 2013) using CSS normalization with metagenomeSeq and phyloseq package (McMurdie and Holmes, 2013; Paulson et al., 2013; Weiss et al., 2017). Amplicon sequence variants were annotated using q2-feature-classifier plugin and gg_13_8_otus database (Bokulich et al., 2018).
Statistical Analysis
All analyses were conducted in R (v1.0.153; R Core Team, 2013), Shannon's and Simpson's indexes (Shannon, 1948; Simpson, 1949), 2-way full factorial permutational multivariate ANOVA (PERMANOVA; Anderson, 2001) on binary, and abundance Jaccard dissimilarities was calculated using the “vegan” R package (Oksanen et al., 2018). Indicator species of cloacal samples were identified by using the indicspecies package (ver. 1.7.1; Dufrêne and Legendre, 1997). Association between paired samples was investigated calculating pairwise binary and nonbinary Jaccard distances in vegan and relative abundance of taxa in cloaca and cecum were compared using Pearson's (Kirch, 2008) and Spearman's correlation coefficient (Hollander and Wolfe, 1973).
Ethics Approval and Consent to Participate
No birds were sacrificed specifically for this study as all samples were collected from animals that were selected for routine gut health husbandry assessment checks. All samples were taken by an authorized Veterinarian. The protocol was approved by the Committee on the Ethics of the University of Lincoln (project number CoSREC342). All efforts were made to minimize suffering. Research on animals met the guidelines approved by the Institutional Animal Care and Use Committee.
Availability of Data and Material
The raw amplicon sequencing data from this study is publicly available in the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra/) under the BioProject ID: PRJNA559084.
Results
A total of 8,396,974 16S rDNA sequences were obtained from 44 cecal and 47 cloacal samples (library construction failed for 3 cecal samples; Supplementary Table 1) with an average of 92,274 ± 22,693 sequences per sample. The total number of DNA sequence reads did not differ between cecal and cloacal samples (Kruskal-Wallis test, chi-square = 0.250, P = 0.616).
Bacterial Community Composition
Analyses with QIIME 2 revealed a total of 693 ASV that clustered with 97% similarity, which we refer to as taxa. Overall, 19 phyla, 34 classes, 58 orders, and 76 families were identified. In terms of taxa numbers, most belong to Firmicutes (74.75%), and other phyla had approximately 10-fold lower taxa numbers: Proteobacteria (6.64%); Tenericutes (5.63%); Actinobacteria (4.62%); and Bacteroidetes (2.74%). Only 1 taxon was identified as belonging to Archea (Methanomassiliicoccaceae family, 0.14%). Of the 518 Firmicutes taxa, 473 belonged to class Clostridia (68.25%) of which 469 were Clostridiales (67.68%). Of Clostridiales, 192 taxa belonged to the family Ruminococcaceae (27.71%) and 69 to Lachnospiraceae (9.96%). No human pathogens (Campylobacter sp. or Salmonella sp.) were detected, and the complete ASV breakdown is supplied in Supplementary Table 2. If taxa abundances are approximated by read abundances, then Firmicutes was the most abundant phylum (94.75%), and other phyla only accounted for a small portion of the abundance: Proteobacteria (2.90%) and Actinobacteria (1.83%) and the remaining phyla accounted for only 0.51% of abundance. Despite the high taxa numbers, Clostridiales accounted for only 13.46% of abundance. Despite the low number of Bacilli taxa, these accounted for 81.22% of the abundance in the 91 samples. Plots representing the bacterial communities at phylum, class, order, and family levels are reported in Supplementary Data file.
Effect of Flock, Bird Age, and Geographic Origin on Biodiversity
Bird age, location, and flock (farm) origin had no significant effect of the numbers, types, or abundances of bacteria when data from cecal and cloacal samples from the same bird were combined (P between 0.70 and 0.98, Kruskal-Wallis; Supplementary Table 3). When cloacal and cecal samples were analyzed separately, there was still no significant effect of any of these factors on the numbers of taxa (P between 0.09 and 0.83, Kruskal-Wallis; Supplementary Table 3). However, there was a significant difference in taxa abundance in both cloaca and ceca by flock origin (R2 = 0.050, P = 0.0062 and R2 = 0.047, P = 0.0006 respectively, PERMANOVA; Supplementary Table 3), and there was a significant difference in the types and abundance of taxa in cecal samples by all factors (R2 between 0.06 and 0.04 and P between 0.007 and 0.00001, PERMANOVA; Supplementary Table 3). However, where there were significant differences by age, location, or flock, the size of these effects is very small: an average of just 5% of the variance in microbiomes is explained by these factors (mean R2 = 0.04983; Supplementary Table 3).
