Abstract
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is caused by mutations in TYMP, the gene encoding the enzyme thymidine phosphorylase (TP). TP dysfunction results in systemic accumulation of the noxious TP substrates thymidine and deoxyuridine. Gene therapy using either a lentiviral vector or adeno-associated vector (AAV) has proven to be a feasible strategy, as both vectors restore biochemical homeostasis in a murine model of the disease. This study shows that the effect of an AAV containing the TYMP coding sequence transcriptionally targeted to the liver persists long term in mice. Although the vector copy number was diluted and AAV-mediated liver TP activity eventually reduced or lost after 21 months at the lowest vector doses, the effect was sustained (with a negligible decrease in TP activity) and fully effective on nucleoside homeostasis for at least 21 months at a dose of 2 × 1012 vg/kg. Macroscopic visual inspection of the animals' organs at completion of the study showed no adverse effects associated with the treatment. These results further support the feasibility of gene therapy for MNGIE.
Keywords: : AAV, liver/metabolic, MNGIE, thymidine phosphorylase, mitochondria
Introduction
Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE) is a mitochondrial disorder caused by autosomal recessive mutations in the nuclear gene TYMP, which encodes the cytosolic enzyme thymidine phosphorylase (TP).1–3 MNGIE patients lack TP activity, and this deficit leads to systemic accumulation of TP substrates, the nucleosides thymidine (dThd) and deoxyuridine (dUrd), which are precursors of the salvage pathway of deoxyribonucleoside triphosphates (dNTPs), used for DNA synthesis. This expanded nucleoside concentration changes the mitochondrial dNTP pool and interferes with proper mitochondrial DNA (mtDNA) replication. As a result of the dNTP imbalance, MNGIE patients develop mtDNA depletion, multiple deletions, and point mutations in certain tissues, which leads to mitochondrial dysfunction4,5 (see Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/hum). MNGIE is a fatal disease, clinically characterized by gastrointestinal dysmotility, progressive external ophthalmoplegia, peripheral neuropathy, diffuse leukoencephalopathy on brain magnetic resonance imaging, and mitochondrial dysfunction. The average age of disease onset is 18 years, but most patients experience their first symptoms in childhood. The condition progressively degenerates until death ensues, usually due to the gastrointestinal complications, at a mean age of 37 years.6
All the hitherto proposed therapeutic approaches for MNGIE have been focused on reducing dThd and dUrd concentrations to normal levels. The systemic overload of these two compounds is the actual biochemical cause of the disease. The noxious effect of dThd excess on mtDNA replication has been demonstrated in different in vitro and in vivo models,7–10 and therefore these constitute good biomarkers to predict the evolution of the disease. Only treatments that provide a permanent biochemical correction result in clinical improvement. Allogeneic hematopoietic stem-cell transplantation (alloHSCT) is the most widely used therapy for MNGIE, as it has proven to be effective in several patients; reduction of dThd and dUrd levels was associated with clinical stabilization or improvement in MNGIE patients treated with alloHSCT.11–13 In two recent reports, orthotopic liver transplantation was also successfully used to treat MNGIE.14,15 The rationale for these strategies is that both hematopoietic and liver tissue show high TP activity in humans. Therefore, engrafted donor tissue clears the dThd and dUrd excess in recipients. However, both of these options are invasive therapies and require compatible donors, and, in the case of alloHSCT, the procedure is associated with high mortality and morbidity rates in MNGIE patients.12
Previously, it was proposed that gene therapy, involving introduction of a functional copy of the human TYMP coding sequence (hcTYMP) into MNGIE patients' cells, could be a noninvasive alternative therapy that additionally would not require donor tissue. The existence of a murine model of the disease16 has allowed this approach to be tested preclinically. Although this animal model does not recapitulate the clinical phenotype of the disease, probably due to the short life expectancy of mice, in combination with some differences in deoxynulceoside metabolism between mice and humans, it reproduces the biochemical imbalances observed in MNGIE patients, thus constituting a useful tool to investigate the effects of experimental therapies on the biomarkers of this disorder, that is, the systemic levels of dThd and dUrd.
Using this animal model, the feasibility of gene therapy was demonstrated using lentiviral vectors targeting the hematopoietic system. Syngeneic transplantation of transduced hematopoietic progenitors into the Tymp/Upp1–/– double knockout (KO) MNGIE model was able to correct the biochemical derangement occurring in the disease.17 However, on long-term follow-up of treated mice, survival was low due to the transplantation procedure, which included total body irradiation of recipient animals before progenitor cell infusion.18 An alternative approach, using an adeno-associated virus (AAV) vector targeting the liver, also prevented biochemical imbalances in mice. Intravenous administration of the AAV vector containing hcTYMP at doses as low as 2 × 1011 genome copies/kg led to a permanent reduction in systemic dThd and dUrd levels to normal values in about 50% of treated mice, and higher doses reduced nucleoside levels in virtually all treated mice.19 This study reports that the therapeutic effect achieved with the AAV vector persists throughout the life of the murine model with no signs of adverse effects.
Materials and Methods
Vector construction, production, and titration
The AAV2/8 vector containing hcTYMP under the control of the tyroxine-binding globulin (TBG) promoter was constructed, produced by triple transfection of 293 cells, and titrated by quantitative polymerase chain reaction (qPCR) and dot blot analysis, as indicated in a previous study.19
Animal procedures
All animal procedures were performed using protocols approved by the authors' Institutional Review Board and committee on animal care and use. Eight- to 12-week-old Tymp/Upp1−/− mice16 were treated with a single intravenous injection (tail vein) of AAV2/8-TBG-hcTYMP. Three groups of animals were treated with three different doses: 2 × 1011, 1012, and 2 × 1012 vector genomes (vg)/kg. Blood samples (EDTA) were collected from the saphenous vein 2 weeks before treatment and every 2–4 weeks after treatment, starting from week 1 and continuing for 28 weeks. Additional blood samples were collected 65, 75, 80, and 88 weeks after treatment. All the animals included in this study were males, except a subgroup of female mice used for the study on the influence of sex on the effect of the treatment.
When mice showed signs of pain, suffering, distress, or lasting harm, they were euthanized before the planned end of the study for humanitarian reasons, according to the following endpoint rule based on a scoring system included in the European Directive 2010/63/UE (Annex III 3.1 b). The parameters that were monitored were body weight, body condition, external appearance, and behavior. Each parameter was scored from 0 to 3 based on different indicators, and euthanasia was practiced in those mice whose score reached 3 in a single parameter or 5 in the sum of the different parameters.