The Similarity Between Cecal and Cloacal Bacterial Communities
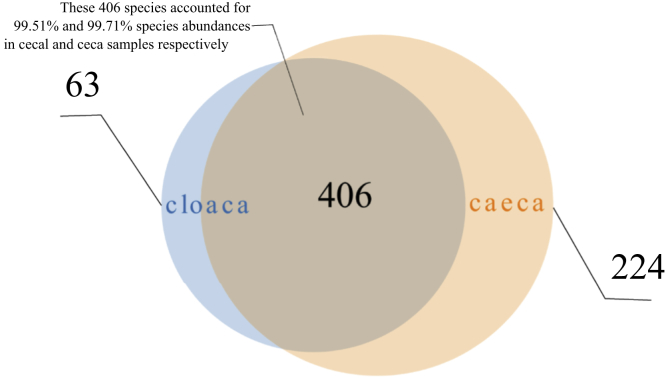
The main hypothesis under test is whether microbiomes from cloacal samples reflect cecal microbiomes to any reasonable degree. There were significantly greater numbers of taxa in cecal than cloacal samples (Kruskal-Wallis test, chi-square = 56.752, P = 5 × 10−14), and the variance in taxa number from cloacal samples was significantly greater than in ceca (F-test, F = 3.3207, P = 1 × 10−4). Ceca had an average of 225 ± 36 and cloaca an average of 68 ± 65 taxa; however, 58.6% of taxa were common to both ceca and cloaca (Figure 1). Simple counts of taxa presence/absence do not consider abundance, and very rare taxa have equal weight as very abundant taxa. Shannon's and Simpson's diversity indexes do account for differential abundances but in a simplistic way by the shape of taxa distributions, and these also show significant differences between cloacal and cecal communities (1.84 and 3.71 and 0.73 and 0.93 for cloacal and cecal communities respectively; Kruskal-Wallis test, chi-square = 64.341, P = 1 × 10−15 and chi-square = 60.947, P = 1 × 10−15, respectively).
Figure 1.
Total number of taxa in cloacal and cecal samples. Venn diagram showing the total and overlap of species in cloaca and ceca.
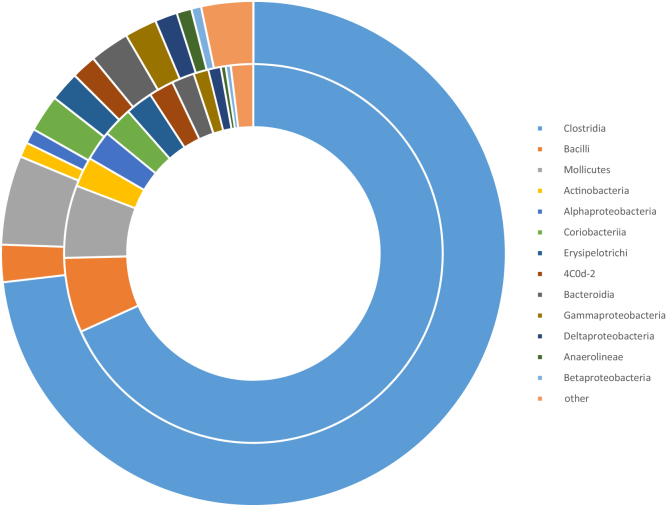
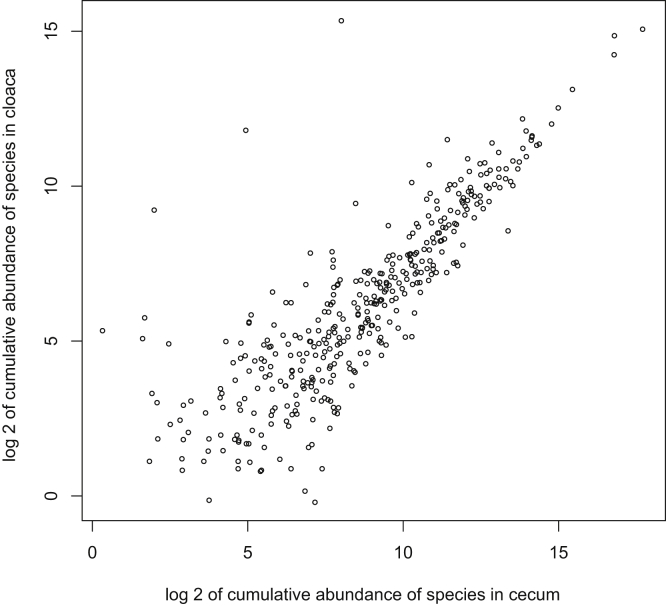
However, more sophisticated analysis that accounts for the differential abundances of specific taxa revealed cecal and cloacal community abundances overlap by >99%: the 224 of 693 taxa discovered in ceca but not cloaca comprised just 0.51% of the cecal community abundance. Figure 2 shows the community composition of cloaca and ceca at class level appears reasonably similar. Moreover, analysis of cumulative taxa abundances in cloaca and ceca reveal these were significantly positively correlated (Spearman's test, Rho = 0.66, P = 2 × 10−16); that is, taxa that are abundant in ceca are also abundant in cloaca (Figure 3).
Figure 2.
Main bacterial classes recorded in cloacal and cecal samples. Pie chart showing the similarity in number of species in cloaca (internal circle) and ceca (external circle) differentiated at class level. Classes representing less than 0.5% of the total number of species are collapsed to the “other” category.
Figure 3.
Correlation between cumulative abundances of taxa in cloacal and cecal samples. The plot represents the correlations between cumulative abundances of cecal and cloacal samples. Indicator species of cloacal samples are not included in the present picture. Pearson's cor = 0.76, P ≤ 2 × 10−16, Spearman's Rho = 0.72, P ≤ 2 × 10−16.
Despite these significant microbiome similarities, especially for abundance, standard relatively sensitive PERMANOVA community ecology tests report both the types (PERMANOVA, R2 = 29.469, P < 0.001) and abundances (PERMANOVA, R2 = 27.009, P < 0.001) of taxa significantly differ between cecal and cloacal samples, and this is presumably driven by difference in the rarer taxa. Table 1 summarizes components of the bacterial communities that have greatest and least similarity in terms of relative abundance in cloaca and ceca at different taxonomic levels. A full description of the difference in relative abundance at multiple taxonomic levels and tests for difference between cloacal and ceca are reported in Supplementary Table 4.
Table 1.
Taxonomic groups having similar and different relative abundances in cloacal and cecal samples.
| Taxonomic level taxonomic group | Cloacal |
Cecal |
Kruskal-Wallis P(fdr) | ||||
|---|---|---|---|---|---|---|---|
| Avg relative abundance | SD | Avg relative abundance | SD | ||||
| Taxonomic groups with most similar abundance (fdr <5 × 10−3) | |||||||
| Phylum | Verrucomicrobia | 0.064 | 0.440 | 0.310 | 1.665 | 0.41199 | 0.54962 |
| Class | Flavobacteriia | 0.367 | 2.515 | 0.113 | 0.638 | 0.37969 | 0.54962 |
| Alphaproteobacteria | 16.433 | 29.062 | 7.859 | 12.786 | 0.50254 | 0.51139 | |
| Actinobacteria | 1741.869 | 1828.78 | 2656.528 | 2575.064 | 1.3261 | 0.35835 | |
| Betaproteobacteria | 37.560 | 100.039 | 44.557 | 102.307 | 1.7064 | 0.35835 | |
| Order | Myxococcales | 0.717 | 4.917 | 0.093914 | 0.623 | 0.00097786 | 0.9751 |
| Xanthomonadales | 0.030 | 0.203 | 0.152 | 1.008 | 0.0039115 | 0.96043 | |
| Desulfuromonadales | 0.653 | 4.474 | 0.128 | 0.604 | 0.37969 | 0.54962 | |
| Flavobacteriales | 0.367 | 2.515 | 0.113 | 0.638 | 0.37969 | 0.54962 | |
| SHA-98 | 0.734 | 5.030 | 0.185 | 0.881 | 0.37969 | 0.54962 | |
| Turicibacterales | 0.175 | 1.201 | 0.099 | 0.484 | 0.37969 | 0.54962 | |
| Taxonomic groups with least similar abundance (fdr <5 × 10−3) | |||||||
| Taxonomic groups with greater relative abundance in cloaca (fdr <5 × 10−3) | |||||||
| Phylum | Firmicutes | 229452.800 | 183758.400 | 24987.86 | 6029.915 | 65.107 | 3 × 10−14 |
| Proteobacteria | 7579.893 | 16863.260 | 180.677 | 327.818 | 33.336 | 5 × 10−8 | |
| Class | Bacilli | 215675.700 | 186524.900 | 1134.072 | 871.034 | 67.435 | <2 × 10−16 |
| Unknown species∗ | 93.831 | 273.191 | 0 | 0 | 27.71 | 8 × 10−7 | |
| Gammaproteobacteria | 7521.226 | 16858.15 | 113.708 | 294.592 | 39.581 | 3 × 10−9 | |
| Order | Actinomycetales | 1097.602 | 1258.386 | 0.827 | 2.390 | 72.291 | <2 × 10−16 |
| Bacillales | 3134.243 | 6656.603 | 56.747 | 81.855 | 58.492 | 5 × 10−13 | |
| Enterobacteriales | 7520.116 | 16858.330 | 113.130 | 294.686 | 39.681 | 3 × 10−9 | |
| Lactobacillales | 212541.3 | 187219.9 | 1077.226 | 886.561 | 67.435 | <2 × 10−16 | |
| Rickettsiales | 8.009 | 21.666 | 0 | 0 | 10.356 | 4 × 10−3 | |
| Streptophyta | 90.845 | 272.048 | 0 | 0 | 26.192 | 2 × 10−6 | |
| Taxonomic groups with greater relative abundance in ceca (fdr <5 × 10−3) | |||||||
| Phylum | Bacteroidetes | 118.768 | 148.397 | 318.616 | 289.758 | 20.894 | 2 × 10−5 |
| Cyanobacteria | 178.086 | 281.062 | 680.720 | 864.613 | 20.467 | 3 × 10−5 | |