TP activity and nucleoside determination
Plasma dThd and dUrd concentrations were analyzed by high-performance liquid chromatography with ultraviolet detection, as previously described.17 TP activity and tissue nucleoside concentrations were measured in mice 34 weeks after treatment and in untreated, age-matched mice. After killing the mice by cervical dislocation, tissues were collected and immediately frozen in liquid nitrogen and stored at −80°C until analysis. Frozen samples were homogenized in lysis buffer (50 mM of Tris-HCl, pH 7.2, 10 mL/L of Triton X-100, 2 mM of phenylmethylsulfonyl fluoride, 0.2 mL/L of 2-mercaptoethanol) in a Potter homogenizer. The homogenates were centrifuged at 20,000 g for 30 min at 4°C, and supernatants were separated into two aliquots. As described elsewhere, one aliquot was used for protein determination20 and TP activity determination.21 The other aliquot was frozen and later used to measure nucleosides by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). To prevent in vitro degradation of dThd and dUrd during the homogenization procedure, for samples with TP activities >100 nmol Thy/h/mg prot, a second piece of tissue was homogenized in the presence of 100 μM of 5-bromouracil (TP inhibitor) for the determination of these nucleosides. Thawed supernatants were centrifuged at 20,000 g for 10 min at 4°C to eliminate any remaining particles, and clean supernatants were deproteinized by ultrafiltration (10 kDa Amicon Ultra filters; Merck Millipore, Billerica, MA) at 14,000 g for 30 min at 4°C. Five microliters of deproteinized homogenate were injected into an Acquity UPLC-MS/MS instrument (Acquity UPLC-Xevo TQ Mass Spectrometer; Waters, Milford, MA) using an Acquity UPLC BEH C18 column (100 × 2.1 mm, 130 Å pore, 1.7 μm particle; Waters). The components of the sample were resolved and detected, as previously described.19 Calibration curves made with aqueous standards were processed in parallel, and concentrations were obtained from interpolation of the peak areas.
Vector copy number
DNA was extracted from liver with the QIAamp DNA mini kit (Qiagen, Hilden, Germany). Detection and quantification of vector genome copies per cell was performed by quantitative PCR (qPCR) in the ABI PRISM 7900 sequence detection system (Applied Biosystems, Foster City, CA). hcTYMP DNA was quantified using the pre-designed TaqMan MGB gene expression assay Hs00157317_m1 (Applied Biosystems), and was referred to the single copy nuclear gene Ang1 using the predesigned TaqMan MGB gene expression assay Mm00833184_s1 (Applied Biosystems). The quantifications were based on a standard curve prepared with different dilutions of vectors containing hcTYMP DNA or a specific region of the Ang1 gene.
Liver mitochondrial dNTP quantification
Liver mitochondria were isolated, as previously described.8 Isolated mitochondria (0.5 mg of protein) were treated with trichloroacetic acid (final concentration 0.5 M) and centrifuged at 20,000 g for 5 min at 4°C. Supernatants were neutralized with 1.5 volumes of 0.5 M tri-N-octylamine in Freon (1,1,2-trichlorotrifluoroethane) and centrifuged for 10 min at 10,000 g at 4°C. Half the aqueous upper phase was recovered and dried under a speed vacuum. Dry dNTP extracts were dissolved in 40 μL of 40 mM Tris-HCl (pH 7.4) and stored at −80°C until measurement. For mitochondrial dNTP quantification, the previously described polymerase-based assay8 was used with some modifications. Briefly, 10 μL of reaction mixture contained 5 μL of dNTP extract in 40 mM of Tris-HCl (pH 7.4), 10 mM of MgCl2, 5 mM of dithiothreitol, 0.25 μM of oligoprimer, 0.75 μM of [8-3H]dATP, 12–21 Ci/mmol, and 0.25 units of Sequencing Taq DNA Polymerase (Bioron, Newtown Square, PA). Reaction mixtures with aqueous dNTP standards were processed in parallel. After incubation at 48°C for 60 min, 9 μL of the mix was spotted onto one position of a DEAE Filtermat glass fiber filter (PerkinElmer, Waltham, MA) and left to dry. The filters were washed six times for 10 min with 5% Na2HPO4, once with water, and once with absolute ethanol, and left to dry again. The filter was covered with a Melt-on Scintillator sheet (PerkinElmer), and the retained radioactivity was determined by scintillation counting in a MicroBeta2 Microplate counter (PerkinElmer). dNTP amounts were calculated from interpolation on the calibration curves. To ensure the reliability of the results, triplicates of two different dilutions of each dNTP extract (usually 1:3 and 1:10 water-diluted) were processed in each independent experiment.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA). The tests used are indicated in the figure legends. For statistical purposes, undetectable values were considered as zero.
Results
Treatment with AAV2/8-TBG-hcTYMP provides a dose-dependent, permanent nucleoside reduction
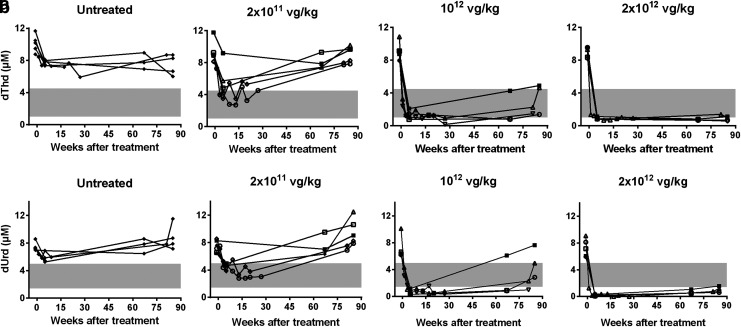
Blood samples were collected 2 weeks before treatment (baseline values) and at several time points over 88 weeks after treatment to measure nucleoside levels (Fig. 1). As was observed in a short-term study with the same vector,19 the lowest dose (2 × 1011 vg/kg) progressively reduced plasma dThd and dUrd concentrations, which reached normal levels 5 weeks after injection in most mice. However, this positive effect was gradually lost in all mice treated with this dose, and plasma dThd and dUrd concentrations returned to KO levels at the end of the study (88 weeks after treatment). Higher doses (1012 and 2 × 1012 vg/kg) produced faster reductions in plasma nucleoside concentrations, which reached wild-type (wt) levels or below in all mice 1 week after vector injection. The effect was maintained over the entire monitoring time, although there was a slight rebound at the last analysis (88 weeks) with the intermediate dose (1012 vg/kg). Nonetheless, nucleoside levels in three of four mice treated with the intermediate dose remained within the wt range at that time.
Figure 1.
Plasma thymidine (dThd) and deoxyuridine (dUrd) monitoring. dThd (a) and dUrd (b) concentrations in plasma obtained from untreated (n = 5) and adeno-associated virus (AAV)-treated knockout (KO) mice (n = 6 for 2 × 1011 and 1012 vector genomes [vg]/kg, and n = 5 for 2 × 1012 vg/kg) over 88 weeks after treatment (time 0). Gray area indicates the plasma dThd and dUrd concentration range in wild-type (wt) mice (n = 11). To facilitate matching the results of each mouse between different figures, the same symbol identifies a particular mouse in Figs. 1, 3, and 4.
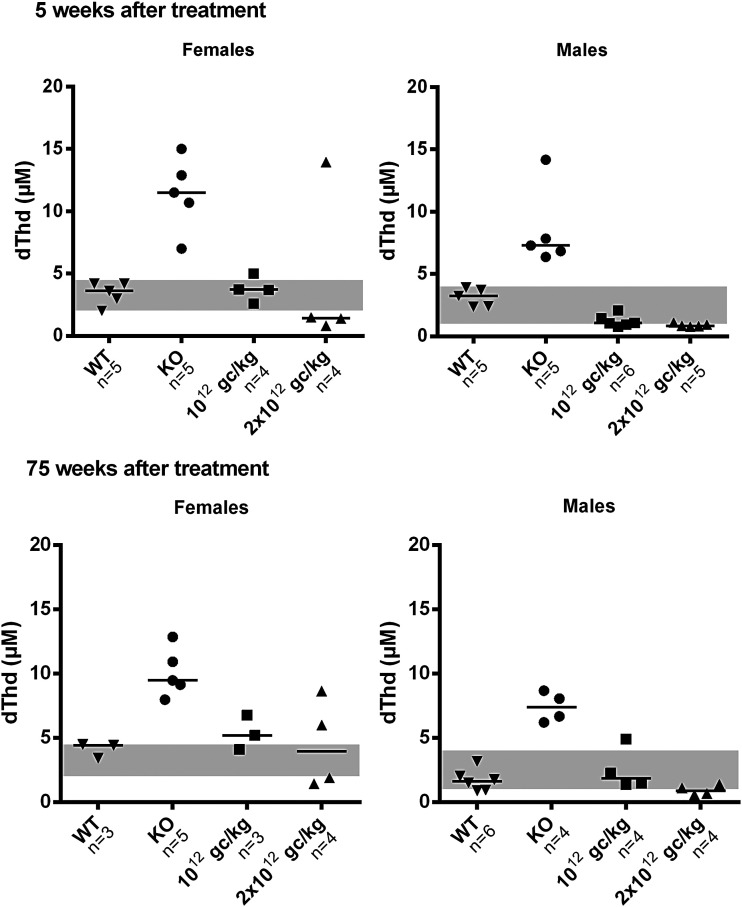
Previous AAV studies in MNGIE have been performed in male mice, as sex-biased AAV transduction efficiency has been observed in rodents (higher in male mice than female mice).22 Therefore, the study tested whether the treatment was also effective in females using two vector doses: 1012 and 2 × 1012 vg/kg. Although the small size of the sample would have compromised a reliable statistical analysis of the data, inspection of the results suggests that the vector was effective in reducing systemic nucleoside levels in females, although the effect was quantitatively less pronounced than in males (Fig. 2). At 5 weeks following treatment, circulating dThd concentrations in female mice were lowered to wt levels with both doses (three of four mice for each dose). This reduction was maintained in females 75 weeks after treatment, but dThd levels persisted within the wt range in only one of three mice (1012 vg/kg) and two of four mice (2 × 1012 vg/kg). In addition, there were reductions in plasma dUrd levels (data not shown). Parallel experiments conducted in male mice showed similar results, but nucleoside reductions were slightly greater in males than females.
Figure 2.
Plasma dThd clearance in male versus female mice. Plasma dThd concentrations in male and female KO mice 5 and 75 weeks after treatment. Doses are indicated on the x-axis (vg/kg). Horizontal lines represent medians. Gray areas indicate the plasma dThd concentration range in wt female (n = 8) and male (n = 11) mice.
AAV treatment restores dNTP balance in liver mitochondria
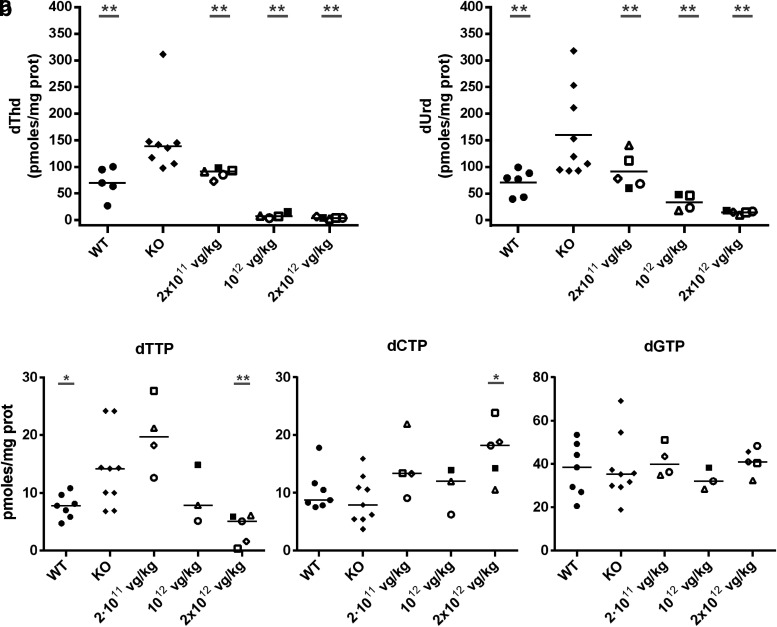
At 96 weeks of age, untreated KO mice showed significantly higher liver dThd and dUrd levels than those of their wt counterparts (Fig. 3), findings consistent with the previously reported results for younger animals (42 weeks).17 These increases were found to be significantly lowered in a dose-dependent manner by treatment with the vector, given at 8 weeks of age. At a vector dose of 2 × 1011 vg/kg, nucleoside levels in the liver had decreased to wt values, and at higher doses, nucleosides had dropped to below normal values.