| Tenericutes | 25.273 | 53.891 | 97.700 | 95.285 | 34.38 | 3 × 10−8 | |
| Thermotogae | 0 | 0 | 0.899 | 1.816 | 13.147 | 1 × 10−5 | |
| Class | 4C0d-2 | 84.255 | 125.320 | 680.720 | 864.613 | 39.102 | 4 × 10−9 |
| Bacteroidia | 118.401 | 148.675 | 318.503 | 289.775 | 20.917 | 2 × 10−5 | |
| Clostridia | 13716.210 | 11504.970 | 23707.880 | 5901.790 | 25.835 | 2 × 10−6 | |
| Coriobacteriia | 74.321 | 125.680 | 630.425 | 557.199 | 57.63 | 6 × 10−13 | |
| Deltaproteobacteria | 4.674 | 11.466 | 14.486 | 15.543 | 23.181 | 7 × 10−6 | |
| Erysipelotrichi | 60.904 | 138.151 | 145.905 | 98.019 | 35.693 | 2 × 10−8 | |
| Mollicutes | 24.665 | 53.320 | 96.493 | 95.143 | 34.958 | 3 × 10−8 | |
| Thermoplasmata | 0 | 0 | 0.899 | 1.816 | 13.147 | 3 × 10−3 | |
| Order | Bacteroidales | 118.401 | 148.675 | 318.503 | 289.775 | 20.917 | 2 × 10−5 |
| Bifidobacteriales | 644.267 | 1139.626 | 2655.702 | 2574.645 | 19.152 | 5 × 10−5 | |
| Clostridiales | 13715.480 | 11503.070 | 23707.530 | 5901.867 | 25.835 | 2 × 10−6 | |
| Coriobacteriales | 74.321 | 125.680 | 630.425 | 557.199 | 57.63 | 6 × 10−13 | |
| Desulfovibrionales | 3.304 | 9.873 | 13.722 | 15.738 | 24.648 | 4 × 10−6 | |
| Erysipelotrichales | 60.904 | 138.151 | 145.905 | 98.018 | 35.693 | 3 × 10−8 | |
| RF39 | 24.199 | 52.812 | 95.481 | 95.419 | 34.764 | 3 × 10−8 | |
| Thermotogales | 0 | 0 | 0.899 | 1.816 | 13.147 | 1 × 10−3 | |
| YS2 | 84.255 | 125.320 | 680.720 | 864.613 | 39.102 | 4 × 10−9 | |
The table summarizes the taxonomic groups with the most and least similar relative abundances between cloaca and ceca, and the groups with greater abundance either in cloaca or in ceca. Average abundance in the cloacal and cecal samples, SD, Benjamini-Hochberg false discovery rate (fdr) corrections are included.
unknown species belonging to phylum Cyanobacteria.
Within and Between Bird Analyses
Thus far, the analyses have evaluated overall cecal and cloacal microbiomes but have not accounted for the fact that specific paired cecal and cloacal samples come from the same bird. We therefore investigated the associations between cecal and cloacal samples from the same animal to those from all other animals using pairwise binary (presence/absence) and nonbinary (abundance) Jaccard distances. Analyses of these similarity ranks show cloaca communities are on average significantly more similar to the cecal communities from the same bird than to cecal communities from other birds for both the types and abundance of taxa (Kruskal-Wallis test, chi-square = 12.02, P = 5 × 10−4 and chi-square = 8.4659, P = 4 × 10−3 respectively). Thirteen and 12 of the 44 comparisons ranked the cloacal to cecal communities from the same bird highest for presence/absence and abundance, respectively, and the probability of recovering this by chance if there were no correlation between cloacal and cecal microbiomes from the same animal is ∼4.3−20. Another way to understand this is to ask what the probability of being able to identify the correct paired cecal sample given a cloacal sample is. Given that 12 to 13 of the 44 comparisons had the correct pair ranked highest (for both types and abundances), this corresponds to a ∼28% chance of being able to identify the correct cecal microbiome given a cloacal microbiome sample, and this rises to ∼50% when the correct pairs are ranked in the top 5. Thus, the considerable bird-to-bird variance in microbiomes, especially for cloaca, partially masks the signal for ceca:cloaca correlation when the combined data are analyzed, but the analyses of paired ceca:cloaca microbiomes from the same bird reveals reasonable correlations.
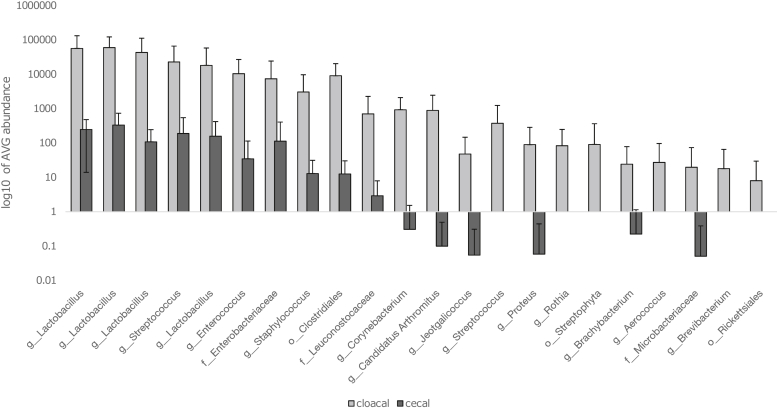
Having demonstrated that there is a detectable correlation between cecal and cloacal bacterial communities in the same bird, we next focus on those components of the community that differed the most to understand discrepancies better. Twenty-two taxa had significantly greater abundance in cloacal compared with cecal communities (and are thus classed as “indicator species” at Benjimni-Hochberg corrected P < 0.005). Figure 4 reports these indicator species and their abundances and indicates that some Firmicutes and Proteobacteria may be relatively more abundant in the cloaca. However, the 10 taxa most overrepresented in cloaca from indicator taxa analysis were also above average abundance in cecal samples (Figure 4). When the abundance of taxa that were present in both ceca and cloaca from the same bird were analyzed, samples from 30 of 44 birds had positive correlations, sample pairs with the lowest P-values (P < 0.0001) all had positive correlations, and all but 2 of the 19 correlations that were significant (P < 0.05) were positive (Supplementary Table 5). There was also a positive correlation between Spearman's Rho values and number of taxa recorded in the cloacal samples (Spearman's Rho = 0.506, P = 4.6 × 10−4) revealing that cloacal samples with greater numbers of taxa are better estimates of cecal samples from the same individual.
Figure 4.
Cloacal indicator taxa. Mean (±SD) log abundance of the indicator species (at adjusted P < 0.005) of cloacal samples (light gray) to abundances in cecal samples (dark gray).
Discussion
Several studies have described the bacterial community compositions of broiler GIT (e.g., Oakley et al., 2014; Kumar et al., 2018), but the focus of the present study was whether there is any value in sampling cloaca to provide an estimate of the cecal microbiomes for cost and animal welfare reasons. We are aware of only 1 previous study that has compared cloacal to cecal microbial communities in broiler poultry, and the findings here are in-line with those reported in Stanley et al. (2015) in that there are correlations in terms of types of species between ceca and cloaca. This study estimates ∼60% of the taxa identified in ceca were also in cloaca, which is less than inferred by Stanley et al. (2015), who reported that ∼90% of the sequences were shared in broilers in Australia. However, the nonshared taxa accounted for a very low percentage of the total biodiversity in both studies: 0.49% of the present study and 0.75% in Stanley et al. (2015). The additional analyses conducted here, including paired cecal-cloacal analyses, show there is a significant correlation between taxa abundance in ceca and cloaca (Spearman's test, Rho = 0.66, P = 2 × 10−16), and that taxa that are abundant in ceca are also in cloaca. Rare taxa in ceca were less well represented in cloaca; we did not evaluate the potential functions of these taxa and so cannot comment on whether the differences in rare taxa between cloaca and ceca may or may not lead to inaccurate inferences of cecal microbiota function.
Several factors are known to influence the composition of bacterial communities in chicken GIT including diet and food additives (Forte et al., 2018; Grant et al., 2018), sex and body weight (Lee et al., 2017), age (Lu et al., 2003), geography (Videnska et al., 2014; Zhou et al., 2016), as extensively reviewed in Kers et al. (2018). Here we found that there was no overall effect of bird location, age or flock origin, on numbers, type or abundance of taxa, but that when ceca were analyzed separately, there was a significant but small (5%) effect of these factors on the types and abundance of taxa. One source of this variance may have been due to the difference in feed in 2 of the flocks (D and F were in grower and the rest finisher feed). Another influence may have been because it was difficult to collect the same volume of sample from both ceca and cloaca. However, rarefaction (discovery) curves analysis of samples show all to plateau which suggest the various sample volumes were all efficient at estimating both cloacal and cecal taxa despite differential sample volumes (Supplementary Figure 1). Cecal communities were more variable than cloacal ones, and some of the variance may be because of the more cecal specific age and location effects. However, the within bird analyses will have controlled for this to some extent. A recent study by Jurburg et al. (2019) demonstrated that age of birds significantly predicted the taxa richness of fecal samples and that this stabilizes after 14 d of age. The age of the animal investigated in the present study ranged between 19 and 31 d posthatch, and thus, our results are in-line with Jurburg et al. (2019) as we found no significant effect of animal age on taxa richness in either cecal or cloacal communities. Ceca had significantly greater microbial biodiversity than cloaca, which is in line with several studies showing ceca has the greatest biodiversity along the chicken gut (Apajalahti et al., 2004; Gong et al., 2007; Grant et al., 2018).
Family Clostridiaceae was significantly more abundant in cecum samples: members of this family are not only well-known chicken pathogens (such as C. perfringens, Van Immerseel et al., 2004) but also some beneficial groups of bacteria (such as Clostridium clusters IV and XIV; Frank et al., 2007; Sokol et al., 2009). Overall, the microbial composition of the GIT revealed here reflects previous reports. Firmicutes represented nearly 75% of the bacterial taxa and ∼95% of the microbial community in terms of abundance which is in line with several studies (e.g., Oakley et al., 2014; Kumar et al., 2018). However, the data here do show small difference with some avian studies; for example, Mohd Shaufi et al. (2015) reported class Bacteroidia to represent 17 to 22% of biodiversity in male commercial Cobb 500 ceca, whereas our data showed only about 1%.