Figure 3.
Nucleoside concentrations and mitochondrial nucleotide levels in the liver after long-term transgene expression. (a) dThd and dUrd concentrations in wt (n = 5), untreated KO (n = 9), and AAV-treated KO mice with 2 × 1011 (n = 5), 1012 (n = 4), and 2 × 1012 vg/kg (n = 5) 21 months (88 weeks) after treatment. (b) Mitochondrial deoxythymidine triphosphate (dTTP), deoxycytidine triphosphate (dCTP), and deoxyguanosine triphosphate levels in wt (n = 7), untreated KO (n = 9), and AAV-treated KO mice with 2 × 1011 (n = 5), 1012 (n = 4), and 2 × 1012 vg/kg (n = 5) 21 months after treatment. Horizontal lines represent medians. Gray underlined asterisks indicate statistical differences compared to untreated KO mice (*p < 0.05; **p < 0.01, Mann-Whitney U-test). The group of mice treated with the higher vector dose had lower dTTP levels (p < 0.05) and higher dCTP levels (p < 0.05) than those of wt mice. To facilitate matching the results of each mouse between different figures, the same symbol identifies a particular mouse in Figs. 1, 3, and 4.
Nucleosides are dNTP precursors within the salvage pathway. As a consequence of this biochemical linkage, dThd and dUrd overload causes a deoxythymidine triphosphate (dTTP) increase and secondary deoxycytidine triphosphate (dCTP) decrease in mitochondria of some tissues, including the liver, and so on.8,18,19,23 These imbalances were partially or totally prevented by the treatment. Mitochondrial dCTP levels increased at all vector doses tested, whereas the mitochondrial dTTP increase was prevented, with reductions down to wt levels or below at doses of 1012 and 2 × 1012 vg/kg. dTTP remained unchanged at the lowest dose of 2 × 1011 vg/kg (Fig. 3). Mitochondrial deoxyguanosine triphosphate (dGTP) levels did not change with the treatment. dATP levels could not be measured in liver mitochondria because the values obtained were below the lower limit of quantification.
Transgene copy number is diluted over time
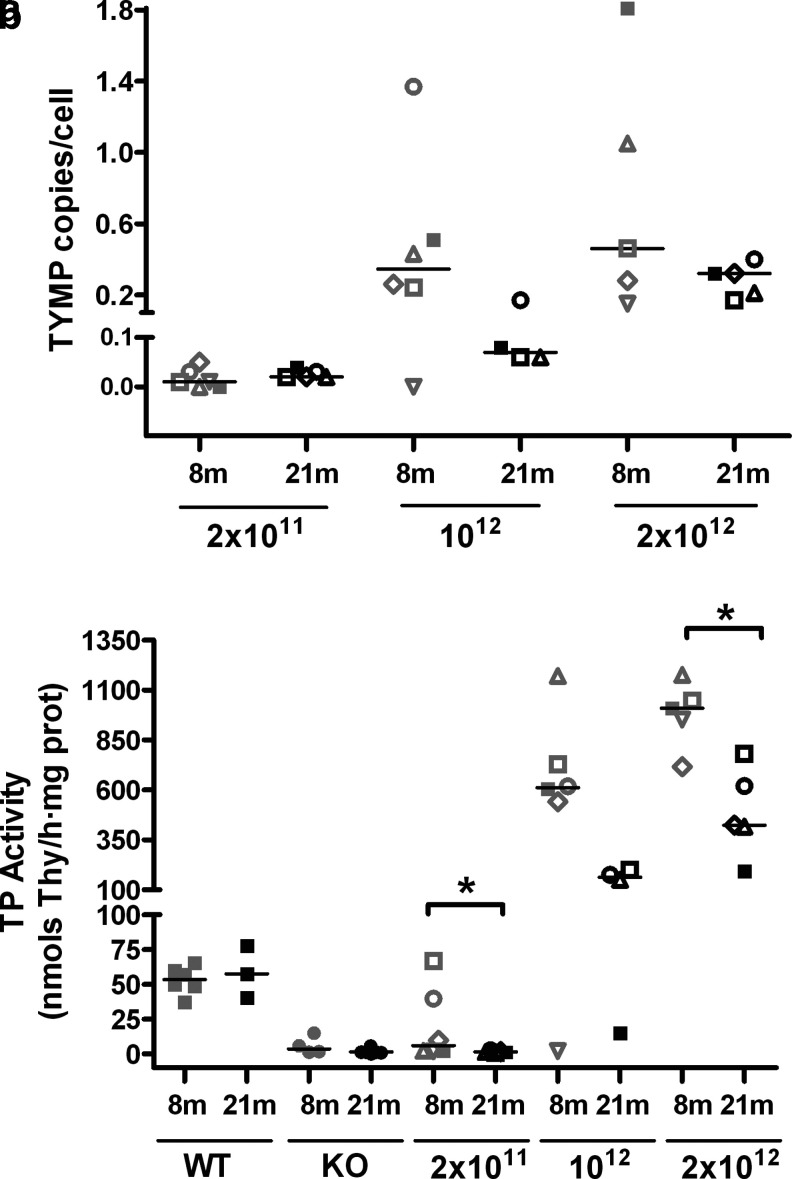
Quantitative PCR analysis of DNA in the liver of mice treated with the AAV2/8-TBG-hcTYMP vector showed a small but noticeable dilution of the vector genomes per cell over time (Fig. 4). At the lowest vector dose, vector copy number per cell was already negligible at 8 months after treatment. At the two higher doses, vector copy number 8 months after treatment ranged from 0.1 to 1.8 vg/cell in 10 of 11 treated mice (median 0.35 and 0.46 vg/cell at doses of 1012 and 2 × 1012 vg/kg, respectively). At 21 months after treatment, vector copy number had decreased (median 0.04 and 0.32 vg/cell at 1012 and 2 × 1012 vg/kg, respectively). This loss of vector copy number correlated with partial loss of TP activity over time. Eight months after treatment, KO mice gained TP activity in the liver in a dose-dependent manner, reaching wt values in two of six mice treated with the lowest dose, and approximate values of 600 to 1,000 nmol/h/mg of protein in mice treated with higher doses. Thirteen months later (21 months after treatment), TP activity had been lost in all mice treated with the lowest dose (consistent with the systemic nucleoside rebound observed in this group; Fig. 1), and reduced in those treated with the higher doses. It should be noted, however, that in most mice treated with higher doses (seven of eight) TP activities were maintained between 200 and 800 nmol/h/mg prot (fourfold increased or higher compared to wt values) at 21 months. The correlation between vector copy number per cell and TP activity in treated mice was statistically significant (p < 0.0001).