Overall, there was a positive correlation between taxa types and abundances in cloacal and cecal bacterial communities, and when we conducted an analysis that compared samples from ceca and cloaca from the same bird, we discovered that these were more similar than expected by chance given the rest of the samples in the study. This indicates that cloacal microbiomes reflect cecal microbiomes from the same animal to a first approximation, especially for abundant taxa, and thus, sampling cloaca is a reasonable way to estimate cecal microbiomes. However, sampling cloaca does not allow a deep insight into cecal microbiota, and the correlation with less abundant taxa is poorer. That cloacal microbiomes reflect but do not perfectly represent cecal microbiomes is an aspect that needs to be balanced with animal welfare when choosing a study design. Well-designed studies which include appropriate controls will likely be able to estimate the relative major changes in cecal microbiomes via cloacal samples. This approach may be less valuable for studies that demand very precisely estimates from a few animals, but it will be much more valuable for population level larger scale studies tracking the general response of flocks to changes in husbandry. Together, findings from Europe and Australasia demonstrate that sampling cloaca shows promise as a method to evaluate gut cecal microbiomes from live broiler poultry, and this will reduce cost and increase livestock welfare for both husbandry and research purposes.
Acknowledgments
The authors would like to thank St. David's Poultry Vets, and Abbie Graham and Megan Nelson from Devenish Nutrition for sampling.
Conflict of Interest Statement: The authors did not provide any conflict of interest statement
Footnotes
Supplementary data associated with this article can be found in the online version at https://doi.org/10.1016/j.psj.2020.08.015.
Supplementary data
References
- Amerah A.M., Péron A., Zaefarian F., Ravindran V. Influence of whole wheat inclusion and a blend of essential oils on the performance, nutrient utilisation, digestive tract development and ileal microbiota profile of broiler chickens. Br. Poult. Sci. 2011;52:124–132. doi: 10.1080/00071668.2010.548791. [DOI] [PubMed] [Google Scholar]
- Anderson M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc Accessed Sep. 2017.
- Apajalahti J., Kettunen A., Graham H. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. Worlds Poult. Sci. J. 2004;60:223–232. [Google Scholar]
- Baldwin S., Hughes R.J., Hao Van T.T., Moore R.J., Stanley D. At-hatch administration of probiotic to chickens can introduce beneficial changes in gut microbiota. PLoS One. 2018;13:e0194825. doi: 10.1371/journal.pone.0194825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa A., Balagué V., Valera F., Martínez A., Benzal J., Motas M., Diaz J.I., Mira A., Pedrós-Alió C. Age-related differences in the gastrointestinal microbiota of Chinstrap penguins (Pygoscelis Antarctica) PLoS One. 2016;11:e0153215. doi: 10.1371/journal.pone.0153215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.E., Thomas R., Williams M., Zalasiewicz J., Edgeworth M., Miller H., Coles B., Foster A., Burton E.J., Marume U. The broiler chicken as a signal of a human reconfigured biosphere. R. Soc. Open Sci. 2018;5:180325. doi: 10.1098/rsos.180325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasato I., De Marco M., Rotolo L., Renna M., Lussiana C., Dabbou S., Capucchio M.T., Biasibetti E., Costa P., Gai F., Pozzo L., Dezzutto D., Bergagna S., Martínez S., Tarantola M., Gasco L., Schiavone A. Effects of dietary Tenebrio molitor meal inclusion in free-range chickens. J. Anim. Physiol. Anim. Nutr. (Berl). 2016;100:1104–1112. doi: 10.1111/jpn.12487. [DOI] [PubMed] [Google Scholar]
- Bokulich N.A., Kaehler B.D., Rideout J.R., Dillon M., Bolyen E., Knight R., Huttley G.A., Caporaso J.G. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2's q2-feature-classifier plugin. Microbiome. 2018;6:90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C., Al-Ghalith G.A. Qiime 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints. 2018;6:e27295v2. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavijo V., Flórez M.J.V. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult. Sci. 2018;97:1006–1021. doi: 10.3382/ps/pex359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clench M.H., Mathias J.R. The avian cecum: a review. Wilson Bull. 1995;107:93–121. [Google Scholar]
- Díaz Carrasco J.M., Redondo E.A., Pin Viso N.D., Redondo L.M., Farber M.D., Fernández Miyakawa M.E. Tannins and bacitracin differentially modulate gut microbiota of broiler chickens. Biomed. Res. Int. 2018;2018:1879168. doi: 10.1155/2018/1879168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drovetski S.V., O'Mahoney M., Ransome E.J., Matterson K.O., Lim H.C., Chesser R.T., Graves G.R. Spatial organization of the gastrointestinal microbiota in urban Canada geese. Sci. Rep. 2018;8:3713. doi: 10.1038/s41598-018-21892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne M., Legendre P. Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol. Monogr. 1997;67:345–366. [Google Scholar]