Figure 4.
Vector genomes and thymidine phosphorylase (TP) activity in the liver. Quantitative polymerase chain reaction–assessed AAV genome copies in the liver from treated KO mice using a human TYMP cDNA probe (a), and TP activity in the liver of wt, untreated, and treated KO mice 8 and 21 months (88 weeks) after treatment (b). The number of animals in consecutive groups of panel (a) are, from left to right, n = 6, 5, 6, 4, 5, and 5. The number of animals in consecutive groups of panel (b) are, from left to right, n = 6, 3, 4, 11, 6, 5, 6, 4, 5, and 5. Horizontal lines represent medians. Doses are indicated on the x-axis (vg/kg). Asterisks indicate statistical differences between groups (*p < 0.05, Mann-Whitney U-test). To facilitate matching the results of each mouse between different figures, the same symbol identifies a particular mouse in Figs. 1, 3, and 4.
Treatment with AAV2/8-TBG-hcTYMP did not increase tumorigenesis or mortality rates
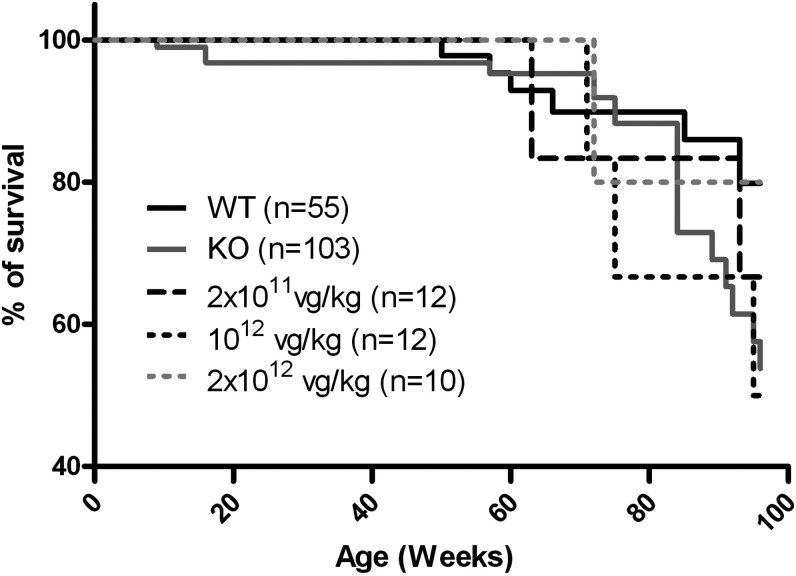
To investigate whether survival was affected in treated mice, mortality was followed up to the end of the study (96 weeks) in wt untreated KO and AAV-treated KO mice at all doses (Fig. 5 and Supplementary Table S1). Kaplan–Meyer analysis showed that median survival was not significantly different between wt and untreated KO mice (although a lower percentage of KO mice than wt mice were alive at the end of the study). The treatment did not affect survival of the animals at any dose, either positively or negatively (i.e., median survival in mice treated with each vector dose did not significantly differ from that of untreated KO mice).
Figure 5.
Survival curve. Kaplan–Meier survival representation of wt, untreated KO, and treated KO mice.
To detect the development of tumors, the organs of a group of mice killed at the end of the study or following the euthanasia recommendation according to the endpoint rule were carefully examined (see Materials and Methods). This analysis only included macroscopic visualization of organs at necropsies, without organ dissection or microscopic analyses. The findings of this examination are summarized in Table 1. The limited number of treated animals examined and the variability of the tumors found precludes definitive conclusions, but the incidence of liver tumors (target tissue of the study) was not higher in AAV-treated KO mice than in wt or untreated KO mice in the overall data analysis.
Table 1.
Tumorigenesis study
| Group/dose | Total studied | Number of mice with tumorsa | Tumors detected |
|---|---|---|---|
| WT | 16 | 3 (19%) | 1 (spleen, liver, and lung), 2 (spleen) |
| KO | 30 | 6 (20%) | 2 (lung), 2 (small intestine), 1 (liver), 1 (spleen, kidney, and liver) |
| 2 × 1011 vg/kg | 6 | 2 (33%) | 1 (lung), 1 (small intestine and lung) |
| 1012 vg/kg | 6 | 1 (17%) | 1 (lung) |
| 2 × 1012 vg/kg | 5 | 2 (40%) | 1 (liver), 1 (small intestine and lung) |
Specific subsets of animals belonging to different groups were selected at the beginning of the study to investigate the occurrence of tumors at the end of the study (when they were killed at the end of the study, or before the end of the study following the endpoint rule for humanitarian reasons).
Tumors found in necropsies at the end of the study (age 96 weeks) or after euthanasia.
wt, wild type; KO, knockout; vg, vector genomes.
Discussion
MNGIE is caused by systemic accumulation of dThd and dUrd, which interfere with mtDNA replication and maintenance in certain tissues that are particularly susceptible to injury by increased nucleoside levels. Consequently, current MNGIE therapies have focused on preventing dThd and dUrd overload in several target tissues, as has been reviewed.24 Past experience has shown that MNGIE therapies are effective only when the systemic nucleoside reduction is sustained over time,12–15 and preclinical evidence indicates that this objective can be achieved using gene therapy. Gene therapy strategies based on a lentiviral vector targeting the hematopoietic system or an AAV targeting the liver have shown biochemical efficacy in a murine model of the disease.17–19 In the case of hematopoietic tissue transduced with a lentiviral vector, the effect is maintained over the life of treated mice, but the procedure itself is associated with a reduction in the animals' life span. The present study reports that treatment with AAV2/8-TBG-hcTYMP targeting the liver in a murine model also provides a permanent biochemical correction, but without adverse effects.