- Forte C., Manuali E., Abbate Y., Papa P., Vieceli L., Tentellini M., Trabalza-Marinucci M., Moscati L. Dietary Lactobacillus acidophilus positively influences growth performance, gut morphology, and gut microbiology in rurally reared chickens. Poult. Sci. 2018;97:930–936. doi: 10.3382/ps/pex396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganz H.H., Doroud L., Firl A.J., Hird S.M., Eisen J.A., Boyce W.M. Community-level differences in the microbiome of healthy wild mallards and those infected by influenza A viruses. mSystems. 2017;2 doi: 10.1128/mSystems.00188-16. e00188-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J., Si W., Forster R.J., Huang R., Yu H., Yin Y., Yang C., Han Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol. Ecol. 2007;59:147–157. doi: 10.1111/j.1574-6941.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- Grant A., Gay C.G., Lillehoj H.S. Bacillus spp. as direct-fed microbial antibiotic alternatives to enhance growth, immunity, and gut health in poultry. Avian Pathol. 2018;47:339–351. doi: 10.1080/03079457.2018.1464117. [DOI] [PubMed] [Google Scholar]
- Grond K., Sandercock B.K., Jumpponen A., Zeglin L.H. The avian gut microbiota: community, physiology and function in wild birds. J. Avian Biol. 2018;49:e01788. [Google Scholar]
- Hang J., Desai V., Zavaljevski N., Yang Y., Lin X., Satya R.V., Martinez L.J., Blaylock J.M., Jarman R.G., Thomas S.J., Kuschner R.A. 16S rRNA gene pyrosequencing of reference and clinical samples and investigation of the temperature stability of microbiome profiles. Microbiome. 2014;2:31. doi: 10.1186/2049-2618-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander M., Wolfe D.A. John Wiley & Sons; New York: 1973. Pages 185–194 in Nonparametric Statistical Methods. (Kendall and Spearman tests) [Google Scholar]
- Jurburg S.D., Brouwer M.S.M., Ceccarelli D., Goot J., Jansman A.J.M., Bossers A. Patterns of community assembly in the developing chicken microbiome reveal rapid primary succession. MicrobiologyOpen. 2019;8:e821. doi: 10.1002/mbo3.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kers J.G., Velkers F.C., Fischer E.A.J., Hermes G.D.A., Stegeman J.A., Smidt H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018;9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirch W. Springer; Dordrecht: 2008. Pearson’s Correlation Coefficient. Encyclopedia of Public Health. [Google Scholar]
- Kumar S., Chen C., Indugu N., Werlang G.O., Singh M., Kim W.K., Thippareddi H. Effect of antibiotic withdrawal in feed on chicken gut microbial dynamics, immunity, growth performance and prevalence of foodborne pathogens. PLoS One. 2018;13:e0192450. doi: 10.1371/journal.pone.0192450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.C., Kil D.Y., Sul W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017;55:939–945. doi: 10.1007/s12275-017-7202-0. [DOI] [PubMed] [Google Scholar]
- Lu J., Idris U., Harmon B., Hofacre C., Maurer J.J., Lee M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003;69:6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie P.J., Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkeviciene L., Ruzauskaite N., Klimiene I., Siugzdiniene R., Dailidaviciene J., Virgailis M., Mockeliunas R., Ruzauskas M. Microbiome and antimicrobial resistance genes in microbiota of cloacal samples from European herring gulls (Larus argentatus) J. Vet. Res. 2017;61:27–35. doi: 10.1515/jvetres-2017-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Shaufi M.A., Sieo C.C., Chong C.W., Gan H.M., Ho Y.W. Deciphering chicken gut microbial dynamics based on high-throughput 16S rRNA metagenomics analyses. Gut Pathog. 2015;7:4. doi: 10.1186/s13099-015-0051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muletz Wolz C.R., Yarwood S.A., Campbell Grant E.H., Fleischer R.C., Lips K.R. Effects of host species and environment on the skin microbiome of Plethodontid salamanders. J. Anim. Ecol. 2018;87:341–353. doi: 10.1111/1365-2656.12726. [DOI] [PubMed] [Google Scholar]
- Nakphaichit M., Thanomwongwattana S., Phraephaisarn C., Sakamoto N., Keawsompong S., Nakayama J., Nitisinprasert S. The effect of including Lactobacillus reuteri KUB-AC5 during post-hatch feeding on the growth and ileum microbiota of broiler chickens. Poult. Sci. 2011;90:2753–2765. doi: 10.3382/ps.2011-01637. [DOI] [PubMed] [Google Scholar]
- Oakley B.B., Buhr R.J., Ritz C.W., Kiepper B.H., Berrang M.E., Seal B.S., Cox N.A. Successional changes in the chicken cecal microbiome during 42 days of growth are independent of organic acid feed additives. BMC Vet. Res. 2014;10:282. doi: 10.1186/s12917-014-0282-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Szoecs E., Wagner H. Vegan: Community Ecology. Package. R package version 2.5-2. 2018. https://CRAN.R-project.org/package=vegan Accessed Sep. 2020.
- Panasevich M.R., Wankhade U.D., Chintapalli S.V., Shankar K., Rector R.S. Cecal versus fecal microbiota in Ossabaw swine and implications for obesity. Physiol. Genomics. 2018;50:355–368. doi: 10.1152/physiolgenomics.00110.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang W., Vogensen F.K., Nielsen D.S., Hansen A.K. Faecal and caecal microbiota profiles of mice do not cluster in the same way. Lab. Anim. 2012;46:231–236. doi: 10.1258/la.2012.011128. [DOI] [PubMed] [Google Scholar]
- Paulson J.N., Stine O.C., Bravo H.C., Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R.E., Jr. Bacterial enteritides of poultry. Poult. Sci. 1998;77:1159–1165. doi: 10.1093/ps/77.8.1159. Review. [DOI] [PubMed] [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;18:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M., Naveed M., BiBi J., Kamboh A.A., Arain M.A., Shah Q.A., Alagawany M., El-Hack M.E.A., Abdel-Latif M.A., Yatoo M.I., Tiwari R., Chakraborty S., Dhama K. The promising pharmacological effects and therapeutic/medicinal applications of Punica granatum L. (pomegranate) as a functional food in humans and animals. Recent Pat Inflamm. Allergy Drug Discov. 2018;12:24–38. doi: 10.2174/1872213X12666180221154713. [DOI] [PubMed] [Google Scholar]
- Shang Y., Kumar S., Oakley B., Kim W.K. Chicken gut microbiota: importance and detection technology. Front. Vet. Sci. 2018;5:254. doi: 10.3389/fvets.2018.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communications. Bell Syst. Tech. J. 1948;27:379–423. [Google Scholar]
- Shapiro S.K., Sarles W.B. Microorganisms in the intestinal tract of normal chickens. J. Bacteriol. 1949;58:531–544. doi: 10.1128/jb.58.4.531-544.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E.H. The measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Sokol H., Seksik P., Furet J.P., Firmesse O., Nion-Larmurier I., Beaugerie L., Cosnes J., Corthier G., Marteau P., Doré J. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm. Bowel Dis. 2009;15:1183–1189. doi: 10.1002/ibd.20903. [DOI] [PubMed] [Google Scholar]
- Stanley D., Geier M.S., Chen H., Hughes R.J., Moore R.J. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. 2015;15:51. doi: 10.1186/s12866-015-0388-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X., Rehman M.U., Huang S., Jiang X., Zhang H., Li J. Comparative analysis of gut microbial community in healthy and tibial dyschondroplasia affected chickens by high throughput sequencing. Microb. Pathog. 2018;118:133–139. doi: 10.1016/j.micpath.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Van Immerseel F., De Buck J., Pasmans F., Huyghebaert G., Haesebrouck F., Ducatelle R. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. 2004;33:537–549. doi: 10.1080/03079450400013162. Review. [DOI] [PubMed] [Google Scholar]
- Videnska P., Rahman M.M., Faldynova M., Babak V., Matulova M.E., Prukner-Radovcic E., Krizek I., Smole-Mozina S., Kovac J., Szmolka A., Nagy B., Sedlar K., Cejkova D., Rychlik I. Characterization of egg laying hen and broiler fecal microbiota in poultry farms in Croatia, Czech Republic, Hungary and Slovenia. PLoS One. 2014;9:e110076. doi: 10.1371/journal.pone.0110076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Nesengani L.T., Gong Y., Yang Y., Lu W. 16s Rrna gene sequencing reveals effects of photoperiod on cecal microbiota of broiler roosters. PeerJ. 2018;6:e4390. doi: 10.7717/peerj.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S., Xu Z.Z., Peddada S., Amir A., Bittinger K., Gonzalez A., Lozupone C., Zaneveld J.R., Vázquez-Baeza Y., Birmingham A., Hyde E.R., Knight R. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yegani M., Korver D.R. Factors affecting intestinal health in poultry. Poult. Sci. 2008;87:2052–2063. doi: 10.3382/ps.2008-00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Simon S.E., Johnson J.A., Allen M.S. Spatial microbial composition along the gastrointestinal tract of Captive Attwater's Prairie chicken. Microb. Ecol. 2017;73:966–977. doi: 10.1007/s00248-016-0870-1. [DOI] [PubMed] [Google Scholar]
- Zhou X., Jiang X., Yang C., Ma B., Lei C., Xu C., Zhang A., Yang X., Xiong Q., Zhang P., Men S., Xiang R., Wang H. Cecal microbiota of Tibetan Chickens from five geographic regions were determined by 16S rRNA sequencing. MicrobiologyOpen. 2016;5:753–762. doi: 10.1002/mbo3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw amplicon sequencing data from this study is publicly available in the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra/) under the BioProject ID: PRJNA559084.