A previous report found that various doses of this vector lowered dThd and dUrd in the blood and several tissues for at least 34 weeks after treatment.19 In this study, it was observed that the biochemical correction is fully maintained over the entire life of most mice treated with 1012 vg/kg and all mice treated with 2 × 1012 vg/kg. At the lowest dose tested (2 × 1011 vg/kg), the biochemical effect persisted for the first 30 weeks (consistent with the results of a previous study) but was lost thereafter in all animals. This loss of effect coincided and correlated with a reduction in vector genomes and TP activity in the liver. Some dilution effect was expected for AAVs because they mainly remain as episomes in the nucleus of transduced cells.25 The average hepatocyte turnover time in uninjured liver of adult mice is 150 days.26 Therefore, an 85% vg/cell reduction would be expected in 350 days (between 8 and 21 months, the times at which this endpoint was assessed). Accurate quantification of this reduction was difficult in this study because vector copy number in the liver cannot be assessed twice in the same mouse (liver biopsies stimulate hepatocyte proliferation, leading to overestimation of the dilution effect), and the wide dispersion of copy number data (illustrated in Fig. 4a) makes precise copy number assessment difficult in different mice at different times. However, the data seem to indicate that the reductions were less marked than was expected, especially for the highest vector dose. At the lowest dose, vector copy number was already barely detectable 8 months after treatment.
The dilution effect was also less pronounced than expected when assessed in terms of liver TP activity. Among the factors that could account for retention of copy number and TP activity, a small but relevant proportion of AAV may have been integrated in the genome of hepatocytes25 and replicated in parallel with cell division. A dilution effect is not expected (and is not observed) when a lentiviral vector is used because these vectors integrate in the genome and are not diluted when cells divide.17,18 In humans, the turnover time of mature hepatocytes in a fully developed, uninjured liver ranges from 200 to 300 days.27 Hence, dilution of the AAV therapeutic effect along time is also expected. Data from the single related clinical trial reported to date, in which hemophilia B was treated with a scAAV8 targeting the liver, have shown that factor IX expression (product of the transgene in this clinical study) is stable for >5 years.28,29 These results indicate that the dilution effect in humans may be slow enough to be undetectable in this moderate time frame. However, huge efforts in the field of AAV gene therapy are aimed at developing new capsid vectors compatible in terms of immunogenicity with a second vector infusion in order to overcome issues derived from the long-term dilution effect.
An impact of sex on liver transduction with AAVs is well recognized in the literature, with greater transduction efficiency reported in males than females.22,30–33 Previous studies in other murine models have proposed various explanations for the higher transgenic expression in male mice, such as an interaction between the AAV ITR and testosterone-dependent host nuclear regulatory proteins22 and contact between AAV vectors and inhibitory molecules present in the blood.30 Only male mice were used in the authors' first study on AAV,19 but the experience was expanded to both male and female mice in the present study. The results indicate that AAV treatment is effective in both sexes, although the reduction in plasma nucleosides is more pronounced in males than it is in females. Since a single intravenous injection was used, both the above-mentioned explanations could account for the differences between sexes. Nevertheless, it is important to note that evidence from studies in nonhuman primates and patients suffices to affirm that this difference only pertains to rodents.34 Therefore, the sex-related differences observed here are not relevant when considering the use of this approach to treat MNGIE patients.
The liver is an advantageous target organ for gene correction in MNGIE patients, not only because of its immunological tolerance, which limits the immune response against the vector capsid and TP protein, but also because it is one of the richest organs in TP activity in humans,21,35 and a particularly suitable tissue to host systemic clearance of metabolites.
TP activity was found to be 3- to 14-fold higher in the murine model than in wt mice at 21 months after treatment with the highest vector dose. This high activity resulted in excessively reduced nucleoside levels in the blood, which could be a concern, as this could lead to dTTP depletion in quiescent hepatocytes. Since de novo dTTP synthesis is cell-cycle dependent and downregulated in nondividing cells, hepatocyte dTTP levels mainly depend on the salvage pathway and would be affected by abnormally low dThd levels. It was found that the dThd over-reduction observed at the highest vector dose was associated with slight mitochondrial depletion of dTTP (below wt values) and expansion of dCTP (above wt values). Hence, it is concluded that the dose-dependent effect on blood nucleoside levels is reflected downstream in the mitochondrial salvage pathway by inducing a dose-dependent dTTP reduction and dCTP expansion, although the above-mentioned over-corrections point out the difficulty of finely adjusting the vector dose to reach the wt dNTP levels. Of note, 21 months after treatment with the lowest vector dose, dTTP remained at KO levels, as also occurred with blood dThd levels in the same group and at the same time. By contrast, liver dThd levels were normal. The reason for these inconsistent findings is unknown, and some bias derived from the low number of available samples from old animals cannot be ruled out. In contrast to the changes observed in pyrimidine deoxynucleotides, mitochondrial dGTP levels were not affected by the treatment.
The effect of the treatment on nucleoside and mitochondrial nucleotide levels underlines the importance of adjusting the vector dose in patients so that excessive TP activity will not change the dNTP pool beyond the objectives of the treatment. However, it is unlikely that the doses tested here will result in liver TP activities higher than normal in humans. Liver TP activities in mice treated with the highest dose were still around four times lower than the normal values in human liver.21 Even considering that we were testing a human gene under a human promoter in mice, this observation suggests that if use of this vector were translated to patients, the concern would not be TYMP overexpression but instead the inability to reach TP levels high enough to normalize systemic nucleoside concentrations. The fact that AAV8 transduction efficiency in rodents is higher than in humans36,37 reinforces this viewpoint. Which is the best serotype to target human liver is a relevant and controversial issue. Some studies in humanized mouse models suggest that AAV3-derived engineered serotypes infected human cells much more efficiently than AAV8,36–38 while other studies do not support this conclusion, showing instead similar efficiencies.39,40
There is considerable controversy regarding the oncogenic potential of AAV use in gene therapy, and several studies have been reported with rather conflicting results. In one experimental study in mice, hepatocellular carcinoma developed after injection of a therapeutic AAV due to integration of the vector into a genomic region encoding numerous regulatory RNAs.41 Furthermore, AAV2 infection has been associated with oncogenic insertional mutagenesis in human hepatocellular carcinoma.42 However, other authors have found no associated increase in tumorigenesis with AAV use in mice,43,44 and a recent study reported an absence of hepatic genotoxicity in relation to systemic administration of an AAV2/5 in nonhuman primates and acute intermittent porphyria patients.45 In another study, random AAV integration in several loci, including mtDNA, was reported,46,47 but the findings were considered largely artefactual by others.48 These discrepancies have been attributed to differing experimental conditions.49 It has also been hypothesized that the enhancer–promoter encoded by the vector can influence its genotoxicity, as it can lead to overexpression of the genes located close to the integration site.49 This study used the TBG promoter, which has been associated with insertional mutagenesis.49 Nonetheless, in this study, the incidence of liver tumors in AAV-treated mice was similar to the spontaneous hepatocellular carcinoma incidence in wt and untreated KO mice and to the rates described in other studies.49–51 The small size of the cohort of treated mice does not allow an increased incidence of tumors to be ruled out. A dedicated study with higher numbers of mice is needed to address this potential issue, but this objective is beyond the scope of this study. Of note, this study treated adult mice and not newborns, and the doses administered were 50 times lower than those resulting in tumorigenesis.49 Thus, at the doses used, it seems that the genotoxic potential of the vector is low, but the therapeutic benefit is retained. To verify the safety of the vector further, a Kaplan–Meier analysis was performed, which found no association of the treatment with higher mortality in this study.
This extended study provides further evidence that gene therapy using an AAV targeting the liver is a feasible therapeutic option for MNGIE treatment. Among the variables that can influence the success of this therapy, selection of the appropriate vector dose is one of the most important to be considered. A vector dose range is described that allows long-term correction and did not show adverse effects derived from transgene overexpression or vector genotoxicity, although more detailed and expanded studies are needed to ensure the safety of the treatment. This information may be of value for designing and implementing future clinical trials with MNGIE patients.
Supplementary Material
Acknowledgments
This work was supported by the Instituto de Salud Carlos III (Grant PI15/00465 to R.M., co-funded with FEDER funds), and the French Muscular Dystrophy Association–Téléthon (AFM Téléthon Postdoctoral Grant 18247 to J.T.).
Author Disclosure
No competing financial interests exist.
References
- 1.Hirano M, Garcia-de-Yebenes J, Jones AC, et al. Mitochondrial neurogastrointestinal encephalomyopathy syndrome maps to chromosome 22q13.32-qter. Am J Hum Genet 1998;63:526–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirano M, Nishigaki Y, Marti R. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): a disease of two genomes. Neurologist 2004;10:8–17 [DOI] [PubMed] [Google Scholar]
- 3.Hirano M, Silvestri G, Blake DM, et al. Mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): clinical, biochemical, and genetic features of an autosomal recessive mitochondrial disorder. Neurology 1994;44:721–727 [DOI] [PubMed] [Google Scholar]
- 4.Marti R, Nishigaki Y, Hirano M. Elevated plasma deoxyuridine in patients with thymidine phosphorylase deficiency. Biochem Biophys Res Commun 2003;303:14–18 [DOI] [PubMed] [Google Scholar]
- 5.Marti R, Nishigaki Y, Vila MR, et al. Alteration of nucleotide metabolism: a new mechanism for mitochondrial disorders. Clin Chem Lab Med 2003;41:845–851 [DOI] [PubMed] [Google Scholar]
- 6.Garone C, Tadesse S, Hirano M. Clinical and genetic spectrum of mitochondrial neurogastrointestinal encephalomyopathy. Brain 2011;134:3326–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Diaz B, Garone C, Barca E, et al. Deoxynucleoside stress exacerbates the phenotype of a mouse model of mitochondrial neurogastrointestinal encephalopathy. Brain 2014;137:1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Vioque E, Torres-Torronteras J, Andreu AL, et al. Limited dCTP availability accounts for mitochondrial DNA depletion in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). PLoS Genet 2011;7:e1002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pontarin G, Ferraro P, Valentino ML, et al. Mitochondrial DNA depletion and thymidine phosphate pool dynamics in a cellular model of mitochondrial neurogastrointestinal encephalomyopathy. J Biol Chem 2006;281:22720–22728 [DOI] [PubMed] [Google Scholar]
- 10.Song S, Wheeler LJ, Mathews CK. Deoxyribonucleotide pool imbalance stimulates deletions in HeLa cell mitochondrial DNA. J Biol Chem 2003;278:43893–43896 [DOI] [PubMed] [Google Scholar]
- 11.Halter J, Schupbach WM, Casali C, et al. Allogeneic hematopoietic SCT as treatment option for patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE): a consensus conference proposal for a standardized approach. Bone Marrow Transplant 2011;46:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halter JP, Michael W, Schupbach M, et al. Allogeneic haematopoietic stem cell transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Brain 2015;138:2847–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirano M, Marti R, Casali C, et al. Allogeneic stem cell transplantation corrects biochemical derangements in MNGIE. Neurology 2006;67:1458–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D'Angelo R, Rinaldi R, Pironi L, et al. Liver transplant reverses biochemical imbalance in mitochondrial neurogastrointestinal encephalomyopathy. Mitochondrion 2017;34:101–102 [DOI] [PubMed] [Google Scholar]
- 15.De Giorgio R, Pironi L, Rinaldi R, et al. Liver transplantation for mitochondrial neurogastrointestinal encephalomyopathy. Ann Neurol 2016;80:448–455 [DOI] [PubMed] [Google Scholar]
- 16.Lopez LC, Akman HO, Garcia-Cazorla A, et al. Unbalanced deoxynucleotide pools cause mitochondrial DNA instability in thymidine phosphorylase-deficient mice. Hum Mol Genet 2009;18:714–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres-Torronteras J, Gomez A, Eixarch H, et al. Hematopoietic gene therapy restores thymidine phosphorylase activity in a cell culture and a murine model of MNGIE. Gene Ther 2011;18:795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres-Torronteras J, Cabrera-Perez R, Barba I, et al. Long-term restoration of thymidine phosphorylase function and nucleoside homeostasis using hematopoietic gene therapy in a murine model of mitochondrial neurogastrointestinal encephalomyopathy. Hum Gene Ther 2016;27:656–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres-Torronteras J, Viscomi C, Cabrera-Perez R, et al. Gene therapy using a liver-targeted AAV vector restores nucleoside and nucleotide homeostasis in a murine model of MNGIE. Mol Ther 2014;22:901–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72:248–254 [DOI] [PubMed] [Google Scholar]
- 21.Valentino ML, Marti R, Tadesse S, et al. Thymidine and deoxyuridine accumulate in tissues of patients with mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). FEBS Lett 2007;581:3410–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davidoff AM, Ng CY, Zhou J, et al. Sex significantly influences transduction of murine liver by recombinant adeno-associated viral vectors through an androgen-dependent pathway. Blood 2003;102:480–488 [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Estevez S, Ferrer G, Torres-Torronteras J, et al. Thymidine phosphorylase is both a therapeutic and a suicide gene in a murine model of mitochondrial neurogastrointestinal encephalomyopathy. Gene Ther 2014;21:673–681 [DOI] [PubMed] [Google Scholar]
- 24.Cabrera-Pérez R, Torres-Torronteras J, Vila-Julià F, et al. Prospective therapeutic approaches in mitochondrial neurogastrointestinal encephalomyopathy (MNGIE). Expert Opin Orphan Drugs 2015;3:1167–1182 [Google Scholar]
- 25.Nakai H, Yant SR, Storm TA, et al. Extrachromosomal recombinant adeno-associated virus vector genomes are primarily responsible for stable liver transduction in vivo. J Virol 2001;75:6969–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magami Y, Azuma T, Inokuchi H, et al. Cell proliferation and renewal of normal hepatocytes and bile duct cells in adult mouse liver. Liver 2002;22:419–425 [DOI] [PubMed] [Google Scholar]
- 27.Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology 2009;137:466–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nathwani AC, Reiss UM, Tuddenham EG, et al. Long-term safety and efficacy of factor IX gene therapy in hemophilia B. N Engl J Med 2014;371:1994–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nienhuis AW, Nathwani AC, Davidoff AM. Gene therapy for hemophilia. Hum Gene Ther 2016;27:305–308 [DOI] [PubMed] [Google Scholar]
- 30.Berraondo P, Crettaz J, Ochoa L, et al. Intrahepatic injection of recombinant adeno-associated virus serotype 2 overcomes gender-related differences in liver transduction. Hum Gene Ther 2006;17:601–610 [DOI] [PubMed] [Google Scholar]
- 31.Dane AP, Cunningham SC, Graf NS, et al. Sexually dimorphic patterns of episomal rAAV genome persistence in the adult mouse liver and correlation with hepatocellular proliferation. Mol Ther 2009;17:1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paneda A, Vanrell L, Mauleon I, et al. Effect of adeno-associated virus serotype and genomic structure on liver transduction and biodistribution in mice of both genders. Hum Gene Ther 2009;20:908–917 [DOI] [PubMed] [Google Scholar]
- 33.Sun B, Zhang H, Franco LM, et al. Correction of glycogen storage disease type II by an adeno-associated virus vector containing a muscle-specific promoter. Mol Ther 2005;11:889–898 [DOI] [PubMed] [Google Scholar]
- 34.Pañeda A, Lopez-Franco E, Kaeppel C, et al. Safety and liver transduction efficacy of rAAV5-cohPBGD in nonhuman primates: a potential therapy for acute intermittent porphyria. Hum Gene Ther 2013;24:1007–1017 [DOI] [PubMed] [Google Scholar]
- 35.Boschetti E, D'Alessandro R, Bianco F, et al. Liver as a source for thymidine phosphorylase replacement in mitochondrial neurogastrointestinal encephalomyopathy. PLoS One 2014;9:e96692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lisowski L, Dane AP, Chu K, et al. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 2014;506:382–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vercauteren K, Hoffman BE, Zolotukhin I, et al. Superior in vivo transduction of human hepatocytes using engineered AAV3 capsid. Mol Ther 2016;24:1042–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kay MA. Selecting the best AAV capsid for human studies. Mol Ther 2015;23:1800–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Ling C, Zhong L, et al. Efficient and targeted transduction of nonhuman primate liver with systemically delivered optimized AAV3B vectors. Mol Ther 2015;23:1867–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Bell P, Somanathan S, et al. Comparative study of liver gene transfer with AAV vectors based on natural and engineered AAV capsids. Mol Ther 2015;23:1877–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donsante A, Miller DG, Li Y, et al. AAV vector integration sites in mouse hepatocellular carcinoma. Science 2007;317:477. [DOI] [PubMed] [Google Scholar]
- 42.Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet 2015;47:1187–1193 [DOI] [PubMed] [Google Scholar]
- 43.Bell P, Wang L, Lebherz C, et al. No evidence for tumorigenesis of AAV vectors in a large-scale study in mice. Mol Ther 2005;12:299–306 [DOI] [PubMed] [Google Scholar]
- 44.Li H, Malani N, Hamilton SR, et al. Assessing the potential for AAV vector genotoxicity in a murine model. Blood 2011;117:3311–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gil-Farina I, Fronza R, Kaeppel C, et al. Recombinant AAV integration is not associated with hepatic genotoxicity in nonhuman primates and patients. Mol Ther 2016;24:1100–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaeppel C, Beattie SG, Fronza R, et al. A largely random AAV integration profile after LPLD gene therapy. Nat Med 2013;19:889–891 [DOI] [PubMed] [Google Scholar]
- 47.Kaeppel C, Beattie SG, Fronza R, et al. Reply to: NGS library preparation may generate artifactual integration sites of AAV vectors. Nat Med 2014;20:578–579 [DOI] [PubMed] [Google Scholar]
- 48.Cogne B, Snyder R, Lindenbaum P, et al. NGS library preparation may generate artifactual integration sites of AAV vectors. Nat Med 2014;20:577–578 [DOI] [PubMed] [Google Scholar]
- 49.Chandler RJ, LaFave MC, Varshney GK, et al. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J Clin Invest 2015;125:870–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeganathan K, Malureanu L, Baker DJ, et al. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol 2007;179:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mahler JF, Stokes W, Mann PC, et al. Spontaneous lesions in aging FVB/N mice. Toxicol Pathol 1996;24:710–716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.